Abstract
Recently four new hypertrophic cardiomyopathy mutations in cardiac troponin C (cTnC) (A8V, C84Y, E134D, and D145E) were reported, and their effects on the Ca2+ sensitivity of force development were evaluated (Landstrom, A. P., Parvatiyar, M. S., Pinto, J. R., Marquardt, M. L., Bos, J. M., Tester, D. J., Ommen, S. R., Potter, J. D., and Ackerman, M. J. (2008) J. Mol. Cell. Cardiol. 45, 281–288). We performed actomyosin ATPase and spectroscopic solution studies to investigate the molecular properties of these mutations. Actomyosin ATPase activity was measured as a function of [Ca2+] utilizing reconstituted thin filaments (TFs) with 50% mutant and 50% wild type (WT) and 100% mutant cardiac troponin (cTn) complexes: A8V, C84Y, and D145E increased the Ca2+ sensitivity with only A8V demonstrating lowered Ca2+ sensitization at the 50% ratio when compared with 100%; E134D was the same as WT at both ratios. Of these four mutants, only D145E showed increased ATPase activation in the presence of Ca2+. None of the mutants affected ATPase inhibition or the binding of cTn to the TF measured by co-sedimentation. Only D145E increased the Ca2+ affinity of site II measured by 2-(4′-(2″-iodoacetamido)phenyl)aminonaphthalene-6-sulfonic acid fluorescence in isolated cTnC or the cTn complex. In the presence of the TF, only A8V was further sensitized to Ca2+. Circular dichroism measurements in different metal-bound states of the isolated cTnCs showed changes in the secondary structure of A8V, C84Y, and D145E, whereas E134D was the same as WT. PyMol modeling of each cTnC mutant within the cTn complex revealed potential for local changes in the tertiary structure of A8V, C84Y, and D145E. Our results indicate that 1) three of the hypertrophic cardiomyopathy cTnC mutants increased the Ca2+ sensitivity of the myofilament; 2) the effects of the mutations on the Ca2+ affinity of isolated cTnC, cTn, and TF are not sufficient to explain the large Ca2+ sensitivity changes seen in reconstituted and fiber assays; and 3) changes in the secondary structure of the cTnC mutants may contribute to modified protein-protein interactions along the sarcomere lattice disrupting the coupling between the cross-bridge and Ca2+ binding to cTnC.
Hypertrophic cardiomyopathy (HCM)3 is typically inherited as an autosomal dominant disease that is caused by mutations in sarcomeric genes and is the most prevalent cause of sudden death in athletes and young people (1, 2). The clinical hallmark of HCM is an increased thickness of the left ventricular wall. Myocyte disarray, fibrosis, septal hypertrophy, and abnormal diastolic function can also be present in HCM patients (3). HCM mutations have been reported in 13 myofilament-related genes; however, the cardiac troponin C (cTnC) gene remained excluded from this list (4–7). The clinical and functional phenotypes may vary according to the gene and the location of the mutation (8). Recently our group has reported evidence that brings cTnC into focus as an HCM susceptibility gene (9). Interestingly the prevalence for cTnC HCM mutations was the same as other well characterized genes (i.e. actin and tropomyosin) (6). To date, prior to our recent report, only one mutation in cTnC (L29Q) had been linked to HCM (10). In vitro and in situ studies demonstrating changes in the functional parameters of cardiac muscle regulation suggest that this mutation is causative of the disease (11, 12).
Analysis of a cohort of 1025 HCM patients from the Mayo Clinic revealed four new cTnC mutations (A8V, C84Y, E134D, and D145E) (9). The clinical records showed that the patients displayed left ventricle hypertrophy and significant left ventricular outflow obstruction managed by surgical myectomy. Symptoms such as dyspnea, syncope, and chest pain were also present. A8V, C84Y, and E134D patients did not present a familial history of HCM indicating that these were likely sporadic de novo mutations. The D145E mutation was observed in six family members suggesting genetic linkage. Functional analysis performed in skinned fibers showed increased Ca2+ sensitivity of force development (a characteristic of troponin (Tn) mutations related to HCM) for three of the four mutations. Additionally the A8V and D145E mutations that are located in different domains caused increases in maximal force in this system. These data strongly suggest that HCM mutations in distinct regions of cTnC can result in a similar functional phenotype (9).
In cardiac muscle, the tropomyosin (Tm)·Tn complex, located in the thin filament, is responsible for muscle regulation (13, 14). Three Tn subunits are involved in this process: troponin T (TnT), which connects the Tn complex to the thin filament and is responsible for actomyosin ATPase activation in the presence of Ca2+ (8, 15); troponin I (TnI) is the subunit that binds to both TnT and TnC, inhibits muscle contraction, and is also implicated in HCM and restrictive cardiomyopathy (16); and TnC, a subunit that plays a crucial function in muscle regulation triggering contraction upon binding Ca2+ and is also considered an important intracellular Ca2+ buffer (17, 18). In the absence of Ca2+ binding to site II of cTnC, its N terminus is detached from the C terminus of cTnI, which under these conditions is bound to actin and inhibits muscle contraction. As Ca2+ binds to site II of cTnC, its N terminus binds to the C terminus of cTnI causing it to dissociate from actin. This is accompanied by the movement of cardiac Tm out of its inhibitory position on actin, thus relieving the inhibition of contraction (19–21). The dynamics of the interactions between Tn subunits and the thin filament that regulate contraction have been extensively studied (22–24).
TnC consists of two globular regions that are connected by a long central helix (25). It is well known that cTnC has two EF-hands containing high affinity Ca2+ binding sites III and IV (∼107 m−1) in the C terminus and only one functional low affinity Ca2+ binding site II (∼105 m−1) in the N terminus (18). An additional feature of helix-loop-helix Ca2+-binding proteins is the presence of short segments of antiparallel β-sheets between the Ca2+ binding loops of each domain (25, 26). The C-terminal domain of cTnC can also bind Mg2+ competitively (∼103 m−1) and is termed the structural domain because it is essential to keep it bound to the thin filament. The N terminus is considered the regulatory domain because Ca2+ binding to site II initiates muscle contraction. When TnC is in the Tn complex, the Ca2+ binding affinity at all sites is increased by ∼10-fold (18, 27, 28). Several studies have shown that there is coupling between TnC and actomyosin ATPase. For example, bepridil and calmidazolium, two known Ca2+ sensitizers that bind to cTnC and enhance its Ca2+ binding affinity, also stimulate myofibrillar ATPase activity (29, 30). In addition, deletion of the N-helix of the TnC N-domain diminishes activation of regulated actomyosin ATPase activity (31, 32).
The purpose of this study was to determine the functional effects of the four newly discovered HCM cTnC mutations not previously addressed and to investigate possible changes in their structure and Ca2+ binding properties. To answer these questions we performed reconstituted ATPase activity, co-sedimentation, and spectroscopy assays. In the presence of 100% HCM mutant or wild type (WT) cTnC, the ATPase activity rate measured by increasing the Ca2+ concentration in an actomyosin·Tm·Tn reconstituted complex showed increases in Ca2+ sensitivity similar to those obtained previously with cardiac skinned fibers (9). At a ratio of 50% mutant to 50% WT, only A8V had a diminished Ca2+ sensitivity. We also evaluated the ability of the Tn HCM mutants to activate and inhibit the ATPase activity in the presence and absence of Ca2+. Only cTnC-D145E showed higher levels of ATPase activation. Co-sedimentation did not show changes in the ability of the Tn complex containing the cTnC mutants to bind to actin·Tm. The Ca2+ binding properties of the regulatory site II of cTnC as estimated from fluorescence and measured at cTnC and cTn levels did not match the apparent affinity of this site in the fiber and reconstituted filaments. However, D145E showed increased Ca2+ affinity in the isolated and cTn states that was minimally affected in the presence of the thin filament (TF). In the presence of the TF, A8V was the only mutant that showed an increase in Ca2+ affinity that more closely approached the Ca2+ sensitivity measured in the fiber. However, the circular dichroism (CD) measurements suggest that significant structural changes exist in the secondary structure of the cTnC mutants A8V, C84Y, and D145E compared with wild type. All of these results considered together with the PyMol illustrations suggest that structural changes are present in at least three TnC HCM mutants that are likely to be crucial for protein-protein interactions but unable to affect the Ca2+ binding properties of TnC at the different levels of TF complexity. Here we show for the first time that the thick filament is probably essential to completely recreate the increased Ca2+ sensitivity produced by HCM TnCs and observed in ATPase and skinned fiber assays.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of Human Cardiac Troponin T, Troponin I, and Troponin C Mutants
cDNAs cloned in our laboratory from human cardiac tissue were used for the expression and purification of cTnI (33) and cTnT (34). Human cTnC (HcTnC) was used as the template for PCR using primers designed to introduce the mutations of interest that result in the TnC HCM mutants (HcTnC-A8V, HcTnC-C84Y, HcTnC-E134D, HcTnC-D145E, HcTnC-C35S, HcTnC-A8V/C35S, HcTnC-E134D/C35S, and HcTnC-D145E/C35S). All subcloned DNA sequences were inserted into the pET3d expression plasmid and sequenced for verification prior to expression and purification. Standard methods previously used in this laboratory were utilized for expression and purification of the HcTnCs (35). Briefly the cTnC WT and cTnC HCM mutants were expressed in Escherichia coli BL21 (DE3) CodonPlus bacterial cells (Stratagene). The bacterially expressed TnCs were passed over a Q-Sepharose column and eluted with a linear gradient of 0–0.6 m KCl (4 °C) in a buffer containing 50 mm Tris-HCl (pH 7.8), 6 m urea, 1 mm EDTA, 1 mm DTT. The purest TnC fractions were dialyzed into 50 mm Tris-HCl (pH 7.5), 1 mm CaCl2, 1 mm MgCl2, 50 mm NaCl, 1 mm DTT; loaded onto a phenyl-Sepharose column after the addition of 0.5 m (NH4)2SO4; and eluted with Ca2+-free buffer containing 50 mm Tris-HCl (pH 7.5), 1 mm EDTA, 1 mm DTT at room temperature. Collected fractions were resolved by SDS-PAGE, and the fractions containing the purest, most concentrated fractions were pooled yielding >97% pure protein, dialyzed exhaustively against 5 mm NH4HCO3, and then lyophilized. All steps were performed at 4 °C unless otherwise indicated.
Formation of Troponin Complexes
The individual purified troponin subunits were first dialyzed against 3 m urea, 1 m KCl, 10 mm MOPS, 1 mm DTT, 0.1 mm phenylmethanesulfonyl fluoride and then twice against the same buffer excluding urea. The protein concentration of the individual subunits was determined using the Coomassie Plus kit (Pierce), and then the subunits were combined in a 1.3:1.3:1 TnT:TnI:TnC molar ratio. After 1 h, the complexes were successively dialyzed against decreasing concentrations of KCl (0.7, 0.5, 0.3, 0.1, 0.05, and 0.025 m). The excess precipitated TnT and TnI that formed during complex formation was removed by centrifugation. Proper stoichiometry was verified by SDS-PAGE before storage of troponin complexes at −80 °C.
Actin·Tm-activated Myosin ATPase Assays
Minimum and Maximum
Porcine cardiac myosin, rabbit skeletal F-actin, and porcine cardiac Tm were prepared as described previously (36). The protein concentrations used for actomyosin ATPase assays were 0.6 μm porcine cardiac myosin, 3.5 μm rabbit skeletal F-actin, 1 μm porcine cardiac tropomyosin, and 0–2 μm preformed Tn complexes as described above. The buffer conditions for the proteins were as follows: myosin in 10 mm MOPS (pH 7.0), 0.4 m KCl, 1 mm DTT; actin in 10 mm MOPS (pH 7.0), 40 mm KCl; tropomyosin in 10 mm (pH 7.0), 300 mm KCl, 1 mm DTT. The final ionic strength of the reactions was ∼75 mm when considering the ionic contributions from the various protein buffers. The ATPase inhibitory assay was performed in a 0.1-ml reaction mixture of 3.4 mm MgCl2, 0.13 μm CaCl2, 1.5 mm EGTA, 3.5 mm ATP, 1 mm DTT, 11.5 mm MOPS (pH 7.0) at 25 °C. The ATPase activation assay was performed using the same 0.1-ml buffer mixture with 3.3 mm MgCl2 and 1.7 mm CaCl2. The ATPase reaction was initiated with the addition of ATP and quenched after 20 min using trichloroacetic acid to a final concentration of 35%. The precipitated assay proteins were removed by centrifugation, and the inorganic phosphate concentration in the supernatant that was released by ATP hydrolysis was determined according to the method of Fiske and Subbarow (37).
pCa Curves
pCa curves were performed using 1 μm preformed Tn complex with the same concentrations of myosin, actin, and tropomyosin as described above. For the experiments of 50% mutant and 50% WT the preformed Tn complexes were combined to a final concentration of 1 μm. The conditions of the assay (11.55 mm MOPS, 2 mm EGTA, 1 mm NTA, 3.7 mm MgCl2 (pH 7.0 at 25 °C)) changed slightly in varied pCa solutions: pCa 8.0 (0.113 mm CaCl2), pCa 7.0 (0.8997 mm CaCl2), pCa 6.5 (1.915 mm CaCl2), pCa 6.0 (2.982 mm CaCl2), pCa 5.5 (3.639 mm CaCl2), pCa 5.0 (3.971 mm CaCl2), pCa 4.5 (4.278 mm CaCl2), and pCa 4.0 (4.913 mm CaCl2) calculated according to methods established in our laboratory (38).
Co-sedimentation
The thin filament constituents, F-actin at a concentration of 20 μm, and Tn/Tm at a concentration of 2.86 μm were used to obtain a molar ratio of 7:1:1 and were co-sedimented using an air-driven ultracentrifuge (Beckman Airfuge; rotor A100) for 30 min at 28 p.s.i. (∼120,000 × g). The buffer conditions were 20 mm imidazole (pH 7.0), 60 mm NaCl, 3 mm MgCl2, 0.5 mm CaCl2, 2 mm β-mercaptoethanol.
Fluorescence Labeling of HCM cTnCs
For isolated cTnC and cTn measurements the cTnC WT and mutants were labeled with IAANS at Cys-35 and Cys-84 except for cTnC-C84Y, which was only labeled at Cys-35. C84S was used as a control for the C84Y mutant. For the thin filament measurements the cTnC WT and mutants A8V, E134D, and D145E had Cys-35 mutated to Ser and were labeled with IAANS only at Cys-84. IAANS was obtained from Molecular Probes, Plano, TX. Fluorescent labeling and purification of cTnC were performed according to established methods (27, 28).
Determination of Apparent Ca2+ Affinities by Fluorescence
Isolated IAANS-labeled HcTnCs (HcTnC mutants and WT) were dialyzed into fluorescence buffer containing 2 mm EGTA, 5 mm nitrilotriacetic acid, 120 mm MOPS, 90 mm KCl. Before each titration 1.25 mm MgCl2 and 1 mm freshly prepared DTT were added. cTn complexes were made as described above (“Formation of Troponin Complexes”); however, the complexes underwent final dialysis in the fluorescence buffer containing 1.25 mm MgCl2. Thin filaments were constructed using the protocol established in our laboratory (39). The cTnCs used in the thin filaments were mutated at Cys-35 to Ser and were labeled with IAANS at Cys-84. For C84Y the label was placed at Cys-35; however, the thin filament did not show spectral changes from pCa 8 to 4. Steady state fluorescence measurements were performed in a Jasco 6500 spectrofluorometer where IAANS fluorescence was excited at 330 nm and emission was detected at 450 nm. The protein concentrations used for isolated cTnC, cTn, and thin filaments were 10 μm, 0.5 μm, and 0.05 mg/ml, respectively. Fluorescence spectral changes were recorded during the titration of microliter amounts of CaCl2. The concentration of free Ca2+ and amounts of titrated Ca2+ were calculated using the pCa calculator program developed by our laboratory (38). The program made corrections for dilution effects that occur during titration of Ca2+. The data were fitted to a version of the Hill equation that accounted for the spectral changes that occur at a low Ca2+ concentration and plotted using SigmaPlot 10.0. The apparent Ca2+ affinities are reported as pCa50 values ±S.E.
Circular Dichroism Measurements
Far-UV CD spectra were collected using a 1-mm-path quartz cell in a Jasco J-720 spectropolarimeter. Spectra were recorded at 195–250 nm with a bandwidth of 1 nm at a speed of 50 nm/min at a resolution of 0.5 nm at room temperature (∼20 °C). Ten scans were averaged, and no numerical smoothing was applied. The optical activity of the buffer was subtracted from relevant protein spectra. Mean residue ellipticity ([θ]MRE in degree·cm2/dmol) for the spectra were calculated utilizing the same Jasco system software using the following equation: [θ]MRE = [θ]/(10 × Cr × l) where [θ] is the measured ellipticity in millidegrees, Cr is the mean residue molar concentration, and l is the path length in cm (40). Protein concentrations were determined by the biuret reaction using TnC as a standard that was calibrated by determining its protein nitrogen content. The CD experiments were performed using three different conditions: for apo, 1 mm EGTA, 20 mm MOPS, 100 mm KCl (pH 7.0); for Mg2+, 1 mm EGTA, 20 mm MOPS, 100 mm KCl, 2.075 mm MgCl2 (pH 7.0) yielding a free [Mg2+] of 2 mm); and for Ca2+, 1 mm EGTA, 20 mm MOPS, 100 mm KCl, 2.075 mm MgCl2, 1.096 mm CaCl2 (pH 7.0) yielding a free [Ca2+] of 10−4 m and free [Mg2+] of 2 mm). The experimental protein concentration for the WT and each mutation was 0.2 mg/ml.
PyMol Analysis
PyMol is an open source molecular visualization program that allows the user to download Protein Data Bank files that contain molecular coordinates from x-ray crystallography- or nuclear magnetic resonance (NMR)-solved structures that were deposited in the Protein Data Bank. The program allows the known structures to be manipulated for example mutagenesis of selected residues, details of potential side chain interactions, and potential for polar contacts such as hydrogen bonding due to the nature and proximity of the side chains.
Statistical Analysis
The experimental results are reported as mean ± S.E. and were analyzed for significance using Student's t test at p < 0.05 (paired or unpaired depending on the experimental design).
RESULTS
Actomyosin ATPase pCa Curves Using Reconstituted Troponin Complexes Containing HCM cTnC Mutants
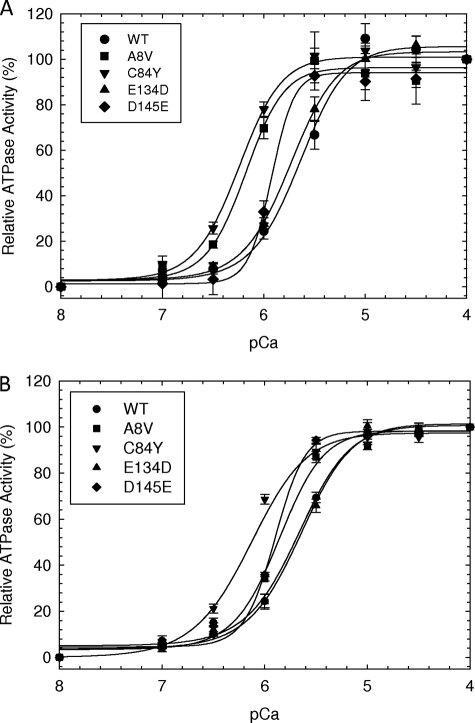
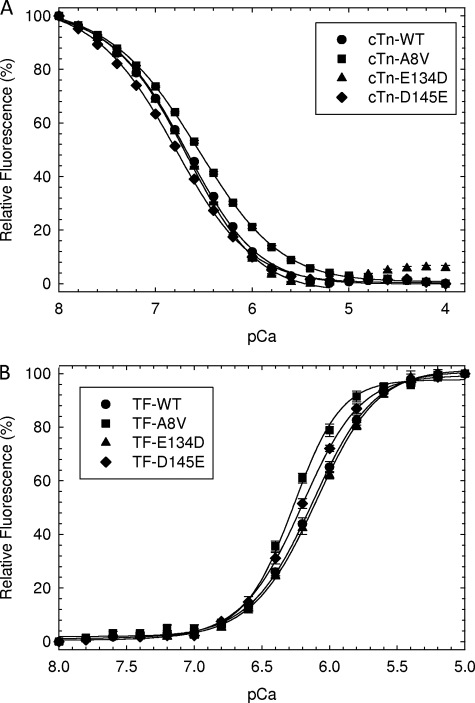
To assess the Ca2+ sensitivity of the newly discovered HCM cTnC mutants at a 100 and 50% ratio in a reconstituted system, actomyosin ATPase assays were evaluated as a function of increasing Ca2+ concentration. This system is more sensitive than the skinned fiber assays because of the lack of organization between thin and thick filaments; however, the Ca2+ sensitivity measured from the actomyosin ATPase correlates to the changes found by isometric tension (41, 42). At 100% mutant reconstitution, A8V and C84Y generated the largest increase in Ca2+ sensitivity of the reconstituted myofilament with a leftward shift of +0.51 (pCa50 = 6.20 ± 0.03) and +0.56 (pCa50 = 6.25 ± 0.03) pCa units compared with the WT (pCa50 = 5.69 ± 0.05), respectively (Fig. 1A). D145E also generated a leftward shift with a ΔpCa50 of +0.25. At the 50% ratio only A8V showed a corresponding decrease in pCa50 compared with the amount of mutant protein present (Fig. 1B). In skinned fibers the leftward shift was ∼0.4 and ∼0.3 pCa units for A8V and C84Y, respectively (9). The D145E mutant (pCa50 = 5.94 ± 0.03) showed an intermediate increase in Ca2+ sensitivity with a 0.25 pCa unit leftward shift compared with WT in both ATPase and force measurements. At both ratios, the E134D (pCa50 = 5.72 ± 0.03 at 100%) mutant showed no significant change compared with WT (ΔpCa50 = 0.02), which was statistically the same in the cardiac skinned fiber results reported previously (Fig. 1, A and B) (9). In addition, D145E displayed the highest degree of cooperativity with the nHill = 3.99 ± 0.25 compared with nHill = 2.14 ± 0.23 for the WT.
FIGURE 1.
The actin-Tm-activated myosin ATPase activity of HCM cTnC mutants as a function of pCa. A, HcTnC mutant or WT at 100%. B, HcTnC mutants and WT at a 50% to 50% ratio. The experiments were performed using 0.6 μm myosin, 3.5 μm actin, 1 μm Tm, and 1 μm preformed Tn complex. The buffer conditions were as described under “Experimental Procedures.” The basal ATPase activity at pCa 8.0 was considered 0%. The intermediate points were normalized to the maximal ATPase activity at pCa 4.0 and were considered 100%. The specific ATPase activity at pCa 8.0 was 0.15, 0.19, 0.11, 0.19, and 0.18 mol of Pi × mol of myosin−1 × s−1 for WT, A8V, C84Y, E134D, and D145E, respectively. Each curve represents an average of six to seven experiments, and error is reported as mean ± S.E.
Actomyosin ATPase Assays Using Reconstituted Troponin Complexes Containing HCM cTnC Mutants (Minimum and Maximum)
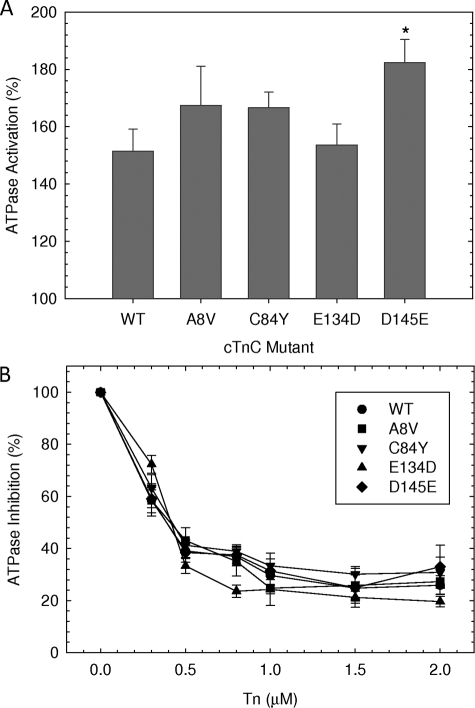
These experiments show the capacity of the cTn complex to activate or inhibit actomyosin ATPase activity in the presence or absence of Ca2+, respectively. Concentrations of preformed cTn complexes ranging from 0.3 to 2 μm were utilized, and the greatest activation was seen at 1 μm. The actomyosin ATPase activity in the absence of Tn complex was considered to be the basal activity level for both activation and inhibition and was set as 100%. Fig. 2A indicates that the cTn complex containing HcTnC-D145E was able to significantly increase the activation of the reconstituted thin filament to 182.3 ± 8.1%, whereas activation for WT complex was 151.4 ± 7.8%. The activation values for cTn complexes containing A8V, C84Y, and E134D were 167.4 ± 13.6, 166.6 ± 5.5, and 153.6 ± 7.3%, respectively, and were not significantly different. The inhibition of ATPase activity as a function of Tn concentration in the absence of Ca2+ did not show any significant changes between the Tn complexes containing either mutants or WT (Fig. 2B).
FIGURE 2.
The activation and inhibition of the actin-Tm-activated myosin ATPase activity by different HCM cTnC mutants in the presence and absence of Ca2+. A, activation of actomyosin ATPase at 1 μm Tn complex in the presence of Ca2+. B, inhibition of preformed actomyosin ATPase activity by HCM cTnC mutants at increasing ratios of Tn complex as indicated on the abscissa. The assay conditions were as follows: 3.5 μm actin, 1 μm Tm, and 0.6 μm myosin were dissolved in 75 mm KCl, 3.3 mm MgCl2, 1.7 mm CaCl2 (for activation assay) or 0.13 μm CaCl2 (for inhibition assay), 1.5 mm EGTA, 3.5 mm ATP, 1 mm DTT, 11.5 mm MOPS, pH 7.0. The myosin ATPase activity that occurs in the absence of cTn complex is considered 100% ATPase activity. The specific ATPase activity in the absence of troponin complexes was measured as 0.35 mol of Pi × mol of myosin−1 × s−1. Each point represents an average of six experiments performed in triplicate and is expressed as mean ± S.E. *, p < 0.05 compared with WT.
Co-sedimentation of the Tn Complex with the Thin Filament
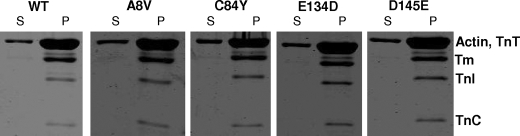
This experiment allowed us to investigate whether the cTnC mutations interfered with the ability of Tn to interact with actin·Tm. All the thin filament proteins were mixed in a ratio of 7:1:1 actin:Tm:Tn and co-sedimented in an air-driven ultracentrifuge (Beckman Airfuge) as detailed under “Experimental Procedures.” Fig. 3 shows no discernable changes in the ability of the different Tn complexes (WT and mutants) to bind to actin·Tm. No changes were observed in the protein content in the pellet between Tn complexes containing cTnC WT or mutants. Also no detectable alterations in the protein content of the supernatant between Tn complexes were seen.
FIGURE 3.
Co-sedimentation of actin, tropomyosin, and Tn complexes containing HCM cTnC mutants. The experimental design and buffer conditions were as described under “Experimental Procedures.” Samples were separated by 15% SDS-PAGE. S, supernatant; P, pellet.
IAANS Fluorescence of Isolated HCM cTnC Mutants
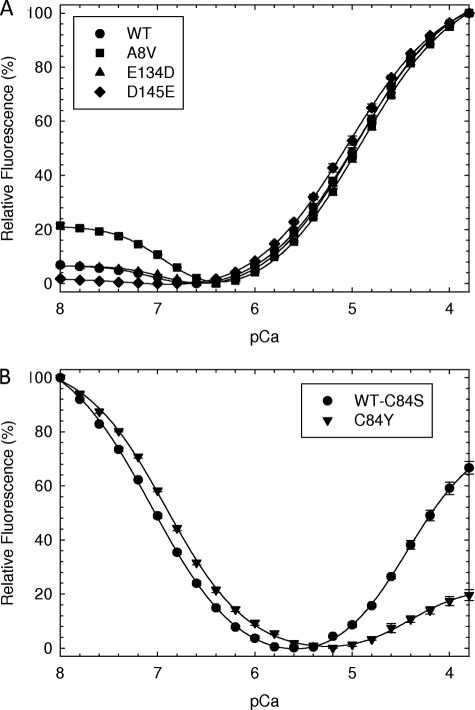
To determine whether these HCM cTnC mutations had an effect on the apparent Ca2+ binding affinity of the regulatory domain site II, we performed Ca2+ titrations and monitored Ca2+ binding-induced conformational changes using IAANS fluorescence (27). The HCM cTnC proteins were doubly labeled at the two native cysteines (Cys-35 and Cys-84) in all mutants except for C84Y. In the isolated HCM cTnC mutants, A8V and E134D showed no significant difference in apparent Ca2+ affinity at site II compared with WT (Fig. 4A and Table 1). However, D145E showed a statistically significant increase in Ca2+ affinity (pCa50 = 5.02 ± 0.02) compared with WT (pCa50 = 4.91 ± 0.01) (Fig. 4A). Although there was a change in the apparent Ca2+ affinity measured by fluorescence (ΔpCa50 = 0.11), it is not sufficient to explain the large changes in Ca2+ sensitivity seen in the skinned fibers (ΔpCa50 = 0.25) (9). Interestingly the doubly IAANS-labeled cTnC was also able to detect the effects of the mutations on the binding of Ca2+ at pCa 8–6.5 to the Ca2+/Mg2+ sites in the C terminus. A8V showed the largest spectral change in the C terminus, whereas D145E, which is present in site IV at the Ca2+-coordinating +Z position, showed no spectral alterations at low Ca2+ concentrations (pCa 8–6.5) (Fig. 4A). Additionally A8V and E134D showed a statistically significant reduction of ∼0.1 pCa unit of the Ca2+ affinity of the C-terminal domain compared with WT (Table 1). Because C84Y has one of the cysteine IAANS binding sites mutated, we used C84S, with cysteine in position 84 that was mutated to serine, as a control for this mutant (27). In this case both proteins (C84Y and C84S) were monolabeled with IAANS only at Cys-35. The fluorescence spectrum of C84Y showed a decrease in fluorescence intensity from pCa 5.5 to 3.8 compared with WT. However, the pCa50 of the low Ca2+ affinity site II was otherwise unchanged from the WT value (Fig. 4B and Table 1). Also C84Y showed a statistically significant decrease, according to the Student's t test, in the apparent affinity for Ca2+ at sites III and IV (pCa50 = 6.92) compared with the WT (pCa50 = 7.06) (Table 1).
FIGURE 4.
Determination of apparent Ca2+ affinities of isolated HCM cTnC mutants by fluorescence. Steady state fluorescence measurements excited the IAANS probe at 330 nm, and emission was detected at 450 nm. A, HcTnCs doubly labeled at cysteines 35 and 84: WTIAANS35IAANS84, A8VIAANS35IAANS84, E134DIAANS35IAANS84, and D145EIAANS35IAANS84. B, HcTnCs labeled at cysteine 35: C84SIAANS35 (control) and C84YIAANS35. Fluorescence spectral changes were recorded during the titration of microliter amounts of CaCl2 to a 2-ml experimental volume. The concentration of free Ca2+ and amounts of titrated Ca2+ were calculated using the pCa calculator program developed by our laboratory (see “Experimental Procedures”). The program made corrections for dilution effects that occur during titration of Ca2+. The data were fitted to a version of the Hill equation that accounted for the spectral changes occurring at low Ca2+ concentration. The Ca2+ affinities are reported in Table 1 as pCa50 and nHill values ±S.E. Each point represents an average of four to five experiments and is expressed as mean ± S.E.
TABLE 1.
Summary of the IAANS fluorescence results of HCM cTnC mutants
The pCa50 and nHill were calculated for both classes of sites from independent Ca2+ titrations, and error is reported as ±S.E.
| TnC | High affinity sites |
Low affinity site |
||
|---|---|---|---|---|
| pCa50 | nHill | pCa50 | nHill | |
| WT | 7.03 ± 0.01 | 1.85 ± 0.02 | 4.91 ± 0.01 | 0.90 ± 0.01 |
| A8V | 6.90 ± 0.01a | 1.56 ± 0.05a | 4.89 ± 0.03 | 0.91 ± 0.02 |
| E134D | 6.91 ± 0.01a | 1.90 ± 0.06 | 4.92 ± 0.02 | 0.94 ± 0.03 |
| D145E | NDb | ND | 5.02 ± 0.02a | 0.88 ± 0.01 |
| WT-C84S | 7.06 ± 0.01 | 0.94 ± 0.03 | 4.44 ± 0.01 | 1.15 ± 0.04 |
| C84Y | 6.92 ± 0.01a | 1.02 ± 0.02 | 4.43 ± 0.02 | 1.42 ± 0.06 |
ap < 0.05 compared with their respective WT.
b ND, not determined.
IAANS Fluorescence of cTn- and TF-containing HCM cTnC Mutants
The changes in Ca2+ affinity of the isolated cTnC (Fig. 4) and within the cTn complex (Fig. 5A) were not sufficient to explain the Ca2+ sensitivity changes that occurred in the actomyosin ATPase assays and skinned fibers. Therefore, it was examined whether the TF was able to increase the apparent affinity of the HCM mutant cTnC to Ca2+ (Fig. 5B). It appears that two mutants, A8V and D145E, for which data were obtained were affected differently by the presence of the TF. For example, A8V showed a decrease in Ca2+ affinity at the low affinity sites of −0.02 and −0.11, respectively, in both isolated cTnC and cTn; however, Ca2+ affinity increased to +0.14 in the presence of the TF (Table 2). This was actually an increase of +0.25 when comparing the Ca2+ affinity of cTn-A8V versus the value obtained in the presence of the TF. D145E on the other hand showed a different trend where the mutation caused an increase in Ca2+ affinity of the isolated cTnC and cTn complex of +0.11 and +0.12, respectively, compared with cTnC WT, and the presence of the TF increased the Ca2+ affinity of cTnC-D145E by a small degree (+0.08) compared with the TF WT (Table 2). The mutant cTnC-E134D had the same Ca2+ affinity as WT in the presence of the TF, and data were not obtained for C84Y because the IAANS label could not be placed at Cys-84. We attempted to measure the Ca2+ affinity of cTnC-C84Y in the TF with IAANS placed at Cys-35; however; there was not a measurable change in Ca2+-dependent fluorescence to take these measurements.
FIGURE 5.
Determination of apparent Ca2+ affinities of cTn and thin filament HCM mutants by fluorescence. Steady state fluorescence measurements excited the IAANS probe at 330 nm, and emission was detected at 450 nm. A, cTn complexes containing HCM HcTnC mutants that were doubly labeled at cysteines 35 and 84. B, TF-containing HCM HcTnC mutants that were singly labeled at cysteine 35. The WT, A8V, E134D, and D145E had the cysteine 84 mutated allowing labeling at a single site. Fluorescence spectral changes were recorded during the titration of microliter amounts of CaCl2 to a 2-ml experimental volume. The data were fitted to a version of the Hill equation that accounted for the spectral changes occurring at low Ca2+ concentration. The Ca2+ affinities are reported in Table 2 as pCa50 values ±S.E. Each point represents an average of four to five experiments and is expressed as mean ± S.E.
TABLE 2.
Summary of pCa50 of Ca2+ affinity or Ca2+ sensitivity measured by various experimental assays at different levels of complexity of the thin filament
| TnC | pCa50 |
||||
|---|---|---|---|---|---|
| cTn | TF | ATPase 100% | ATPase 50%a | Skinned fiberb | |
| WT | 6.69 ± 0.01 | 6.13 ± 0.02 | 5.69 ± 0.05 | 5.68 ± 0.02 | 5.66 ± 0.01 |
| A8V | 6.58 ± 0.01c | 6.27 ± 0.02c | 6.20 ± 0.03c | 5.87 ± 0.03c | 6.02 ± 0.01c |
| E134D | 6.69 ± 0.01 | 6.11 ± 0.02 | 5.72 ± 0.03 | 5.66 ± 0.04 | 5.68 ± 0.01 |
| D145E | 6.81 ± 0.01c | 6.21 ± 0.02c | 5.94 ± 0.03c | 5.90 ± 0.02c | 5.90 ± 0.01c |
| C84S | 6.77 ± 0.01 | NDd | ND | ND | ND |
| C84Y | 6.76 ± 0.01 | ND | 6.25 ± 0.03c | 6.20 ± 0.03 | 5.93 ± 0.01c |
a WT in ATPase 50% was measured at 100% ratio and compared with the mutants at a 50% ratio.
b Skinned fiber values were obtained from Ref. (9).
cp < 0.05 compared with their respective WT.
d ND, not determined.
Circular Dichroism of HCM cTnC Mutants
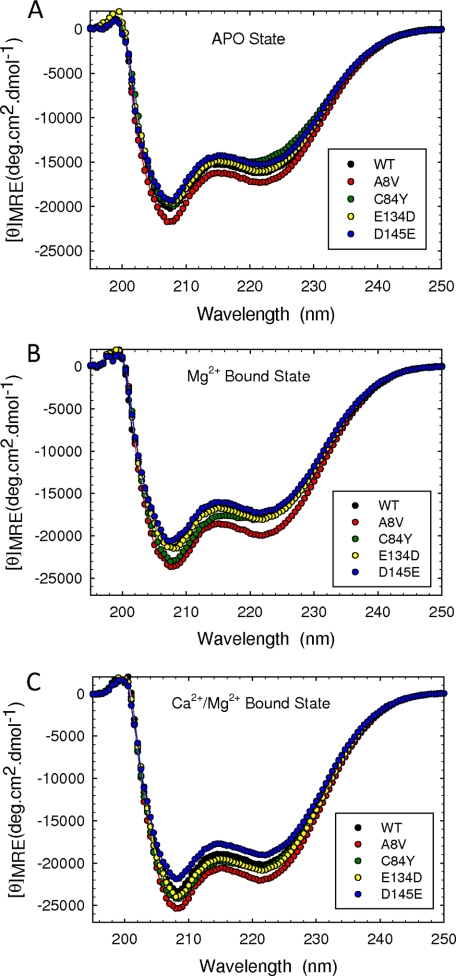
In the next set of experiments we addressed whether the HCM mutations affect the secondary structure of cTnC. In the apo state (absence of Ca2+ and Mg2+) the A8V, C84Y, and D145E mutations modified the total amount of secondary structure of the cTnC compared with WT (Fig. 6A and Table 3). In the presence of Mg2+, only the A8V mutant showed structural differences compared with WT. In the presence of both Ca2+ and Mg2+, A8V and D145E were the only mutants that showed differences from WT (Fig. 6, B and C). Note that the alterations observed in the presence of Ca2+ and Mg2+ were different for the A8V and D145E mutants. The mean residue ellipticity (MRE) at 222 nm (one of the peaks used to evaluate the α-helical content in proteins) was −21,840.0 and −19,064.7 for A8V and D145E, respectively, compared with −20,124.2 for the WT (Table 3). The A8V mutant showed an increase in the α-helical content, whereas the D145E mutant showed a decrease.
FIGURE 6.
Circular dichroism spectra of wild type and HCM cTnC mutations. Far-UV CD spectra were performed in apo, Mg2+-bound, and Ca2+/Mg2+-bound states. Spectra were recorded at 195–250 nm utilizing a 1-mm-path quartz cell in a Jasco-720 spectropolarimeter at room temperature (20 °C). The spectra shown are the average of three independent measurements. For each independent measurement 10 scans were averaged, and no numerical smoothing was applied. The optical activity of the buffer was subtracted from relevant protein spectra. The buffer conditions were as described under “Experimental Procedures.” Mean residue ellipticity ([θ]MRE in degree·cm2/dmol) for the spectra was calculated utilizing the same Jasco system software and the following equation: [θ]MRE = [θ]/(10 × Cr × l) where [θ] is the measured ellipticity in millidegrees, Cr is the mean residue molar concentration, and l is the path length in cm. deg, degrees.
TABLE 3.
Summary of circular dichroism results for HCM cTnC mutants
The CD spectrum is reported for apo (without divalent cations), Mg2+-bound, and Ca2+/Mg2+-bound states, and error is reported as ± S.E. deg, degrees.
| TnC | [θ]222 nm |
n | ||
|---|---|---|---|---|
| Apo | Mg2+ | Ca2+/Mg2+ | ||
| deg·cm2/dmol | ||||
| WT | −16,178.9 ± 178.9 | −17,945.4 ± 327.5 | −20,124.2 ± 212.2 | 3 |
| A8V | −17,276.2 ± 168.0a | −19,952.6 ± 37.2a | −21,848.0 ± 199.2a | 3 |
| C84Y | −14,786.7 ± 73.6a | −17,892.0 ± 136.9 | −20,545.5 ± 98.7 | 3 |
| E134D | −16,027.7 ± 177.5 | −18,077.7 ± 233.9 | −20,788.3 ± 196.0 | 3 |
| D145E | −15,246.2 ± 105.9a | −17,203.6 ± 202.5 | −19,064.7 ± 169.5a | 3 |
a p < 0.05 compared with the WT at same condition.
DISCUSSION
Here we assessed the functional and biochemical properties of four newly described HcTnC mutations linked to HCM. The approach we used was to look first at the mutations in the context of a reconstituted actomyosin system and then at the level of isolated HcTnC, cTn, and TF. We present data that details the effects of these mutations on the ability of the thin filament to inhibit or activate actomyosin ATPase activity. Fluorescence experiments were performed to determine the effects of these HCM mutations on the Ca2+ affinity of TnC. Circular dichroism was used to assess general structural changes in cTnC. From these data we determined whether perturbations in Ca2+ binding to HcTnC are responsible for the altered Ca2+ sensitivity of force development seen in cardiac skinned fibers in our previous study (9) or actomyosin ATPase activity as reported here.
The actomyosin ATPase experiments demonstrated results that may in part explain the HCM phenotype of diastolic dysfunction and cardiac hypertrophy found in patients. ATPase activation, in the presence of Ca2+, demonstrated that D145E, located in the C-terminal domain, displayed a significant increase in the ability to activate actomyosin ATPase. Recently it was shown that different types of cross-bridges can modulate TnC affinity for the thin filament demonstrating the influence of myosin on the C-terminal domain of TnC (43). Here the C-terminal domain HCM mutation D145E strengthens this concept as it shows the influence of this domain on actomyosin function. The mutations A8V and C84Y showed a tendency to increase ATPase activity; however, these differences were not statistically significant compared with WT. The inhibition of actomyosin ATPase activity by the HCM HcTnC mutations, in the absence of Ca2+, was unaltered. With regard to actomyosin ATPase activity as a function of pCa, the results recapitulated the skinned fiber findings (9). We measured the Ca2+ sensitivity of the myofilament by ATPase activity using two different amounts of cTnC mutants incorporated into the lattice. At 100% mutant reconstitution into the thin filament cTnC-A8V and -C84Y showed the largest increase in Ca2+ sensitivity in the actomyosin ATPase as seen in the skinned fibers. D145E displayed an intermediate increase in Ca2+ sensitivity in both systems, whereas E134D showed no significant change in this parameter, also consistent with previously performed skinned fiber experiments. To better mimic what is seen in patients that normally exist in the heterozygous state we decided to investigate the effects of the cTnC mutants reconstituted into the thin filament at a ratio of 50% mutant to 50% WT. At the 50% A8V to 50% WT ratio there was a decrease in the pCa50, whereas C84Y, E134D, and D145E did not show any significant change of pCa50 when compared with 100% mutant protein. This suggests that for some of the mutants the ratio of mutant protein to WT protein did not appear to have an effect: the mutant had a dominant effect even when present in a smaller amount. However, it appears that not all mutations behave in the same manner in the reconstituted thin filament since A8V showed a decrease in pCa50 when only 50% of the mutant was present. The increased Ca2+ sensitivity (A8V, C84Y, and D145E) seen using two different systems and increased ATPase activity (at least for D145E) could be associated with cardiac hypertrophy. Because cTnC Ca2+ affinity was increased it suggests that these mutants bind Ca2+ more tightly and consequently dissociate more slowly from cTnC. This delay in Ca2+ dissociation from cTnC maintains the heart in systole longer and is implicated in diastolic dysfunction (44). We hypothesize that changes in the Ca2+ affinity of cTnC would change the homeostasis of the cardiac cell triggering Ca2+-regulated pathways that lead to sudden cardiac death (45) and/or cardiac hypertrophy (46).
Co-sedimentation analysis using actin·Tm·Tn was performed to determine whether any of the HCM HcTnC mutants had altered the affinity of cTn for the components of the thin filament. The results (Fig. 3) suggest that the cTnC mutations either do not interfere with the association between cTn and the thin filament or that this method may not be sensitive enough to detect small affinity changes. The other possibility is that the perturbations caused by the cTnC mutations occur locally at the level of TnC·TnI, TnC·TnT, or TnC·TnI·TnT, and the physiological changes we measured were not due to altered affinity of cTn for the thin filament. Further studies may help to elucidate whether any modifications of the thin filament occur in the presence of HCM TnC mutations.
We investigated whether the changes in Ca2+ sensitivity in the reconstituted actomyosin ATPase and skinned fibers were due in part to altered Ca2+ affinity of HcTnC. To address this question we performed fluorescence experiments. The Ca2+ affinity measurements of the mutant HCM cTnC in the presence of the cTn complex and the thin filament indicated that the increased Ca2+ sensitivity seen in ATPase pCa curves and skinned fibers is achieved through a different mechanism for A8V and D145E (Table 2). The cTnC-A8V appears to be sensitized by the addition of tropomyosin and actin meaning that important interactions are occurring further down the TF. It does not appear that Ca2+ affinity of cTnC-A8V is intrinsically increased because the fluorescence was actually decreased in isolated cTnC and cTn. Therefore it is expected that the indirect mechanism of increased Ca2+ sensitivity is due to altered protein-protein interactions that occur in the TF that either may result from increased association of cTnI with cTnC or dictate more strongly binding cross-bridges that would in turn further increase the Ca2+ affinity of HCM cTnC. The cTnC-D145E mutation caused an increase in the apparent Ca2+ affinity of cTnC and the cTn complex that was not modified upon addition of the members of the TF. It appears that myosin cross-bridges may display a stronger role in sensitizing the myofilament containing cTnC-D145E compared with cTnC-A8V. These results strongly indicate that the mechanism of altered Ca2+ sensitivity of force development is far more complex than simply alterations in the Ca2+ affinity of the TF. It was reported that most of the mutations related to hypertrophic or dilated cardiomyopathy in tropomyosin, TnT, or TnI need the presence of all the thin filament proteins (actin·Tm·Tn) to reproduce the Ca2+ sensitivity changes observed in fibers or reconstituted systems (47). Here we show for the first time that mutations in cTnC related to HCM that affect the function N-terminal domain in fibers need additional components of the sarcomere and not only the proteins of the thin filament.
To assess whether HcTnC secondary structure was perturbed by the presence of the HCM mutations, CD was performed to determine the extent that the mutants changed the α-helical (θ = 222) and β-sheet content (θ = 208). The spectra of HcTnC in an apo state, Mg2+-bound state, and Mg2+ + Ca2+-saturating conditions were collected. Generally the Ca2+-bound states of the isolated skeletal TnC N and C termini have similar conformations, whereas the apo state of each displays striking differences. The N terminus possesses a higher percentage of α-helical content, and the C terminus has a larger percentage of random coil (48). The binding of metals to the high affinity sites III and IV greatly increases the α-helical content of the C-terminal domain and induces the skeletal TnC C-terminal domain to adopt a more compact globular conformation (27, 49). However, metal binding to the N terminus induces smaller conformational changes in the N terminus as indicated by tyrosine fluorescence and CD measurements (50).
The previous CD structural studies suggest that the large increase in α-helical content of the N-terminal mutation cTnC-A8V, under all conditions, compared with WT (see Table 3) is due to effects on the C terminus. Smith et al. (51) reported that the major effect of the N-helix mutations they studied including R11C in the N terminus of cTnC was on Ca2+ binding to the C-terminal domain. It is interesting to note that cTnC-A8V also showed the largest spectral change in IAANS fluorescence from probes located in the N terminus at the low Ca2+ levels sufficient to saturate only the C terminus. Therefore, in the apo state, cTnC-A8V may have increased α-helical content that consequently stabilizes the N terminus. Additional structural studies are essential to determine how this mutation increases the α-helical content of cTnC in the Ca2+- and Mg2+-bound state especially because physiological Mg2+ binding occurs at sites III and IV (52). Also the β-sheet content was greatly increased in the apo and divalent cation-bound states (Fig. 6C). In the absence of Ca2+ these short β-strands located between the Ca2+ binding sites normally do not form or form loosely between Ca2+ binding sites III and IV (25, 53). NMR studies have also shown that the short β-strands that are derived from the helix-loop-helix motif of Ca2+ binding sites I and II are present even in the apo state (54). This would suggest that much of the increase in β-strand content due to the A8V mutation might be derived from conformational changes within the N terminus.
The N-helix also makes contacts with helices A and D through hydrophobic and electrostatic interactions (25, 55, 56). PyMol modeling of A8V into the Takeda et al. (21) crystal structure, Protein Data Bank code 1J1E, showed that substitution of alanine to valine with a larger side chain could change the relationship between the N-helix and D-helix. This mutation may affect the overall N-terminal structure (see supplemental Fig. 1, A and B), thereby altering TnC and TnI interactions as well as influencing the C-terminal domain (31, 51). There is evidence that the N-helix and the C-terminal domain are physically close in three-dimensional space (51, 57). The effect of this mutation on the C terminus is also confirmed by studies in which deletion of the first 11–14 amino acids of the N terminus of troponin C reduces the affinity of the high affinity Ca2+ binding sites (31, 58).
C84Y, in contrast to A8V, had a significant reduction of these secondary structural elements in the apo state as seen from the CD experiments (Table 3). The HCM mutation C84Y had reduced α-helical content in the apo state compared with WT; however, the binding of Mg2+ and Ca2+ appears to stabilize the structure of TnC-C84Y. The cTnC-C84Y mutation is located at the end of the N terminus near the beginning of the flexible linker in the region that makes contact with the cTnI regulatory region (residues 147–162) as seen from modeling of this mutation into Protein Data Bank structure 1J1E (see supplemental Fig. 1, C and D). Cysteine 84 normally makes a hydrogen bond with the backbone amide of cTnI Ala-151, which is maintained in the mutant cTnC-C84Y. However, introduction of the bulky tyrosine side group decreases the space between these two interacting regions. Introduction of a tyrosine may create a wedge effect between interacting portions of cTnC and cTnI. Also previous results have attributed the stability of the N terminus to important interactions between the N-helix and residues of apo chicken skeletal TnC that are C-terminal to residue 85 (in helix D) that correspond to residue 83 in HcTnC (58).
E134D is located in the region of the cTnC C terminus where the cTnI N terminus interacts and protein kinase C phosphorylated residues 43/45 perturb the structure of TnC helix G in the vicinity of Glu-134 (59). Possibly the mutation alters transmission of the phosphorylation signal to cTnC and thereby affects the Ca2+ affinity of cTnC. Also a portion of TnT (residues 160–193) binds in this region as well, and cTnC residues near Glu-134 are perturbed upon TnT binding and are located on the outer surfaces of helices F and G and exposed to the solvent (60). PyMol modeling of cTnC-E134D into Protein Data Bank structure 1JIE did not show any potential structural changes in the vicinity of the mutation (data not shown), leading to speculation that other physiological processes such as phosphorylation of cTnI by protein kinase C or even other kinases might be affected (11, 61).
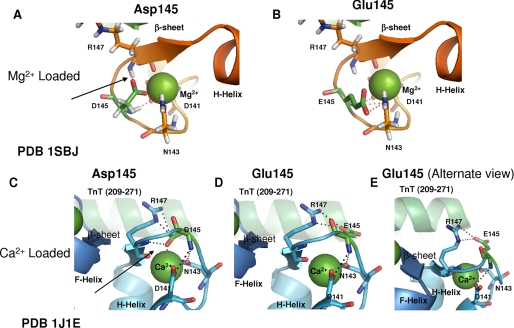
D145E showed no fluorescence changes attributed to the C terminus that may indicate that the C-terminal domain of cTnC is not properly binding Ca2+. The mutation is located in the +Z position of Ca2+ binding site IV that has been shown to be strictly conserved among species and EF-hand proteins; therefore it is likely that substitution at this position may have a deleterious effect. Previous studies have shown that the mutation of Asp to Ala in skeletal TnC at the X-coordinated position of the Ca2+ binding loop disrupts binding of Ca2+ to sites III and/or IV and results in an increase in the Ca2+ sensitivity of force development as seen in the D145E mutation (62). The cTnC HCM mutation D145E had a decrease in α-helicity in the Ca2+-bound state as seen by CD that may be due to the effects of the mutation on the helix-loop-helix structure of the EF-hand as Ca2+ binding stabilizes the adjacent α-helices (25). The Mg2+-saturated structure of cTnC-(81–161) in complex with the N-domain of cTnI-(33–80) shows that Mg2+ ligation causes partial closure of the cTnI binding hydrophobic cleft surrounding binding site IV and is consistent with condensation of the C-terminal portion of site IV around the smaller Mg2+ ion (63). The structure of the Ca2+/Mg2+ binding loops is maintained through a network of hydrogen bonding interactions (64). Any alterations in hydrogen bonding may cause a slight increase in flexibility of site IV and affect divalent cation binding (Fig. 7, A and B). The D145E mutation was modeled into Protein Data Bank structure 1SCV using PyMol in the presence of Ca2+ (Fig. 7, C, D, and E). In addition, a loss of β-sheet content was seen in the Ca2+-saturated D145E indicating that the mutation may cause destabilizing effects on the C-terminal β-sheet structure (65). The effects of the D145E mutation are likely due to an altered ability of site IV to coordinate Ca2+. It is possible that loss of cooperativity between these sites might affect α-helical content.
FIGURE 7.
Mapping and modeling of D145E HCM mutation in HcTnC in Mg2+-loaded C terminus and Ca2+-saturated states. cTnC HCM mutation D145E modeled into the NMR structure of the Mg2+-loaded C terminus of cTnC-(81–161) complexed with cTnI-(33–80) (Protein Data Bank code 1SBJ (63)) and Ca2+-saturated cTn complex crystal structure (Protein Data Bank (PDB) code 1J1E (21)) is shown. A, location of the residue Asp-145 in the Mg2+-bound state and H-bonding shown with red dotted lines. B, E145 modeled into Protein Data Bank structure 1SBJ showing H-bonding contacts. C, the residue Asp-145 shown in the Ca2+-bound cTn structure (Protein Data Bank code 1J1E). Helices in cTnC are indicated, and the interacting portion of cTnT is labeled. D, the mutation D145E modeled into the Ca2+-bound structure (Protein Data Bank code 1J1E) showing residues that make contact through H-bonding. E, different orientation of modeled mutation D145E.
The results indicate that the mechanism of increased Ca2+ sensitivity is different between the HCM mutations A8V and D145E. However, it appears that D145E may need the presence of the thick filament to fully explain how this mutation increased the Ca2+ sensitivity seen in ATPase and skinned fibers. A8V showed increased Ca2+ affinity upon addition of the TF; however, the myosin cross-bridge may be less involved with its Ca2+ sensitization. The structural illustrations presented here are unable to make assumptions of whether the mutations have global effects on the troponin complex. Structural studies are required to reveal the overall changes at the protein level. To this end we have presented results from the new HCM TnC mutations that support the idea that other thin and thick filament proteins are required to recapitulate the functional changes observed, i.e. increased Ca2+ sensitivity. The structural data lend credence to the idea that crucial intra- and interchemical interactions may reveal the possible mechanism of these new mutations in disrupting the myofilament Ca2+ binding function.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants HL-67415 and HL-42325 (to J. D. P.). This work was also supported by the Windland Smith Rice Comprehensive Sudden Cardiac Death Program (to M. A. J.) and American Heart Association Postdoctoral Fellowship AHA 0825368E (to J. R. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- HCM
- hypertrophic cardiomyopathy
- DTT
- dithiothreitol
- IAANS
- 2-(4′-(2″-iodoacetamido)phenyl)aminonaphthalene-6-sulfonic acid
- MOPS
- 3-(N-morpholino)propanesulfonic acid
- WT
- wild type
- cTn
- cardiac troponin
- Tn
- troponin
- Tm
- tropomyosin
- CD
- circular dichroism
- HcTnC
- human cTnC
- MRE
- mean residue ellipticity.
REFERENCES
- 1.Marian A. J., Roberts R. (1998) J. Cardiovasc. Electrophysiol. 9, 88–99 [DOI] [PubMed] [Google Scholar]
- 2.Maron B. J. (2002) Card. Electrophysiol. Rev. 6, 100–103 [DOI] [PubMed] [Google Scholar]
- 3.Maron B. J. (2002) J. Am. Med. Assoc. 287, 1308–1320 [Google Scholar]
- 4.Towbin J. A., Bowles N. E. (2002) Nature 415, 227–233 [DOI] [PubMed] [Google Scholar]
- 5.Liew C. C., Dzau V. J. (2004) Nat. Rev. Genet. 5, 811–825 [DOI] [PubMed] [Google Scholar]
- 6.Van Driest S. L., Ellsworth E. G., Ommen S. R., Tajik A. J., Gersh B. J., Ackerman M. J. (2003) Circulation 108, 445–451 [DOI] [PubMed] [Google Scholar]
- 7.Alcalai R., Seidman J. G., Seidman C. E. (2008) J. Cardiovasc. Electrophysiol. 19, 104–110 [DOI] [PubMed] [Google Scholar]
- 8.Gomes A. V., Potter J. D. (2004) Ann. N.Y. Acad. Sci. 1015, 214–224 [DOI] [PubMed] [Google Scholar]
- 9.Landstrom A. P., Parvatiyar M. S., Pinto J. R., Marquardt M. L., Bos J. M., Tester D. J., Ommen S. R., Potter J. D., Ackerman M. J. (2008) J. Mol. Cell. Cardiol. 45, 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann B., Schmidt-Traub H., Perrot A., Osterziel K. J., Gessner R. (2001) Hum. Mutat. 17, 524. [DOI] [PubMed] [Google Scholar]
- 11.Schmidtmann A., Lindow C., Villard S., Heuser A., Mügge A., Gessner R., Granier C., Jaquet K. (2005) FEBS J. 272, 6087–6097 [DOI] [PubMed] [Google Scholar]
- 12.Liang B., Chung F., Qu Y., Pavlov D., Gillis T. E., Tikunova S. B., Davis J. P., Tibbits G. F. (2008) Physiol. Genomics 33, 257–266 [DOI] [PubMed] [Google Scholar]
- 13.Gordon A. M., Homsher E., Regnier M. (2000) Physiol. Rev. 80, 853–924 [DOI] [PubMed] [Google Scholar]
- 14.Farah C. S., Reinach F. C. (1995) FASEB J. 9, 755–767 [DOI] [PubMed] [Google Scholar]
- 15.Potter J. D., Sheng Z., Pan B. S., Zhao J. (1995) J. Biol. Chem. 270, 2557–2562 [DOI] [PubMed] [Google Scholar]
- 16.Gomes A. V., Potter J. D. (2004) Mol. Cell. Biochem. 263, 99–114 [DOI] [PubMed] [Google Scholar]
- 17.Bers D. M. (2001) in Excitation-Contraction Coupling and Cardiac Contractile Force, 2nd Ed., pp. 39–56, Kluwer Academic Publishers, London [Google Scholar]
- 18.Holroyde M. J., Robertson S. P., Johnson J. D., Solaro R. J., Potter J. D. (1980) J. Biol. Chem. 255, 11688–11693 [PubMed] [Google Scholar]
- 19.Gordon A. M., Regnier M., Homsher E. (2001) News Physiol. Sci. 16, 49–55 [PubMed] [Google Scholar]
- 20.Vibert P., Craig R., Lehman W. (1997) J. Mol. Biol. 266, 8–14 [DOI] [PubMed] [Google Scholar]
- 21.Takeda S., Yamashita A., Maeda K., Maéda Y. (2003) Nature 424, 35–41 [DOI] [PubMed] [Google Scholar]
- 22.Lindhout D. A., Boyko R. F., Corson D. C., Li M. X., Sykes B. D. (2005) Biochemistry 44, 14750–14759 [DOI] [PubMed] [Google Scholar]
- 23.Hoffman R. M., Blumenschein T. M., Sykes B. D. (2006) J. Mol. Biol. 361, 625–633 [DOI] [PubMed] [Google Scholar]
- 24.Murakami K., Yumoto F., Ohki S. Y., Yasunaga T., Tanokura M., Wakabayashi T. (2005) J. Mol. Biol. 352, 178–201 [DOI] [PubMed] [Google Scholar]
- 25.Herzberg O., James M. N. (1985) Nature 313, 653–659 [DOI] [PubMed] [Google Scholar]
- 26.Moews P. C., Kretsinger R. H. (1975) J. Mol. Biol. 91, 201–225 [DOI] [PubMed] [Google Scholar]
- 27.Putkey J. A., Liu W., Lin X., Ahmed S., Zhang M., Potter J. D., Kerrick W. G. (1997) Biochemistry 36, 970–978 [DOI] [PubMed] [Google Scholar]
- 28.Johnson J. D., Collins J. H., Robertson S. P., Potter J. D. (1980) J. Biol. Chem. 255, 9635–9640 [PubMed] [Google Scholar]
- 29.Solaro R. J., Bousquet P., Johnson J. D. (1986) J. Pharmacol. Exp. Ther. 238, 502–507 [PubMed] [Google Scholar]
- 30.el-Saleh S. C., Solaro R. J. (1987) J. Biol. Chem. 262, 17240–17246 [PubMed] [Google Scholar]
- 31.Chandra M., da Silva E. F., Sorenson M. M., Ferro J. A., Pearlstone J. R., Nash B. E., Borgford T., Kay C. M., Smillie L. B. (1994) J. Biol. Chem. 269, 14988–14994 [PubMed] [Google Scholar]
- 32.Smith L., Greenfield N. J., Hitchcock-DeGregori S. E. (1994) J. Biol. Chem. 269, 9857–9863 [PubMed] [Google Scholar]
- 33.Zhang R., Zhao J., Potter J. D. (1995) J. Biol. Chem. 270, 30773–30780 [DOI] [PubMed] [Google Scholar]
- 34.Szczesna D., Zhang R., Zhao J., Jones M., Guzman G., Potter J. D. (2000) J. Biol. Chem. 275, 624–630 [DOI] [PubMed] [Google Scholar]
- 35.Pinto J. R., Parvatiyar M. S., Jones M. A., Liang J., Potter J. D. (2008) J. Biol. Chem. 283, 2156–2166 [DOI] [PubMed] [Google Scholar]
- 36.Gomes A. V., Guzman G., Zhao J., Potter J. D. (2002) J. Biol. Chem. 277, 35341–35349 [DOI] [PubMed] [Google Scholar]
- 37.Fiske C. H., Subbarow Y. (1925) J. Biol. Chem. 66, 375–400 [Google Scholar]
- 38.Dweck D., Reyes-Alfonso A., Jr., Potter J. D. (2005) Anal. Biochem. 347, 303–315 [DOI] [PubMed] [Google Scholar]
- 39.Dweck D., Hus N., Potter J. D. (2008) J. Biol. Chem. 283, 33119–33128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szczesna D., Ghosh D., Li Q., Gomes A. V., Guzman G., Arana C., Zhi G., Stull J. T., Potter J. D. (2001) J. Biol. Chem. 276, 7086–7092 [DOI] [PubMed] [Google Scholar]
- 41.Zot A. S., Potter J. D. (1989) Biochemistry 28, 6751–6756 [DOI] [PubMed] [Google Scholar]
- 42.Güth K., Potter J. D. (1987) J. Biol. Chem. 262, 13627–13635 [PubMed] [Google Scholar]
- 43.Pinto J. R., Veltri T., Sorenson M. M. (2008) Pflugers Arch. 456, 1177–1187 [DOI] [PubMed] [Google Scholar]
- 44.Wen Y., Pinto J. R., Gomes A. V., Xu Y., Wang Y., Wang Y., Potter J. D., Kerrick W. G. (2008) J. Biol. Chem. 283, 20484–20494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baudenbacher F., Schober T., Pinto J. R., Sidorov V. Y., Hilliard F., Solaro R. J., Potter J. D., Knollmann B. C. (2008) J. Clin. Investig. 118, 3893–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heineke J., Molkentin J. D. (2006) Nat. Rev. Mol. Cell Biol. 7, 589–600 [DOI] [PubMed] [Google Scholar]
- 47.Robinson P., Griffiths P. J., Watkins H., Redwood C. S. (2007) Circ. Res. 101, 1266–1273 [DOI] [PubMed] [Google Scholar]
- 48.Negele J. C., Dotson D. G., Liu W., Sweeney H. L., Putkey J. A. (1992) J. Biol. Chem. 267, 825–831 [PubMed] [Google Scholar]
- 49.Leavis P. C., Kraft E. L. (1978) Arch. Biochem. Biophys. 186, 411–415 [DOI] [PubMed] [Google Scholar]
- 50.Johnson J. D., Potter J. D. (1978) J. Biol. Chem. 253, 3775–3777 [PubMed] [Google Scholar]
- 51.Smith L., Greenfield N. J., Hitchcock-DeGregori S. E. (1999) Biophys. J. 76, 400–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zot H. G., Potter J. D. (1982) J. Biol. Chem. 257, 7678–7683 [PubMed] [Google Scholar]
- 53.Brito R. M., Krudy G. A., Negele J. C., Putkey J. A., Rosevear P. R. (1993) J. Biol. Chem. 268, 20966–20973 [PubMed] [Google Scholar]
- 54.Brito R. M., Putkey J. A., Strynadka N. C., James M. N., Rosevear P. R. (1991) Biochemistry 30, 10236–10245 [DOI] [PubMed] [Google Scholar]
- 55.Satyshur K. A., Rao S. T., Pyzalska D., Drendel W., Greaser M., Sundaralingam M. (1988) J. Biol. Chem. 263, 1628–1647 [PubMed] [Google Scholar]
- 56.Strynadka N. C., Cherney M., Sielecki A. R., Li M. X., Smillie L. B., James M. N. (1997) J. Mol. Biol. 273, 238–255 [DOI] [PubMed] [Google Scholar]
- 57.Vassylyev D. G., Takeda S., Wakatsuki S., Maeda K., Maéda Y. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4847–4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fredricksen R. S., Swenson C. A. (1996) Biochemistry 35, 14012–14026 [DOI] [PubMed] [Google Scholar]
- 59.Finley N. L., Rosevear P. R. (2004) J. Biol. Chem. 279, 54833–54840 [DOI] [PubMed] [Google Scholar]
- 60.Blumenschein T. M., Tripet B. P., Hodges R. S., Sykes B. D. (2001) J. Biol. Chem. 276, 36606–36612 [DOI] [PubMed] [Google Scholar]
- 61.Biesiadecki B. J., Kobayashi T., Walker J. S., John Solaro R., de Tombe P. P. (2007) Circ. Res. 100, 1486–1493 [DOI] [PubMed] [Google Scholar]
- 62.Szczesna D., Guzman G., Miller T., Zhao J., Farokhi K., Ellemberger H., Potter J. D. (1996) J. Biol. Chem. 271, 8381–8386 [DOI] [PubMed] [Google Scholar]
- 63.Finley N. L., Howarth J. W., Rosevear P. R. (2004) Biochemistry 43, 11371–11379 [DOI] [PubMed] [Google Scholar]
- 64.Strynadka N. C., James M. N. (1989) Annu. Rev. Biochem. 58, 951–998 [DOI] [PubMed] [Google Scholar]
- 65.Krudy G. A., Brito R. M., Putkey J. A., Rosevear P. R. (1992) Biochemistry 31, 1595–1602 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.