Abstract
The cystic fibrosis transmembrane conductance regulator (CFTR), a member of the ABC transporter superfamily, is a cyclic AMP-regulated chloride channel and a regulator of other ion channels and transporters. In epithelial cells CFTR is rapidly endocytosed from the apical plasma membrane and efficiently recycles back to the plasma membrane. Because ubiquitination targets endocytosed CFTR for degradation in the lysosome, deubiquitinating enzymes (DUBs) are likely to facilitate CFTR recycling. Accordingly, the aim of this study was to identify DUBs that regulate the post-endocytic sorting of CFTR. Using an activity-based chemical screen to identify active DUBs in human airway epithelial cells, we demonstrated that Ubiquitin Specific Protease-10 (USP10) is located in early endosomes and regulates the deubiquitination of CFTR and its trafficking in the post-endocytic compartment. small interference RNA-mediated knockdown of USP10 increased the amount of ubiquitinated CFTR and its degradation in lysosomes, and reduced both apical membrane CFTR and CFTR-mediated chloride secretion. Moreover, a dominant negative USP10 (USP10-C424A) increased the amount of ubiquitinated CFTR and its degradation, whereas overexpression of wt-USP10 decreased the amount of ubiquitinated CFTR and increased the abundance of CFTR. These studies demonstrate a novel function for USP10 in facilitating the deubiquitination of CFTR in early endosomes and thereby enhancing the endocytic recycling of CFTR.
The endocytosis, endocytic recycling, and endosomal sorting of numerous transport proteins and receptors are regulated by ubiquitination (1–6). Ubiquitin, an 8-kDa protein, is conjugated to target proteins via a series of steps that includes ubiquitin-activating enzymes (E1),2 ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3) (1). Proteins that are ubiquitinated in the plasma membrane are internalized and are either deubiquitinated and recycle back to the plasma membrane or, via interactions with the endosomal sorting complexes required for transport machinery, are delivered to the lysosome for degradation (1–7). Sorting of ubiquitinated plasma membrane proteins for either the lysosomal pathway or for the recycling pathway is regulated, in part, by the removal of ubiquitin by deubiquitinating enzymes (DUBs) (1–6). Thus, the balance between ubiquitination and deubiquitination regulates the plasma membrane abundance of several membrane proteins, including the epithelial sodium channel (ENaC), the epidermal growth factor receptor, the transforming growth factor-β receptor, and the cytokine receptor γ-c (8–14).
CFTR is rapidly endocytosed from the plasma membrane and undergoes rapid and efficient recycling back to the plasma membrane in human airway epithelial cells, with >75% of endocytosed wild-type CFTR recycling back to the plasma membrane (15–18). A study published several years ago demonstrated that, although ubiquitination did not regulate CFTR endocytosis, ubiquitination reduced the plasma membrane abundance of CFTR in BHK cells by redirecting CFTR from recycling endosomes to lysosomes for degradation (19). However, neither the E3 ubiquitin ligase(s) responsible for the ubiquitination of CFTR nor the DUB(s) responsible for the deubiquitination of CFTR in the endocytic pathway have been identified in any cell type. Moreover, the effect of the ubiquitin status of CFTR on its endocytic sorting in human airway epithelial cells has not been reported. Thus, the goals of this study were to determine if the ubiquitin status regulates the post-endocytic sorting of CFTR in polarized airway epithelial cells, and to identify the DUBs that deubiquitinate CFTR.
Approximately 100 DUBs have been identified in the human genome and are classified into five families based on sequence similarity and mechanism of action (1–6, 20, 21). To identify DUBs that regulate the deubiquitination of CFTR from this large class of enzymes, we chose an activity-based, chemical probe screening approach developed by Dr. Hidde Ploegh (4, 21, 22). This approach utilizes a hemagglutinin (HA)-tagged ubiquitin probe engineered with a C-terminal modification incorporating a thiol-reactive group that forms an irreversible, covalent bond with active DUBs. Using this approach we demonstrated in polarized human airway epithelial cells that ubiquitin-specific protease-10 (USP10) is located in early endosomes and regulates the deubiquitination of CFTR and thus its trafficking in the post-endocytic compartment. These studies demonstrate a novel function for USP10 in promoting the deubiquitination of CFTR in early endosomes and thereby enhancing the endocytic recycling of CFTR.
EXPERIMENTAL PROCEDURES
Cell Culture
The role of DUBs in the intracellular trafficking of CFTR was studied in human airway epithelial cells (CFBE41o− cells, homozygous for the ΔF508 mutation) stably expressing wt-CFTR. Details on the stable transfection and characterization of CFBE41o− cells expressing wt-CFTR (hereafter called CFBE cells) have been described in detail by several laboratories (17, 18, 23). CFBE cells between passages 18 and 27 were maintained in minimal essential medium supplemented with 50 μg/ml penicillin, 50 μg/ml streptomycin, 2 mm l-glutamine, 10% fetal bovine serum, 2 μg/ml puromycin, and 5 μg/ml plasmocin in a 5% CO2-95% air incubator at 37 °C. To establish confluent, polarized monolayers, 1 × 106 cells were seeded onto 24-mm Transwell permeable supports (0.4-μm pore size, Corning, Corning, NY) coated with Vitrogen plating medium containing human fibronectin (10 μg/ml, Collaborative Biomedical Products, Bedford, MA), Vitrogen 100 (1%, Collagen, Palo Alto, CA), and bovine serum albumin (10 μg/ml, Sigma-Aldrich) and grown in an air-liquid interface culture at 37 °C for 6–9 days, as described (17).
Identification of Active DUBs
To identify active DUBs in CFBE cells we used a chemical probe screening approach described in detail by Dr. Hidde Ploegh (4, 21, 22). The chemical structure of the probe utilized in this study is presented in Fig. 1 by Borodovsky et al. (22). Briefly, cells were lysed in radioimmunoprecipitation assay buffer (25 mm Tris-HCl, pH 7.6, 10 mm NaCl, 1% Nonidet P-40 (IGEPAL), 1% sodium deoxycholate, 0.1% SDS), and 0.1 μg of the HA-UbVME probe was added to 20 μg of the post-nuclear supernatant obtained by low speed (10,000 × g) centrifugation of cell lysates or to early endosomal fractions (isolated as described below) isolated from CFBE cells. The HA-UbVME probe forms an irreversible, covalent bond with active DUBs. Identification of DUBs covalently linked to the HA-UbVME probe was achieved by immunoprecipitation of the HA-UbVME·DUB complex(s) using an anti-HA monoclonal antibody (Santa Cruz Biotechnology) followed by SDS-PAGE and Western blot analysis using specific anti-DUB antibodies (see “Antibodies and Reagents”). The specificity of the HA-UbVME probe for active DUBs was confirmed with the addition of N-ethylmaleimide (10 μm), which inhibits cysteine protease DUBs, during the labeling reaction (4, 21, 22).
FIGURE 1.
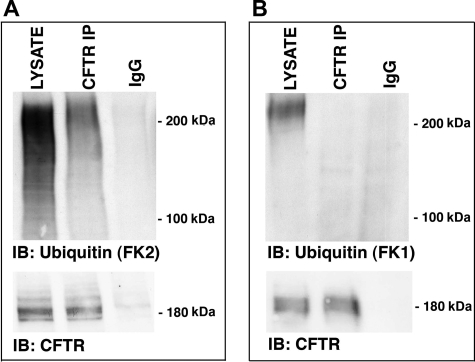
Ubiquitinated CFTR is present in early endosomes. CFBE cells were lysed, early endosomes were purified, CFTR was immunoprecipitated using a monoclonal CFTR antibody (clone M3A7, Upstate Biotechnology), and ubiquitinated CFTR was detected via Western blot analysis (IB) using either an anti-ubiquitin antibody that recognizes: mono-, multi-, and polyubiquitin additions (FK2) (A) or polyubiquitin additions (FK1) (B). LYSATE, early endosomal lysate; CFTR IP, immunoprecipitated CFTR; IgG, immunoprecipitation using a non-immune IgG antibody. In the bottom panels of A and B, immunoprecipitated CFTR was blotted with a CFTR monoclonal antibody (clone 24-1, R&D Systems). Similar amounts of CFTR were immunoprecipitated in all cases: thus, differences in the amount of polyubiquitinated CFTR detected was not due to differences in the efficiency of the immunoprecipitation step for CFTR. Gels were cut between the stacking and running gel interface. Experiments performed three times. Representative blots are shown.
Isolation of Early Endosomes
To determine if USP10 is expressed in early endosomes, differential centrifugation and fractionation techniques were used to isolate early endosomes from CFBE cells using a protocol adapted from Butterworth et al. (9). Briefly, polarized CFBE cells, grown on 24-mm permeable membrane supports, were scraped into phosphate-buffered saline, pelleted, and resuspended in 600 μl of HEPES buffer (250 mm sucrose, 10 mm HEPES, 0.5 mm EDTA at pH 7.4 containing protease inhibitors (Roche Applied Science)). The cells were homogenized with a Dounce homogenizer and passed through a 22-gauge needle 20 times. Following a low speed spin (3000 × g), the post-nuclear supernatant was diluted 1:1 with 62% sucrose in HEPES buffer and placed at the bottom of a 4.4-ml ultracentrifuge tube (Sorvall, Ashville, NC). 1.5 ml of 35% sucrose in HEPES buffer was layered on top followed by 1.5 ml of 25% sucrose in HEPES buffer and 0.5 ml of HEPES buffer. The gradients were centrifuged in a TH-660 rotor at 167,000 × g for 75 min at 4 °C, and the interfaces were collected to isolate the early endosomal fractions. Western blot analysis for various Rab GTPases was used to confirm purity of the early endosomal fraction. Rab5a and early endosome antigen-1 served as markers for the early endosomal fraction, whereas LAMP-1 and actin served as negative controls.
Ubiquitination Assay
To assess the amount of ubiquitinated CFTR in CFBE cells, a protocol was adapted from Urbe et al. (24). Briefly, polarized CFBE cells were lysed in boiling lysis buffer (2% SDS, 1 mm EDTA, 50 mm sodium fluoride, and Complete Protease Inhibitor Mixture (Roche Applied Science)) preheated to 100 °C. The lysates were transferred to screw-cap tubes, incubated for 10 min at 100 °C, and cooled to room temperature, and the lysates were diluted by the addition of four volumes of the dilution buffer (2.5% Triton X-100, 12.5 mm Tris, pH 7.5, 187.5 mm NaCl, and Complete Protease Inhibitor Mixture (Roche Applied Science)). After pelleting cell debris by low speed centrifugation (3000 × g), the lysates were immunoprecipitated overnight at 4 °C with 5 μg of anti-CFTR antibody (clone M3A7, Upstate Biotechnology) complexed with Protein G-agarose. Immunoprecipitated complexes were washed three times with dilution buffer (2% Triton X-100, 0.4% SDS, 10 mm Tris, pH 7.5, 150 mm NaCl), once with a high salt wash buffer (200 mm NaCl, 400 mm NaOAc), and once more with the dilution buffer before preparation for SDS-PAGE and Western blot analysis using a ubiquitin antibody that recognizes mono-, multi-, and polyubiquitin additions (FK2 ubiquitin clone, Biomol) or a ubiquitin antibody that recognizes only polyubiquitin additions (FK1 ubiquitin clone, Biomol). The quantitation of ubiquitinated CFTR was calculated as the signal obtained with the ubiquitin antibody normalized for immunoprecipitated CFTR detected with the CFTR antibody (clone 24-1, R&D Systems).
Immunoprecipitation
To determine if CFTR interacts with USP10 in the early endosomal fractions, USP10 was immunoprecipitated from early endosomal fractions isolated from the CFBE cell lysate by methods described previously in detail (18). Briefly, CFBE cells were lysed in immunoprecipitation buffer containing 150 mm NaCl, 50 mm Tris (pH 7.2), 0.1% IGEPAL (Sigma), 5 mm MgCl2, 5 mm EDTA, 1 mm EGTA, 30 mm NaF, 1 mm Na3VO4, and Complete Protease Inhibitor Mixture (Roche Applied Science), and USP10 was immunoprecipitated by incubation with 5 μg of a polyclonal USP10 antibody (Bethyl Laboratories) and protein A-agarose complex. Immunoprecipitated proteins were eluted from the protein A-agarose complexes by incubation at 100 °C for 3 min in Laemmli sample buffer (Bio-Rad) containing 80 mm dithiothreitol. Immunoprecipitated proteins were separated by SDS-PAGE using 15% gels (Bio-Rad) and analyzed by Western blot analysis.
Biochemical Determination of the Apical Membrane CFTR
The biochemical determination of apical membrane CFTR was performed by domain selective cell surface biotinylation using EZ-LinkTM Sulfo-NHS-LC-Biotin (Pierce), as described previously in detail (25, 26).
RNA Isolation and Reverse Transcription-PCR
Reverse transcription (RT)-PCR studies were conducted to examine the endogenous expression of USP10 in CFBE cells, as previously described in detail (18, 27). For USP10, triplicate reactions of each cDNA sample were incubated at 95° C for 10 min, followed by 35 cycles of 15 s at 95° C and 1 min at 60° C using TaqMan Gene Expression Assay primers (Applied Biosystems) for human USP10. RT-PCR products were run on an low melting point agarose gel to confirm product size, subcloned into pCR4-TOPO (Invitrogen), and submitted for sequence analysis to confirm the identity of the products.
Q-RT-PCR
Q-RT-PCR studies were conducted to examine the effect of siUSP10 on USP10 mRNA expression using a protocol published previously in detail (18). Predesigned TaqMan Gene Expression Assay Q-RT-PCR primers (Applied Biosystems) for human USP10 were employed. The cDNA generated during RT was quantified (NanoDrop, NanoDrop Technologies), and data were expressed as percent change in USP10 mRNA expression.
Plasmids and Transient Transfections
Plasmids containing GFP-wt-USP10 and GFP-USP10 (C424A) were a generous gift from Dr. Susanna Chiocca (European Institute of Oncology, IFOM-IEO campus (28)). All constructs were sequence verified upon receipt by ABI PRISM dye terminator cycle sequencing (Applied Biosystems, Foster City, CA). Transient transfections of CFBE cells with GFP-wt-USP10 and USP10-C424A were conducted using Effectene (Qiagen, Valencia, CA) according to the manufacturer's instructions.
RNA-mediated Interference
USP10 expression was selectively reduced using siRNA for USP10 purchased from Qiagen, by methods described previously (18, 29). In brief, CFBE cells were seeded at 0.1 × 106 on 24-mm Transwell permeable membrane supports and cultured for 3 days. On day 4, post-seeding, cells were transfected with either 5, 15, or 50 nm siRNA for USP10 or a scrambled, control siRNA with HiPerfect transfection reagent according to the manufacturer's protocol (Qiagen). Sequences for siRNAs are: siUSP10 sense (5′-CACAGCUUCUGUUGACUCUTT-3′) and antisense (5′-AGAGUCAACAGAAGCUGUGTT-3′), and siNegative scrambled sense (5′-UUCUCCGAACGUGUCACGU-3′) and antisense (5′-ACGUGACACGUUCGGAGAA-3′). Cells were studied on day 8 post-seeding (i.e. 4 days after transfection with siRNA).
Confocal Microscopy
Co-localization studies were conducted to confirm Western blot studies demonstrating that endogenous USP10 is expressed in early endosomes. Briefly, CFBE cells seeded at 0.1 × 106 on collagen-coated, glass-bottom Mat-Tek dishes, were infected 24 h after seeding with a baculovirus expressing a eGFP-Rab5a-eGFP plasmid (Organelle LightsTM Endosomes-GFP, Molecular Probes, Invitrogen), according to the manufacturer's instructions, and fixed for immunolabeling 96 h post-infection, as described previously (30). USP10 was visualized by indirect immunofluorescence using a polyclonal antibody for USP10 (Bethyl Laboratories, Montgomery, TX) followed by an Alexa 586-labeled secondary antibody. Z-stack images (0.4-μm sections) of labeled cells were acquired with a Nikon Sweptfield confocal microscope (Apo TIRF 100× oil immersion 1.49 numerical aperture objective) fitted with a QuantEM:512sc camera (Photometrics, Tucson, AZ) and Elements 2.2 software (Nikon, Inc.) to reconstruct and render three-dimensional images. Experiments were repeated three times, with five fields imaged for each experiment.
Ussing Chamber Measurements
Ussing chamber measurements of CFTR-mediated chloride secretion were performed as described previously (31).
Endocytosis and Recycling Assays
Endocytic and recycling assays were performed in CFBE cells as described previously (17, 18, 31). For both assays, the plasma membrane proteins were first biotinylated at 4 °C using EZ-LinkTM Sulfo-NHS-SS-Biotin (Pierce). For the endocytic assay, cells were warmed to 37 °C for 5 min after biotinylation, and GSH was applied at 4 °C to reduce the disulfide bonds between proteins labeled with Sulfo-NHS-SS-Biotin in the plasma membrane. Biotinylated proteins that were endocytosed during the 5-min period at 37 °C are not reduced by GSH, which is impermeant to the plasma membrane, and thus reside in the endosomal compartment. Cells were lysed, and biotinylated proteins were isolated using streptavidin-agarose beads, eluted into SDS sample buffer, and separated by 7.5% SDS-PAGE. For recycling assays, cells were warmed to 37 °C for 5 min after biotinylation to load endocytic vesicles with biotinylated proteins. Cells were then cooled immediately to 4 °C, and the disulfide bonds on Sulfo-NHS-SS-Biotin-labeled proteins remaining in the plasma membrane were reduced by GSH at 4 °C. Subsequently, cells were either lysed or warmed again to 37 °C for 5 min (to allow endocytosed and biotinylated CFTR to recycle to the plasma membrane). Cells were then cooled again to 4 °C, and the disulfide bonds on the proteins biotinylated with Sulfo-NHS-SS-Biotin remaining in the plasma membrane were reduced with GSH. The recycling of endocytosed CFTR was calculated as the difference between the amount of biotinylated CFTR after the first and second GSH treatments.
Antibodies and Reagents
The antibodies used were: mouse anti-human CFTR C terminus antibody (clone 24-1, R&D systems, Minneapolis, MN); mouse anti-CFTR antibody (clone M3A7, Upstate Biotechnology, Lake Placid, NY); mouse anti-EEA1 antibody, mouse anti-ezrin antibody, mouse anti-Rab5 antibody, mouse anti-LAMP-1 antibody, mouse anti-actin antibody, mouse anti-GFP antibody (BD Biosciences, San Jose, CA); mouse anti-HA antibody (Santa Cruz Biotechnology, Santa Cruz, CA); mouse anti-ubiquitin (clones FK2 and FK1) antibodies (Biomol, Plymouth Meeting, PA); rabbit anti-USP10 antibody, rabbit anti-USP34 antibody, and rabbit anti-USP8 (Bethyl Laboratories, Montgomery, TX); and horseradish peroxidase-conjugated goat anti-mouse and goat anti-rabbit secondary antibodies (Bio-Rad). All antibodies and reagents were used at the concentrations recommended by the manufacturers or as indicated in the figure legends.
Data Analysis and Statistics
Statistical analysis of the data was performed using Prism version 4.0a for Macintosh (GraphPad, San Diego, CA). Means were compared using a t test or analysis of variance followed by a Tukey test, as appropriate. p < 0.05 was considered significant. Data are expressed as the mean ± S.E.
RESULTS
Ubiquitinated CFTR Is Located in Early Endosomes
If ubiquitination regulates CFTR sorting in the endocytic pathway in human airway epithelial cells, we reasoned that CFTR in early endosomes should be ubiquitinated. Thus, early endosomes were isolated from polarized human bronchial epithelial cells (CFBEs), CFTR was immunoprecipitated and probed using an antibody that detects mono-, multi-, and polyubiquitinated proteins (FK2). SDS-PAGE followed by Western blotting with the FK2 antibody revealed that ubiquitinated CFTR is present in early endosomes (Fig. 1A). However, an antibody specific for polyubiquitinated proteins (FK1) did not detect polyubiquitinated CFTR in early endosomes (Fig. 1B). Thus, CFTR in early endosomes is mono- and/or multiubiquitinated. Because most of the ubiquitinated CFTR was ∼200 kDa (Fig. 1A), and CFTR detected by Western blot was ∼180 kDa (Fig. 1A), and a single ubiquitin has a molecular mass of 8 kDa, we infer that most of the ubiquitinated CFTR in early endosomes is multiubiquitinated, a signal that targets proteins in early endosomes for lysosomal degradation (32–35).
The DUB USP10 Is Active in Airway Epithelial Cells
To identify active DUBs in polarized airway epithelial cells we used a chemical probe screening approach developed by Dr. Hidde Ploegh (4, 21, 22). CFBE cells were lysed, and an HA-tagged ubiquitin-vinyl methyl ester probe (HA-UbVME) was added to the lysates. The HA-UbVME probe forms an irreversible, covalent bond with active DUBs. Subsequently one or more HA-UbVME·DUB complexes were immunoprecipitated with an anti-HA antibody, and one or more immunoprecipitated complexes were analyzed by SDS-PAGE followed by Western blotting using an anti-HA antibody (Fig. 2A). Using this chemical approach we identified an active DUB with a molecular mass of ∼110 kDa. N-Ethylmaleimide, which inhibits cysteine proteases (i.e. DUBs), significantly reduced the interaction between HA-UbVME and the 110-kDa DUB (Fig. 2A).
FIGURE 2.
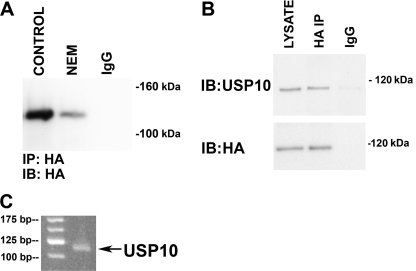
USP10 is expressed and active in airway epithelial cells. CFBE cells were lysed and incubated with the HA-UbVME probe in the presence and absence of N-ethylmaleimide (10 μm) to identify active DUBs. N-Ethylmaleimide (NEM) inhibits cysteine proteases and therefore eliminates the covalent linkage between DUBs and the HA-UbVME probe. The HA-UbVME·DUB complex was immunoprecipitated with an anti-HA antibody, and the immunoprecipitated complex was analyzed by SDS-PAGE followed by Western blot analysis (IB) using an anti-HA antibody (A) or an anti-USP10 antibody (B). A, the anti-HA antibody identified an N-ethylmaleimide-sensitive band at a molecular mass of 110 kDa. IgG, immunoprecipitation using a non-immune IgG was used as a negative control. B, the anti-USP10 antibody detected a 110-kDa protein in cell lysates (LYSATE) and in the lane containing the immunoprecipitated HA-UbVME·DUB. The molecular mass of HA-UbVME is ∼9 kDa, thus USP10 detected in the center lane (i.e. HA-UbVME-USP10) runs ∼9 kDa larger than the USP10 in the lane on the left (i.e. most of the USP10 detected in cell lysates is most likely not covalently attached to HA-UbVME). A non-immune IgG was used as a negative control. C, RT-PCR detection of USP10 mRNA expression in polarized CFBE cells. Experiments were performed three times. Representative blots or gels are shown.
A search of the literature revealed that USP10 has a molecular mass of 110 kDa (28). Thus, to determine if the 110-kDa protein is USP10, the HA-UbVME·DUB complex was immunoprecipitated with an anti-HA antibody, and the immunoprecipitated complex was analyzed by SDS-PAGE followed by Western blotting using an anti-USP10 antibody (Fig. 2B). The 110-kDa complex was recognized by the USP10 antibody. In addition, RT-PCR and sequencing of the amplicon verified mRNA expression of USP10 in CFBE cells (Fig. 2C). Taken together, these studies identify USP10 as an active DUB in human airway epithelial cells. Additional studies, described below, were conducted to determine if USP10 is expressed and active in early endosomes and regulates the deubiquitination of CFTR.
USP10 Is Located in Early Endosomes
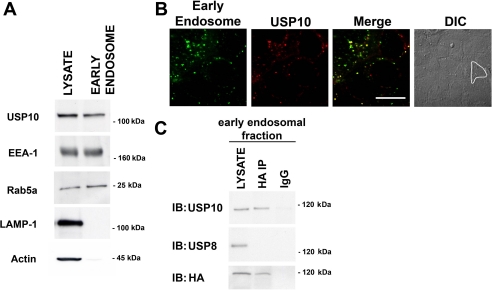
The presence of USP10 in early endosomes was established by two independent methods: biochemical and confocal microscopy. First, early endosomes were isolated as described under “Experimental Procedures,” and USP10 was identified in the early endosomal fraction by SDS-PAGE followed by Western blot analysis (Fig. 3A). The isolation of early endosomes was confirmed by the presence of early endosome antigen-1 and Rab5a (Fig. 3A), and by the absence of actin and lysosome-associated protein-1 (LAMP-1), which served as negative controls (Fig. 3A). In addition, immunofluorescence microscopy confirmed the expression of USP10 in early endosomes (Fig. 3B). The co-localization between USP10 and eGFP-labeled Rab5a, an early endosomal protein, was quantified by intensity correlation analysis using Nikon Elements Software. Pearson's correlation and Mander's overlap coefficients, measured on images obtained on a confocal microscope, confirmed a high degree of co-localization of USP10 with early endosomes. Pearson's correlation coefficient is a number between −1 and +1 that measures the degree of pattern similarity between two fluorochromes (+1 indicates a complete positive correlation and −1 for a negative correlation, with 0 indicating no correlation). A value of +0.654 ± 0.021 indicates a strong positive correlation between USP10 (red channel) and Rab5a (green channel). Mander's overlap coefficient ranges from 0 to 1, with 0 indicating low co-localization and 1 indicating high co-localization. Mander's coefficient values are independent of the pixel intensities within respective channels. A Mander's overlap coefficient of +0.862 ± 0.019 indicates a high level of overlap between USP10 (red channel) and Rab5a (green channel). Most of the USP10 was localized to early endosomes, although a very small amount also appeared to be localized to other subcellular compartments. Taken together, our biochemical and immunolocalization studies demonstrate that USP10 is localized primarily to early endosomes in human airway epithelial cells.
FIGURE 3.
USP10 is active and expressed in early endosomes of airway epithelial cells. A, early endosomes were isolated from CFBE cells using a sucrose gradient. The isolation and purification of early endosomes was confirmed by Western blot analysis. The isolated fractions contained the early endosomal antigen-1 (EEA-1) and Rab5a, which are located in early endosomes, but did not contain LAMP-1, a lysosomal protein, or actin, a cytoplasmic protein. The early endosome fraction was positive for USP10. Lysates (LYSATE), which contain cytoplasm, lysosomes, and early endosomes, were positive for USP10, early endosome antigen-1, Rab5, LAMP-1, and actin. Rab11a and Rab7 protein were not detected in the early endosomal fraction (data not shown). B, representative confocal images of CFBE cells infected with a baculovirus Rab5a-eGFP construct, which is expressed in early endosomes (green). USP10 was immunolocalized using an anti-USP10 antibody and an Alexa-568 secondary antibody (red). DIC, differential interference contrast image of cells imaged for Rab5a and USP10. A single cell is outlined in white in the polarized monolayer of airway cells. Infection with the baculovirus expressing the empty vector had no effect on cellular morphology, and fluorescence in the green channel in cells infected with this baculovirus was similar to background (data not shown). Scale bar, equals 10 μm. C, early endosomes were isolated and incubated with the HA-UbVME probe to identify active DUBs. The HA-UbVME·DUB complex was immunoprecipitated with an anti-HA antibody, and the immunoprecipitated complex was analyzed by SDS-PAGE followed by Western blot analysis using an anti-USP10 antibody, an anti-USP8 antibody or an anti-HA antibody (lane labeled HA-IP). The lane labeled LYSATE represent lysates of early endosomes blotted with the anti-USP10, the anti-USP8, or the anti-HA antibody. Although USP10 and USP8 were expressed in early endosomes, only USP10 was active as determined by the chemical probe technique. The non-immune IgG did not immunoprecipitate USP10-, USP8-, or HA-labeled complexes, and thus served as a negative control. Experiments were performed three times. Representative blots are shown.
USP10 Is Active in Early Endosomes
To determine if USP10 is active in early endosomes we used the chemical probe screening approach described above. Early endosomes were isolated as described above, the HA-UbVME probe was added to early endosomes, the HA-UbVME·DUB complex was immunoprecipitated with an anti-HA antibody, and Western blots of the immunoprecipitated complex were blotted with an anti-USP10 antibody. The USP10 antibody detected a 110-kDa protein in the early endosomal fraction, demonstrating that USP10 is active in early endosomes (Fig. 3C). Although USP8 (also known as UBPY) was also identified in lysates of early endosomes, USP8 was not active as determined by the chemical probe assay and Western blotting (Fig. 3C). The possibility exists that lysis of airway epithelial cells releases an inhibitor of USP10 activity, resulting in the activation of USP10 activity, as determined using the chemical probe assay. However, experiments described later in this manuscript using siRNA knockdown and mutant USP10 constructs demonstrate that USP10 is active in intact cells.
USP10 Interacts with CFTR in Early Endosomes
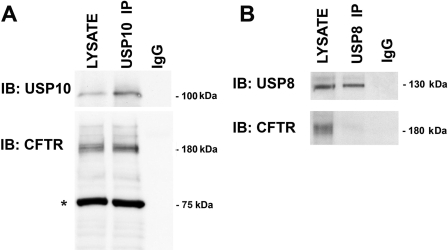
The ability of USP10 to deubiquitinate proteins has been shown to require interaction between USP10 and its substrate (8, 28). Thus, if USP10 regulates the deubiquitination of CFTR, we predict that USP10 will interact with CFTR in CFBE cells. Accordingly, USP10 was immunoprecipitated from the early endosomal fraction isolated from CFBE cells, the immunoprecipitated proteins were separated by SDS-PAGE, and Western blots were probed for CFTR. Western blot analysis demonstrated that CFTR immunoprecipitates with USP10, but not with USP8 (Fig. 4, A and B). Thus, USP10 and CFTR interact in the early endosomal compartment of polarized human airway epithelial cells.
FIGURE 4.
USP10 immunoprecipitates CFTR. A, CFBE cells were lysed, USP10 was immunoprecipitated using an anti-USP10 antibody, and Western blot analysis was performed for USP10 and CFTR. LYSATE, cell lysates (2.5% of lysate run on gel). USP10 IP indicates proteins that were immunoprecipitated using the USP10 antibody. The non-immune IgG did not immunoprecipitate USP10 or CFTR, and thus served as a negative control. *, denotes ezrin, a protein used to quantitate protein loading. B, USP8 does not immunoprecipitate with CFTR. CFBE cells were lysed, USP8 was immunoprecipitated using an anti-USP8 antibody, and Western blot analysis was performed for USP8 and CFTR. LYSATE, cell lysates (2.5% of lysate run on gel). USP8 IP indicates proteins that were immunoprecipitated using the USP8 antibody. The non-immune IgG did not immunoprecipitate USP8 or CFTR, and thus served as a negative control. Experiments were performed three times. Representative blots are shown.
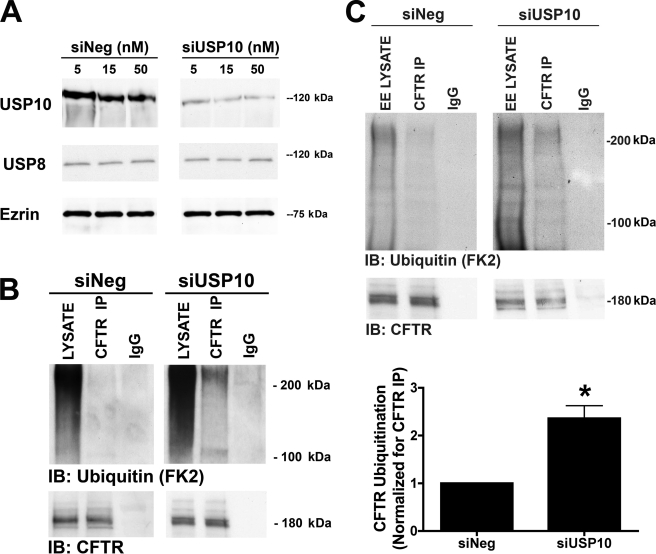
siRNA-mediated Silencing of USP10 Increases the Amount of Ubiquitinated CFTR and Its Degradation in the Lysosome
The abundance of many ubiquitinated proteins is determined by the balance between the rate of ubiquitination, mediated by an E3 ligase, and the rate of deubiquitination, mediated by a DUB. If USP10 deubiquitinates CFTR, then siRNA-mediated knockdown of USP10 should increase the amount of ubiquitinated CFTR and increase the degradation of ubiquitinated CFTR in the lysosome. Preliminary studies were conducted to reduce USP10 protein abundance in polarized CFBE cells using a siRNA approach. All concentrations of siUSP10 used (5, 15, and 50 nm) reduced USP10 protein abundance, without altering the abundance of USP8 and ezrin (Fig. 5A) and decreased USP10 mRNA levels. The lowest siRNA concentration tested (5 nm) reduced USP10 protein abundance by 76 ± 4% (p < 0.05) and mRNA levels by 54 ± 5% (p < 0.05). Thus, 5 nm was used for the remainder of the siUSP10 studies. The effect of siUSP10 on the amount of ubiquitinated CFTR is illustrated in Fig. 5B. Immunoprecipitation of CFTR, followed by SDS-PAGE and Western blot analysis of ubiquitinated CFTR with the FK2 antibody, revealed that siUSP10 increased the amount of ubiquitinated CFTR by 258 ± 17% (Fig. 5B). The siUSP10-mediated increase in CFTR ubiquitination was confirmed in an early endosomal fraction from CFBE cells with USP10 knockdown (Fig. 5C).
FIGURE 5.
siRNA-mediated knockdown of USP10 increases ubiquitinated CFTR. CFBE cells were transfected with scrambled, negative control siRNA (siNeg) or siRNA specific for USP10 (siUSP10). A, Western blot analysis was performed to examine the effect of siUSP10 on the expression of USP10, USP8, and ezrin. siUSP10 (5 nm) reduced USP10 protein abundance by 76 ± 4% (p < 0.05), but had no effect on USP8 or ezrin abundance. 5 nm siUSP10 reduced USP10 mRNA by 54 ± 5%. B, CFBE cells were transfected with scrambled, negative control siRNA (siNeg), or siUSP10 (5 nm). CFTR was immunoprecipitated from whole cell lysate using a monoclonal CFTR antibody (clone M3A7, Upstate Biotechnology), and ubiquitinated CFTR was detected via Western blot analysis (IB) using an anti-ubiquitin antibody (FK2, Biomol). C, CFBE cells were transfected with scrambled, negative control siRNA (siNeg), or siUSP10 (5 nm). CFTR was immunoprecipitated from the early endosome fraction using a monoclonal CFTR antibody (clone M3A7, Upstate Biotechnology), and ubiquitinated CFTR was detected via Western blot analysis (IB) using an anti-ubiquitin antibody (FK2, Biomol). LYSATE, cell lysates. EE LYSATE, lysate from early endosomal preparation. CFTR IP, immunoprecipitated CFTR. IgG, immunoprecipitation using a non-immune IgG antibody as a negative control. In the bottom panels (B and C) immunoprecipitated CFTR was blotted (IB) with a CFTR monoclonal antibody: similar amounts of CFTR were immunoprecipitated in all cases: thus, differences in the amount of ubiquitinated CFTR detected were not due to differences in the efficiency of the immunoprecipitation step for CFTR. Representative blots are shown. The panel on the right is a quantitation of ubiquitinated CFTR in siNeg and siUSP10 experiments. Data normalized for the amount of CFTR immunoprecipitated. Experiments were performed four times. *, p < 0.05 versus siNeg.
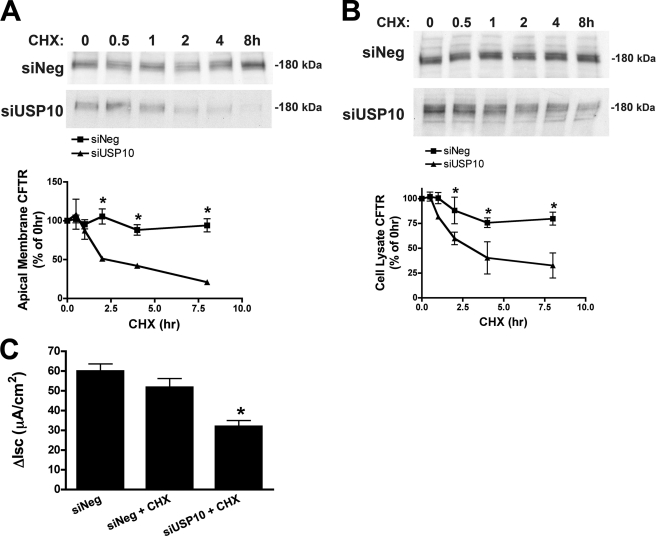
An increase in the amount of ubiquitinated CFTR, induced by siUSP10, would be expected to enhance the trafficking of CFTR to lysosomes for degradation, and thereby decrease the recycling of CFTR from early endosomes to the plasma membrane and subsequently decrease the plasma membrane abundance of CFTR. To specifically assess the effect of USP10 on CFTR trafficking, the following experiments were performed in the presence of cycloheximide. Fig. 6A demonstrates that siUSP10, but not a control siRNA (siNeg), produced a time-dependent decrease in apical plasma membrane CFTR. The siUSP10-mediated reduction in apical membrane CFTR was accompanied by a decrease in the abundance of CFTR in the cell lysate (Fig. 6B). siUSP10 also reduced CFTR-mediated chloride secretion across polarized CFBE cells (Fig. 6C).
FIGURE 6.
siUSP10 reduces CFTR abundance and CFTR-mediated chloride currents. Polarized CFBE cells were transfected with scrambled siRNA (siNeg) or siUSP10. A, the effect of siUSP10 on the amount of CFTR in the apical plasma membrane was determined by cell surface biotinylation and Western blot analysis as a function of time after cycloheximide (CHX, 5 μg/ml), to inhibit CFTR translation. *, p < 0.05 versus siNeg at the same time point. B, the effect of siUSP10 on the amount of CFTR in cell lysates was determined by Western blot analysis as a function of time after cycloheximide (CHX, 5 μg/ml), which inhibits CFTR translation. *, p < 0.05 versus siNeg at the same time point. C, the effect of siUSP10 on CFTR-mediated chloride currents across CFBE cells. Polarized CFBE cells were transfected with scrambled siRNA (siNeg) or siUSP10 and treated with vehicle or cycloheximide (CHX, 5 μg/ml for 4 h before Ussing chamber studies) to inhibit CFTR translation. Data reported as the short circuit current inhibited by the CFTR channel blocker CFTR-172 in forskolin (10 μm) treated cells (ΔIsc). Experiments performed four times; representative blots are shown. *, p < 0.05 versus siNeg and siNeg + CHX.
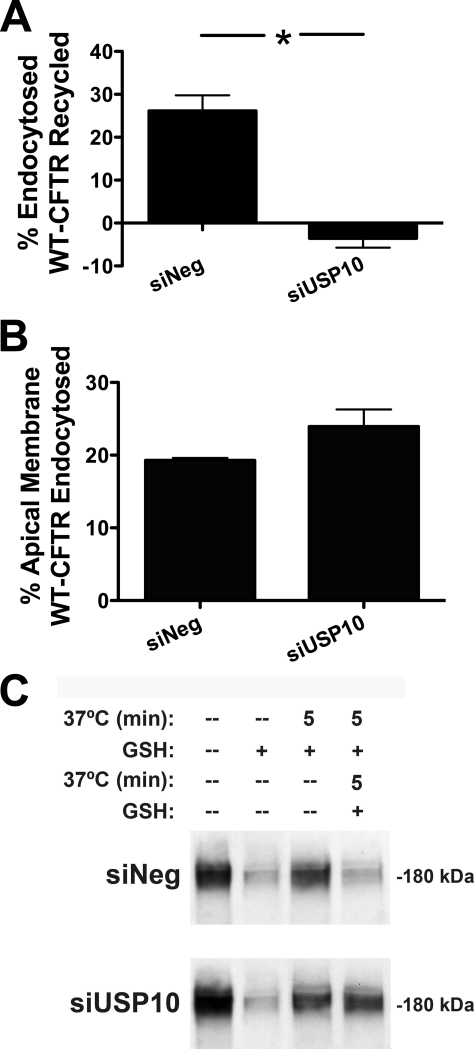
To assess if the reduction in apical membrane CFTR abundance and function in the presence of USP10 knockdown was due to an alteration in the endocytic recycling of CFTR, we performed endocytosis and recycling assays on CFBE cells in the presence and absence of USP10 knockdown. Fig. 7 demonstrates that siRNA-mediated reduction in USP10 expression did not alter the endocytosis of CFTR in CFBE cells (Fig. 7B), but dramatically inhibited the endocytic recycling of CFTR (Fig. 7A). Therefore, USP10 enhances the expression and function of CFTR at the apical membrane by increasing the post-endocytic recycling of the chloride channel.
FIGURE 7.
siUSP10 inhibits the endocytic recycling of CFTR. Polarized CFBE cells were transfected with scrambled siRNA (siNeg) or siUSP10 and endocytosis and endocytic recycling of CFTR were measured as described under “Experimental Procedures.” A, siRNA-mediated knockdown of USP10 inhibits the endocytic recycling of CFTR. B, USP10 knockdown by siRNA does not alter the endocytosis of CFTR. C, representative blots from endocytic and recycling assays. Cells from all four lanes were cooled to 4 °C and biotinylated. The amount of biotinylated CFTR remaining in the plasma membrane after GSH treatment at 4 °C (lane 2) was subtracted from the amount of CFTR remaining biotinylated after warming to 37 °C and GSH treatment (lane 3) to determine the amount of endocytosed CFTR. Cells from lane 3 were then warmed at 37 °C to allow recycling of endocytosed CFTR before a second GSH treatment. CFTR recycling was calculated as the difference between the amount of biotinylated CFTR after the first (lane 3) and second (lane 4) GSH treatments. Experiments performed three times. *, p < 0.05.
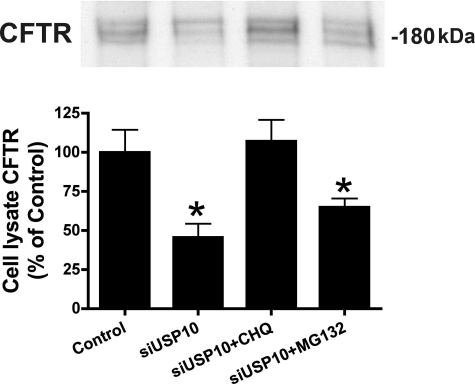
To determine if siRNA knockdown of USP10 reduced the abundance of CFTR by enhancing its targeting and degradation in lysosomes or proteasomes, cells were treated with siRNA for USP10 and either chloroquine, a lysosomal inhibitor, or MG132, a proteasomal inhibitor. siUSP10 reduced CFTR in cell lysates, and chloroquine, but not MG132, blocked the siUSP10-mediated decrease in CFTR (Fig. 8). These studies are consistent with the view that siUSP10, by reducing USP10, increases the amount of ubiquitinated CFTR, which is degraded in the lysosome.
FIGURE 8.
siUSP10 enhances the lysosomal degradation of CFTR. CFBE cells were transfected with siNeg or siUSP10 and CFTR in cell lysates was detected by Western blot analysis. siUSP10 reduced the amount of CFTR, confirming studies reported in Fig. 7B. Chloroquine (CHQ: 200 μm), an inhibitor of the lysosomal degradation of proteins, blocked the siUSP10-induced decrease in CFTR abundance. By contrast, MG132 (50 μm), an inhibitor of the proteasomal degradation of proteins, had no effect on the siUSP10-induced decrease in CFTR abundance. Experiments performed four times; representative blots are shown. *, p < 0.05 versus Control (siNeg) and siUSP10 + CHQ.
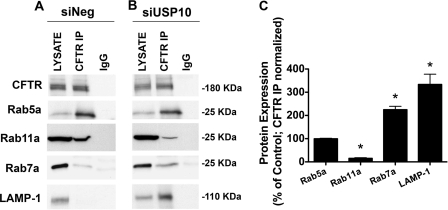
siRNA-mediated Knockdown of USP10 Redirects CFTR from the Recycling Pathway to the Lysosomal Pathway
To provide additional support for the view that knockdown of USP10 increases the ubiquitination of CFTR and redirects CFTR from recycling endosomes to lysosomes, studies were conducted to examine the effect of siUSP10 on the subcellular distribution of CFTR. Previous studies have shown that co-immunoprecipitation of CFTR with a variety of Rab GTPases can be used to identify the subcellular localization of transporters and ion channels, including CFTR (16–18, 36, 37). In control cells (siNeg), CFTR immunoprecipitated with Rab5a, a marker of early endosomes, and Rab11a, a marker for recycling endosomes, but not Rab7a, a marker for late endosomes, and LAMP-1, a marker for lysosomes (16, 17, 19) (Fig. 9A). These observations are consistent with previous studies demonstrating that CFTR is efficiently recycled from early endosomes to recycling endosomes, to the plasma membrane (15–18). siRNA knockdown of USP10 decreased CFTR immunoprecipitation with Rab11a, increased CFTR immunoprecipitation with Rab7a and LAMP-1, and had no effect on CFTR immunoprecipitation with Rab5a (Fig. 9B). These results are consistent with the view that siUSP10, by increasing the amount of ubiquitinated CFTR, redirects CFTR from recycling endosomes to late endosomes and lysosomes.
FIGURE 9.
siUSP10 redirects CFTR from the recycling pathway to the lysosomal pathway. CFBE cells were transfected with siNeg or siUSP10. Subsequently, co-immunoprecipitation studies were conducted to determine the subcellular location of CFTR in cells treated with siNeg (A) and with siUSP10 (B). C, summary of data for Rab GTPase immunoprecipitation with CFTR in the presence and absence of siUSP10, normalized for the amount of CFTR immunoprecipitation. *, p < 0.05 versus control (100%). CFTR was immunoprecipitated using a monoclonal CFTR antibody (clone M3A7, Upstate) and interacting proteins were determined by SDS-PAGE and Western blot analysis. Rab5a, a marker of early endosomes; Rab11a, a marker of recycling endosomes; Rab7a, a marker of late endosomes; and LAMP-1, a marker of lysosomes. Comparison of panels A and B reveals that siUSP10 reduced CFTR-Rab11a interaction, and increased CFTR and Rab7a and LAMP-1 interaction. By contrast siUSP10 had no effect on CFTR-Rab5a interaction. These experiments indicate that siUSP10 redirects CFTR from recycling endosomes to lysosomes and that siUSP10 has no effect on the amount of CFTR in early endosomes. Experiments performed three times; representative blots are shown.
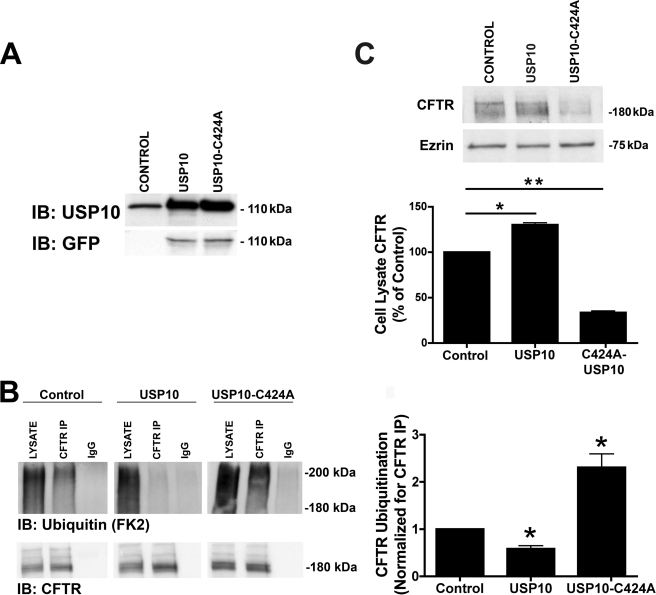
wt-USP10 Decreases, and a Dominant Negative USP10 Increases, the Ubiquitination and Degradation of CFTR
To provide additional support for a role of USP10 in regulating the deubiquitination status of CFTR, CFBE cells were transfected with either wt-CFTR or a dominant negative, catalytically inactive USP10 (USP10-C424A), and the amount of ubiquitinated CFTR was measured as described above. We predicted that overexpression of wt-USP10 would decrease the amount of ubiquitinated CFTR, and that expression of a dominant-negative USP10 would increase the amount of ubiquitinated CFTR. Western blot analysis confirmed that transfection increased the amount of wt-USP10 and USP10-C424A in CFBE cells by ∼9-fold compared with endogenous USP10 (Fig. 10A). Whereas overexpression of wt-USP10 decreased the amount of ubiquitinated CFTR by 58 ± 7% (Fig. 10B), USP10-C424A increased the amount of ubiquitinated CFTR by 231 ± 3% (Fig. 10B). Overexpression of wt-USP10 also increased the amount of CFTR in cell lysates (Fig. 10C), whereas USP10-C424A reduced the amount of CFTR in cells lysates (Fig. 10C). These results support the conclusion that USP10 deubiquitinates CFTR.
FIGURE 10.
wt-USP10 reduces, and USP10-C424A increases, the amount of ubiquitinated CFTR and the amount of CFTR that is degraded. CFBE cells were transiently transfected with either wild-type GFP-USP10 or a dominant-negative GFP-USP10 (USP-C424A). A, USP10 expression in control cells and in cells transfected with GFP-USP10 and GFP-USP10-C424A was determined by Western blot analysis using either an anti-USP10 antibody or an anti-GFP antibody. B, cells were incubated in 200 μm chloroquine for 4 h and lysed, and CFTR was immunoprecipitated (IP) using a monoclonal CFTR antibody (clone M3A7, Upstate). Ubiquitinated CFTR was detected via Western blot analysis (IB) using an anti-ubiquitin antibody (FK2). LYSATE, cell lysates. CFTR IP, immunoprecipitated CFTR using an anti-CFTR monoclonal antibody (M3A7). IgG, immunoprecipitation using a non-immune IgG antibody. In the bottom panels immunoprecipitated CFTR was blotted with a CFTR monoclonal antibody (clone 24-1, R&D Systems): similar amounts of CFTR were immunoprecipitated in all cases; thus, differences in the amount of ubiquitinated CFTR were not due to differences in the efficiency of the immunoprecipitation step for CFTR. Experiments performed three times. Representative blots are shown. The panel on the right is a summary of the data in Control, USP10, and USP10-C424A experiments. Data normalized for the amount of CFTR immunoprecipitated. *, p < 0.05 versus control. C, Western blot analysis of CFTR and ezrin demonstrating the effect wt-USP10 and USP10-C424A on CFTR in cell lysates. The top panel is a representative blot with quantitation of the results in the bottom panel. Experiments performed four times. Representative blots are shown. *, p < 0.05; **, p < 0.01.
DISCUSSION
The studies in this report demonstrate that ubiquitination regulates the endocytic sorting of CFTR in human airway epithelial cells and identifies a novel function for USP10 in facilitating the deubiquitination of CFTR in early endosomes, an effect that enhances the endocytic recycling of CFTR back to the plasma membrane, and thereby increases the cell surface expression of CFTR. Using an activity-based chemical screen to identify active DUBs in human airway epithelial cells, we demonstrated that USP10 is located in early endosomes and regulates the deubiquitination of CFTR and trafficking in the post-endocytic compartment. siRNA-mediated knockdown of USP10 increased the amount of ubiquitinated CFTR and its degradation in lysosomes, and reduced both apical membrane CFTR and CFTR-mediated chloride secretion. Moreover, a dominant negative USP10 (USP10-C424A) increased the amount of ubiquitinated CFTR and its degradation, while overexpression of wt-USP10 decreased the amount of ubiquitinated CFTR and increased the abundance of CFTR. We offer a model that incorporates the data in the preset study with data in the literature on CFTR trafficking, and the role of ubiquitination in regulating the endocytic trafficking of cell surface receptors (1–6, 15–18, 26). We propose that CFTR is ubiquitinated by an unidentified E3 ligase and that ubiquitinated CFTR is removed from the plasma membrane by endocytosis (19). The majority of endocytosed CFTR is deubiquitinated by USP10 in early endosomes, and deubiquitinated CFTR recycles back to the plasma membrane. The CFTR that is not deubiquitinated in early endosomes is sorted to multivesicular bodies, to late endosomes, and ultimately, to lysosomes for degradation. The highly efficient endocytic recycling of CFTR, as a result of the deubiquitination of CFTR by USP10, accounts for the long half-life of CFTR (∼3–24 h in polarized human airway cells) in the plasma membrane (15–17, 38). Outstanding issues that need to be addressed by additional studies include the identification of the E3 ligase(s) that ubiquitinate CFTR, and the identification of other DUBS in the endocytic pathway that may deubiquitinate CFTR.
Several years ago Sharma et al. (19) demonstrated that ubiquitination reduced the plasma membrane abundance of CFTR in cells heterologously expressing CFTR (i.e. baby hamster kidney cells) by redirecting CFTR from recycling endosomes to lysosomes for degradation. Our studies are in agreement with those of Sharma et al. and extend their observations by demonstrating that the ubiquitination of CFTR also redirects CFTR from recycling endosomes to lysosomes for degradation in polarized human airway epithelial cells. In addition, the present studies provide the novel observation that USP10 deubiquitinates CFTR in early endosomes, thereby facilitating the endocytic recycling of CFTR. Although we did not characterize all DUBs identified using the HA-UbVME probe, we did detect other active DUBs in the airway epithelial cells. It is possible that some of these DUBs may also regulate the deubiquitination of CFTR. Identification and characterization of these DUBs, as well as the identification of DUBS from other classes, require further study and the use of different chemical probes that react with other classes of DUBs in this large family (>100) of enzymes (4).
In kidney cortical collecting duct cells vasopressin up-regulates the expression of USP10, which deubiquitinates and stabilizes sorting nexin3 that, by an unknown mechanism, increases the cell surface expression of ENaC (8). Interestingly, vasopressin also stimulates CFTR-mediated chloride secretion by dog kidney (Madin-Darby canine kidney) epithelial cells (39, 40), raising the possibility that vasopressin-mediated stimulation of CFTR chloride secretion may be mediated by up-regulation of USP10, deubiquitination of CFTR, and enhanced endocytic recycling of CFTR to the cell surface. Additional studies, beyond the scope of this study, are required to examine this hypothesis.
Our biochemical and imaging studies revealed that in airway epithelial cells UPS10 is localized primarily in early endosomes, consistent with our data identifying USP10 in regulating the post-endocytic sorting of CFTR. A recent study identified USP10 in a diffuse intracellular location (8), consistent with the expression in early endosomes. Although our studies clearly demonstrate that USP10 deubiquitinates CFTR in early endosomes, it is possible that USP10 may deubiquitinate CFTR in other subcellular compartments, as well. Each DUB is typically expressed in several subcellular sites and has multiple substrates/binding partners, thus a single DUB may regulate the function of several different proteins or the same protein in several different subcellular sites, depending on the substrates/binding partners. For example, the observation that USP10 regulates trafficking between the endoplasmic reticulum and Golgi in yeast (41, 42) raises the interesting possibility that USP10 may also regulate CFTR trafficking between the endoplasmic reticulum and the Golgi in epithelial cells. However, our observation that the proteasomal inhibitor MG132 did not affect CFTR degradation in cells treated with siUSP10 (see Fig. 8) suggests that USP10 does not regulate CFTR deubiquitination in the endoplasmic reticulum.
Several other DUBs, in addition to USP10, have been shown to deubiquitinate receptors and channels. Deubiquitination of the epidermal growth factor receptor and the yeast ABC transporter Ste6, by AMSH and Ubp1, respectively, increase their cell surface expression (12, 43, 44). UCH-L3 and USP2–45 deubiquitinate ENaC in a cortical collecting duct cell line and enhance ENaC cell surface expression (9, 10). Thus, it is becoming increasingly clear that DUB-mediated deubiquitination of ion channels is an important mechanism regulating channel cell surface expression.
In conclusion, our data provide direct evidence that, in polarized human airway epithelial cells, deubiquitination of CFTR by USP10 regulates CFTR-dependent chloride secretion by facilitating the endocytic recycling of CFTR from early endosomes to the apical plasma membrane.
Acknowledgments
We thank Dr. Hidde Ploegh for his generous gift of the HA-UbVME probe for the DUB activity assays, Bonita Coutermarsh for her technical assistance with the Ussing chamber experiments, Dr. Susanna Chiocca for the USP10 plasmid constructs, and Dr. J. P. Clancy for the CFBE cells. We also thank Dr. Dean Madden for his critical analysis of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants 5T32DK007301 and 5R01DK45581 (to B. A. S.). This work was also supported by Cystic Fibrosis Foundation Grants BOMBER08F0 (to J. M. B.) and STANTO07R0 (to B. A. S.).
- E1
- ubiquitin-activating enzyme
- E2
- ubiquitin-conjugating enzyme
- E3
- ubiquitin ligase
- CFTR
- cystic fibrosis transmembrane conductance regulator
- USP10
- ubiquitin-specific protease-10
- DUB
- deubiquitinating enzyme
- ENaC
- epithelial sodium channel
- ABC
- ATP-binding cassette
- HA
- hemagglutinin
- RT
- reverse transcription
- siRNA
- small interference RNA
- UbVME
- ubiquitin-vinyl methyl ester
- LAMP-1
- lysosome-associated protein-1.
REFERENCES
- 1.Hicke L., Dunn R. (2003) Annu. Rev. Cell Dev. Biol. 19, 141–172 [DOI] [PubMed] [Google Scholar]
- 2.Clague M. J., Urbé S. (2006) Trends Cell Biol. 16, 551–559 [DOI] [PubMed] [Google Scholar]
- 3.Mukhopadhyay D., Riezman H. (2007) Science 315, 201–205 [DOI] [PubMed] [Google Scholar]
- 4.Love K. R., Catic A., Schlieker C., Ploegh H. L. (2007) Nat. Chem. Biol. 3, 697–705 [DOI] [PubMed] [Google Scholar]
- 5.Williams R. L., Urbé S. (2007) Nat. Rev. 8, 355–368 [DOI] [PubMed] [Google Scholar]
- 6.Millard S. M., Wood S. A. (2006) J. Cell Biol. 173, 463–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katzmann D. J., Babst M., Emr S. D. (2001) Cell 106, 145–155 [DOI] [PubMed] [Google Scholar]
- 8.Boulkroun S., Ruffieux-Daidie D., Vitagliano J. J., Poirot O., Charles R. P., Lagnaz D., Firsov D., Kellenberger S., Staub O. (2008) Am. J. Physiol. Renal Physiol. 295, F889–F900 [DOI] [PubMed] [Google Scholar]
- 9.Butterworth M. B., Edinger R. S., Ovaa H., Burg D., Johnson J. P., Frizzell R. A. (2007) J. Biol. Chem. 282, 37885–37893 [DOI] [PubMed] [Google Scholar]
- 10.Fakitsas P., Adam G., Daidié D., van Bemmelen M. X., Fouladkou F., Patrignani A., Wagner U., Warth R., Camargo S. M., Staub O., Verrey F. (2007) J. Am. Soc. Nephrol. 18, 1084–1092 [DOI] [PubMed] [Google Scholar]
- 11.Gesbert F., Malardé V., Dautry-Varsat A. (2005) Biochem. Biophys. Res. Commun. 334, 474–480 [DOI] [PubMed] [Google Scholar]
- 12.Urbé S., McCullough J., Row P., Prior I. A., Welchman R., Clague M. J. (2006) Biochem. Soc. Trans. 34, 754–756 [DOI] [PubMed] [Google Scholar]
- 13.Wicks S. J., Grocott T., Haros K., Maillard M., ten Dijke P., Chantry A. (2006) Biochem. Soc. Trans. 34, 761–763 [DOI] [PubMed] [Google Scholar]
- 14.Wicks S. J., Haros K., Maillard M., Song L., Cohen R. E., Dijke P. T., Chantry A. (2005) Oncogene 24, 8080–8084 [DOI] [PubMed] [Google Scholar]
- 15.Ameen N., Silvis M., Bradbury N. A. (2007) J. Cyst. Fibros. 6, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gentzsch M., Chang X. B., Cui L., Wu Y., Ozols V. V., Choudhury A., Pagano R. E., Riordan J. R. (2004) Mol. Biol. Cell 15, 2684–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swiatecka-Urban A., Brown A., Moreau-Marquis S., Renuka J., Coutermarsh B., Barnaby R., Karlson K. H., Flotte T. R., Fukuda M., Langford G. M., Stanton B. A. (2005) J. Biol. Chem. 280, 36762–36772 [DOI] [PubMed] [Google Scholar]
- 18.Swiatecka-Urban A., Talebian L., Kanno E., Moreau-Marquis S., Coutermarsh B., Hansen K., Karlson K. H., Barnaby R., Cheney R. E., Langford G. M., Fukuda M., Stanton B. A. (2007) J. Biol. Chem. 282, 23725–23736 [DOI] [PubMed] [Google Scholar]
- 19.Sharma M., Pampinella F., Nemes C., Benharouga M., So J., Du K., Bache K. G., Papsin B., Zerangue N., Stenmark H., Lukacs G. L. (2004) J. Cell Biol. 164, 923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorenzo M. E., Jung J. U., Ploegh H. L. (2002) J. Virol. 76, 5522–5531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlieker C., Korbel G. A., Kattenhorn L. M., Ploegh H. L. (2005) J. Virol. 79, 15582–15585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borodovsky A., Ovaa H., Kolli N., Gan-Erdene T., Wilkinson K. D., Ploegh H. L., Kessler B. M. (2002) Chem. Biol. 9, 1149–1159 [DOI] [PubMed] [Google Scholar]
- 23.Bebok Z., Collawn J. F., Wakefield J., Parker W., Li Y., Varga K., Sorscher E. J., Clancy J. P. (2005) J. Physiol. 569, 601–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Urbé S., Sachse M., Row P. E., Preisinger C., Barr F. A., Strous G., Klumperman J., Clague M. J. (2003) J. Cell Sci. 116, 4169–4179 [DOI] [PubMed] [Google Scholar]
- 25.Moyer B. D., Loffing J., Schwiebert E. M., Loffing-Cueni D., Halpin P. A., Karlson K. H., Ismailov II, Guggino W. B., Langford G. M., Stanton B. A. (1998) J. Biol. Chem. 273, 21759–21768 [DOI] [PubMed] [Google Scholar]
- 26.Swiatecka-Urban A., Duhaime M., Coutermarsh B., Karlson K. H., Collawn J., Milewski M., Cutting G. R., Guggino W. B., Langford G., Stanton B. A. (2002) J. Biol. Chem. 277, 40099–40105 [DOI] [PubMed] [Google Scholar]
- 27.Swiatecka-Urban A., Boyd C., Coutermarsh B., Karlson K. H., Barnaby R., Aschenbrenner L., Langford G. M., Hasson T., Stanton B. A. (2004) J. Biol. Chem. 279, 38025–38031 [DOI] [PubMed] [Google Scholar]
- 28.Soncini C., Berdo I., Draetta G. (2001) Oncogene 20, 3869–3879 [DOI] [PubMed] [Google Scholar]
- 29.Wolde M., Fellows A., Cheng J., Kivenson A., Coutermarsh B., Talebian L., Karlson K., Piserchio A., Mierke D. F., Stanton B. A., Guggino W. B., Madden D. R. (2007) J. Biol. Chem. 282, 8099–8109 [DOI] [PubMed] [Google Scholar]
- 30.Bomberger J. M., Parameswaran N., Hall C. S., Aiyar N., Spielman W. S. (2005) J. Biol. Chem. 280, 9297–9307 [DOI] [PubMed] [Google Scholar]
- 31.Swiatecka-Urban A., Moreau-Marquis S., Maceachran D. P., Connolly J. P., Stanton C. R., Su J. R., Barnaby R., O'Toole G. A., Stanton B. A. (2006) Am. J. Physiol. Cell Physiol. 290, C862–C872 [DOI] [PubMed] [Google Scholar]
- 32.Haglund K., Di Fiore P. P., Dikic I. (2003) Trends Biochem. Sci. 28, 598–603 [DOI] [PubMed] [Google Scholar]
- 33.Haglund K., Sigismund S., Polo S., Szymkiewicz I., Di Fiore P. P., Dikic I. (2003) Nat. Cell Biol. 5, 461–466 [DOI] [PubMed] [Google Scholar]
- 34.Monami G., Emiliozzi V., Morrione A. (2008) J. Cell. Physiol. 216, 426–437 [DOI] [PubMed] [Google Scholar]
- 35.Mosesson Y., Shtiegman K., Katz M., Zwang Y., Vereb G., Szollosi J., Yarden Y. (2003) J. Biol. Chem. 278, 21323–21326 [DOI] [PubMed] [Google Scholar]
- 36.Saxena S., Singh M., Engisch K., Fukuda M., Kaur S. (2005) Biochem. Biophys. Res. Commun. 337, 1219–1223 [DOI] [PubMed] [Google Scholar]
- 37.Saxena S. K., Kaur S. (2006) Biochem. Biophys. Res. Commun. 346, 259–267 [DOI] [PubMed] [Google Scholar]
- 38.Lukacs G. L., Chang X. B., Bear C., Kartner N., Mohamed A., Riordan J. R., Grinstein S. (1993) J. Biol. Chem. 268, 21592–21598 [PubMed] [Google Scholar]
- 39.Chang C. T., Bens M., Hummler E., Boulkroun S., Schild L., Teulon J., Rossier B. C., Vandewalle A. (2005) J. Physiol. 562, 271–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lahr T. F., Record R. D., Hoover D. K., Hughes C. L., Blazer-Yost B. L. (2000) Pflugers Arch. 439, 610–617 [DOI] [PubMed] [Google Scholar]
- 41.Cohen M., Stutz F., Belgareh N., Haguenauer-Tsapis R., Dargemont C. (2003) Nat. Cell Biol. 5, 661–667 [DOI] [PubMed] [Google Scholar]
- 42.Cohen M., Stutz F., Dargemont C. (2003) J. Biol. Chem. 278, 51989–51992 [DOI] [PubMed] [Google Scholar]
- 43.Schmitz C., Kinner A., Kölling R. (2005) Mol. Biol. Cell 16, 1319–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCullough J., Clague M. J., Urbé S. (2004) J. Cell Biol. 166, 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]