Abstract
Bacterioferritin (BFR) is a bacterial member of the ferritin family that functions in iron metabolism and protects against oxidative stress. BFR differs from the mammalian protein in that it is comprised of 24 identical subunits and is able to bind 12 equivalents of heme at sites located between adjacent pairs of subunits. The mechanism by which iron enters the protein to form the dinuclear (ferroxidase) catalytic site present in every subunit and the mineralized iron core housed within the 24-mer is not well understood. To address this issue, the properties of a catalytically functional assembly variant (E128R/E135R) of Escherichia coli BFR are characterized by a combination of crystallography, site-directed mutagenesis, and kinetics. The three-dimensional structure of the protein (1.8 Å resolution) includes two ethylene glycol molecules located on either side of the dinuclear iron site. One of these ethylene glycol molecules is integrated into the surface of the protein that would normally be exposed to solvent, and the other is integrated into the surface of the protein that would normally face the iron core where it is surrounded by the anionic residues Glu47, Asp50, and Asp126. We propose that the sites occupied by these ethylene glycol molecules define regions where iron interacts with the protein, and, in keeping with this proposal, ferroxidase activity decreases significantly when they are replaced with the corresponding amides.
Bacterioferritin (BFR)4 is a prokaryotic form of ferritin that has been identified in a number of bacteria (1–3). Despite low sequence similarity with eukaryotic ferritins, the three-dimensional structures and functional properties of BFRs from Escherichia coli, Rhodobacter capsulatus, Desulfovibrio desulfuricans, and Azotobacter vinelandii (4–10) are remarkably reminiscent of those reported for mammalian ferritins. For example, BFRs are oligomeric proteins comprised of 24 subunits (∼18 kDa each) that catalyze oxidation of Fe2+ by dioxygen (ferroxidase activity) to promote formation of a mineralized iron core that can contain as many as 2700 iron atoms/ 24-meric molecule (11). On the other hand, those BFRs that have been characterized differ from the mammalian proteins in that the 24 subunits are identical, and each possesses a catalytic dinuclear iron center that is referred to as the ferroxidase site (in mammalian ferritins, only the H-chains possess such catalytic sites). The pairwise arrangement of BFR monomers within the 24-mer creates 12 binding sites for heme, commonly protoheme IX but iron-coproporphyrin III in D. desulfuricans BFR, in which a methionyl residue on the surface of adjacent BFR monomers provides an axial ligand to create a b-type heme-binding site with bismethionine axial coordination (12, 13). Although a functional role for the heme of BFR has not been identified, the functional role of BFR is believed to be in iron storage and detoxification (14), thereby protecting against oxidative stress (15).
The subunits of BFR are arranged to form eight 3-fold channels and six 4-fold channels. These channels have been proposed as possible entry and exit routes for iron incorporation into or release from the central iron core. For human ferritin, the 3-fold channel plays a significant role in the transport of iron into the iron core (16), but a similar role for this channel in BFR has not been demonstrated.
The dinuclear ferroxidase site located within each subunit binds two iron atoms. Coordination of these iron atoms involves Glu51 and Glu127 as bridging ligands for both irons, Glu18 and His54 as ligands for FE1, and Glu94 and His130 as ligands for FE2. Previous studies of E. coli BFR have demonstrated that the ferroxidase center is essential for core formation and that core formation involves at least three kinetically distinguishable phases (11, 17). Phase 1 involves the very rapid reversible binding of two Fe2+ ions to each of the 24 dinuclear ferroxidase centers and can be studied by monitoring small changes in the spectrum of the bound heme. Phase 2 occurs in the presence of dioxygen (or an alternative oxidant such as hydrogen peroxide) and involves the rapid oxidation of each di-Fe2+ center to form an intermediate that is probably an oxo- or hydroxo-bridged di-Fe3+ center. In the presence of Fe2+ exceeding the amount required to saturate the ferroxidase centers, a slower reaction, Phase 3, is observed in which a large ferric oxyhydroxo mineral is synthesized within the protein cavity. The change in absorbance at 340 nm that is observed during aerobic addition of Fe2+ to apo-BFR results from Phases 2 and 3 but is influenced by the kinetics of Phase 1. Although Phases 1 and 2 are well characterized, less is known about Phase 3. This phase probably involves the interaction of Fe2+ (or Fe3+) with amino acid residues on the inner surface of the ferritin oligomer, as part of a complex and poorly defined process known as nucleation. Further information on this phase of core formation is now required.
An assembly variant of E. coli BFR (E128R/E135R) has been shown previously to form stable subunit dimers that bind one equivalent of protoheme IX and not to form higher order oligomers (18). Each monomer in this minimal functional unit can form a dinuclear iron center that catalyzes the formation of a minimal iron core comprised of four to six iron atoms before precipitating (18, 19). The overall kinetics of Fe2+ oxidation observed on addition of Fe2+ to this variant are similar to those observed for wild-type BFR but have not been reported in detail. Nevertheless, the properties of this variant are of interest because the minimal structural unit that it forms constitutes a potentially important experimental model for evaluating detailed mechanistic features of BFR function. Simplification of the oligomeric structure of the protein as represented by this variant form of BFR makes the inner surface of the protein as accessible to bulk solvent as the outer surface, thereby removing any kinetic influences of the channels present in the 24-mer protein.
The present paper reports detailed kinetic and structural studies that validate this dimeric variant of BFR as a model of the minimal functional unit of wild-type BFR. In addition, the crystallographic structural data suggest a likely functional role of acidic inner surface residues in iron core formation that led to construction of a family of variants of the stable subunit dimer involving replacement of Glu47, Asp50, and Asp126 by site-directed mutagenesis. Kinetic studies of these additional variants confirm a functional role for these residues and lead to the proposal of a model of BFR action.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The E128R/E135R assembly variant of E. coli BFR was expressed in E. coli MAK96 (BL21 ΔBFR) (19) transformed with the pALN18 plasmid (18). Expression was induced with 1 mm isopropyl-β-d-thiogalactopyranoside when cultures achieved OD600 = 1.0, and the cells were harvested after growth for 10 h at 37 °C with shaking. Following harvesting by centrifugation, the cells were resuspended in potassium phosphate buffer (pH 7.2, 50 mm) containing phenylmethanesulfonyl fluoride (1 mm; Sigma-Aldrich) and lysed by incubating with lysozyme (125 mg/100 ml) and subjected to freeze thaw cycles. The cell-free extract was centrifuged to remove insoluble material and applied to a DEAE-Sepharose (GE Bioscience) anion exchange column (2.75 × 10 cm; ambient temperature) that had been equilibrated with potassium phosphate buffer (pH 7.2, 50 mm; ambient temperature). BFR was eluted with a linear NaCl gradient (0–0.5 m, formed by mixing 300 ml of buffer lacking NaCl with 300 ml of buffer with 0.5 m NaCl). The fractions containing BFR were pooled and concentrated to ∼5 ml by centrifugal ultrafiltration (Amicon Ultra) and then loaded onto a Sephadex G-75 (GE Bioscience) gel filtration column (2.6 × 90 cm) that was equilibrated and developed at ambient temperature with potassium phosphate buffer (pH 7.2, 50 mm). The resulting purified protein was dialyzed exhaustively against MES buffer (pH 6.5, 0.2 m) that also contained a dialysis bag containing Amberlite IRC-718 chelating resin (ICN Biomedical). Protein purified in this manner was used for the preparation of crystals for structure determination.
Kinetic measurements and other experiments were performed with the E128R/E135R BFR variant expressed from a pET32b+ plasmid (Novagen) following insertion of the gene encoding the E128R/E135 variant of BFR by PCR integration (20). This plasmid was further modified by introduction of a coding sequence for the tobacco etch virus (TEV) cleavage site (ENLYFQM) immediately after the sequence encoding S tag such that the N-terminal methionyl residue of BFR remains following cleavage with TEV (21, 22). Site-directed mutagenesis was performed with the Stratagene method and the following primer pairs: E47Q, 5′-GTGGAGTATCATCAGTCCATTGATGAGATGAAACACGCCG-3′ and 5′-CGGCGTGTTTCATCTCATCAATGGACTGATGATACTCCAC-3′; D50N, 5′-CCATTAATGAGATGAAACACGCCGATC-3′ and 5′- GATCGGCGTGTTTCATCTCATTAATGG-3′; D126N, 5′-GAAATTTTGCGTAATGAACGAGGCC-3′ and 5′-GGCCTCGTTCATTACGCAAAATTTC-3′; E47Q/D50N, 5′-GTGGAGTATCATCAGTCCATTAATGAGATGAAACACGCCGATCG-3′ and 5′-CGATCGGCGTGTTTCATCTCATTAATGGACTGATGATACTCCAC-3′ (underlined nucleotides indicate the mutations). The validity of all mutant genes produced in this work was verified by DNA sequence analysis. The TEV protease variant His-TEV(S219V)-Arg expressed from the plasmid pRK793 and purified as described by Kapust et al. (21) was the kind gift of Dr. Susanne Ludwiczek.
E. coli MAK96 cells transformed with the resulting plasmid were grown and processed initially as described above. Harvested cells were resuspended in potassium phosphate buffer (pH 7.2, 50 mm) containing 1 mm phenylmethanesulfonyl fluoride, 0.5 m NaCl, and 1 mm imidazole (Fisher) prior to lysis with an Avestin EmulsiFlex-C5 high pressure homogenizer. The cell-free extract was applied to a Ni2+-chelate column (2.75 × 2.5 cm, Chelating Sepharose FF; Amersham Biosciences) and washed with 50 mm potassium phosphate buffer (pH 7.2) containing 0.5 m NaCl and 50 mm imidazole. Protein was eluted with 50 ml of 50 mm potassium phosphate buffer (pH 7.2) containing 0.5 m NaCl and 300 mm imidazole. Fractions containing His6-tagged BFR were incubated overnight at room temperature with TEV protease (100:1 BFR/TEV protease A280 ratio) in potassium phosphate buffer (pH 7.2, 50 mm) containing 1 mm dithiothreitol and 1 mm EDTA. The resulting reaction mixture was dialyzed against 50 mm potassium phosphate buffer (pH 7.2) containing 0.5 m NaCl and eluted over a Ni2+-chelate column to remove uncleaved BFR, the cleaved tag sequence, and the TEV protease. The purified BFR assembly variant lacking the N-terminal tag was eluted in the void volume with 50 mm potassium phosphate buffer (pH 7.2) containing 25 mm imidazole.
Recombinant protein purified by either protocol was obtained as a mixture of protein with only partial heme occupancy. Full occupancy of heme-binding sites was achieved by the addition of hemin chloride (prepared by dissolution in 0.1 m sodium hydroxide and dilution into MES buffer (pH 6.5)) to a final 1.1:1 (heme:subunit dimer) ratio. Free and adventitiously bound heme were removed by elution over PD-10 desalting columns (GE Biosciences). Iron and other metal ions were removed from BFR samples prepared for kinetics analysis by repeated dilution and concentration by centrifugal ultrafiltration (Amicon Ultra) of the protein in 50 mm MES buffer (pH 6.5) containing 1 mm dithiothreitol, 10 mm EDTA, and 5 mm 2–2′-bipyridyl. Removal of iron from samples prepared in this manner was confirmed colorimetrically with ferrozine (Sigma) (23), and removal of zinc was confirmed colorimetrically with 4-(2-pyridylazo)resorcinol (Aldrich) (24). The concentration of protein with heme bound was determined from the absorbance at 280 nm (3.3 × 104 m−1 cm−1 subunit (25)) or 418 nm (1.09 × 105 m−1 cm−1 (26)) with the molar absorptivities indicated. All of the experiments reported here were performed with the dimeric E128R/E135R variant or with variants of this protein.
Crystallization and X-ray Diffraction Analysis
The purified E128R/E135R variant of E. coli BFR was crystallized by hanging drop vapor diffusion and microseeding in 20% polyethylene glycol 4000, 0.2 m Li2SO4, and 0.1 m Tris·HCl buffer (pH 8.5) at 30 °C by mixing equal volumes of protein (0.2 mm in 0.2 m MES buffer (pH 6.5)) and precipitant solutions. The resulting crystals were then soaked in a series of crystallization solutions (0.1 m Tris·HCl buffer (pH 8.5) containing 20% polyethylene glycol 4000 and 0.2 m Li2SO4) for 5 min each. These solutions were of identical composition except with ethylene glycol added at concentrations starting at 5% and increasing to 30% at increments of 5%. X-ray diffraction data were collected at the Stanford Synchrotron Radiation Laboratory (Palo Alto, CA) on Beam-line 1-5 using 0.979440 Å x-ray radiation and on Beam-line 9-2 using 1.30505 Å. These data were then indexed, integrated, and scaled with the program HKL2000 (27). The structure was solved by molecular replacement with MOLREP (28) using a pair of subunits from a structure (Protein Data Bank accession number 1BCF) of wild-type E. coli BFR from which the metal ions were removed as the search model. A single solution was obtained clearly separated from the alternatives. Refinement was carried out with Refmac5 (29) from the CCP4 suite (30) and with Swiss-Pdb Viewer (31). The final refined model for data collected at 0.979440 Å was used to provide phases for the anomalous difference maps computed at both wavelengths. The graphics figures were constructed with Swiss-Pdb Viewer v 3.7 and POV-Ray v 3.6.
Spectroscopy
Electronic absorption spectra were recorded with a Cary 6000i or 300 spectrophotometer. Circular dichroism spectra were recorded with a Jasco J-720 spectropolarimeter fitted with a circulating water bath and thermostatted cell holder (2-mm path length).
Kinetics
The kinetics of iron oxidation was studied with a Varian Cary 6000i spectrophotometer or with a Bio-Logic SFM-400 stopped flow spectrometer. Fe2+ solutions were prepared by dissolving ferrous ammonium sulfate in HCl (0.1 m) shortly before addition to apo-BFR for kinetics measurements. For stopped flow mixing experiments, these solutions were prepared anaerobically in HCl (6 mm). For experiments involving zinc, ZnSO4 solution was added to the BFR solution at least 5 min prior to the addition of Fe2+ used to initiate the kinetics measurements.
RESULTS
X-ray Structure Determination
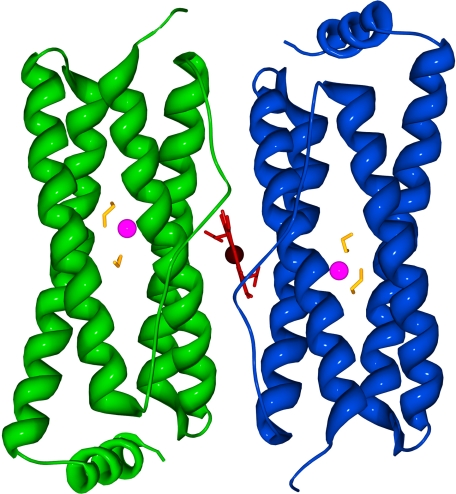
The E128R/E135R variant crystallizes in the space group P212121 with a = 33.64 Å, b = 91.09 Å, and c = 102.08 Å and contains two subunits in the asymmetric unit (Fig. 1). As observed in the structure of the wild-type protein, each subunit consists of four anti-parallel α-helices with a shorter C-terminal α-helix. The binding of heme is symmetric as observed in the structures of wild-type BFR from several species (e.g. E. coli, D. desulfuricans, R. capsulatus, and A. vinelandii) and is modeled with 50% occupancy for each of the two orientations (4, 8–10). After modeling all the residues into the electron density, two alternate conformations were found for Asp96 and Asp132 in the A subunit and for Asn17 for both subunits. Each subunit possesses one metal ion bound in the ferroxidase site, one sulfate ion, and two ethylene glycol molecules located nearby, all of which were modeled with 100% occupancy. Notably, one ethylene glycol molecule is located in an area of the protein related to the ferroxidase pore that has been identified in the structure of bacterioferritin from D. desulfuricans (8), and the other is located near the entrance at the inner surface.
FIGURE 1.
Ribbon diagram of the structure determined for the E128R/E135R variant of E. coli BFR. The ethylene glycol molecules, zinc, and heme are colored yellow, magenta, and red, respectively. The heme group is located between the two subunits. The model is viewed from what would be the outer surface of the 24-mer.
Electron density representing the presence of a metal ion is observed at one location of each dinuclear iron site in a position that is consistent with coordination by Glu18, His54, Glu51, and Glu127 in a tetrahedral geometry. This density was modeled as a Zn2+ ion, and metal ligand bonds were not restrained. The presence of zinc was confirmed by observing a decrease in the anomalous scattering of the metal when shifting the wavelength from 0.97944 Å, which is above the K-shell absorption edge of zinc, to 1.30505 Å, immediately below the K-edge of zinc (Table 1 and supplemental Fig. S1). Anomalous scattering would not decrease if manganese, iron, cobalt, nickel, or copper were located at the site in high occupancy because both wavelengths used are above the K-edges of these metals. The peak heights at the metal sites in the anomalous difference maps (supplemental Table S1) were scaled by comparison with the peak height at the heme iron. The presence of zinc and the absence of iron in the protein sample used to prepare these crystals was subsequently confirmed by the colorimetric methods described above.
TABLE 1.
Crystallographic data collection and refinement statistics
| P212121 space group |
|||
|---|---|---|---|
| Diffraction (0.97944 Å) | Anomalous scattering |
||
| 0.97944 Å | 1.30505 Å | ||
| Cell dimensions (Å) | a = 33.6, b = 91.1, c = 102.1 | a = 33.6, b = 91.1, c = 102.1 | a = 33.5, b = 91.3, c = 102.0 |
| Resolution (Å) | 45.5-1.80 (1.86-1.80)a | 45.5-1.80 (1.86-1.80)a | 44.5-2.02 (2.09-2.02)a |
| Rmerge (%) | 4.8 (25.7)a | 5.0 (26.0)a | 6.0 (19.7)a |
| I/σ(I) | 45.3 (6.7)a | 45.2 (6.7)a | 37.5 (16.3)a |
| Completeness (%) | 99.6 (98.7)a | 99.6 (98.7)a | 100 (100)a |
| No. total reflections | 339,676 | 339,912 | 562,393 |
| No. unique reflections | 29,997 | 29,982 | 21,335 |
| Wilson B factor (Å2) | 24.2 | ||
| Working R factor (%) | 20.2 | ||
| Free R factor (%) | 22.8 | ||
| B factors (Å2) | |||
| All atoms | 25.2 | ||
| Heme | 14.3 | ||
| Zinc | 19.9 | ||
| Ethylene glycol | 27.1 | ||
| Water molecules | 228 | ||
| Model geometry | |||
| Root mean square deviation bond length (Å) | 0.009 | ||
| Root mean square deviation bond angle (°) | 1.063 | ||
| Ramachandran plot | |||
| Most favored region (%) | 96.6 | ||
| Additionally allowed region (%) | 3.4 | ||
a The numbers in parentheses correspond to the highest resolution shell.
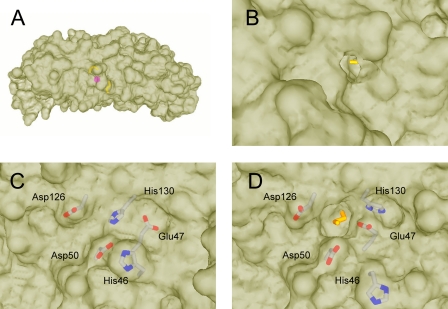
The other half of each dinuclear site consisting of the ligands Glu94, His130, Glu51, and Glu127 is unoccupied (Fig. 2). Interestingly, His130 is rotated away from the ferroxidase site as observed previously in the structures of a uranyl derivative of E. coli BFR (32) and A. vinelandii BFR with a partially occupied ferroxidase site (7, 9). For each subunit, one ethylene glycol molecule is located between the bound metal ion and the “outer surface,” and the other ethylene glycol molecule is located between the empty site and the “inner surface” (Fig. 3). The outer ethylene glycol (Fig. 3B) is bound in a pocket surrounded by the residues Leu14, Asn17, Glu18, Tyr58, Gly97, Glu127, and either waters 10 and 16 in subunit A or waters 28 and 78 in subunit B. One end of this ethylene glycol molecule appears to form an H-bond with Tyr58 and Glu127, and the other appears to form H-bonds with Glu127, Asn17, Glu18 as well as two water molecules. One hydroxyl end of the inner ethylene glycol is buried in the empty dinuclear iron site and is bound by Glu51, Glu94, and Glu127, whereas the other hydroxyl group is not bound by the protein but points toward the opening that is surrounded by residues Glu47, Asp50, and Asp126. Interestingly, the positions of Glu47 and His46 adjacent to the ferroxidase site on the inner surface of the protein (Fig. 3, C and D) differ from their positions in the structure of fully occupied E. coli BFR (5, 6, 10).
FIGURE 2.
Images of the partially occupied dinuclear iron site. A, 2Fo − Fc representative electron density of the dinuclear iron site contoured at 1 σ (blue) and 5 σ (green). B, the metal ion (magenta) and ethylene glycol (yellow) are highlighted. His130 is directed away from the vacant site where the iron is normally coordinated by His130.
FIGURE 3.
Molecular surfaces showing the ferroxidase pore openings at the inner and outer surface. A, side view showing the relative positions of the ethylene glycol molecules and metal ion inside the protein. B, outer surface of BFR subunit dimer showing ethylene glycol molecule colored yellow in outer pore. C, inner surface of wild-type BFR showing previously observed conformations of His46, Glu47, and His130 in the closed state. D, inner surface of BFR subunit dimer showing ethylene glycol molecule in inner pore and alternate conformations of His46, Glu47, and His130 in the open state.
As described under “Experimental Procedures,” the BFR used in the crystallization studies was freshly prepared subunit-dimer variant that had been repeatedly exchanged against buffer containing a dialysis bag of chelating resin to remove metal ions. Thus, the mechanistic implications of the presence of Zn2+ in only one part of the ferroxidase site are uncertain, although it does seem apparent that the two metal ion-binding sites are inequivalent insofar as zinc binding is concerned. The notable result here is that Zn2+ has been found at the ferroxidase site, as it was (as a mixture with iron) in a recent report of the structure of the E. coli 24-mer protein (10). This observation provides strong evidence in favor of our suggestion that Zn2+ inhibits BFR by binding at its ferroxidase center (see below).
Kinetics of Fe2+ Oxidation by the E128R/E135R Variant
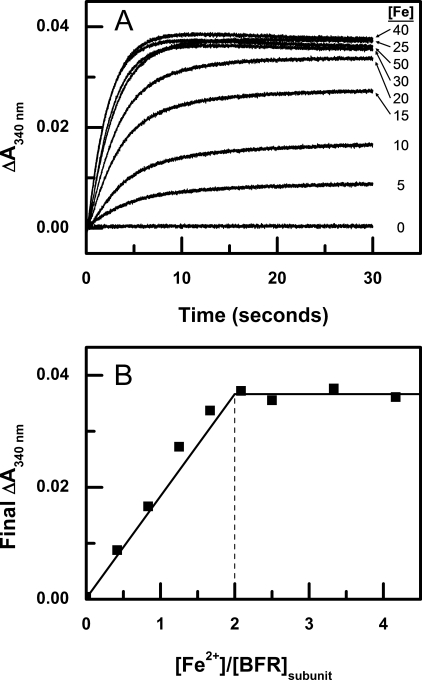
On addition of Fe2+ to the subunit dimer variant, a biphasic increase in absorbance at 340 nm is observed. The initial burst phase of oxidation, the so-called Phase 2, is clearly distinct from the slower Phase 3. The kinetics of Phase 2 were studied by stopped flow mixing experiments in which the iron-free protein was mixed with varying concentrations of Fe2+, and the resulting absorbance change was monitored at 340 nm (Fig. 4A). For iron loadings of 25 μm and greater, absorbance at 340 nm decreases slightly after completion of Phase 2. This effect has been observed in wild-type bacterioferritin in the presence of phosphate (33). As can be seen, the change in absorbance at 340 nm reaches a maximum when ∼2 equivalents of Fe2+ are added for each BFR subunit. This amount of iron corresponds to the number of iron atoms required to occupy the dinuclear iron sites completely (Fig. 4B). These data provide the first clear demonstration that the ferroxidase centers of the subunit dimer variant exhibit activity similar to that of the wild-type 24-mer, because the earlier kinetic study of the subunit dimer variant (18) did not show this. Consequently, these results provide critical validation of the subunit dimer variant as a functional model that is relevant to the intact 24-mer wild-type protein.
FIGURE 4.
Stopped flow kinetic measurements of iron oxidation after addition of Fe2+ to apo-BFR at 25 °C. A, Fe2+ was added to final concentrations of 0, 5, 10, 15, 20, 25, 30, 40, and 50 μm to subunit dimer (6 μm BFR) in 0.1 m MES buffer (pH 6.5). B, final absorbance change after 30 s of iron oxidation at various ratios of Fe2+/BFR subunit concentration.
Zinc Inhibition of Fe2+ Oxidation by E128R/E135R BFR
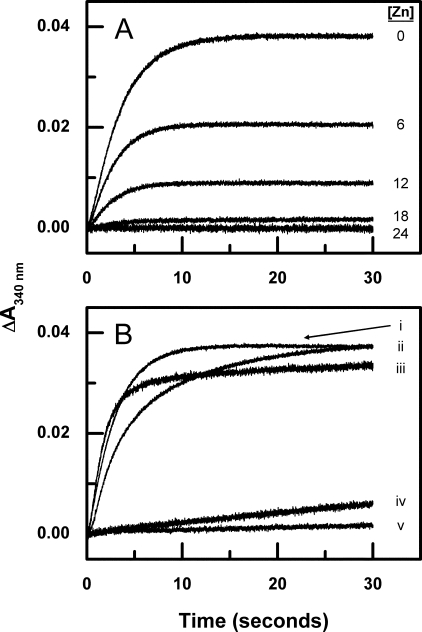
Zinc has been shown previously to inhibit the ferroxidase activity of wild-type BFR. Although the mechanism of this inhibition has been proposed to involve binding of Zn2+ to the ferroxidase sites (25, 26, 34), there is no direct evidence for this. Thus, it remains possible that Zn2+ inhibits 24-mer BFR by binding to the channels located between protein subunits, thereby preventing access of Fe2+ to the central cavity. Because the latter mechanism is not possible with the variant used in the current work, the effect of Zn2+ on the ferroxidase activity of the subunit dimer variant was evaluated by stopped flow experiments in which increasing concentrations of Zn2+ were added to the BFR prior to mixing with Fe2+. As reported for wild-type BFR (25), the addition of Zn2+ to the apo-BFR subunit dimer inhibits oxidation of Fe2+ (Fig. 5A) in a manner that is not linearly dependent on [Zn2+] and that is essentially complete following the addition of two equiv of Zn2+ for each BFR subunit. Thus, the effect of Zn2+ on the kinetics of Fe2+ oxidation does not require the presence of channels that are present in the structure of the wild-type 24-mer protein, consistent with Zn2+ inhibiting Fe2+ oxidation by binding to the ferroxidase centers.
FIGURE 5.
Stopped flow kinetic measurements of iron oxidation after addition of Fe2+ to zinc bound BFR and apo-BFR nucleation site mutants. A, inhibition of Phase 2 iron oxidation by various zinc concentrations was measured by stopped flow mixing at 25 °C. 25 μm Fe2+ was added to 6 μm BFR dimer in 0.1 m MES buffer (pH 6.5) with [Zn2+] = 0, 6, 12, 18, and 24 μm. B, 25 μm Fe2+ was added to (i) BFR subunit dimer, (ii) subunit dimer D50N, (iii) subunit dimer D126N, (iv) subunit dimer E47Q, and (v) 6 μm subunit dimer E47Q/D50N/D126N in 0.1 m MES buffer (pH 6.5).
E47Q, D50N, and D126N Variants of E128R/E135R BFR
Three acidic residues, Glu47, Asp50, and Asp126, have been noted previously to occur on the inner surface of the protein in proximity to the ferroxidase center (11, 18). In the structure shown in Fig. 3D, these acidic residues are also notable because they form a negative electrostatic surface close to one of the ethylene glycol molecules that is partially exposed on the inner surface of the protein. The proximity of these residues to this bound ethylene glycol molecule reinforces the previous suggestion that this region of the protein provides a possible site for nucleation of the iron core. To evaluate this possibility, site-directed mutagenesis was used to replace Glu47, Asp50, and Asp126 with the corresponding amidated residues, individually and in combination with each other, for kinetics analysis. The far-UV CD spectra of the BFR subunit dimer and its E47Q, D50N and D126N variants are essentially identical to each other (Fig. 6A). Similarly, the electronic absorption spectra of these proteins (Fig. 6B) are identical to that exhibited by the wild-type 24-mer (33). These spectroscopic data provide evidence that the heme-binding sites, the high content of helical structure, and the three-dimensional structure of the subunit dimer in general are not perturbed to any significant extent by the replacement of the surface carboxylates by amides.
FIGURE 6.
Comparison of far-UV circular dichroism and electronic absorption spectra of the wild-type BFR dimer and its variants. A, CD spectra of BFR subunit dimer (solid line), subunit dimer E47Q (· · ·), subunit dimer D50N (- · - ·), subunit dimer D126N (- · ·), and subunit dimer E47Q/D50N/D126N (- - -) in 20 mm MES buffer (pH 6.5). B, electronic spectra of BFR subunit dimer (solid line), subunit dimer E47Q (· · ·), subunit dimer D50N (- · - ·), subunit dimer D126N (- · ·), and subunit dimer E47Q/D50N/D126N (- - -) in 100 mm MES buffer (pH 6.5).
Stopped flow kinetic measurements demonstrate the effect of the single E47Q, D50N, and D126N mutations on Phase 2 iron oxidation activities relative to the activity of the parent subunit dimer (Fig. 5B). The D50N and D126N variants exhibit small differences with a slight reduction in rate for the D50N variant, the E47Q variant exhibiting a large reduction in activity and the E47Q/D50N/D126N triple variant exhibiting the most significantly reduced activity. Presumably, the influence exhibited by replacement of Glu47, which was greater than the other acidic residues of this group, results from the shorter distance between this residue and FE2 (6.3 as opposed to 7.6 and 8.0 for Asp50 and Asp126, respectively) as measured from the side chain carboxyl carbon in the structure of van Eerde et al. (10), but clearly the ferroxidase centers of the D50N and D126N variants function in a manner that is similar to that of the parent dimer.
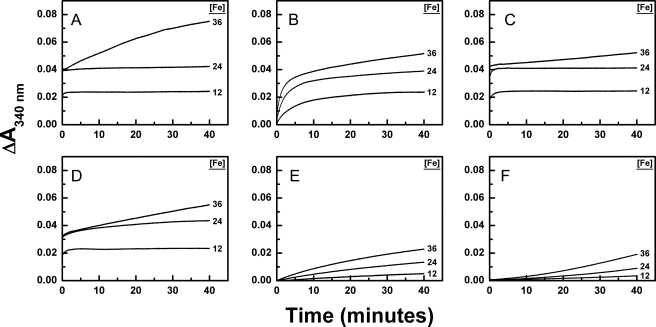
Measurements were performed to compare the kinetics of Phase 3 iron oxidation by the dimeric variant of BFR with that of its amidated variants (Fig. 7). Under the conditions of these experiments, full occupancy of the dinuclear iron sites is expected to occur at an iron concentration of 24 μm. The D50N and D126N variants exhibited similar iron oxidation activity up to 24 μm of iron added and diminished Phase 3 activity relative to the unmodified dimeric BFR variant (Fig. 7, compare A with C and D). As observed for the Phase 2 kinetics, the E47Q and E47Q/D50N/D126N variants exhibit reduced activity at all iron concentrations, but their activity is greater than that observed for iron autooxidation alone. Once again, the triple variant demonstrates the most dramatically diminished ferroxidase activity Fe2+ (Fig. 7, B and E).
FIGURE 7.
Kinetic measurements of iron oxidation after addition of Fe2+ to apo-BFR (6 μm) (25 °C). Fe2+ was added to final concentrations of 12, 24, and 36 μm to 6 μm (A) BFR subunit dimer, (B) subunit dimer E47Q, (C) subunit dimer D50N, (D) subunit dimer D126N, and (E) subunit dimer E47Q/D50N/D126N in 0.1 m MES buffer (pH 6.5). F, auto-oxidation of Fe2+ was measured in 0.1 m MES buffer (pH 6.5) in the absence of protein.
DISCUSSION
The three-dimensional structure of the E128R/E135R variant unambiguously confirms the dimeric structure of the BFR subunit dimer and establishes that the three-dimensional structures of the individual monomers are nearly identical to those of monomers in wild-type BFR (10) (root mean square deviation = 1.43 Å over all 158 Cα atoms). One notable difference, however, can be observed in the position of the small C-terminal α-helix, which forms the intersubunit 4-fold channels in the wild-type protein; removal of the intersubunit contact leads to a small repositioning of this helix relative to the four-helix bundle. Omitting 15 residues from the C-terminal α-helix and using only residues 1–143 resulted in a root mean square deviation of 0.48 Å. The remarkable structural similarity between the stabilized subunit dimer and a subunit dimer from the wild-type 24-mer protein indicates that the main features of the structure of the subunit dimer do not require the higher order assembly of the wild-type protein.
Cryoprotection of crystals with ethylene glycol serendipitously resulted in integration of one ethylene glycol molecule into a pocket that opens on the outer surface of the monomer and provides access to one-half of the dinuclear center. A second ethylene glycol molecule is integrated into a corresponding pocket that is accessed from the inner surface of the monomer and provides access to the other half. The ethylene glycol molecule integrated into the outer surface of the protein is located in a position that corresponds to that of a so-called ferroxidase pore that has been observed in the structure of D. desulfuricans (8) and that was suggested to be the route by which iron enters the ferroxidase site and iron core of that protein. Thus, ethylene glycol appears to be a suitable probe for hydrated Fe2+ despite their differing shape and size. In the current structure, residue Asn17 is located close to the ethylene glycol on the inner surface, so the two positions described above for this residue probably result from the presence of the ethylene glycol molecule.
Iron has been proposed previously to be translocated from the dinuclear iron site to the iron core by movements of residues His59 and Glu131 in D. desulfuricans BFR and residues Glu47 and His130 in A. vinelandii BFR (7, 9). The observation of a similar conformational change by residues Glu47 and His130 in the current structure suggests that a structural change of this type upon metal ion release from, or partial occupancy of, the ferroxidase center is characteristic of this family of proteins. Frolow and Kalb (32) have reported that the uranyl derivative of E. coli BFR exhibits a similar movement of Glu47 and His130 for the binding of uranyl in the dinuclear iron site. A significant body of mechanistic data on the E. coli BFR protein indicates that following the Phase 2 oxidation of the di-Fe2+ ferroxidase center, a stable bridged di-Fe3+ center is generated (17, 19). Further high resolution structural data on iron-bound forms of the ferroxidase center are now required to further address the fate of Fe3+ following oxidation at the ferroxidase center.
The ethylene glycol on the inner surface of the E128R/E135R variant is close to the negatively charged residues Glu47, Asp50, and Asp126 that have been speculated previously as participating either in guiding of Fe2+ to the unoccupied dinuclear iron center or nucleation of the iron core (3, 11, 18), although clear evidence that these residues are involved in such processes has not been reported until now. To evaluate the potential functional role of these residues in the E128R/E135R variant, the ferroxidase kinetics of variants in which residues Glu47, Asp50, and Asp126 were replaced individually and in combination by the corresponding amides were investigated. Stopped flow measurements established that the E47Q and E47Q/D50N/D126N variants possess markedly decreased Phase 2 iron oxidation activity relative to the wild-type protein and the D50N and D126N variants. These results clearly support a significant role for Glu47 in the ferroxidase activity of BFR. Because Glu47 is not a ferroxidase center ligand, the mechanistic origin of this effect is not clear. For example, Glu47 could play a role in guiding iron to the ferroxidase center, which may result simply from the closer proximity of this residue to FE2 than is the case for Asp50 or Asp126 as noted above, although the difference in orientation of this residue in the current structure and in protein with both irons present (10) may also be a factor. Both D50N and D126N exhibit diminished reactivity during Phase 3 of Fe2+ oxidation. This result supports a role for Asp50 and Asp126 in the nucleation step of BFR iron core formation. Notably, these modifications of the electrostatic properties of the inner surface of the dimeric variant are sufficient to impede oxidation of Fe2+ in the absence of changes to the corresponding outer surface of the protein adjacent to the outer ferroxidase pore.
To evaluate the possibility that the acidic residues studied in the current work might participate similarly in the ferroxidase activity of other bacterioferritins, the sequences of several members of this protein family are aligned (35) in Fig. 8. From this analysis, it is clear that each of these residues is highly but not absolutely conserved, so it seems highly likely that the functional contributions of these residues observed in the present study would also be observed for these other species of the protein. In those unusual cases where one of these acidic residues is not conserved, an adjacent acidic residue may occupy the same stereochemical position, but confirmation of this possibility must await detailed characterization of these other proteins.
FIGURE 8.
Alignment of amino acid sequences of several bacterioferritins. The acidic residues replaced in the current work are indicated by arrows. The intensity of the background shading is proportional to the extent of conservation.
Supplementary Material
Acknowledgments
We thank Dr. David S. Waugh (National Cancer Institute-Frederic Cancer Research and Development Center) for the plasmid for expression of the TEV protease and Dr. Susanne Ludwiczek for samples of the purified TEV protease.
This work was supported by a Canadian Institutes of Health Research/Canadian Blood Services partnership grant (to A. G. M.), Canadian Institutes of Health Research Grant MOP-49597 (to M. E. P. M.), and Grants 83/B14704 and BB/D001943/1 from the Biotechnology and Biological Sciences Research Council (to G. R. M. and N. E. L.). This work was also supported in part by a grant from the Canadian Foundation for Innovation to the University of British Columbia Laboratory of Molecular Biophysics, an infrastructure support grant from the Michael Smith Foundation for Health Sciences to the University of British Columbia Centre for Blood Research and the University of British Columbia Laboratory of Molecular Biophysics, and funds from the United States Department of Energy Office of Basic Energy Sciences and Office of Biological and Environmental Research, the National Institutes of Health, the National Center for Research Resources, the Biomedical Technology Program, and the National Institute of General Medical Sciences.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
The atomic coordinates and structure factors (code 3E2C) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- BFR
- bacterioferritin
- MES
- 4-morpholineethanesulfonic acid
- TEV
- tobacco etch virus.
REFERENCES
- 1.Stiefel E. I., Watt G. D. (1979) Nature 279, 81–83 [DOI] [PubMed] [Google Scholar]
- 2.Stiefel E. I., Grossman M. J., Hinton S. M., Minak-Bernero V., George G. N., Prince R. C., Bare R. E., Watt G. D. (1994) Adv. Exp. Med. Biol. 356, 157–164 [DOI] [PubMed] [Google Scholar]
- 3.Lewin A., Moore G. R., Le Brun N. E. (2005) Dalton Trans. 22, 3597–3610 [DOI] [PubMed] [Google Scholar]
- 4.Cobessi D., Huang L. S., Ban M., Pon N. G., Daldal F., Berry E. A. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dautant A., Meyer J. B., Yariv J., Précigoux G., Sweet R. M., Kalb A. J., Frolow F. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 16–24 [DOI] [PubMed] [Google Scholar]
- 6.Frolow F., Kalb A. J., Yariv J. (1994) Nat. Struct. Biol. 1, 453–560 [DOI] [PubMed] [Google Scholar]
- 7.Liu H. L., Zhou H. N., Xing W. M., Zhao J. F., Li S. X., Huang J. F., Bi R. C. (2004) FEBS Lett. 573, 93–98 [DOI] [PubMed] [Google Scholar]
- 8.Macedo S., Romão C. V., Mitchell E., Matias P. M., Liu M. Y., Xavier A. V., LeGall J., Teixeira M., Lindley P., Carrondo M. A. (2003) Nat. Struct. Biol. 10, 285–290 [DOI] [PubMed] [Google Scholar]
- 9.Swartz L., Kuchinskas M., Li H., Poulos T. L., Lanzilotta W. N. (2006) Biochemistry 45, 4421–4428 [DOI] [PubMed] [Google Scholar]
- 10.van Eerde A., Wolterink-van Loo S., van der Oost J., Dijkstra B. W. (2006) Acta Crystallograph. Sect. F Struct. Biol. Cryst. Commun. 62, 1061–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baaghil S., Lewin A., Moore G. R., Le Brun N. E. (2003) Biochemistry 42, 14047–14056 [DOI] [PubMed] [Google Scholar]
- 12.Andrews S. C., Le Brun N. E., Barynin V., Thomson A. J., Moore G. R., Guest J. R., Harrison P. M. (1995) J. Biol. Chem. 270, 23268–23274 [DOI] [PubMed] [Google Scholar]
- 13.Cheesman M. R., Thomson A. J., Greenwood C., Moore G. R., Kadir F. (1990) Nature 346, 771–773 [DOI] [PubMed] [Google Scholar]
- 14.Bou-Abdallah F., Lewin A. C., Le Brun N. E., Moore G. R., Chasteen N. D. (2002) J. Biol. Chem. 277, 37064–37069 [DOI] [PubMed] [Google Scholar]
- 15.Vasil M. L., Ochsner U. A. (1999) Mol. Microbiol. 34, 399–413 [DOI] [PubMed] [Google Scholar]
- 16.Levi S., Santambrogio P., Corsi B., Cozzi A., Arosio P. (1996) Biochem. J. 317, 467–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Brun N. E., Wilson M. T., Andrews S. C., Guest J. R., Harrison P. M., Thomson A. J., Moore G. R. (1993) FEBS Lett. 333, 197–202 [DOI] [PubMed] [Google Scholar]
- 18.Malone S. A., Lewin A., Kilic M. A., Svistunenko D. A., Cooper C. E., Wilson M. T., Le Brun N. E., Spiro S., Moore G. R. (2004) J. Am. Chem. Soc. 126, 496–504 [DOI] [PubMed] [Google Scholar]
- 19.Kilic M. A., Spiro S., Moore G. R. (2003) Protein Sci. 12, 1663–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geiser M., Cèbe R., Drewello D., Schmitz R. (2001) BioTechniques 31, 88–90, 92 [DOI] [PubMed] [Google Scholar]
- 21.Kapust R. B., Tözsér J., Fox J. D., Anderson D. E., Cherry S., Copeland T. D., Waugh D. S. (2001) Protein Eng. 14, 993–1000 [DOI] [PubMed] [Google Scholar]
- 22.Kapust R. B., Tözsér J., Copeland T. D., Waugh D. S. (2002) Biochem. Biophys. Res. Commun. 294, 949–955 [DOI] [PubMed] [Google Scholar]
- 23.Stookey L. L. (1970) Anal. Chem. 42, 779–781 [Google Scholar]
- 24.Liu J. L., Rigolet P., Dou S. X., Wang P. Y., Xi X. G. (2004) J. Biol. Chem. 279, 42794–42802 [DOI] [PubMed] [Google Scholar]
- 25.Yang X., Le Brun N. E., Thomson A. J., Moore G. R., Chasteen N. D. (2000) Biochemistry 39, 4915–4923 [DOI] [PubMed] [Google Scholar]
- 26.Le Brun N. E., Andrews S. C., Guest J. R., Harrison P. M., Moore G. R., Thomson A. J. (1995) Biochem. J. 312, 385–392 [PMC free article] [PubMed] [Google Scholar]
- 27.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 28.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 29.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 30.Collaborative Computational Project (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 31.Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 32.Frolow F., Kalb A. J. (2001) in Handbook of Metalloproteins (Messerschmidt A. ed) pp. 782–790, Wiley, New York [Google Scholar]
- 33.Aitken-Rogers H., Singleton C., Lewin A., Taylor-Gee A., Moore G. R., Le Brun N. E. (2004) J. Biol. Inorg. Chem. 9, 161–170 [DOI] [PubMed] [Google Scholar]
- 34.Le Brun N. E., Keech A. M., Mauk M. R., Mauk A. G., Andrews S. C., Thomson A. J., Moore G. R. (1996) FEBS Lett. 397, 159–163 [DOI] [PubMed] [Google Scholar]
- 35.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.