Abstract
Cytochrome c oxidase (COX) is the terminal enzyme of the electron transport chain composed of 13 subunits; three are mitochondria-encoded, and 10 are nucleus-inscribed on nine different chromosomes within the mammalian genome. The transcriptional regulation of such a multisubunit, multichromosomal, and bigenomic enzyme is mechanistically challenging. Transcription factories have been proposed as one mechanism by which genes from different genomic loci congregate to transcribe functionally related genes, and chromosome conformation capture (3C) is a means by which such interactions can be revealed. Thus far, however, only loci from the same chromosome or at most two chromosomes have been co-localized by 3C. The present study used 3C to test our hypothesis that not only the 10 genomic loci from nine chromosomes encoding the 10 nuclear subunits of COX, but also genes from three chromosomes encoding mitochondrial transcription factors A and B (Tfam, Tfb1m, and Tfb2m) critical for the transcription of the three mitochondria-encoded COX subunit genes all occupy common intranuclear sites in the murine neuronal nuclei. The pairing of various COX subunit genes and Tf genes indicates that interactions are present among all of them. On the other hand, genes for a non-mitochondrial protein (calreticulin) as well as a mitochondrial enzyme (citrate synthase) did not interact with COX genes. Furthermore, interactions between COX subunit and Tf genes were up-regulated by depolarizing stimulation and down-regulated by impulse blockade in primary neurons. Thus, a viable mechanism is in place for a synchronized, coordinated transcriptional regulation of this multisubunit, bigenomic COX enzyme in neurons.
Cytochrome c oxidase (COX)2 or complex IV of the mitochondrial electron transport chain is made up of three subunits encoded in the mitochondrial genome and 10 nucleus-encoded subunits located on nine different chromosomes (1–3). The 10 nucleus-encoded COX subunits are 4, 5a, 5b, 6a, 6b, 6c, 7a, 7b, 7c, and 8a. Subunits 4, 6a, 6b, 7a, and 8a each has a ubiquitous isoform and a tissue-specific isoform. Transcription of all ubiquitously expressed subunits in neurons are coordinately regulated according to the cell's ATP requirement (4). Transcriptional regulators of COX in rats, mice, and humans include NRF-1 and NRF-2 (nuclear respiratory factors 1 and 2) (5–8). In particular, both NRF-1 and NRF-2 activate the transcription of all 10 nucleus-encoded COX subunits and modulate the level of transcription in response to changing cellular energy demands in neurons (9–14). Additionally, NRF-1 and NRF-2 indirectly activate the three mitochondria-encoded COX subunit genes by regulating Tfam, Tfb1m, and Tfb2m (mitochondrial transcription factors A, B1, and B2) critical for mitochondrial DNA transcription and replication (15–18). Transcriptional coactivators, such as PGC-1α (peroxisome proliferator-activated receptor γ-coactivator 1α) (19, 20), also play an important role in stimulating COX transcription in the presence of upstream signals in neurons (5, 21, 22). Thus, regulation of these 13 COX subunits has been credited to a small set of transcription factors and coactivators (21). Nevertheless, it remains unknown if all 10 nuclear subunits and mitochondrial transcription factors from different chromosomes are transcribed simultaneously within the nucleus. Such a synchronous process would enhance the efficiency of coordination as well as ensure that the 1:1 stoichiometry of the 13 subunits in the holoenzyme is maintained.
Transcription of nuclear genes is known to occur in discrete sites within the nucleus. Known as transcription factories, these sites of active transcription are thought to contain several RNA polymerase II molecules surrounded by transcription factors and loops of specific chromatin encoding actively expressing genes (23–25). Furthermore, loci on one or two different chromosomes seem to co-localize to the same transcription factory (26–28). Such long range interactions among the genes can be studied using chromosome conformation capture (3C), a recently developed technique that can convert chromatin conformation and physical interactions in vivo into a specific ligation product demonstrable with PCR (29, 30). However, only loci from the same chromosome or at most two chromosomes have thus far been co-localized by 3C.
The present study employed 3C to test our hypothesis that all 10 nuclear subunits of COX encoded on nine different chromosomes as well as Tfam, Tfb1m, and Tfb2m genes, located on three chromosomes and vital for the transcription of the three mitochondria-encoded COX subunit genes, all reside in the same intranuclear locale in neuronal nuclei. Pairing of various COX subunit genes and mitochondrial transcription factor genes indicates physical interactions among all of them. Thus, our results are consistent with a viable mechanism by which multiple subunit-coding genes from different chromosomes as well as transcription factor genes are simultaneously recruited to a common transcription factory for the coordinated expression of a multisubunit, multichromosomal bigenomic enzyme in neurons.
EXPERIMENTAL PROCEDURES
In Silico Analysis of Mouse COX Subunit Gene Sequence
Genomic sequence of the 10 murine nucleus-encoded COX subunit genes, including chromosomal positions and surrounding non-coding sequences, were obtained from the mouse genome data base (3). Each COX subunit gene was independently verified by comparing coding sequence with known COX cDNA sequence and confirming the presence of intron-exon structure. DNA sequence corresponding to each COX gene and its surrounding 10,000-bp region was analyzed for the presence of BglII restriction sites.
Cell Culture
Mouse N2a neuroblastoma cells (ATCC, Manassaa, VA) were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT). Cells were passaged every 2–3 days and harvested at 80–90% confluence for 3C experiments.
Primary Cortical Neuronal Cultures and Treatment with KCl or TTX
All experiments were carried out in accordance with National Institutes of Health guidelines (43) and Medical College of Wisconsin regulations. All efforts were made to minimize the number of animals and their suffering.
Primary visual cortical neurons were prepared as described previously (9). Briefly, 1–2-day-old Sprague-Dawley rat pups were anesthetized with CO2 and killed by decapitation. The brains were removed and cleansed of meninges and surface blood vessels, and the cortices were dissected, trypsinized, and triturated to dissociate individual neurons. Cells were plated onto a poly-l-lysine (Sigma)-coated 100-mm dish at a density of ∼5 × 106 cells/ml in Dulbecco's minimal essential medium (Invitrogen) with 10% fetal bovine serum (Hyclone) for 3 h. The medium was changed to neurobasal/B27 supplement (Invitrogen), and cultures were maintained at 37 °C with 5% CO2 in a humidified incubator. Cytosine arabinoside (Sigma) was added on the second day of plating neurons to inhibit the replication of non-neuronal cells. Neuronal cultures were maintained by replacing half of the medium every 5 days. Cells were cultured for 7–15 days before exposure to 20 mm KCl for 5 h. TTX (Sigma) at a final concentration of 0.4 μm was added to the culture medium from the 12th day after plating. Cultures were exposed to TTX for 3 days. All cells were harvested on the same day for each experiment, and 3C samples were prepared for each group.
Control cultures were neither depolarized with KCl nor inactivated by TTX. Real time quantitative PCRs were carried out in a Cepheid Smart Cycler detection system (Cepheid, Sunnyvale, CA). SyBr Green (Biowhittaker Molecular Application, Rockland, ME) and EX Taq real time quantitative PCR hot start polymerase (Takara Mirus Bio, Madison, WI) were used following the manufacturer's protocols and as described previously (11).
Chromosome Conformation Capture
3C experiments were performed as described previously (31) with minor modifications. Briefly, N2a cells and primary neurons were harvested by trypsinization and resuspended in fresh culture medium with 10% fetal bovine serum. To cross-link chromatin, 1 × 107 N2a cells or 5 × 106 primary neurons in 10 ml of cell culture medium were treated with 2% formaldehyde for 10 min at room temperature. Cross-linking was stopped by quenching with 0.125 m glycine for 5 min at room temperature. Nuclei were isolated by incubating cells in a lysis buffer (10 mm Tris, pH 8.0, 10 mm NaCl, 0.2% Nonidet P-40) on ice with agitation for 30 min. Chromatin was subsequently released by treating isolated nuclei with 0.3% SDS and digested with 400 units of BglII (Fermentas, Hanover, MD) overnight. Afterward, restriction enzyme was heat-inactivated, and chromatin was diluted in T4 ligation buffer to achieve a low genomic DNA concentration of 2.5 ng/μl to facilitate intermolecular religation. Chromatin religation was performed by incubating diluted chromatin with 100 Weiss units of T4 ligase (Fermentas) for 4 h at 16 °C. Following RNase A (Roche Applied Science) digestion to remove RNA contaminant, the 3C chromatin was purified by phenol/chloroform extraction.
Generation of Control Templates
To generate individual PCR template controls for comparison with 3C product, all 10 COX subunit genes were paired, as were the pairing of other genes or gene loci, as shown in the control lanes (left columns) of Figs. 1–3. For example, to generate the control template for COX4i1/COX5a, we used PCR to amplify the regions spanning a single BglII site of interest with 100–150 bp on either side for each subunit gene. Approximately 30 μg of each PCR product were digested with 300 units of BglII overnight at 37 °C. DNA was extracted using phenol chloroform and ethanol precipitation. Equimolar amounts of digested product (COX4i1 and COX5a) were ligated at a high concentration (∼10 ng/μl for each product) to ensure formation of intermolecular cross-linked product. One primer of a given restriction fragment (COX4i1) was combined with a primer of a second (COX5a) in all pairwise combinations of ligated orientations (e.g. forward primer of COX4i1 with forward primer of COX5a, reverse primer of COX4i1 with forward primer of COX5a, etc.). For all other pairing of COX genes, the same steps were used to generate a large number of pairings among subunits and were used as control templates for comparison with 3C samples. The final chosen control template for each pairing had the same orientation as the 3C-generated product. By comparing the signals obtained by quantitative PCR for the 3C cross-linked templates versus the control templates, we corrected for the differences in amplification efficiency between primer sets and also for differences in signal intensities due to the size of the PCR products (since all of the primers were designed to give PCR products of 150–300 bp) (supplemental Table 1). All primers were designed to have an annealing temperature of 58–60 °C, and they all yielded a product when used with the positive control templates. These control templates and 3C samples for each primer pair were performed during the same PCR run.
FIGURE 1.
Positive and negative controls for chromosome conformation capture. All possible restriction fragments after BglII digestions were present in equimolar amounts. A, two regions of calreticulin gene, CalR2.8 and CalR4.4, were cross-linked and represented a positive control. On the other hand, Cox6a1 and CalR were two functionally unrelated genes, and they did not interact in our 3C reactions. Likewise, a mitochondrial enzyme citrate synthase also failed to interact with COX4i1, COX6a1, and COX8a in our 3C reactions. B, CalR2.8 and CalR4.4, COX4i1, and COX8a as well as Tfam did not interact in the absence of a cross-linking agent (formaldehyde) or T4 ligase. Primers far beyond the BglII sites for COX4i1 and COX8a did not yield any interaction between COX subunits. Experiments were performed in triplicates.
FIGURE 2.
3C interactions among all 10 nucleus-encoded COX subunit genes. All possible interactive pairs among COX4i1, -5a, -5b, -6a1, -6b, -6c, -7a2, -7b, -7c, and -8a were tested by PCR targeting their cross-linking products. COX4i1 and COX8a consistently interacted with each of the remaining nine subunits of COX genes. Pairing of the remaining eight COX subunits with other randomly selected COX genes all yielded positive hybrid product (see also Table 2). All experiments were performed in triplicates.
FIGURE 3.
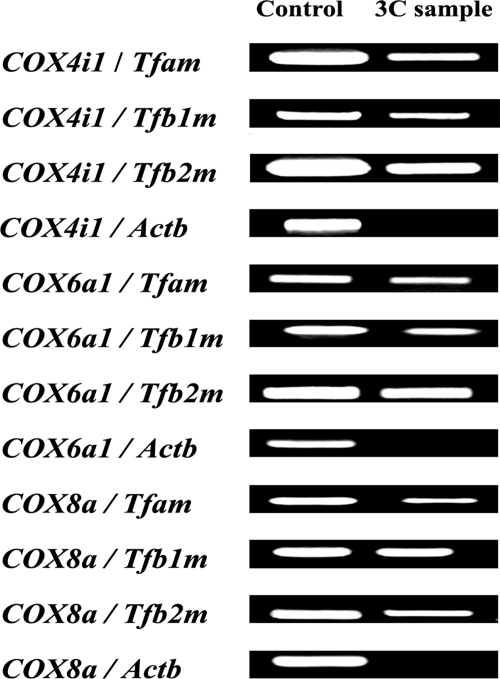
Co-localization of COX subunits with Tfam, Tfb1m, and Tfb2m. Cross-linked products between each of three nucleus-encoded COX subunit genes and Tfam, Tfb1m, and Tfb2m, respectively, were consistently detected by 3C reactions. β-Actin (Actb) was used as a negative control, and it did not cross-react with any of the three COX subunits tested.
A positive 3C control targeted an “internal” cross-linking product between two BglII sites located 1.6 kb apart within the coding region of the CalR (calreticulin) gene (+2.8 and +4.4 kb relative to the CalR transcriptional start point) (31) (Fig. 1A).
Five sets of 3C negative controls were employed: (a) possible cross-linking between the CalR 4.4-kb BglII site and another BglII site on COX6a1; (b) possible cross-linking between the BglII site of Cs (citrate synthase) and those of COX4i1, COX6a1, and COX8a, respectively; (c) no cross-linking agent (formaldehyde); (d) no ligase; and (e) possible cross-linking for regions far beyond the BglII sites for COX4i1 and COX8a (Fig. 1, A and B).
PCR Assays
The PCR conditions used were very stringent in order to detect only specific signals. PCR cycles used were as follows: initial denaturing step for 2 min at 94 °C followed by 36 repeats of a cycle with 30 s at 94 °C, 15 s at 59 °C, and 15 s at 72 °C, followed by a final step of 1 min at 72 °C. Thirty-six PCR cycles were sufficient to detect the specific PCR fragments generated after digestion and ligation that captured the rare molecular phenomenon occurring in the nucleus. Cross-linking among COX subunit gene loci in 3C samples was detected using semiquantitative PCR. Specifically, a pair of primers targeting two different COX subunit genes was used to amplify cross-linked DNA generated from 3C reactions. Each primer targeted a DNA region 100–150 bp from the BglII site and generated 200–300-bp amplicons. Primers were designed for all 10 nucleus-encoded COX genes, as shown in supplemental Table 1. The linear range of amplification was determined for 3C samples by serial dilution, as described previously (28, 30). An appropriate amount (150 μg) of 3C DNA within the linear range was added to each PCR. PCR product was run on 2% agarose gel stained with ethidium bromide. A large number of possible interactive pairs among 10 COX subunits (4i1, 5a, 5b, 6a1, 6b, 6c, 7a2, 7b, 7c, and 8a) were tested by PCRs targeting their possible cross-linked products (Table 2). All PCRs were performed in triplicates.
TABLE 2.
3C interactions among all 10 nucleus-encoded COX subunit genes
A plus sign indicates positive cross-linked product obtained by PCR between two interacting COX genes.
| Gene | COX4i1 | COX5a | COX5b | COX6a1 | COX6b | COX6c | COX7a2 | COX7b | COX7c | COX8a |
|---|---|---|---|---|---|---|---|---|---|---|
| COX4i1 | + | + | + | + | + | + | + | + | + | |
| COX5a | + | + | + | + | + | |||||
| COX5b | + | + | + | + | + | |||||
| COX6a1 | + | + | + | + | ||||||
| COX6b | + | + | + | + | + | |||||
| COX6c | + | + | + | + | + | + | ||||
| COX7a2 | + | + | + | + | + | + | ||||
| COX7b | + | + | + | + | + | + | ||||
| COX7c | + | + | + | + | + | + | ||||
| COX8a | + | + | + | + | + | + | + | + | + |
RESULTS
Distribution of COX Subunit Genes among Mouse Chromosomes
Chromosomal distributions of 10 nucleus-encoded COX subunit genes and mitochondrial transcription factors (Tfam, Tfb1m, and Tfb2m) in the mouse genome are summarized in Table 1. The ubiquitously expressed COX subunit genes (4i1, 5a, 5b, 6a1, 6b, 6c, 7a2, 7b, 7c, and 8a) are dispersed throughout the mouse genome on different chromosomes (chromosomes 1, 5, 7, 8, 9, 13, 15, 19, and X), with the exception of COX5a and COX7a2, both of which are on chromosome 9 but are 22.23 Mb apart and do not form a gene cluster. COX5a is located on the 9 B locus, whereas COX7a2 is on the 9 E1 locus. All COX heart/skeletal muscle-specific isoforms (COX6a2, -7a1, and -8b) are located on chromosome 7. The COX6b testes isoform is also on chromosome 7. COX4i2, the lung isoform of COX4, is on chromosome 2. The present study focused only on the ubiquitous isoforms present in neurons.
TABLE 1.
Chromosomal location of murine nucleus-encoded COX subunit genes arranged by chromosomal number
Genes encoding liver/ubiquitous and tissue-specific isoforms are shown in their respective columns.
| Chromosome | Ubiquitous isoforms | Tissue-specific isoforms |
|---|---|---|
| 1 | COX5b, Tfb2m | |
| 2 | COX4i2 (lung) | |
| 5 | COX6a1 | |
| 7 | COX8b (heart/muscle) | |
| 7 | COX6a2 (heart/muscle) | |
| 7 | COX6b1 | |
| 7 | COX6b2 (testes) | |
| 7 | COX7a1 (heart/muscle) | |
| 8 | COX4i1 | |
| 9 | COX5a | |
| 9 | COX7a2 | |
| 10 | Tfam | |
| 13 | COX7c | |
| 15 | COX6c | |
| 17 | Tfb1m | |
| 19 | COX8a | |
| X | COX7b |
Co-localization of Nucleus-encoded COX Subunit Genes
To make certain our ability to identify interactions between two non-contiguous DNA regions, a positive control PCR was achieved using primers targeting an “internal” cross-linkable product between two BglII sites located 1.6 kb apart within the coding region of the CalR gene (Fig. 1A). CalR and COX6a1 are unrelated genes and would co-localize only at a background frequency. As Fig. 1A illustrates, the internal cross-linking within the CalR gene was detected by PCR of our 3C samples. In contrast, any chance cross-linking event between CalR and COX6a1 was not detected by our PCRs. PCRs were also performed to target for possible cross-linking product between the BglII site of another mitochondrial enzyme, Cs, and those of COX4i1, COX6a1, or COX8a, respectively. Again, no background cross-linking between citrate synthase and all three COX subunits was detectable in our PCR product (Fig. 1A). In the absence of a cross-linking agent (formaldehyde) or T4 ligase, PCR products were not detected among CalR, COX4i1, COX8a, and Tfam genes (Fig. 1B). Moreover, primers designed for regions beyond BglII sites for COX4i1 and COX8a did not yield any cross-linking interactions (Fig. 1B).
A large number of possible interactive pairs among the 10 COX subunits were tested by PCR. Initially, COX4i1 and COX8a were individually documented to form a cross-linking pair with each of the remaining nine subunits of COX genes (Fig. 2 and Table 2). Each of the remaining eight COX subunit genes was then tested for, and their interactions were confirmed with a few other COX subunit genes (Fig. 2 and Table 2). Thus, all possible interactions that were tested were consistently detectable by 3C in our PCR product.
3C Interactions between Mitochondrial Transcription Factors and Nucleus-encoded COX Subunit Genes
Possible interactions between mitochondrial transcription factors A and B (Tfam, Tfb1m, and Tfb2m) derived from three different chromosomes and COX subunit genes were tested using PCR primers for Tfam, Tfb1m, Tfb2m, COX4i1, COX6a1, and COX8a (supplemental Table 1). β-Actin (Actb) served as a negative control, and PCR was performed, targeting possible cross-linking product between β-actin BglII site and COX4i1, COX6a1, and COX8a BglII sites, respectively. Any chance background cross-linking between β-actin and all three COX subunits was not detectable in our PCR product (Fig. 3). On the other hand, cross-linked products between the three nucleus-encoded COX subunit genes and mitochondrial transcription factors were consistently detected in our 3C-reacted PCR samples (Fig. 3).
Effect of KCl Stimulation and TTX Inactivation on 3C Interactions in Primary Neurons
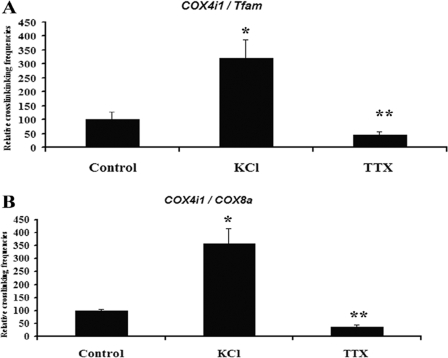
To determine the effect of functional perturbations on COX gene interactions, primary neurons were subjected to depolarizing stimulation with 20 mm KCl for 5 h or impulse blockade with TTX for 3 days. As shown in Fig. 4, A and B, KCl treatment induced a 219–259% increase (p < 0.05), whereas TTX resulted in a 55–65% decrease (p < 0.001) in the interactions between COX4i1 and Tfam or COX4i1 and COX8a, as monitored by real time quantitative PCR. KCl and TTX experiments were also performed on N2a cells, and comparable results were obtained (data not shown).
FIGURE 4.
3C interactions in primary neurons exposed to KCl stimulation or TTX impulse blockade. KCl depolarization induced an up-regulation of cross-linking frequencies between COX4i1 and Tfam (A) and between COX4i1 and COX8a (B), whereas TTX exposure for 3 days down-regulated the interactions of these subunits. *, p < 0.05; **, p < 0.001.
DISCUSSION
The present study documents for the first time that a multisubunit, multichromosomal bigenomic enzyme can be precisely regulated at the transcriptional level by having all relevant nucleus-encoded subunit genes and mitochondrial transcription factors be simultaneously located at the same nuclear site, enabling them to be transcribed concurrently in the same presumed transcription factory in mammalian neurons (Fig. 5).
FIGURE 5.
Schematic representation of co-localization of COX subunit genes and mitochondrial transcription factor genes in a presumed transcription factory. Loops of all 10 genomic loci for the 10 nucleus-encoded COX subunit genes and three genomic loci for mitochondrial transcription factors A and B (Tfam, Tfb1m, and Tfb2m) plus RNA polymerase II and the relevant transcription factors (NRF-1, NRF-2, and others?) are schematically depicted as occupying the same transcription factory in the murine neuronal nuclei.
Chromosome conformation capture was devised to capitalize on the assumption that genes that are actively co-expressed are physically localized in a “transcription factory” that contains copies of active RNA polymerases, transcription factors, and associated chromatin undergoing transcription (32). Physical interactions between chromatin within such a factory can be immobilized, extracted, digested with a restriction enzyme for common restriction sites, religated at very low DNA concentrations, reverse cross-linked, and subjected to PCR to yield a hybrid product encompassing nucleotides from the two interacting chromatin or genomic loci. As shown in the present study, such interactions are not random but are highly specific. Genes that do not share common functions did not interact, and even if their gene products target the same organelle (such as citrate synthase and COX), there was no evidence that their chromatin interacted either (Figs. 1 and 3). The original 3C method was applied to the analysis of chromosomal conformation in yeast (29), but the approach was found useful to reveal folding of complex gene loci and active chromatin hub in mammalian cells as well (23–25). Thus far, however, 3C has uncovered interactions only between genomic loci of the same chromosome or at most of two different chromosomes (26–28), and no report has dealt with a multisubunit protein involving 11 chromosomes or 13 genomic loci, let alone a bigenomic enzyme.
What are the advantages of such precise transcriptional regulation of all 13 subunits from the two genomes for COX? First and foremost, COX is a vital enzyme, without which oxidative metabolism cannot be carried to completion and mitochondrial ATP cannot be generated (33, 34). Dysfunction of this enzyme jeopardizes health, and absence of this enzyme is incompatible with life (35, 36). COX is a housekeeping enzyme present in almost every eukaryotic cell (with a few exceptions, such as mature, short lived red blood cells lacking in both nuclei and mitochondria). In highly oxidative organs, such as the heart, kidney, liver, skeletal muscles, and especially the brain, COX is a workhorse that is constitutively active and is exquisitely regulated by the functional demands of the organ, such as the brain (33, 34, 37). Functional COX holoenzyme is composed of the 13 subunits in a 1:1 stoichiometry (1, 38), hence the need to coordinate the transcription of all subunits.
In neurons, all 13 subunit mRNAs of COX have been found to be coordinately up-regulated by depolarizing activity in vitro. With reduced or blockage of neuronal activity in vivo or in vitro, all 13 subunit transcripts were significantly down-regulated (4, 39). The present study extends these findings with the 3C approach and documents that the occurrence of COX subunit genes in the same transcription factory is up-regulated by KCl stimulation and down-regulated by TTX.
To coordinate the transcriptional regulation of all 13 subunits from the two genomes, transcription factor candidates are needed. Both nuclear respiratory factors 1 and 2 (NRF-1 and NRF-2) proved to be viable candidates, since they each regulate the expressions of mitochondrial transcription factors A, B1, and B2 (15–18), which are responsible for the transcription and replication of mitochondrial DNA (16–18) and hence the three mitochondria-encoded COX subunits, and each also regulates all 10 nucleus-encoded subunit genes of COX in neurons (9–11). An important coactivator of NRF-1 and NRF-2, known as PGC-1α (peroxisome proliferator-activated receptor γ coactivator 1α) (19, 20, 40), is also involved in the transcriptional regulation of COX in neurons (5, 22). Thus, a transcription factory for COX gene expression is likely to include all of these factors, in addition to the necessary RNA polymerase II. Such a factory is expected to be in an active state most of the time for functionally active neurons. However, the relevant chromatin loops are likely to be migrating dynamically into and out of transcription factories throughout a 24-h period in response to the functional demands of the cells. As the present study documents, depolarizing stimulation or impulse blockade readily adjusts the frequency of such occurrence in the same transcription factory.
With our recent findings that NRF-1 co-regulates COX and subunits of glutamatergic NMDA and AMPA receptors (41, 42), the possibility exists that under conditions of sustained glutamatergic neurotransmission, transcription of genes for both COX and the glutamate receptors has to be precisely coordinated to balance the supply and demand sides of energy for optimal neuronal functioning. Such molecular coupling may very well require a common transcription factory. Studies are under way to test this hypothesis.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant R01 EY018441.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- COX
- cytochrome c oxidase
- 3C
- chromosome conformation capture
- TTX
- tetrodotoxin.
REFERENCES
- 1.Kadenbach B., Jarausch J., Hartmann R., Merle P. (1983) Anal. Biochem. 129, 517–521 [DOI] [PubMed] [Google Scholar]
- 2.Attardi G., Schatz G. (1988) Annu. Rev. Cell Biol. 4, 289–333 [DOI] [PubMed] [Google Scholar]
- 3.Eppig J. T., Blake J. A., Bult C. J., Kadin J. A., Richardson J. E. (2007) Nucleic Acids Res. 35, D630–D637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang H. L., Ongwijitwat S., Wong-Riley M. T. (2006) Neuroscience 140, 177–190 [DOI] [PubMed] [Google Scholar]
- 5.Liang H. L., Wong-Riley M. T. (2006) Neuroreport 17, 401–405 [DOI] [PubMed] [Google Scholar]
- 6.Scarpulla R. C. (2002) Biochim. Biophys. Acta 1576, 1–14 [DOI] [PubMed] [Google Scholar]
- 7.Scarpulla R. C. (2006) J. Cell. Biochem. 97, 673–683 [DOI] [PubMed] [Google Scholar]
- 8.Scarpulla R. C. (2008) Physiol. Rev. 88, 611–638 [DOI] [PubMed] [Google Scholar]
- 9.Ongwijitwat S., Wong-Riley M. T. T. (2005) Gene 360, 65–77 [DOI] [PubMed] [Google Scholar]
- 10.Ongwijitwat S., Liang H. L., Graboyes E. M., Wong-Riley M. T. T. (2006) Gene 374, 39–49 [DOI] [PubMed] [Google Scholar]
- 11.Dhar S. S., Ongwijitwat S., Wong-Riley M. T. T. (2008) J. Biol. Chem. 283, 3120–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang S. J., Liang H. L., Ning G., Wong-Riley M. T. T. (2004) Eur. J. Neurosci. 19, 1153–1162 [DOI] [PubMed] [Google Scholar]
- 13.Yang S. J., Liang H. L., Wong-Riley M. T. T. (2006) Neuroscience 141, 1181–1192 [DOI] [PubMed] [Google Scholar]
- 14.Nie F., Wong-Riley M. (1999) J. Comp. Neurol. 404, 310–320 [DOI] [PubMed] [Google Scholar]
- 15.Escrivá H., Rodríguez-Peña A., Vallejo C. G. (1999) Biochimie 81, 965–971 [DOI] [PubMed] [Google Scholar]
- 16.Virbasius J. V., Scarpulla R. C. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 1309–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gleyzer N., Vercauteren K., Scarpulla R. C. (2005) Mol. Cell. Biol. 25, 1354–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Topper J. N., Clayton D. A. (1989) Mol. Cell. Biol. 9, 1200–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R. C., Spiegelman B. M. (1999) Cell 98, 115–124 [DOI] [PubMed] [Google Scholar]
- 20.Andersson U., Scarpulla R. C. (2001) Mol. Cell. Biol. 21, 3738–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong-Riley M. T., Liang H. L., Ongwijitwat S. (2008) in Understanding Transcriptional Regulation by Neuronal Activity: To the Nucleus and Back (Dudek S. M. ed) pp. 209–228, Springer Science, New York [Google Scholar]
- 22.Meng H., Liang H. L., Wong-Riley M. (2007) Brain Res. 1175, 10–16 [DOI] [PubMed] [Google Scholar]
- 23.Jackson D. A., Iborra F. J., Manders E. M., Cook P. R. (1998) Mol. Biol. Cell 9, 1523–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osborne C. S., Chakalova L., Brown K. E., Carter D., Horton A., Debrand E., Goyenechea B., Mitchell J. A., Lopes S., Reik W., Fraser P. (2004) Nat. Genet. 36, 1065–1071 [DOI] [PubMed] [Google Scholar]
- 25.Zhou G. L., Xin L., Song W., Di L. J., Liu G., Wu X. S., Liu D. P., Liang C. C. (2006) Mol. Cell. Biol. 26, 5096–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ling J. Q., Li T., Hu J. F., Vu T. H., Chen H. L., Qiu X. W., Cherry A. M., Hoffman A. R. (2006) Science 312, 269–272 [DOI] [PubMed] [Google Scholar]
- 27.Spilianakis C. G., Lalioti M. D., Town T., Lee G. R., Flavell R. A. (2005) Nature 435, 637–645 [DOI] [PubMed] [Google Scholar]
- 28.Lomvardas S., Barnea G., Pisapia D. J., Mendelsohn M., Kirkland J., Axel R. (2006) Cell 126, 403–413 [DOI] [PubMed] [Google Scholar]
- 29.Dekker J., Rippe K., Dekker M., Kleckner N. (2002) Science 295, 1306–1311 [DOI] [PubMed] [Google Scholar]
- 30.Miele A., Dekker J. (2009) Methods Mol. Biol. 464, 105–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tolhuis B., Palstra R. J., Splinter E., Grosveld F., de Laat W. (2002) Mol. Cell 10, 1453–1465 [DOI] [PubMed] [Google Scholar]
- 32.Xu M., Cook P. R. (2008) J. Cell Biol. 181, 615–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong-Riley M. T. (1989) Trends Neurosci. 12, 94–101 [DOI] [PubMed] [Google Scholar]
- 34.Wikström M., Krab K., Saraste M. (1981) Cytochrome Oxidase: A Synthesis, Academic Press, Inc., New York [Google Scholar]
- 35.Péquignot M. O., Dey R., Zeviani M., Tiranti V., Godinot C., Poyau A., Sue C., Di Mauro S., Abitbol M., Marsac C. (2001) Hum. Mutat. 17, 374–381 [DOI] [PubMed] [Google Scholar]
- 36.Servidei S., Zeviani M., Manfredi G., Ricci E., Silvestri G., Bertini E., Gellera C., Di Mauro S., Di Donato S., Tonali P. (1991) Neurology 41, 1053–1059 [DOI] [PubMed] [Google Scholar]
- 37.Wong-Riley M. T. T., Nie F., Hevner R. F., Liu S. (1998) in Cytochrome Oxidase in Neuronal Metabolism and Alzheimer's Disease (González-Lima F. ed) pp. 1–53, Plenum Press, New York [Google Scholar]
- 38.Kadenbach B. (1986) J. Bioenerg. Biomembr. 18, 39–54 [DOI] [PubMed] [Google Scholar]
- 39.Hevner R. F., Wong-Riley M. T. T. (1993) J. Neurosci. 13, 1805–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelly D. P., Scarpulla R. C. (2004) Genes Dev. 18, 357–368 [DOI] [PubMed] [Google Scholar]
- 41.Dhar S. S., Wong-Riley M. T. T. (2009) J. Neurosci. 29, 483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dhar S. S., Liang H. L., Wong-Riley M. T. (2009) J. Neurochem. 108, 1595–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.National Research Council (1996) Guide for the Care and Use of Laboratory Animals, National Academy Press, Washington, DC [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.