Abstract
Mounting evidence from clinical and basic research suggests that estrogen signaling may be altered in the brains of people with schizophrenia. Previously, we found that DNA sequence variation in the estrogen receptor (ER) α gene, lower ERα mRNA levels, and/or blunted ERα signaling is associated with schizophrenia. In this study, we asked whether the naturally occurring truncated ERα isoform, Δ7, which acts as a dominant negative, can attenuate gene expression induced by the wild-type (WT) receptor in an estrogen-dependent manner in neuronal (SHSY5Y) and non-neuronal (CHOK1 and HeLa) cells. In addition, we determined the extent to which ERα interacts with NRG1-ErbB4, a leading schizophrenia susceptibility pathway. Reductions in the transcriptionally active form of ErbB4 comprising the intracytoplasmic domain (ErbB4-ICD) have been found in schizophrenia, and we hypothesized that ERα and ErbB4 may converge to control gene expression. In the present study, we show that truncated Δ7-ERα attenuates WT-ERα-driven gene expression across a wide range of estrogen concentrations in cells that express functional ERα at base line or upon co-transfection of full-length ERα. Furthermore, we find that ErbB4-ICD can potentiate the transcriptional activity of WT-ERα at EREs in two cell lines and that this potentiation effect is abolished by the presence of Δ7-ERα. Immunofluorescence microscopy revealed nuclear co-localization of WT-ERα, Δ7-ERα, and ErbB4-ICD, whereas immunoprecipitation assays showed direct interaction. Our findings demonstrate convergence between ERα and ErbB4-ICD in the transcriptional control of ERα-target gene expression and suggest that this may represent a convergent pathway that may be disrupted in schizophrenia.
Schizophrenia is a devastating mental illness of unknown cause, afflicting 1% of the population worldwide. At present, the molecular processes underlying the development and progression of schizophrenia have not been clearly identified; however, we have recently found evidence to support the hypothesis that schizophrenia arises from a derailment of normal brain maturation during adolescence where the brain fails to respond to the developmental increase in sex steroids (testosterone and estrogen) (1). Testosterone is commonly converted into estrogen by the action of the enzyme aromatase whereby the converted hormone serves as a ligand for estrogen receptors. Schizophrenia affects both men and women occurring at a ratio of ∼3:2, respectively (2). Males typically have an earlier age of onset and more severe symptoms and tend to be less treatment responsive than females with schizophrenia (3). However, women with schizophrenia can have symptom exacerbation at times of low estrogen: during post-partum and menopause (4–6), and schizophrenic symptoms can be ameliorated with estrogen treatment (7–11).
Estrogen affects the developing brain by influencing the genesis, development, migration, plasticity, and survival of neurons (12–16). Estrogen has also been shown to act as a trophic factor promoting the growth and arborization of axons and dendrites in culture (17–19), as well as promoting synapse/spine formation (20–26). In addition, estrogen also regulates the transcription of key enzymes involved in the synthesis and turnover of classic neurotransmitters including noradrenaline, dopamine, serotonin, and acetylcholine (27–30), as well as the transcription of neurotrophins; neuropeptides, including vasopressin and insulin-like growth factor (31–34); and cell surface receptors such as oxytocin receptor (35).
The regulatory effects of estrogen are mediated by two closely related receptors, estrogen receptor α (ERα3 (ESR1, NR3A1)) (36) and ERβ (ESR2, NR3A2) (37). The ERs are ligand-activated transcription factors belonging to the steroid/thyroid hormone superfamily of nuclear receptors (38). In our current study, we focus our attention toward ERα because we have recently shown that sequence variation in the ERα gene, lower ERα mRNA, and/or decreased full-length or wild-type (WT) ERα mRNA is associated with the diagnosis of schizophrenia (1, 39, 40). The ERα gene contains eight coding exons, which translate into three independent but interacting functional domains including an activation domain, a DNA-binding domain, and an estrogen-binding domain (41). The estrogen-binding domain within the C-terminal region is necessary for ligand-dependent receptor dimerization. This region also contains the activation function 2, which promotes transcription by recruiting co-activators upon ligand binding (42–44). This estrogen-binding domain is coded for by exons 6–8 of the ERα gene. We have previously detected as many as 18 distinct ERα splice variants in the human cortex with the exon-skipped Δ7 being the most abundant naturally occurring splice transcript of the ERα expressed in the primate brain (1, 39), in normal breast (45), and in ER-positive mammary carcinoma cell lines (46). We and others have previously shown that Δ7-ERα encodes a truncated protein that can modulate WT-ERα-mediated transcription in yeast and various non-neuronal cell lines by functioning as a dominant negative receptor with the ability to suppress transcriptional activation by WT-ERα (1, 47–49). However, no study to date has investigated the function of Δ7-ERα in neuronal cells. As a primary aim, in this current study we determine whether the truncated ERα, Δ7, can act as a dominant negative receptor in neuronal cells to suppress WT-ERα mediated transcription in response to estrogen.
In schizophrenia, estrogen signaling may also be altered or impacted through convergence with other disease causing pathways, such as neuregulin (NRG1)-ErbB4. The NRG1-ErbB4 pathway is a leading schizophrenia susceptibility pathway (50–54), and the ErbB4 mRNA, protein, and functional coupling has been shown to be altered in the brains of patients with schizophrenia (55–58). ErbB4 is a member of the type I or ErbB receptor tyrosine kinase subfamily that is activated by it's ligand NRG1. Binding of NRG1 stimulates sequential cleavage of the full-length ErbB-4 receptor by tumor neurosis factor α converting enzyme and γ-secretase, which releases the transcriptionally active intracytoplasmic domain (ErbB4-ICD).
Evidence suggests that alterations in cortical ErbB4-ICD and WT-ERα may be involved in the molecular pathology of schizophrenia. Cross-talk between WT-ERα and ErbB4-ICD has been implicated in the development and progression of endrocrine-related cancers (59). However, in neurons, interaction between WT-ERα and ErbB4 has yet to be described. We hypothesize that ERα and ErbB4 may converge to regulate ERα-mediated gene transcription at EREs where ErbB4-ICD may potentiate estrogen signaling by acting as a co-activator. Thus, as a secondary aim, we also determined whether ErbB4-ICD can potentiate transcriptional activity of WT-ERα at EREs in both neuronal and non-neuronal cells.
Although reductions in cortical WT-ERα mRNA and ErbB4-ICD protein have been linked to schizophrenia susceptibility, we have also found possible increases in non-WT ERα transcripts (splice variants) in mental illness (1, 39, 58). Considering that Δ7-ERα is the most abundant naturally occurring ERα splice transcript, as a final aim, we determined whether Δ7-ERα can impact WT-ERα and ErbB4-ICD transcriptional interaction at EREs.
EXPERIMENTAL PROCEDURES
Plasmid Construction
pDsRed-CMV-wtERα, pDsRed-CMV- Δ7ERα, pcDNA3.1V5His-wtERα, pcDNA3.1V5His-Δ7ERα, pZsGreen-ErbB4-ICD, pcDNA6.2-EmGFP-Full-ErbB4, pcDNA6.2- EmGFP-CYT1, and pcDNA6.2-EmGFP-CYT2 were constructed using a method as previously described (1). For construction of plasmids, DNA fragments encoding full-length WT-ERα, Δ7-ERα, and ErbB4-ICD were amplified from cDNA using the following primer sets: ERα1F, CTCGAGACCATGACCCTCCAC; ERα1R, GGTACCTCAGACCGTGGCAGGGA; ErbB4-ICDF: AGATCTATGAGAAGGAAGAGCATCAAA; ErbB4-ICDR, CACCACAGTATTCCGGTGTC; full ErbB4F, ATGAAGCCGGCGACAGGAC; and full ErbB4R, CACCACAGTATTCCGGTGTC.
Briefly, cDNA of WT-ERα, Δ7-ERα, or ErbB4-ICD were ligated into pcDNA3.1-V5-His using the TOPO TA cloning kit (Molecular Probes) according to the manufacturer's instructions. For pcDNA6.2-EmGFP-Full-ErbB4, pcDNA6.2- EmGFP-CYT1, and pcDNA6.2-EmGFP-CYT2, cDNAs of full-length ErbB4, CYT1-ErbB4-ICD, and CYT2-ErbB4-ICD were ligated into pcDNA6.2-EmGFP using the TOPO TA cloning kit (Molecular Probes) according to the manufacturer's instructions. The CYT2 ICD was obtained by mini-prep screening and sequencing for the isoform lacking the 48 bp coding the PI3K-binding domain. For construction of pDsRed-CMV-wtERα, pDsRed-CMV-Δ7ERα, and pZsGreen-ErbB4-ICD, genes of interest were then excised by restriction endonuclease digestion with BamHI and XhoI (for WT-ERα and Δ7-ERα) or BglII and SacII (for ErbB4-ICD) and ligated into pZsGreen1-N1 or pDsRed-Express-C1 (Clontech) using T4 DNA ligase (Fermenats). Positive clones were verified by sequencing.
Cell Culture
All of the cell lines were obtained from the American Type Culture Collection and were grown at 37 °C in a 5% CO2 atmosphere. CHOK1 cells were cultured in Dulbecco's modified Eagle's medium/Ham's F-12 medium (DF12 1:1 mixture) containing 10% (v/v) FBS supplemented with 100 units/100 μg/ml penicillin/streptomycin and 2 mm glutamax. HEK293 cells were cultured in Dulbecco's modified Eagle's medium containing 10% (v/v) FBS supplemented with 100 units/100 μg/ml penicillin/streptomycin and 2 mm glutamax. HeLa and SHSY5Y cells were cultured in RPMI containing 10% (v/v) FBS supplemented with 100 units/100 μg/ml penicillin/streptomycin and 2 mm glutamax. To differentiate SHSY5Y cells, all-trans-retinoic acid (10 μm) was added to the culture medium and incubated for 9 days. Prior to transfection the cells were washed twice in PBS and incubated for 24 h in phenol red-free medium containing 10% (v/v) charcoal-stripped FBS supplemented with glutamax (2 mm). Transfections and treatments were conducted for 24 h in the same medium. All of the treatments were added to cells either in absolute ethanol or dimethyl sulfoxide and compared with vehicle-only controls. No cell toxicity was observed for any treatment at the concentrations employed.
Luciferase Reporter Assay
On day 1, the cells were seeded in 48-well plates (density was 2 × 104 for CHO K1, 3 × 104 for HeLa, and 2 × 105 for SHSY5Y). For SHSY5Y, the cells were differentiated 9 days prior. On day 2, the cells were washed and incubated in phenol red-free medium containing 10% (v/v) charcoal-stripped FBS supplemented with 2 mm glutamax. On day 3, 3×ERE-luc reporter plasmids (a kind gift from Professor Rakesh Kumar, M. D. Anderson Cancer Center, Houston, TX)) (125 ng/well), pZsGreen-ErbB4-ICD (50 ng/well), pDsRed-CMV-wtERα (12.5 ng/well), and/or pDsRed-CMV-Δ7ERα (37.5 ng/well) were transfected into cells for 24 h using Lipofectamine 2000 (0.5 μl/well) or Lipofectamine 2000 combined with magnetofection (0.2 μl/well; 20 min on magnet) (Chemicell) for SHSY5Y. The pRL-TK Renilla internal control plasmid (Promega) (12.5 ng/well) was co-transfected for normalization of transfection efficiency. On day 4, the cells were treated with 0.01 nm or the indicated concentrations of 17β-estradiol for 24 h. On day 5, the cells were washed and resuspended in 50 μl of 1× passive lysis buffer. The luciferase assays were performed using a dual luciferase assay reporter system (Promega) according to the manufacturer's instructions in a FluorStar OPTIMA (BMG). The results were normalized to the Renilla control and expressed as relative fold change in luciferase activity relative to the empty vector vehicle-treated control condition.
Immunofluorescence Staining and Fluorescence Microscopy
On day 1, the cells were seeded in 24-well plates containing sterile 19 × 19-mm glass coverslips (density was 4 × 104 for CHOK1, 6 × 104 for HeLa, and 4 × 105 for SHSY5Y). For SHSY5Y, the cells were differentiated 9 days prior. On day 2, the cells were washed and incubated in phenol red-free medium containing 10% (v/v) charcoal-stripped FBS supplemented with 2 mm glutamax. On day 3, the cells were transfected with 100 ng/well overexpression plasmids for 24 h using Lipofectamine 2000 (1 μl/well) or Lipofectamine 2000 combined with magnetofection (0.2 μl/well; 20 min on magnet) (Chemicell) for SHSY5Y. On day 4, the cells were treated with 0.01 nm 17β-estradiol for 24 h. On day 5, the cells were washed twice with PBS and fixed with 3% (v/v) formaldehyde/PBS for 15 min at room temperature or 37 °C for SHSY5Y. For fluorescence microscopy without immunostaining, the cells were then rinsed twice with PBS and mounted on glass slides using mounting medium containing anti-fading reagent ProLong® Gold antifade reagent with DAPI (Invitrogen). For immunofluorescence microscopy, the cells were rinsed with PBS (3 × 5 min) and then permeabilized with 0.1% (v/v) Triton X-100 in PBS (5 min). After washing with PBS (3 × 5 min), the cells were incubated with 10% FBS (v/v) in PBS for 1 h at room temperature. The cells on the coverslips were then incubated with primary antibody against ERα (Santa Cruz catalog number sc-8005) 1:50 dilution in 10% FBS (v/v) in PBS plus 0.1% (w/v) of saponin) for 16 h at 4 °C. After washing with 10% FBS (v/v) in PBS (3 × 5 min), the cells were incubated with 5 μg/ml of Alexa Fluor 594-conjugated secondary antibody (Molecular Probes catalog number A21203) for 1 h at room temperature. The cells were washed with 10% FBS (v/v) in PBS (3 × 10 min) and then mounted on glass slides using mounting medium containing anti-fading reagent ProLong® Gold antifade reagent with DAPI (Invitrogen). The images were obtained using a Nikon D-Eclipse C1 confocal microscope (Nikon).
Immunoprecipitation and Western Blotting
On day 1, all of the cells were seeded at a density of 2 × 106 cells/dish. HEK293 cells were seeded in poly-l-lysine-coated 60-mm dishes. HeLa and CHOK1 cells were seeded in 60-mm dishes. On day 2, the cells were transfected for 48 h with pcDNA3.1V5His-wtERα or pcDNA3.1V5His-Δ7ERα (2 μg) using Lipofectamine LTX transfection reagent (Invitrogen). On day 4, the cells were washed twice with PBS and then scraped into 1 ml of ice-cold PBS. The cells were pelleted and lysed in 1× Tris-buffered saline with 0.01% Triton X-100 (TBST) containing protease inhibitor mixture (Sigma). The cell lysates were then precleared with 20 μl of protein A/G-agarose plus beads (Santa Cruz) by rotating for 30 min at 4 °C. After centrifugation at 5000 × g for 5 min, precleared lysates (supernatants) were rotated overnight at 4 °C with protein A/G-agarose plus beads prebound with 2 μg of ErbB4 polyclonal antibody (Santa Cruz catalog number sc-283). The beads were pelleted by centrifugation at 2500 × g and washed with 1× TBST + protease inhibitor mixture for 10 min at 4 °C with rotation. This was repeated three times. The beads were then mixed with 5× SDS loading buffer and boiled for 5 min at 95 °C. For SHSY5Y and MCF-7 lysates, 40 and 10 μg, respectively, were mixed with 5× SDS loading buffer and boiled for 5 min at 95 °C. All of the samples were subjected to SDS-PAGE on 12% Bis-Tris gels (Bio-Rad). The proteins were transferred onto nitrocellulose membranes and incubated with blocking solution (5% w/v nonfat milk, 0.1% (v/v) Tween 20 in PBS (PBST) at room temperature for 1 h). The membranes were incubated with a primary antibody for V5 (Invitrogen catalog number R960-25) diluted 1:10000 in blocking solution at 4 °C overnight. For detection of endogenous ERα, the membranes were incubated with a primary antibody for ERα (Novocastra catalog number NCL-ER-6F11) diluted 1:500 in 5% bovine serum albumin-PBST; 4 °C; overnight. The membranes were washed three times for 10 min with PBST and incubated with peroxidase-conjugated affinity-purified secondary antibodies (anti-mouse (for V5 antibody, diluted 1:5000 in blocking solution; for ERα antibody, diluted 1:1000 in 5% bovine serum albumin-PBST) at 4 °C for 1 h) (Chemicon International). After further washing, the bound antibodies were incubated with enhanced chemiluminescence reagent (Millipore) and visualized by autoradiography or by chemiluminescence on a Chemidoc Imaging System (Bio-Rad).
Statistical Analysis
All of the experiments are representative of at least two independent experiments. For luciferase, the data are presented as the means ± S.E. from three replicate cultures representative of two independent experiments. The statistical analyses were conducted using Statistica 7 (StatSoft Inc., 2000, STATISTICA for Windows). One-way analysis of variance (ANOVA) was used to assess significance of increasing 17β-estradiol concentration. The distinct concentration or the type(s) of transfected vectors were used as the independent variable, and relative luciferase activity was used as the dependent variable. ANOVAs were followed up with the Fisher LSD post-hoc analysis to assess the significance among groups. A p value less than 0.05 was considered statistically significant.
RESULTS
Endogenous ERα Expression in Cell Lines
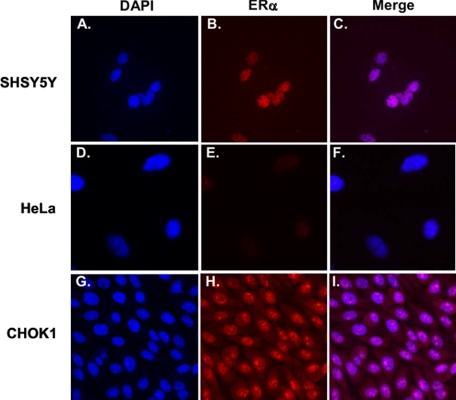
In the present study we used three cell types to assess ERα-driven gene expression. The neuronal cell line SHSY5Y and the non-neuronal cell lines HeLa and CHOK1. By immunofluorescence staining, we observed nuclear localization of ERα in SHSY5Y (Fig. 1, A–C). HeLa cells are reported to be ER-negative (60) and were used as a negative control. We confirmed that no expression of ERα was observed in HeLa cells (Fig. 1, D–F). Conversely, CHO K1 cells are ERα-positive and were used as a positive control. We observed robust staining of ERα in CHO K1 cells, most of which is localized in the nucleus (Fig. 1, G–I).
FIGURE 1.
Photomicrographs from immunocytochemistry of ERα. SHSY5Y (A–C), HeLa (D–F), and CHO K1 (G–I) cells were fixed, immunoprobed for endogenous ERα protein expression (red), and stained for DAPI (blue). Co-localization visualized as purple nuclei can be seen in the merge in C and I but not in F.
ERα Is Required for Estrogen-stimulated Transcription at EREs
To determine the extent to which estrogen influences ERα-mediated transcription in neurons, SHSY5Y cells were treated with increasing concentrations of 17β-estradiol. In the absence of WT-ERα overexpression, 17β-estradiol had no effect on transcriptional activity (ANOVA, F = 0.3, df = 5, 12, p = 0.9) (Fig. 2A) in SHSY5Y cells. When WT-ERα was overexpressed, we observed a robust increase in transcriptional activity (ANOVA, F = 68.7, df = 5, 12, p < 0.000001) (Fig. 2B). When Δ7-ERα was overexpressed, we observed no change in luciferase reporter activity from base line (ANOVA, F = 0.37, df = 5, 12, p = 0.86) (Fig. 2C). Similar to our findings in SHSY5Y cells, we observed no significant change in transcriptional activity in the ERα negative HeLa cells (ANOVA, F = 2.75, df = 5, 12, p = 0.07) (Fig. 2D) unless they were transfected with WT-ERα (ANOVA, F = 10.4, df = 5, 12, p = 0.0005) (Fig. 2E) and not with Δ7-ERα (ANOVA, F = 2.66, df = 5, 12, p = 0.08) (Fig. 2F). But the overall magnitude of the increase in luciferase assay is less in HeLa cells as compared with SHSY5Y cells. In contrast to SHSY5Y and HeLa, CHO K1 cells showed a concentration-dependent increase in transcriptional activity in the absence or presence of WT-ERα overexpression (empty vector ANOVA, F = 16.1, df = 5,12, p = 0.00006 (Fig. 2G); WT-ERα ANOVA, F = 98.2, df = 5, 12, p < 0.00001) (Fig. 2H). Interestingly, when Δ7-ERα was overexpressed in CHO K1 cells, we observed a modest but significant dose-dependent increase in transcription. Treatment with 1 nm 17β-estradiol elicited a 3-fold induction (ANOVA, F = 67.6, df = 5, 12, p < 0.000001) (Fig. 2I). However, when this was compared with the empty vector condition, i.e. in the absence of Δ7-ERα, 1 nm 17β-estradiol stimulated transcription up to 8-fold (Fig. 2G). Because we showed that SHSY5Y cells contained ERα immunoreactivity at base line as do CHO K1 cells, the lack of estrogen responsiveness at base line was surprising.
FIGURE 2.
17β-Estradiol dose response curves. SHSY5Y (top panels) HeLa (middle panels), and CHO K1 cells (bottom panels) were transfected with 3× ERE-luc and empty vector (A, D, and G), WT-ERα (B, E, and H), or Δ7-ERα (C, F, and I) as indicated for 24 h and treated with the indicated concentrations of 17β-estradiol for 24 h. Relative luciferase activity is plotted on the y axis, and the dose of 17β-estradiol is plotted on the x axis.
We determined whether this lack of response was due to the type of ERα expressed in SHSY5Y cells at base line. We found that SHSY5Y cells express both WT-ERα and Δ7-ERα mRNA and protein (supplemental Fig. S1). Endogenous ERα protein was detected by Western blotting using an ERα antibody targeted to an epitope at the N-terminus of the ERα protein between amino acids 1 and 184. We have previously characterized this antibody extensively, demonstrating that it detects WT-ERα and smaller ERα isoforms in both cell lysates and brain tissue (61). Indeed, multiple bands were detected in SHSY5Y and the MCF-7 positive control. Compared with MCF-7, expression of WT-ERα in SHSY5Y is low to negligible. The prominent bands migrating below WT-ERα are putative truncated ERα proteins. Aside from Δ7-ERα, which migrates at ∼45 kDa, we predict that the band migrating at ∼42 kDa to be a possible Δ5-ERα based on the predicted molecular masses as previously reported (60). Although both cell types express similar banding pattern, expression levels between the two differed, consistent with our previous suggestion that expression of ERα isoforms may be cell type-specific (61).
ErbB4-ICD Potentiation of Transcription at EREs
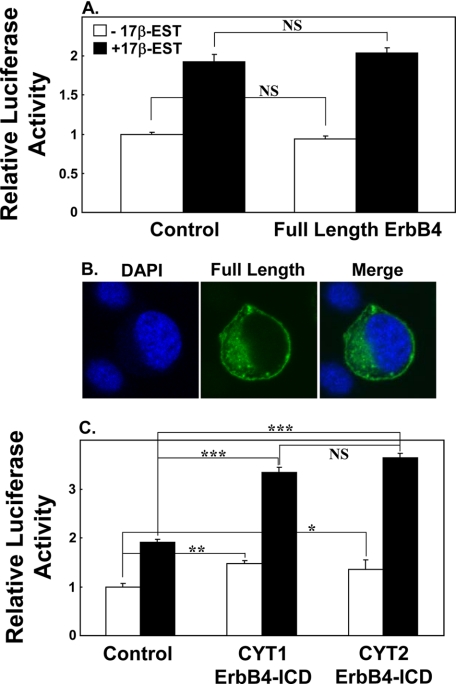
Previous studies have shown that ErbB4-ICD could stimulate ERα transcription at EREs in non-neuronal cell types (59). However, the effect of the full-length ErbB4 receptor has yet to be shown. In our next approach, we determined whether ErbB4 potentiation of ERα could be incited by the membrane-bound form of ErbB4 or whether it was dependent on cleavage of the full-length ErbB4. Transfection of the full-length ErbB4 receptor into CHOK1 cells did not stimulate transcription at EREs beyond the increase that was seen with 17β-estradiol alone (p > 0.1) (Fig. 3A). Analysis of subcellular localization of the GFP-tagged full-length ErbB4 receptor showed only plasma membrane and cytoplasmic localization with no apparent localization to the nucleus (Fig. 3B). Together, this implies that cleavage of the ICD (nuclear form) of ErbB4 is necessary to increase transcription at EREs. Because the ICD domain of ErbB4 can exist as 2 isoforms: one that is PI3K binding (CYT1) and one that lacks the PI3K binding domain (CYT2), we next determined whether the ErbB4-ICD PI3K-binding site influences ERα-mediated transcription. Using CHO K1 cells, we found that both the CYT1-ErbB4-ICD and CYT2-ErbB4-ICD potentiated transcription at EREs to a similar extent. No difference in ERE-driven gene transcription was observed between the two ErbB4-ICD isoforms (p > 0.1) (Fig. 3C). Both CYT1 and CYT2 ErbB4-ICDs were capable of stimulating transcriptional activity at EREs in the absence of estrogen, although this effect was greater in the presence of estrogen. Considering that no difference was observed between the two ErbB4-ICD isoforms, CYT1 ErbB4-ICD was used in subsequent experiments.
FIGURE 3.
Effect of full-length ErbB4, CYT1, and CYT2 on ERE promoter activity. CHOK1 cells were transfected with 3×ERE-luc alone (control condition) or 3×ERE-luc and full-length ErbB4 (A), or 3×ERE-luc and CYT1-ErbB4-ICD or CYT2-ErbB4-ICD (C) for 24 h and treated with 0.01 nm of 17β-estradiol for 24 h. A photomicrograph of CHOK1 cells transfected with fluorescently tagged full-length ErbB4 (green) for 24 h is shown in B with the DAPI nuclear counterstain (blue). Note the predominant fluorescence at the perimeter of the cell, some fluorescence in the cytoplasm, but no fluorescence in the nucleus. Asterisks denote statistical significance. *, p = 0.05; **, p < 0.01; ***, p < 0.000001; NS, not significant.
ErbB4-ICD Potentiates ERα-mediated Transcription at EREs in SHSY5Y Cells
We next determined whether ErbB4-ICD could potentiate estrogen-stimulated transcription at EREs in neurons using two approaches, first at base line (Fig. 4) and then upon transfection with WT-ERα (Fig. 5). Overexpression of ErbB4-ICD in SHSY5Y cells had no effect on ERE-related transcriptional activity at base line (ANOVA, F = 0.82, df = 3, 8, p = 0.52) (Fig. 4A). A similar lack of ErbB4-ICD-related change in transcriptional activity was observed in HeLa cells (ANOVA, F = 0.9, df = 3, 8, p = 0.48) (Fig. 4B). In contrast, overexpression of ErbB4-ICD in CHOK1 cells increased transcriptional activity by over 35% in the absence and presence of 0.01 nm 17β-estradiol (ANOVA, F = 54.8, df = 3, 8, p = 0.00001) (Fig. 4C).
FIGURE 4.
Effect of ErbB4-ICD on ERE promoter activity at base line. SHSY5Y (A), HeLa (B), and CHOK1 (C) cells were transfected with 3×ERE-luc and ErbB4-ICD for 24 h and treated with (black bars) or without (white bars) 0.01 nm of 17β-estradiol for 24 h. Asterisks denotes statistical significance. *, p < 0.05; **, p < 0.005.
FIGURE 5.
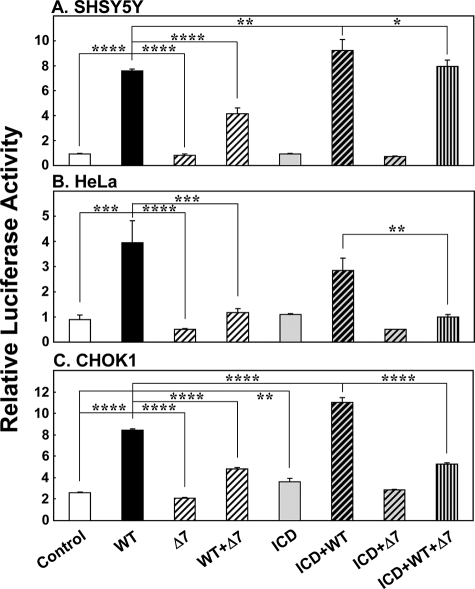
Potentiation of WT-ERα by ErbB4-ICD in neuronal cells is blocked by Δ7-ERα. CHOK1, HeLa and SHSY5Y cells were transfected with 3× ERE alone (white bars), WT-ERα (black bars), Δ7-ERα (hatched bars), ErbB4-ICD (gray bars), and their combinations as indicated for 24 h. The cells were then treated with 0.01 nm of 17β-estradiol for 24 h. A, SHSY5Y. *, p < 0.05; **, p < 0.01; ****, p < 0.00001. B, HeLa. **, p < 0.005; ***, p < 0.0001; ****, p < 0.00001. C, CHOK1. **, p < 0.01; ****, p < 0.00001. Note that ErbB4-ICD potentiates luciferase reporter activity in SHSY5Y and CHOK1 cells in the presence of WT-ERα (A and C, compare second column (WT; black bars) and sixth column (ICD+WT; hatched gray bars).
In the absence of exogenous expression of WT-ERα, ErbB4-ICD failed to stimulate transcription at EREs in SHSY5Y and HeLa cells (p > 0.05) as found in Figs. 4 (A and B) and 5 (A and B, fifth column). Interestingly, in SHSY5Y cells, when ErbB4-ICD was co-expressed with WT-ERα, ErbB4-ICD potentiated WT-ERα-mediated transcription further stimulating luciferase reporter activity greater than in the WT-ERα condition alone (SHSY5Y ANOVA, F = 84.2, df = 7, 16, p < 0.01) (Fig. 5A, second and sixth columns). In CHOK1 cells, ErbB4-ICD potentiated transcriptional activity at EREs in the absence and presence of WT-ERα overexpression (CHOK1 ANOVA, F = 218.5, df = 7, 16, p < 0.0000001) (Fig. 5C, first and fifth columns and second and sixth columns). In HeLa cells, ErbB4-ICD did not potentiate ERα-mediated transcription (p > 0.05) (Fig. 5B, second and sixth columns).
Δ7-ERα Disrupts Synergy in Transcriptional Activation by WT-ERα and ErbB4-ICD at EREs
In our next approach, we overexpressed both WT-ERα and Δ7-ERα. As we previously found, overexpression of WT-ERα increased transcriptional activity at EREs in all three cell types (all p < 0.0001), whereas overexpression of Δ7-ERα did not stimulate transcription (all p < 0.00001) (Fig. 5, second and third columns). Co-expression of WT-ERα together with Δ7-ERα significantly attenuated transcriptional activity at EREs in all three cell types (all p < 0.0001) (Fig. 5, fourth column). These findings confirm what we show in Fig. 2.
When ErbB4-ICD was co-expressed with Δ7-ERα in the absence of WT-ERα overexpression, we observed no significant change in transcriptional activity from the control condition in all three cell types (Fig. 5, seventh column), suggesting that ErbB4-ICD potentiation cannot be mediated through Δ7-ERα. In the presence of WT-ERα overexpression, Δ7-ERα attenuated ErbB4-ICD potentiation of ERα-mediated transcription (all p < 0.05) (Fig. 5, eighth column), suggesting that Δ7-ERα may be interfering with the interaction between WT-ERα and ErbB4-ICD.
Subcellular Localization of WT-ERα, Δ7-ERα, and ErbB4-ICD
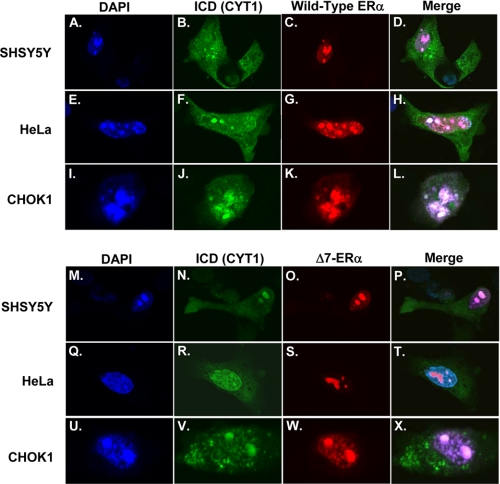
We next determined the subcellular distribution of WT-ERα, Δ7-ERα and ErbB4-ICD. All three cell types were transfected with the combination of fluorescently tagged GFP-ErbB4-ICD (green) (Fig. 6, B, F, and J) and RFP-WT-ERα (red) (Fig. 6, C, G, and K) or the combination of GFP-ErbB4-ICD (green) (Fig. 6, N, R, and V) and RFP-Δ7-ERα (red) (Fig. 6, O, S, and W). In all three cell types, RFP-WT-ERα (Fig. 6, C, G, and K) and RFP-Δ7-ERα (Fig. 6, O, S, and W) showed primarily nuclear localization (similar to what we show in Fig. 1), whereas GFP-ErbB4-ICD showed both nuclear and cytoplasmic localization (Fig. 6, B, F, J, N, R, and V). In co-localization experiments, we observed nuclear co-localization of GFP-ErbB4-ICD with RFP-WT-ERα (Fig. 6, D, H, and L) and ErbB4-ICD with Δ7-ERα (Fig. 6, P, T, and X).
FIGURE 6.
Subcellular localization of transfected ErbB4-ICD, WT-ERα, and Δ7-ERα. CHO K1, HeLa, and SHSY5Y cells were transfected for 24 h with the combination of fluorescently tagged GFP-ErbB4-ICD (green) (B, F, and J) and RFP-WT-ERα (red) (C, G, and K) or GFP-ErbB4-ICD (green) (N, R, and V) and RFP-Δ7-ERα (red) (O, S, and W). The cells were then fixed, stained with DAPI (blue), (A, E, I, M, Q, and U), and mounted. Co-localization of GFP-ErbB4-ICD and RFP-WT-ERα or GFP-ErbB4-ICD and RFP-Δ7-ERα can be seen in the merge as a whitish pink color (D, H, L, P, I, and X).
ErbB4-ICD Interacts with WT-ERα and Δ7-ERα
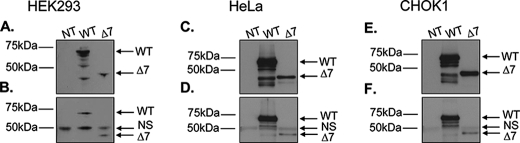
To determine whether ErbB4-ICD physically interacts with WT-ERα or Δ7-ERα, HEK293, HeLa and CHOK1 cells transfected with either V5-tagged WT-ERα or Δ7-ERα (Fig. 7, A, C, and E) were immunoprecipitated for endogenous ErbB4 and immunoprobed via Western blotting for the V5-tagged ERα. In all three cell types, both WT-ERα and Δ7-ERα were found to co-immunoprecipitate with endogenous ErbB4 (Fig. 7, B, D, and F). Consistent with our co-localization data (Fig. 6, D, H, L, P, T, and X), we show that a direct protein-protein interaction between WT-ERα and ErbB4-ICD as well as Δ7-ERα and ErbB4-ICD within three different cell types is possible.
FIGURE 7.
ERα and ErbB4 protein immunoprecipitation. HEK293 (A and B), HeLa (C and D), and CHOK1 cells (E and F) were transfected with V5-tagged WT-ERα or Δ7-ERα for 48 h. Protein from cells was then harvested and immunoprecipitated with ErbB4 antibody. The immunoprecipitants were separated by SDS-PAGE and immunoprobed with V5 antibody. Immunoblotting for V5 in whole cell lysates (A, C, and E) and immunoblotting for V5 after pulldown with ErbB4 c-terminal antibody (B, D, and F) are shown. Whole cell lysates and immunoprecipitated bands (V5-WT-ERα, ∼69 kDa; V5-Δ7-ERα, ∼48 kDa) are shown. NS, nonspecific.
DISCUSSION
In this current study, we present several major findings. 1) We showed that the truncated ERα receptor, Δ7-ERα, can abolish or attenuate estrogen-dependent WT-ERα-mediated transcription in neuronal and non-neuronal cell lines. 2) In addition, we showed that ErbB4-ICD, but not full-length ErbB4, can potentiate the transcriptional activity of WT-ERα at EREs. 3) The positive effect of ErbB4-ICD on WT-ERα-mediated transcription at EREs is not dependent on the CYT form of ErbB4-ICD. 4) Moreover, we show that ErbB4-ICD potentiation of ERα-driven gene expression at EREs can occur in the CHO ovary and SHSY5Y neuronal cell line but not in HeLa cells, suggesting that only certain cell types provide a permissive ERα-responsive environment. 5) Finally, we present two lines of evidence showing direct interaction between ERα and ErbB4-ICD. Both WT-ERα and Δ7-ERα were found to co-localize with ErbB4-ICD in the nucleus of neuronal and non-neuronal cells. Moreover, immunoprecipitation revealed direct protein-protein interaction between ErbB4-ICD and WT-ERα and between ErbB4-ICD and Δ7-ERα.
Interestingly, although treatment with estrogen and/or overexpression of ErbB4-ICD can potentiate ERα-mediated transcription in neuronal cells, this effect was only observed in the presence of exogenous WT-ERα expression but not at base line despite positive ERα immunoreactivity. In SHSY5Y cells, although we detect ERα by immunoprobing, analysis of the predicted sizes for ERα splice variants from Western blotting showed low to negligible levels of the full-length WT-ERα but with relatively higher levels of smaller ERα reactive bands and Δ7-ERα mRNA (supplemental Fig. S1) at base line. This would suggest that SHSY5Y cells may naturally express increased truncated forms of ERα and would thus normally be in a repressed state with regards to ERα-mediated transcription. Previous studies have reported that the relative levels of WT-ERα to Δ7-ERα are critical in determining the effectiveness of WT-ERα transcriptional activity (47, 49). Because Δ7-ERα is naturally expressed with WT-ERα, it has been suggested that the dominant negative function of the ERα splice variants may act to protect tissues from excessive estrogenic signals. This notion is based on a 30-fold higher expression of the Δ3-ERα variant in normal breast cells rather than in breast cancer tissues (62, 63). Moreover, estradiol treatment of human endometrial adenocarcinomas grown in nude mice show decreased WT-ERα and increased Δ7-ERα protein expression (64). Our finding that SHSY5Y cells are capable of robust induction of estrogen-stimulated luciferase reporter activity by exogenous WT-ERα overexpression is consistent with this notion. In the presence of exogenously expressed Δ7-ERα, WT-ERα-mediated transcription at EREs was severely attenuated in the SHSY5Y neuronal cell line.
ErbB4-ICD Potentiation of Transcription at EREs
ErbB4 is thought to be unique among the receptor ErbB tyrosine kinase family members because proteolytic processing of the full-length ErbB4 receptor releases a bioactive ICD. The biological significance of the ErbB4-ICD is not fully understood. In this current study, we showed that it is the ErbB4-ICD but not full-length ErbB4 receptor that is responsible for the stimulation in transcriptional activity. By transient transfection of the full-length ErbB4 receptor, we observed no change in transcriptional activity at EREs or targeting of the full-length receptor to the nucleus of the cell, suggesting that potentiation of ERα-mediated transcription may depend upon cleavage and nuclear localization of ErbB4-ICD. Moreover, this potentiation is not dependent on the availability of the PI3K-binding site, suggesting that coupling to the PI3K/AKT signaling pathway does not affect the ability of the ErbB4-ICD to transclocate to the nucleus and stimulate transcription because both CYT1 and CYT2 ErbB4-ICDs stimulated transcription at EREs to the same extent. This is in contrast to the work of Sundvall et al. (65), who showed that CYT2-ErbB4-ICD was more efficient than CYT1-ErbB4-ICD at stimulating β-casein promoter activity. These differences in transcriptional activity may depend on the cell type and gene promoter to which the ErbB4-ICDs are targeted to. This is likely because ErbB4-ICD has been shown to affect a variety of cellular processes in different settings (66, 67).
ErbB4-ICD and ERα
Although the dominant negative activity of Δ7-ERα has been described (1, 47–49), the mechanism by which this occurs is debatable. Although Δ7-ERα possesses a nuclear localization signal, previous studies have reported nuclear exclusion and primarily cytoplasmic localization of Δ7-ERα (60), suggesting that Δ7-ERα interferes with WT-ERα entering the nucleus. In contrast, we showed nuclear localization of Δ7-ERα, suggesting that interference with WT-ERα occurs within the nucleus. In contrast, ErbB4-ICD is likely to be acting as a co-activator for WT-ERα-mediated transcription in both a ligand-dependent and independent manner because ErbB4-ICD was able to slightly stimulate transcriptional activity in CHOK1 cells in the absence of estrogen stimulation. However, it is possible that there is endogenous production of estrogen by CHOK1 cells, although ErbB4-ICD stimulated transcriptional activity even further in the presence of exogenous estrogen. The potentiation of ErbB4-ICD-WT-ERα-mediated transcription was blunted in the presence of Δ7-ERα. Other studies have shown by in vitro protein-protein interaction that Δ7-ERα is unable to induce co-activator recruitment and binding in the presence of estrogen (49). Thus, our finding that WT-ERα and Δ7-ERα can co-immunoprecipitate with ErbB4 suggests that ErbB4-ICD can directly interact with WT-ERα-Δ7 heterodimers or WT-WT and Δ7-Δ7 homodimers. Our finding of Δ7-ERα repression of WT-ERα-ICD mediated transcription at EREs suggests that the presence of the dominant negative ERα as a dimerization partner suppresses a large transcription complex and renders it less estrogen-responsive.
ERα and ErbB4-ICD in Human Disease
Although the focus of our laboratory is to determine the relationship between ERα and ErbB4 in schizophrenia, the mechanistic data we present in this study suggest that WT-ERα, Δ7-ERα, and ErbB4-ICD may regulate a wide variety of cellular processes that are essential for normal cell function. Indeed, dysregulation of ERα and ErbB4 is implicated in various human diseases. In schizophrenia, we have previously reported decreased frequency of WT-ERα mRNA and increases in splice variant mRNA expression (1). In various cancers including breast, meningiomas, endometrial hyperplasias, and moderate to well differentiated endometrial adenocarcinomas, changed frequencies of WT-ERα and Δ7-ERα mRNA have been reported (68, 69). Similarly, with Alzheimer disease, expression of the both WT-ERα mRNA and Δ7 are substantially increased (70). Collectively our finding that Δ7-ERα can function as a dominant negative receptor to suppress estrogen-dependent ERα-mediated transcription in neuronal and non-neuronal cells suggests that increases in non-wild-type ERα mRNA expression may represent a predisposition to disease.
Using postmortem tissue from the human prefrontal cortex, we and others found elevations in the JMA/CYT1 form of the ErbB4 receptor in schizophrenia (56–58) and decreases in ErbB4-ICD protein (58). Considering that full-length ErbB4 was unable to potentiate WT-ERα-mediated transcription at EREs, this would suggest that increases in full-length ErbB4 and reductions in ErbB4-ICD may alter the appropriate balance of transcriptional versus nontranscriptional ErbB4 signaling in neurons in schizophrenia.
Summary
To date, only one other study in ERα-positive breast cancer cells has shown a convergence in function between WT-ERα and ErbB4-ICD (59). In our current work, we confirm their findings using another ERα-positive cell line, CHOK1, and extend it by showing that convergence between WT-ERα and ErbB4-ICD can be detected in neuronal cells. Moreover, we found that the ability of ErbB4-ICD to potentiate ERα-mediated transcription may be dependent on cell type. Additionally, we show for the first time that Δ7-ERα can not only disrupt the transcriptional effects of WT-ERα but also disrupt the potentiation mediated by ErbB4-ICD in neuronal and non-neuronal cells. The impact of ErbB4-ICD on ERα is likely to be mediated by direct nuclear interaction.
Collectively, our current findings suggest that gene regulation by ERα-ErbB4-ICD may function as a general mechanism of ERα-mediated transcription in various WT-ERα-expressing cell types. In addition, disruption of this interaction by Δ7-ERα may act as an additional transcriptional regulatory mechanism in controlling ERα target gene expression. Thus, our findings demonstrate that molecular alterations of ERα and ErbB4 expression in ERα expressing neuronal and non-neuronal could lead to blunted transcriptional control of DNA promoters regulated by estrogen. In regards to schizophrenia, we suggest that the lack of appropriate transcriptional control of steroid-driven maturation in the brain may represent a faulty regulatory pathway where two potential schizophrenia susceptibility genes, ERα and ErbB4, may converge to cause alterations in neuronal functioning.
Supplementary Material
Acknowledgments
We acknowledge Heng Giap Woon and Shan Yuan Tsai for technical assistance.
This work was supported by the Schizophrenia Research Institute, New South Wales Health, Macquarie Group Foundation, Prince of Wales Medical Research Institute, and the University of New South Wales.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- ER
- estrogen receptor
- ICD
- intracytoplasmic domain
- ERE
- estrogen response element
- WT
- wild-type
- ANOVA
- analysis of variance
- NRG1
- neuregulin
- PI3K
- phosphatidylinositol 3-kinase
- FBS
- fetal bovine serum
- PBS
- phosphate-buffered saline
- DAPI
- 4′,6′-diamino-2-phenylindole
- CHO
- Chinese hamster ovary
- df
- degrees of freedom.
REFERENCES
- 1.Weickert C. S., Miranda-Angulo A. L., Wong J., Perlman W. R., Ward S. E., Radhakrishna V., Straub R. E., Weinberger D. R., Kleinman J. E. (2008) Hum. Mol. Genet. 17, 2293–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGrath J., Saha S., Chant D., Welham J. (2008) Epidemiol. Rev. 30, 67–76 [DOI] [PubMed] [Google Scholar]
- 3.Leung A., Chue P. (2000) Acta Psychiatr. Scand. Suppl. 401, 3–38 [DOI] [PubMed] [Google Scholar]
- 4.Seeman M. V. (1997) Am. J. Psychiatry 154, 1641–1647 [DOI] [PubMed] [Google Scholar]
- 5.Grigoriadis S., Seeman M. V. (2002) Can. J. Psychiatry 47, 437–442 [DOI] [PubMed] [Google Scholar]
- 6.Riecher-Rössler A., Seeman M. V. (2002) Arch. Womens Ment. Health 5, 91–92 [DOI] [PubMed] [Google Scholar]
- 7.Seeman M. V. (1986) Acta Psychiatr. Scand. 73, 609–617 [DOI] [PubMed] [Google Scholar]
- 8.Kulkarni J., de Castella A., Smith D., Taffe J., Keks N., Copolov D. (1996) Schizophr. Res. 20, 247–252 [DOI] [PubMed] [Google Scholar]
- 9.Kulkarni J., Riedel A., de Castella A. R., Fitzgerald P. B., Rolfe T. J., Taffe J., Burger H. (2001) Schizophr. Res. 48, 137–144 [DOI] [PubMed] [Google Scholar]
- 10.Kulkarni J., Riedel A., de Castella A. R., Fitzgerald P. B., Rolfe T. J., Taffe J., Burger H. (2002) Arch. Womens Ment. Health 5, 99–104 [DOI] [PubMed] [Google Scholar]
- 11.Kulkarni J., de Castella A., Fitzgerald P. B., Gurvich C. T., Bailey M., Bartholomeusz C., Burger H. (2008) Arch. Gen. Psychiatry 65, 955–960 [DOI] [PubMed] [Google Scholar]
- 12.Gorski R. A. (1985) J. Anim. Sci. 61, (Suppl. 3) 38–61 [DOI] [PubMed] [Google Scholar]
- 13.Dodson R. E., Shryne J. E., Gorski R. A. (1988) J. Comp. Neurol. 275, 623–629 [DOI] [PubMed] [Google Scholar]
- 14.Arai Y., Sekine Y., Murakami S. (1996) Neurosci. Res. 25, 403–407 [DOI] [PubMed] [Google Scholar]
- 15.McEwen B. S. (2001) J. Appl. Physiol. 91, 2785–2801 [DOI] [PubMed] [Google Scholar]
- 16.Wang L., Andersson S., Warner M., Gustafsson J. A. (2002) Sci STKE 2002, PE29. [DOI] [PubMed] [Google Scholar]
- 17.Toran-Allerand C. D. (1976) Brain Res. 106, 407–412 [DOI] [PubMed] [Google Scholar]
- 18.Beyer C., Karolczak M. (2000) J. Neurosci. Res. 59, 107–116 [PubMed] [Google Scholar]
- 19.Dominguez R., Jalali C., de Lacalle S. (2004) J. Neurosci. 24, 982–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsumoto A., Arai Y. (1976) Cell Tissue Res. 169, 143–156 [DOI] [PubMed] [Google Scholar]
- 21.Woolley C. S., McEwen B. S. (1992) J. Neurosci. 12, 2549–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lustig R. H. (1994) Horm. Behav. 28, 383–395 [DOI] [PubMed] [Google Scholar]
- 23.McEwen B. S., Woolley C. S. (1994) Exp. Gerontol. 29, 431–436 [DOI] [PubMed] [Google Scholar]
- 24.Woolley C. S., Weiland N. G., McEwen B. S., Schwartzkroin P. A. (1997) J. Neurosci. 17, 1848–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hao J., Rapp P. R., Leffler A. E., Leffler S. R., Janssen W. G., Lou W., McKay H., Roberts J. A., Wearne S. L., Hof P. R., Morrison J. H. (2006) J. Neurosci. 26, 2571–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srivastava D. P., Woolfrey K. M., Woolfrey K., Jones K. A., Shum C. Y., Lash L. L., Swanson G. T., Penzes P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14650–14655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liaw J. J., He J. R., Hartman R. D., Barraclough C. A. (1992) Brain Res. Mol. Brain Res. 13, 231–238 [DOI] [PubMed] [Google Scholar]
- 28.Di Paolo T. (1994) Rev. Neurosci. 5, 27–41 [DOI] [PubMed] [Google Scholar]
- 29.Singer C. A., McMillan P. J., Dobie D. J., Dorsa D. M. (1998) Brain Res. 789, 343–346 [DOI] [PubMed] [Google Scholar]
- 30.Osterlund M. K., Halldin C., Hurd Y. L. (2000) Synapse 35, 39–44 [DOI] [PubMed] [Google Scholar]
- 31.Harlan R. E. (1988) Mol. Neurobiol. 2, 183–200 [DOI] [PubMed] [Google Scholar]
- 32.Miranda R. C., Sohrabji F., Toran-Allerand D. (1994) Horm. Behav. 28, 367–375 [DOI] [PubMed] [Google Scholar]
- 33.Sohrabji F., Miranda R. C., Toran-Allerand C. D. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 11110–11114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng C. M., Cohen M., Wang J., Bondy C. A. (2001) FASEB J. 15, 907–915 [DOI] [PubMed] [Google Scholar]
- 35.Fleming J. G., Spencer T. E., Safe S. H., Bazer F. W. (2006) Endocrinology 147, 899–911 [DOI] [PubMed] [Google Scholar]
- 36.Greene G. L., Gilna P., Waterfield M., Baker A., Hort Y., Shine J. (1986) Science 231, 1150–1154 [DOI] [PubMed] [Google Scholar]
- 37.Mosselman S., Polman J., Dijkema R. (1996) FEBS Lett. 392, 49–53 [DOI] [PubMed] [Google Scholar]
- 38.Tsai M. J., O'Malley B. W. (1994) Annu. Rev. Biochem. 63, 451–486 [DOI] [PubMed] [Google Scholar]
- 39.Perlman W. R., Matsumoto M., Beltaifa S., Hyde T. M., Saunders R. C., Webster M. J., Rubinow D. R., Kleinman J. E., Weickert C. S. (2005) Neuroscience 134, 81–95 [DOI] [PubMed] [Google Scholar]
- 40.Perlman W. R., Tomaskovic-Crook E., Montague D. M., Webster M. J., Rubinow D. R., Kleinman J. E., Weickert C. S. (2005) Biol. Psychiatry 58, 812–824 [DOI] [PubMed] [Google Scholar]
- 41.Nilsson S., Mäkelä S., Treuter E., Tujague M., Thomsen J., Andersson G., Enmark E., Pettersson K., Warner M., Gustafsson J. A. (2001) Physiol. Rev. 81, 1535–1565 [DOI] [PubMed] [Google Scholar]
- 42.Kumar V., Green S., Staub A., Chambon P. (1986) EMBO J. 5, 2231–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tora L., White J., Brou C., Tasset D., Webster N., Scheer E., Chambon P. (1989) Cell 59, 477–487 [DOI] [PubMed] [Google Scholar]
- 44.Beato M., Sánchez-Pacheco A. (1996) Endocr. Rev. 17, 587–609 [DOI] [PubMed] [Google Scholar]
- 45.Poola I., Speirs V. (2001) J. Steroid Biochem. Mol. Biol. 78, 459–469 [DOI] [PubMed] [Google Scholar]
- 46.Miksicek R. J., Lei Y., Wang Y. (1993) Breast Cancer Res. Treat. 26, 163–174 [DOI] [PubMed] [Google Scholar]
- 47.Fuqua S. A., Fitzgerald S. D., Allred D. C., Elledge R. M., Nawaz Z., McDonnell D. P., O'Malley B. W., Greene G. L., McGuire W. L. (1992) Cancer Res. 52, 483–486 [PubMed] [Google Scholar]
- 48.Wang H., Zeng X., Khan S. A. (1999) Mol. Cell. Endocrinol. 156, 159–168 [DOI] [PubMed] [Google Scholar]
- 49.García Pedrero J. M., Zuazua P., Martínez-Campa C., Lazo P. S., Ramos S. (2003) Endocrinology 144, 2967–2976 [DOI] [PubMed] [Google Scholar]
- 50.Pulver A. E., Lasseter V. K., Kasch L., Wolyniec P., Nestadt G., Blouin J. L., Kimberland M., Babb R., Vourlis S., Chen H., et al. (1995) Am. J. Med. Genet. 60, 252–260 [DOI] [PubMed] [Google Scholar]
- 51.Stefansson H., Sigurdsson E., Steinthorsdottir V., Bjornsdottir S., Sigmundsson T., Ghosh S., Brynjolfsson J., Gunnarsdottir S., Ivarsson O., Chou T. T., Hjaltason O., Birgisdottir B., Jonsson H., Gudnadottir V. G., Gudmundsdottir E., Bjornsson A., Ingvarsson B., Ingason A., Sigfusson S., Hardardottir H., Harvey R. P., Lai D., Zhou M., Brunner D., Mutel V., Gonzalo A., Lemke G., Sainz J., Johannesson G., Andresson T., Gudbjartsson D., Manolescu A., Frigge M. L., Gurney M. E., Kong A., Gulcher J. R., Petursson H., Stefansson K. (2002) Am. J. Hum. Genet. 71, 877–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stefansson H., Sarginson J., Kong A., Yates P., Steinthorsdottir V., Gudfinnsson E., Gunnarsdottir S., Walker N., Petursson H., Crombie C., Ingason A., Gulcher J. R., Stefansson K., St Clair D. (2003) Am. J. Hum. Genet. 72, 83–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu C. M., Hwu H. G., Fann C. S., Lin C. Y., Liu Y. L., Ou-Yang W. C., Lee S. F. (2005) Am. J. Med. Genet. B Neuropsychiatr. Genet. 134B, 79–83 [DOI] [PubMed] [Google Scholar]
- 54.Harrison P. J., Law A. J. (2006) Biol. Psychiatry 60, 132–140 [DOI] [PubMed] [Google Scholar]
- 55.Hahn C. G., Wang H. Y., Cho D. S., Talbot K., Gur R. E., Berrettini W. H., Bakshi K., Kamins J., Borgmann-Winter K. E., Siegel S. J., Gallop R. J., Arnold S. E. (2006) Nat. Med. 12, 824–828 [DOI] [PubMed] [Google Scholar]
- 56.Silberberg G., Darvasi A., Pinkas-Kramarski R., Navon R. (2006) Am. J. Med. Genet. B Neuropsychiatr. Genet. 141B, 142–148 [DOI] [PubMed] [Google Scholar]
- 57.Law A. J., Kleinman J. E., Weinberger D. R., Weickert C. S. (2007) Hum. Mol. Genet. 16, 129–141 [DOI] [PubMed] [Google Scholar]
- 58.Chong V. Z., Thompson M., Beltaifa S., Webster M. J., Law A. J., Weickert C. S. (2008) Schizophr. Res. 100, 270–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu Y., Sullivan L. L., Nair S. S., Williams C. C., Pandey A. K., Marrero L., Vadlamudi R. K., Jones F. E. (2006) Cancer Res. 66, 7991–7998 [DOI] [PubMed] [Google Scholar]
- 60.Bollig A., Miksicek R. J. (2000) Mol. Endocrinol. 14, 634–649 [DOI] [PubMed] [Google Scholar]
- 61.Montague D., Weickert C. S., Tomaskovic-Crook E., Rothmond D. A., Kleinman J. E., Rubinow D. R. (2008) J. Neuroendocrinol. 20, 893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Erenburg I., Schachter B., Mira y Lopez R., Ossowski L. (1997) Mol. Endocrinol. 11, 2004–2015 [DOI] [PubMed] [Google Scholar]
- 63.Ferro P., Forlani A., Muselli M., Pfeffer U. (2003) Int. J. Mol. Med. 12, 355–363 [PubMed] [Google Scholar]
- 64.Horvath G., Leser G., Helou K., Henriksson M. (2002) Gynecol. Oncol. 84, 271–279 [DOI] [PubMed] [Google Scholar]
- 65.Sundvall M., Peri L., Määttä J. A., Tvorogov D., Paatero I., Savisalo M., Silvennoinen O., Yarden Y., Elenius K. (2007) Oncogene 26, 6905–6914 [DOI] [PubMed] [Google Scholar]
- 66.Williams C. C., Allison J. G., Vidal G. A., Burow M. E., Beckman B. S., Marrero L., Jones F. E. (2004) J. Cell Biol. 167, 469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sardi S. P., Murtie J., Koirala S., Patten B. A., Corfas G. (2006) Cell 127, 185–197 [DOI] [PubMed] [Google Scholar]
- 68.Koehorst S. G., Jacobs H. M., Thijssen J. H., Blankenstein M. A. (1993) J. Steroid Biochem. Mol. Biol. 45, 227–233 [DOI] [PubMed] [Google Scholar]
- 69.Horvath G., Leser G., Hahlin M., Henriksson M. (2000) Int. J. Gynecol. Cancer 10, 128–136 [DOI] [PubMed] [Google Scholar]
- 70.Ishunina T. A., Swaab D. F., Fischer D. F. (2005) J. Clin. Endocrinol. Metab. 90, 3757–3765 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.