Abstract
CAPS (Ca2+-dependent activator protein for secretion) functions in priming Ca2+-dependent vesicle exocytosis, but the regulation of CAPS activity has not been characterized. Here we show that phosphorylation by protein kinase CK2 is required for CAPS activity. Dephosphorylation eliminated CAPS activity in reconstituting Ca2+-dependent vesicle exocytosis in permeable and intact PC12 cells. Ser-5, -6, and -7 and Ser-1281 were identified by mass spectrometry as the major phosphorylation sites in the 1289 residue protein. Ser-5, -6, and -7 but not Ser-1281 to Ala substitutions abolished CAPS activity. Protein kinase CK2 phosphorylated CAPS in vitro at these sites and restored the activity of dephosphorylated CAPS. CK2 is the likely in vivo CAPS protein kinase based on inhibition of phosphorylation by tetrabromo-2-benzotriazole in PC12 cells and by the identity of in vivo and in vitro phosphorylation sites. CAPS phosphorylation by CK2 was constitutive, but the elevation of Ca2+ in synaptosomes increased CAPS Ser-5 and -6 dephosphorylation, which terminates CAPS activity. These results identify a functionally important N-terminal phosphorylation site that regulates CAPS activity in priming vesicle exocytosis.
Regulated neurotransmitter secretion is central to intercellular communication in the nervous system. Two types of secretory vesicles mediate neurotransmitter release; that is, synaptic vesicles that release transmitters such as glutamate at synapses and dense-core vesicles that release modulatory transmitters and neuropeptides at non-synaptic sites. Both types of secretory vesicles are recruited to docking sites on the plasma membrane where they are primed to a ready release state to undergo fusion in response to Ca2+ elevations. Many of the proteins that mediate the targeting, docking, priming, and Ca2+-dependent fusion of vesicles with the plasma membrane function in both synaptic vesicle and dense-core vesicle pathways (1). CAPS-12 (also known as Cadps1) is a 1289-residue protein that reconstitutes Ca2+-triggered dense-core vesicle exocytosis in permeable neuroendocrine cells at a priming step (2–4). CAPS is required for secretion of a subset of transmitters in Caenorhabditis elegans (5) and Drosophila melanogaster (6) and for priming dense-core vesicle exocytosis in neuroendocrine cells (7) and synaptic vesicle exocytosis in neurons (8). Vesicle priming reactions are extensively modulated during physiological demand (9), but mechanisms that regulate CAPS function remain to be identified.
Reversible protein phosphorylation is a major mechanism for the regulation of cellular processes including vesicle exocytosis. Many proteins that function in evoked vesicle exocytosis are phosphoproteins (10, 11). The neuronal SNARE proteins syntaxin 1A, VAMP-2, and SNAP-25 are phosphorylated by several protein kinases in vitro (12–14). Protein kinase C and protein kinase A sites on SNAP-25 affect refilling rates and size, respectively, of the primed pool of vesicles in chromaffin cells (15, 16). Several SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor)-binding proteins such as munc18, RIM1, and rabphilin undergo regulated phosphorylation, but it is not known whether phosphorylation affects function (10, 11, 17).
Because the function of CAPS at a priming step in vesicle exocytosis may be regulated, we determined whether CAPS is phosphorylated. We show that CAPS is a phosphoprotein with functionally essential N-terminal phosphorylated Ser residues. Ser-5, -6, and -7 in CAPS were substrates for protein kinase CK2 in vitro and in vivo as well as for a Ca2+-dependent dephosphorylation mechanism. The results indicate that phosphorylation by protein kinase CK2 is necessary for CAPS activity in priming vesicle exocytosis and that regulated dephosphorylation may constitute a mechanism for terminating CAPS activity.
EXPERIMENTAL PROCEDURES
Cell Labeling, Transfection, and Immunoprecipitation
PC12 cells and COS-1 cells were cultured and transfected by electroporation (18, 19). Cells were incubated in phosphate-free Dulbecco's modified Eagle's medium (Sigma) for 30 min and labeled with 1 mCi of [32P]orthophosphate (GE Healthcare) for 1 h. For CK2 inhibition, 60 μm 4,5,6,7-tetrabromobenzotriazole (Calbiochem) was added to the labeling medium. Cell lysates were prepared in radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Triton X-100, 1 mm EDTA, 0.25% fish gelatin, 0.1% SDS, 50 mm NaF, 1 mm Na4O7P2) with a protease inhibitor mixture (Roche Applied Science) and incubated with CAPS polyclonal antibody for 1 h at 4 °C and with protein A-Sepharose Fast Flow beads for an additional 2 h. Beads were washed with radioimmune precipitation assay buffer and water before boiling in sample buffer. CAPS was detected by Western blotting with a CAPS pleckstrin homology (PH) domain polyclonal antibody, and 32P incorporation was detected by autoradiography. Signals were quantified by densitometry using ImageQuant (GE Healthcare).
Synaptosomes were prepared and stimulated as described previously (20). Mouse brains from animals older than P21 were homogenized in 0.32 m sucrose, 5 mm Hepes-NaOH, pH 7.4, 0.1 mm EDTA with a glass Teflon homogenizer at 900 rpm, homogenates were centrifuged at 900 × g for 10 min, and the supernatant was centrifuged at 14,400 × g for 20min. The synaptosome pellet was resuspended in freshly aerated KHH (Krebs-Henseleit-Hepes) buffer (118 mm NaCl, 3.5 mm KCl, 1.2 mm CaCl2, 1.2 mm MgSO4, 1.2 mm KH2PO4, 25 mm NaHCO3, 10 mm Hepes-NaOH, pH 7.4, 11.5 mm glucose) or high K+ KHH buffer (same as KHH but with 94 mm NaCl, 50 mm KCl) and incubated at 37 °C. Incubations were terminated by centrifugation at 735 × g for 20 min, and the pellet was resuspended in hypotonic lysis buffer (20 mm Hepes, pH 7.4, 10 mm Na4P2O7, 1 mm EDTA, 1 mm EGTA, 50 mm NaF, 1× protease inhibitor mixture) and centrifuged at 200,000 × g for 1 h. Supernatants were analyzed by Western blotting with phosphospecific CAPS antibody and with CAPS PH domain antibody.
Site-directed Mutagenesis of CAPS
To mutagenize Ser-5, -6, and/or -7 to Ala in full-length CAPS cDNA, a fragment containing nucleotides 1–1222 of the coding sequence was excised from pCMVCAPSmyc6xHis (19) using XhoI and SacI sites and cloned into pBluescript II KS− vector (Fermentas) using the same restriction sites. Site-directed mutagenesis was performed on the pBluescript II KS− CAPS XhoI-SacI construct, and the mutations were confirmed by sequencing. The fragment of CAPS flanked by XhoI and AflII sites was placed back into the corresponding site of pCMVCAPSmyc6xHis in place of the wild-type fragment. To mutagenize Ser-1281 to Ala, a fragment containing nucleotides 2326–3868 of the coding sequence was excised from pCMVCAPS-myc6xHis using Eco47III and EcoRI sites and cloned into pBluescript II KS− vector using SmaI and EcoRI sites. The construct pBluescript II KS− CAPS Eco47III-EcoRI was used as a template in site-directed mutagenesis, and the fragment of CAPS flanked by Eco47III and EcoRI with the mutation was placed back into the corresponding site of pCMVCAPSmyc6xHis as above.
Protein and Antibody Preparation
The C-terminal Myc-His6-tagged CAPS proteins used for in vitro dephosphorylation and phosphorylation and mass spectroscopy were expressed in Sf9 insect cells and purified as described (3). The C-terminal Myc-His6-tagged wild-type and mutant CAPS proteins used for secretion assays were purified from transfected COS-1 cell lysates by nickel-nitrilotriacetic acid chromatography. Circular dichroism was performed using the Aviv 62A DS Circular Dichroism Spectrometer (Aviv Associates). Proteins at 1.2 μm were scanned at 25 °C in 20 mm phosphate buffer, pH 7.4. Partial proteinase K digestion was conducted by incubation for 1 h at 25 °C at a CAPS:enzyme mass ratio of 100:1. Reactions were terminated by adding 1 mm 4-(2-aminoethyl)benzenesulfonyl fluoride plus 20 mm phenylmethylsulfonyl fluoride, and samples were analyzed by 13.5% SDS-PAGE and stained with Coomassie Brilliant Blue.
To dephosphorylate Myc-His6-tagged CAPS, the protein was incubated with λ protein phosphatase (New England Biolabs) in 50 mm Tris-HCl, pH 7.5, 1 mm EDTA, 1 mm dithiothreitol, 1 mm MnCl2 for 1 h at 30 °C and purified on nickel-nitrilotriacetic acid-agarose. To phosphorylate CAPS, the dephosphorylated protein was incubated with CK1, CK2, glycogen synthase kinase 3 (New England Biolabs), Ca2+/calmodulin-dependent protein kinase II, protein kinase A (Sigma), or protein kinase C (Calbiochem) in the manufacturers' suggested buffer plus 200 μm ATP and final specific activity of 500 μCi/μmol [32P]ATP (GE Healthcare) for 1 h at 30 °C. Proteins were analyzed on 7.5% SDS-PAGE gels, and gels were exposed to a Storage Phosphor Screen (GE Healthcare). 32P autoradiographic signals were processed on a Typhoon 9410 (GE Healthcare), and densitometry was conducted using ImageQuant.
A rabbit antibody was generated against a 20-mer peptide comprising amino acids 1–19 of CAPS with phospho-Ser at positions 5 and 6 and a C-terminal Cys residue for conjugation to KLH (MLDPpSpSSEEESDEILEEESC, pS is phosphoserine). Phosphopeptides and cognate unphosphorylated peptides were synthesized at the University of Wisconsin Biotechnology Center (Madison, WI), and antisera were generated by Harlan (Madison, WI). Rabbit serum was purified on protein A-Sepharose Fast Flow (Sigma) and applied to a SulfoLink coupling gel (Pierce) affinity column saturated with the non-phosphorylated CAPS-(1–19) peptide. The flow-through was applied to a SulfoLink coupling gel affinity column saturated with CAPS-(1–19) phosphopeptide. The phosphospecific antibody was eluted with 100 mm glycine, pH 3.0, neutralized with 1 m Tris-HCl, pH 8.0, and dialyzed against phosphate-buffered saline.
Permeable PC12 Cell Secretion Assays
Two permeable cell assays were used to detect Ca2+-dependent vesicle exocytosis. These assays effectively measure the priming activity of CAPS (4). To assay for the Ca2+-triggered release of [3H]norepinephrine, PC12 cells were labeled with 0.5 μCi/ml [3H]norepinephrine (GE Healthcare) plus 0.5 mm sodium ascorbate for 16 h at 37 °C and permeabilized by passage through a ball homogenizer (18). The permeable cells were preincubated with 1 mg/ml rat brain cytosol plus 2 mm MgATP and washed. [3H]Norepinephrine release was determined by incubating permeable cells with CAPS plus 10 μm Ca2+ for 3 min at 30 °C. In a second assay, the Ca2+-triggered release of norepinephrine was measured by rotating disc electrode voltammetry as described previously (4). PC12 cells were incubated with 1.5 mm norepinephrine and 0.5 mm sodium ascorbate for 16 h at 37 °C and subsequently processed as in the first assay. Ca2+ plus CAPS-triggered norepinephrine release was measured at 35 °C as norepinephrine oxidized at the surface of the rotating disc electrode.
CAPS Down-regulation and Rescue
CAPS was down-regulated in PC12 cells by transfection with a pSHAG plasmid (21) containing a hairpin sequence targeted to nucleotides 3839–3866 of CAPS mRNA. Rescue constructs were generated in a pcDNA3.1 CAPS plasmid by introducing four silent mutations into the target sequence. Constructs were subcloned into tag red fluorescent protein-containing plasmids to monitor successful re-expression of CAPS by fluorescence. Studies of evoked dense-core vesicle exocytosis were conducted in PC12 cells expressing brain-derived neurotrophic factor-enhanced green fluorescent protein by total internal reflection fluorescence microscopy as previously described (22).
Mass Spectrometry
CAPS peptides resulting from complete tryptic digestion were subjected to Ga(III)-immobilized metal affinity chromatography (23) to enrich for phosphopeptides. Phosphopeptides were incubated for 15 min at 37 °C with or without alkaline phosphatase (Calbiochem) (24). For phosphopeptide enrichment of native dephosphorylated and re-phosphorylated CAPS tryptic peptides, a Ga(III)-based phosphopeptide isolation kit (Pierce) was used as per the manufacturer's instructions. For matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF), samples were prepared with a ZipTip C18 (Millipore) and mixed with matrix (2 mg/ml 2′,4′,6′-trihydroxyacetophenone, 10 mm ammonium citrate, 25% acetonitrile, 0.2% trifluoroacetic acid). Phosphopeptide analysis was performed on a Voyager-STR (Applied Biosystems) in reflectron mode with a mass range from 900–4000 Da at a laser power of 2000–2300 arbitrary units and an acceleration voltage of 25 kV. Tandem mass spectrometry was performed on a Q-STAR QTOF with a MALDI source (Applied Biosystems). For additional sequence confirmation, tryptic peptides were analyzed with capillary reverse phase high performance liquid chromatography in-line with an electrospray ionization LCQ Deca ion-trap mass spectrometer (Thermo Finnigan) using a guided single ion monitoring approach as previously described (25, 26).
RESULTS
Phosphorylation Is Essential for CAPS Activity
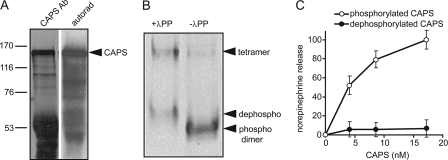
To determine whether CAPS is a phosphoprotein, PC12 cells were labeled with [32P]orthophosphate, and CAPS was immunoprecipitated from cell lysates. Immunoprecipitation of CAPS was specific (not shown), and autoradiography revealed that CAPS was phosphorylated (Fig. 1A). CAPS phosphorylation appeared to be constitutive because cells were not stimulated during 32P-labeling. We determined whether CAPS phosphorylation was required for its activity. CAPS proteins are tested for activity by their ability to restore Ca2+-dependent vesicle exocytosis in permeable PC12 cells (4, 18). A recombinant CAPS protein produced in insect Sf9 cells was purified and dephosphorylated with λ-protein phosphatase. CAPS is a ∼300-kDa dimer (2), and phosphatase-treated CAPS migrated more slowly on native gels due to the loss of phosphate (Fig. 1B). In addition to the mobility shift of the dimer, dephosphorylation increased the amount of a CAPS tetramer. Dephosphorylated CAPS was found to exhibit a complete loss of activity in reconstituting Ca2+-dependent exocytosis in permeable PC12 cells (Fig. 1C).
FIGURE 1.
Phosphorylation is essential for CAPS activity. A, CAPS was immunoprecipitated from extracts of 32P-labeled PC12 cells and analyzed on 7.5% SDS gels for Western blotting with CAPS antibody (Ab, left lane) or by autoradiography for 32P (right lane). B, recombinant CAPS purified from Sf9 cells (right lane) was incubated with λ-protein phosphatase (λPP, left lane) and analyzed by electrophoresis on native 6% polyacrylamide gels and Coomassie staining. Bands from the bottom to top correspond to native phosphorylated dimer (phospho), dephosphorylated dimer (dephospho), and higher molecular weight tetramer. C, native phosphorylated (○) but not dephosphorylated CAPS (●) restores Ca2+-triggered release of [3H]norepinephrine from permeable PC12 cells. Permeable cells were prepared as described under “Experimental Procedures” and incubated for 3 min at 30 °C with 10 μm Ca2+ plus the indicated concentrations of CAPS. Mean values of duplicate determinations with the indicated ranges correspond to the CAPS-dependent component of norepinephrine release with maximal value set at 100% for wild type CAPS.
A loss in the activity of CAPS upon dephosphorylation might be due to an overall structural change in the protein. When we compared phosphorylated and dephosphorylated CAPS proteins by circular dichroism, phosphorylated CAPS was ∼33% α-helical, whereas dephosphorylated CAPS was ∼23% α-helical (not shown). This indicated that dephosphorylation of CAPS results in some reduction in secondary structure. Limited proteolysis studies were used to identify protein conformational changes (27). Limited digestion of CAPS by proteinase K generated a discrete set of 20–35-kDa core fragments from phosphorylated CAPS, and this pattern was essentially identical for dephosphorylated CAPS (not shown). This suggested that CAPS dephosphorylation did not result in major conformational changes. Overall, the results indicated that CAPS dephosphorylation dramatically reduced its activity, promoted its oligomerization, and reduced its secondary structure.
Identification of CAPS Phosphorylation Sites
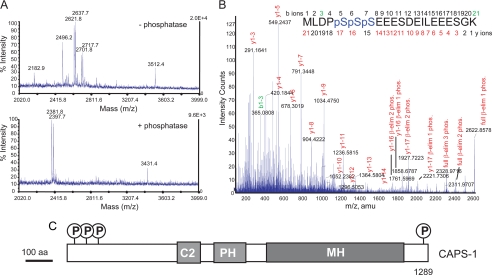
Mass spectrometry was used to map phosphorylation sites on CAPS. Purified CAPS produced in Sf9 insect cells was digested with trypsin, and phosphopeptides were enriched by chromatography on Ga(III)-immobilized metal affinity columns (23). Alkaline phosphatase treatment (24) combined with MALDI-TOF mass spectrometry (MS) was used to identify CAPS phosphopeptides. Several CAPS peptides treated with alkaline phosphatase exhibited mass shifts of −80 Da and multiples of −80 Da (HPO3−) (Fig. 2A). Comparison of the MALDI-TOF MS spectra identified three single-charged peptides that corresponded to dephosphorylated CAPS 1–21 (m/z = 2381.8 and 2397.7) and CAPS 1280–1309 (m/z = 3431.4) (Fig. 3A). CAPS 1280–1309 corresponded to CAPS 1280–1289 with a linker sequence.
FIGURE 2.
CAPS is phosphorylated at N- and C-terminal Ser residues. A, MALDI-TOF spectra of CAPS phosphopeptides. Phosphopeptides enriched from CAPS tryptic peptides by Ga(III)- immobilized metal ion affinity chromatography were incubated without (upper panel) or with (lower panel) alkaline phosphatase before MALDI-TOF to detect mass shifts of −80 Da (HPO3−) or multiples of −80 Da. B, MALDI-QTOF MS/MS spectrum of triply phosphorylated CAPS residues 1–21 (2622.9 atomic mass units (amu)) with a summary of Ser phosphorylations detected by neutral loss of H3PO4 by β-elimination (β-elim). C, schematic figure of CAPS indicating phosphorylation sites relative to other functional domains. Functional domains correspond to C2 (protein kinase C C2-like), PH, and MH (munc 13 homology).
FIGURE 3.
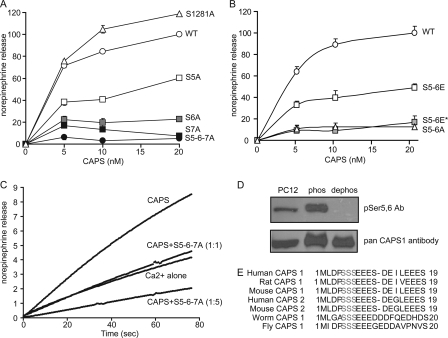
Phosphorylation of Ser-5, -6, and -7 is essential for CAPS activity. A, reconstituting activity of S5A, S6A, S7A, S5A/S6A/S7A, and S1281A CAPS mutants. Each mutant was expressed in COS-1 cells as a C-terminal Myc-His6-tagged protein and purified. [3H]Norepinephrine release from permeable PC12 cells was measured as in Fig. 1C. Maximal release promoted by wild-type CAPS was set as 100%, and release with Ca2+ alone was set as 0%. Values are the means of duplicate determinations with indicated ranges. WT, wild type. B, reconstituting activity of S5E/S6E CAPS was compared with that of the S5A/S6A mutant. Dephosphorylation of S5E/S6E (S5-6E*) reduced activity. C, dominant negative activity of S5A/S6A/S7A mutant CAPS. The effect of adding the mutant CAPS proteins to wild-type CAPS proteins in 1:1 or 5:1 ratio on norepinephrine release from permeable PC12 cells was measured by rotating disk electrode assay. D, detection of phosphorylated CAPS with phosphospecific antibody (Ab). Phosphospecific antibody generated against CAPS 1–19 with phospho-Ser-5 and -6 (upper panel) and a general CAPS antibody (lower panel) were used in Western blotting with PC12 cell lysate (lane 1), recombinant phosphorylated CAPS (lane 2), and dephosphorylated CAPS (lane 3). E, comparison of CAPS N-terminal amino acid sequences indicates conservation of Ser-5, -6, and -7.
CAPS 1–21 was triple or quadruple-phosphorylated, whereas CAPS 1280–1309 was single-phosphorylated (Fig. 2A). CAPS 1–21 had additional N-terminal acetylation or Met residue oxidation modifications as confirmed by liquid chromatography-electrospray ionization-tandem mass spectrometry (MS/MS) using guided single ion monitoring (not shown). There were five possible phosphorylation sites in triple- or quadruple-phosphorylated CAPS 1–21 (MLDPSSSEEESDEILEEESGK). MALDI-QTOF MS/MS analysis of CAPS 1–21 indicated it was phosphorylated at Ser-5, -6, and -7 but not at Ser-11 or Ser-19 (Fig. 2B). The collision-induced dissociation spectrum showed neutral loss of H3PO4 via β elimination only on y-ions containing Ser-5, -6, or -7. By contrast, CAPS 1280–1309 (DSDEEDEEDD+linker) was singly phosphorylated on the one possible phosphorylation site corresponding to Ser-1281 (not shown). These data indicated that CAPS phosphorylation sites were located at the extreme N and C termini of the protein corresponding to Ser-5, -6, and -7 and Ser-1281, respectively (Fig. 2C).
Effect of Ser-5, -6, and -7 Mutation on CAPS Activity
To determine which phosphoacceptor Ser residues were essential for CAPS activity, site-directed mutagenesis was used to generate Ala for Ser substitutions. The nonphosphorylatable Ala mutant proteins expressed in COS-1 cells were purified and tested for reconstituting activity in Ca2+-dependent exocytosis. CAPS activity was impaired when Ser-5, -6, and -7 were individually substituted with Ala (Fig. 3A). A severe loss of activity was observed for the triple S5A/S6A/S7A mutant (Fig. 3A). By contrast, there was no loss of activity for the S1281A mutant (Fig. 3A) or S11A or S19A mutants (not shown). The results indicated that Ser-5, -6 and -7 but not Ser-1281 phosphorylation was essential for CAPS activity. We also generated potential phosphomimetic CAPS mutants in which Ser-5, -6, and -7 were substituted with Glu or Asp residues. The activity of a S5E/S6E mutant was greater than that of the S5A/S6A mutant but less than that of wild-type protein (Fig. 3B). However, activity of S5E/S6E was completely eliminated by dephosphorylation (Fig. 3B). This indicated that Glu (Fig. 3B) and Asp (not shown) residues were not phosphomimetic, presumably due to differences in the charge density or size of their side chains compared with phospho-Ser. Asp and Glu substitutions at Ser-5 and-6 did, however, appear to preserve consensus sites for Ser-7 phosphorylation.
Dephosphorylated CAPS and the nonphosphorylatable S5A/S6A/S7A CAPS mutant were found to not only lack reconstituting activity in Ca2+-dependent exocytosis but also to exert dominant inhibitory effects. The S5A/S6A/S7A CAPS mutant inhibited wild-type CAPS activity completely at a 1:1 molar ratio (Fig. 3C). At a 5:1 molar ratio of S5A/S6A/S7A to wild-type CAPS, Ca2+-triggered secretion was reduced below the level of the Ca2+ alone control (Fig. 3C). This resulted from the inhibition of the activity of endogenous residual CAPS that is present in the permeable cells. The dominant negative activity of S5A/S6A/S7A CAPS may be related to the effect that Ser-5, -6, and -7 dephosphorylation has on the CAPS dimer-tetramer equilibrium (see Fig. 1B).
To determine whether CAPS is phosphorylated at Ser-5 and -6 in neuroendocrine cells, a phosphospecific antibody was generated to CAPS 1–19 (MLDPSpSpSEEESDEILEEES). The phosphospecific antibody reacted with recombinant wild type but not with dephosphorylated CAPS (Fig. 3D). The phosphospecific antibody also reacted with CAPS in PC12 cell lysates, indicating that Ser-5 and/or Ser-6 were phosphorylated in endogenous CAPS. Quantitative immunoblotting studies (not shown) indicated that at least 50% of CAPS in PC12 cell lysates was phosphorylated. We were unable to confirm phosphorylation of CAPS at Ser-7 in PC12 cells because technical limitations precluded generation of an antibody specific for all three phospho-Ser residues. The absolute conservation of the three N-terminal Ser phosphoacceptors across CAPS isoforms in different species (Fig. 3E) is consistent with an essential role for phosphorylatable Ser-5, -6, and -7 in CAPS function.
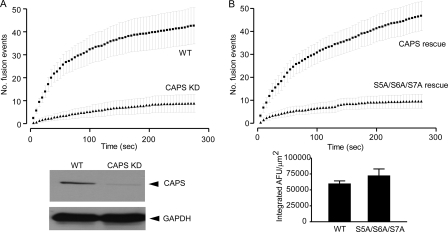
To assess the requirement for Ser-5, -6, and -7 phosphorylation in CAPS activity in vivo, we conducted studies of evoked dense-core vesicle exocytosis in PC12 cells expressing a CAPS short hairpin RNA plasmid. The short hairpin RNA plasmid reduced CAPS protein levels to 5% that of control levels and reduced evoked dense-core vesicle exocytosis to ∼20% that of control levels (Fig. 4A). Re-expression of wild-type CAPS fully restored evoked vesicle exocytosis (Fig. 4B). By contrast, re-expression of S5A/S6A/S7A CAPS was completely unable to restore evoked vesicle exocytosis even though it was expressed at the same level as wild-type CAPS (Fig. 4B). These results confirm the requirement for CAPS in evoked dense-core vesicle exocytosis and reveal that phosphorylatable Ser residues at positions 5, 6, or 7 are essential for CAPS function in vivo.
FIGURE 4.
Phosphoacceptor Ser-5, -6, and -7 residues are essential for in vivo CAPS activity in evoked vesicle exocytosis. A, CAPS was down-regulated in PC12 cells by transfection of an short hairpin RNA plasmid (lower panel) resulting in a 5-fold reduction in the number of exocytic events evoked by 56 mm K+ depolarization (upper panel). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; WT, wild type. B, transfection with a pcDNA3.1-CAPS-tag red fluorescent protein harboring silent mutations enabled re-expression of wild-type or S5A/S6A/S7A CAPS in down-regulated PC12 cells (lower panel). Wild-type CAPS expression restored evoked exocytosis, whereas S5A/S6A/S7A CAPS expression failed to do so (upper panel). Error bars indicate S.E. for n = 13–17 cells. AFM, arbitrary fluorescence units.
Phosphorylation of CAPS by Protein Kinase CK2
The Netphos 2.0 program (28, 29) identified the context of multiple acidic residues near Ser-5, -6, and -7 and Ser-1281 of CAPS as fitting the consensus (S/T)XX(D/E/pS/pY) for sites phosphorylated by protein kinase CK2. This prediction was tested by conducting in vitro phosphorylation studies with protein kinases using dephosphorylated CAPS as substrate. Dephosphorylated CAPS was a substrate for protein kinases CK1 and CK2, whereas no significant phosphorylation of dephosphorylated CAPS by Ca2+-calmodulin-dependent protein kinase II, glycogen synthase kinase 3, or protein kinase A was detected (Fig. 5A). Previous studies (30) showed that native CAPS is a substrate for protein kinase C. We confirmed that native CAPS was phosphorylated by protein kinase C (not shown); however, dephosphorylated CAPS was not a substrate for protein kinase C (Fig. 5A). Phosphorylation of native CAPS by protein kinase C did not affect CAPS activity (not shown).
FIGURE 5.
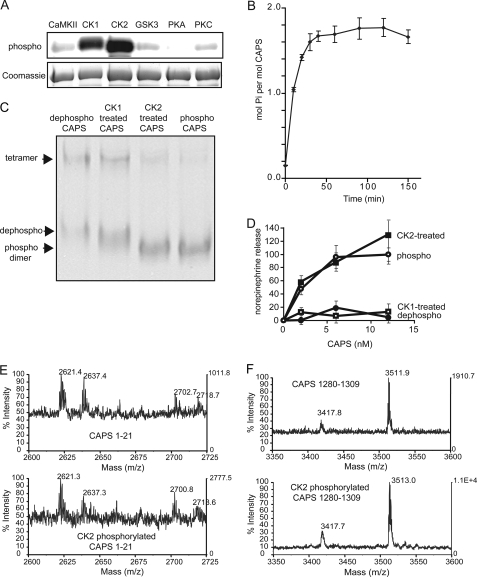
CK2 phosphorylates CAPS in vitro and restores activity. A, in vitro phosphorylation of CAPS by Ca2+/calmodulin-dependent protein kinase II, CK1, CK2, glycogen synthase kinase 3, protein kinase A (PKA), and protein kinase C (PKC). Dephosphorylated CAPS was incubated with the indicated enzymes in the presence of [32P]ATP. Proteins separated by 7.5% SDS-PAGE were analyzed by phosphorimaging (upper panel) and Coomassie G250 staining (lower panel). B, the approximate stoichiometry of phosphorylation of 20 μg of dephospho-CAPS by 50 units of CK2 was determined at the indicated incubation times at 30 °C. C, CAPS proteins were analyzed by 6% acrylamide native gel electrophoresis and staining with Coomassie G250. Phosphorylated dimer, dephosphorylated dimer, and tetramer are indicated by arrowheads from bottom to top. D, activity of CAPS phosphorylated by CK1 or CK2. Dephosphorylated CAPS (●) incubated with either CK1 (□) or CK2 (■) plus MgATP were re-purified and tested for activity in Ca2+-dependent [3H]norepinephrine release from permeable PC12 cells. Maximal release promoted by native phosphorylated CAPS (○) was set as 100%, and release with Ca2+ alone was set as 0%. Values are the mean of duplicate determination with indicated ranges. E and F, native phosphorylated CAPS (upper panels) and dephosphorylated CAPS incubated with CK2 and MgATP (lower panels) were compared by MALDI-TOF after trypsin digestion. MALDI-TOF spectra corresponding to CAPS 1–21 (E) and CAPS 1280–1309 (F) are shown.
The phosphorylation of dephosphorylated CAPS catalyzed by CK2 was more extensive than that by CK1 (Fig. 5A). The stoichiometry of CAPS phosphorylation in vitro by CK2 was estimated to be ∼2 mol of Pi incorporated per mol CAPS (Fig. 5B). Native gel electrophoresis indicated that CK2 phosphorylation of dephosphorylated CAPS fully restored the mobility of the CAPS dimer and reduced CAPS tetramerization (Fig. 5C). By contrast, CK1 promoted only a partial mobility shift of the dimer (Fig. 5C). Mass spectrometry confirmed that CK2 phosphorylated CAPS in vitro at the same sites phosphorylated in native CAPS (Fig. 5, E and F). Both native and CK2-phosphorylated CAPS contained single-charged peptides with masses of 2622, 2638, 2702, 2718 (Fig. 5E), and 3512 (Fig. 5F), which correspond to phosphorylated CAPS 1–21 and CAPS 1280–1289 with linker, respectively. These peptides were absent in dephosphorylated CAPS (not shown). The results indicated that sites phosphorylated by CK2 in vitro correspond to sites phosphorylated in vivo.
Consistent with the ability of CK2 to restore the native properties and phosphorylation of dephosphorylated CAPS, we found that phosphorylation by CK2 fully restored the activity of dephosphorylated CAPS in reconstituting Ca2+-dependent exocytosis (Fig. 5D). This contrasted with CK1-mediated phosphorylation, which failed to restore CAPS activity (Fig. 5D). Collectively, the results indicate that phosphorylation by protein kinase CK2 is both necessary and sufficient for CAPS activity.
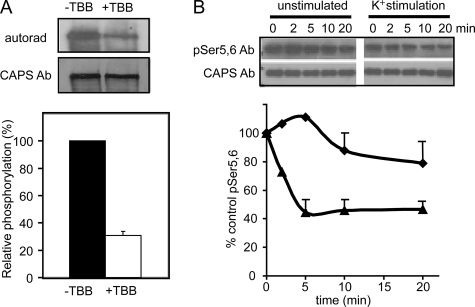
To determine whether CK2 was the major protein kinase that mediates in vivo CAPS phosphorylation, the selective CK2 inhibitor tetrabromo-2-benzotriazole (31, 32) was used in 32P-labeling of PC12 cells. Tetrabromo-2-benzotriazole treatment reduced the 32P-labeling of CAPS by ∼70% (Fig. 6A). This result indicated that CK2 is likely the main protein kinase responsible for CAPS phosphorylation in vivo.
FIGURE 6.
CK2 phosphorylates CAPS in vivo. A, CAPS phosphorylation was inhibited in PC12 cells treated with a specific CK2 inhibitor. PC12 cells were labeled with [32P]orthophosphate for 1 h in the absence or presence of 60 μm tetrabromo-2-benzotriazole (TBB). CAPS immunoprecipitates were analyzed by 7.5% SDS-PAGE and autoradiography or Western blotting with a general CAPS antibody (Ab). Densitometric scanning of autoradiogram and Western blot was used to assess relative phosphorylation. Values are the mean of triplicate determinations with the indicated S.D. Tetrabromo-2-benzotriazole inhibited CAPS phosphorylation by 70%. B, CAPS dephosphorylation was promoted by Ca2+ influx in synaptosomes. Mouse brain synaptosomes were incubated under control conditions (♦) or stimulated with high K+ (▲) for the indicated times. Synaptosomes were hypotonically lysed, and supernatant fractions were analyzed by Western blotting with Ser(P)-5 and -6 phosphospecific antibody or general CAPS antibody. For the graphic representation the signal at the zero time point is set as 100%, and values are the mean of duplicate determination with indicated ranges.
Regulation of CAPS Dephosphorylation
CAPS is an abundant brain protein (2) that was shown to be required for Ca2+-triggered catecholamine secretion from brain synaptosome preparations (33, 34). To determine whether the phosphorylation state of CAPS was altered during Ca2+-triggered exocytosis, we prepared synaptosomes for probing with the Ser(P)-5- and -6-specific CAPS antibody. Synaptosomes were stimulated with depolarizing high K+ buffers to promote Ca2+ influx and neurotransmitter release. CAPS levels, detected with a pan-specific antibody, were maintained during the incubation period in both control and stimulated synaptosomes (Fig. 6B). By contrast, CAPS reactive with the phosphospecific antibody rapidly decreased in depolarized but not in unstimulated synaptosomes (Fig. 6B). The results indicate that CAPS undergoes rapid activity-dependent dephosphorylation in response to depolarization. Because dephosphorylation at the N-terminal Ser residues inactivates CAPS for Ca2+-dependent exocytosis, the dephosphorylation mechanism revealed by these studies suggests possible negative feedback control of CAPS activity in response to strong stimulation.
DISCUSSION
CAPS is part of the machinery required for priming Ca2+-dependent vesicle exocytosis (2, 4, 6, 7). The major conclusion of the current work is that phosphorylation of CAPS at Ser-5, -6, and -7 by protein kinase CK2 is essential for CAPS activity. Dephosphorylation in vitro by λ-protein phosphatase completely abolished CAPS activity in reconstituting Ca2+-dependent vesicle exocytosis in permeable PC12 cells. Moreover, a non-phosphorylatable S5A/S6A/S7A CAPS mutant was incapable of rescuing CAPS function in permeable or intact PC12 cells. Mass spectrometry revealed that active insect cell-generated recombinant CAPS was multiply phosphorylated at Ser-5, -6, and -7 and at Ser-1281. These residues fall within consensus sites for protein kinase CK2, and in vitro studies confirmed that CK2 phosphorylated these sites and fully restored the activity of dephosphorylated CAPS. Ser to Ala substitutions showed that the N-terminal triple-phosphorylated site, but not the C-terminal phosphorylation site, was essential for CAPS activity. In vivo CAPS phosphorylation occurs at sites corresponding to those catalyzed by CK2 in vitro, indicating that CK2 corresponds to the CAPS protein kinase. Collectively, the results show that phosphorylation of CAPS at Ser-5, -6, and -7 by protein kinase CK2 is necessary and sufficient for CAPS function in Ca2+-dependent dense-core vesicle exocytosis.
By contrast, CK2-mediated phosphorylation of Ser-1281 was not essential for CAPS function in priming vesicle exocytosis. However, Ser-1281 phosphorylation may regulate other aspects of CAPS function. C-terminal sequences in CAPS are essential for interactions with dense-core vesicles (19). These could mediate the reported CAPS stimulation of catecholamine loading by vesicular amine transporter in chromaffin cells (35). Intriguingly, the C terminus of vesicular amine transporter is also a substrate for CK2 (36).
Protein kinase CK2 is a Ser/Thr kinase ubiquitously expressed in vertebrate tissues with particularly high levels in brain (37, 38) where CAPS is also most highly expressed (2, 39, 40). Numerous CK2 substrates have been suggested to function in synaptic transmission and synaptic plasticity (37). Proteins required for Ca2+-dependent vesicle exocytosis such as synaptotagmin-1, syntaxin 1A, and VAMP-2 are in vitro substrates of CK2 (12, 14, 41). Of these, only syntaxin 1A was shown to be phosphorylated at a CK2 site in vivo (42), but whether syntaxin 1A phosphorylation by CK2 is required for function in vesicle exocytosis was not determined (43).
CAPS is the first protein involved in vesicle exocytosis whose activity has been shown to be dependent on CK2 phosphorylation. Thus, the regulation of CK2 activity could be an important mechanism to modulate the priming of vesicle exocytosis. Glutamate N-methyl-d-aspartate receptor activation leads to a Ca2+-dependent increase in CK2 activity (44) that may be mediated by Ca2+-calmodulin-dependent protein kinase II phosphorylation of CK2 (45). Neurotrophins such as brain-derived neurotrophic factor also acutely enhance CK2 activity in neurons (46). Increased CK2 activity may enhance CAPS priming activity in evoked vesicle exocytosis. We were, however, unable to identify conditions in which CK2-mediated phosphorylation of CAPS was enhanced or regulated. Instead, at least 50% of CAPS was found to be phosphorylated in PC12 cells under resting conditions, suggesting that CAPS phosphorylation by CK2 may be constitutive rather than regulated.
Recent studies indicated that the size of the primed vesicle pool in insulin-secreting pancreatic β cells was dependent on the level of diphosphoinositol pentakisphosphate (48), which non-enzymatically pyrophosphorylates proteins on phospho-Ser residues within CK2 consensus sites (49). Whether CAPS is further activated by diphosphoinositol pentakisphosphate-mediated pyrophosphorylation to enhance vesicle priming will require further study.
Studies in synaptosomes revealed a regulated dephosphorylation mechanism for CAPS. Several synaptic proteins required for vesicle endocytosis such as dynamin, amphiphysin, synaptojanin, and phosphatidylinositol 5-kinase undergo rapid Ca2+-triggered dephosphorylation in a phosphatase PP2B/calcineurin-dependent mechanism that activates endocytic retrieval (47). The depolarization-evoked dephosphorylation at Ser-5 and -6 detected by the phosphospecific antibody would terminate CAPS activity for regulated exocytosis in parallel with the activation of the endocytic machinery for compensatory endocytosis. Whether PP2B/calcineurin mediates Ca2+-dependent CAPS dephosphorylation requires further study. Preliminary in vitro studies with purified phosphatases showed that the N-terminal phosphorylation sites of CAPS were resistant to dephosphorylation by phosphatases PP1, PP2A, and PP2B/calcineurin.3
Phosphatidylinositol 4,5-diphosphate is essential for priming evoked vesicle exocytosis (50) and serves as a co-factor for CAPS membrane recruitment and activation (4, 19, 51). Protein kinase CK2 was reported to be inhibited by the binding of phosphatidylinositol 4,5-diphosphate (52). CAPS might be locally inactivated after it functions in exocytosis by the simultaneous inhibition of CK2 activity and Ca2+-dependent activation of a protein phosphatase. It will be important to determine whether CAPS phosphorylation is dynamic and whether a phosphorylation/dephosphorylation cycle is coupled to the regulation of its activity in vesicle exocytosis.
In summary, our results indicate that CAPS is phosphorylated by CK2 and that this phosphorylation is essential for CAPS activity. In addition to the pleckstrin homology and C2 domains that have been identified as essential for CAPS function (5, 19, 51), this study characterizes a functionally important N-terminal phosphorylation domain in CAPS.
This work was supported, in whole or in part, by National Institutes of Health Grants DK25861 and DK40428 (to T. F. J. M.). This work was also supported by an American Heart Association fellowship (to M. N.).
- CAPS
- Ca2+-dependent activator protein for secretion
- CK2
- casein kinase 2
- CK1
- casein kinase 1
- PH
- pleckstrin homology
- MALDI-TOF
- matrix-assisted laser desorption/ionization time-of-flight
- QTOF
- quadrupole-time of flight
- MS
- mass spectrometry.
M. Nojiri, unpublished information.
REFERENCES
- 1.Malsam J., Kreye S., Söllner T. H. (2008) Cell. Mol. Life Sci. 65, 2814–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walent J. H., Porter B. W., Martin T. F. (1992) Cell 70, 765–775 [DOI] [PubMed] [Google Scholar]
- 3.Ann K., Kowalchyk J. A., Loyet K. M., Martin T. F. (1997) J. Biol. Chem. 272, 19637–19640 [DOI] [PubMed] [Google Scholar]
- 4.Grishanin R. N., Kowalchyk J. A., Klenchin V. A., Ann K., Earles C. A., Chapman E. R., Gerona R. R., Martin T. F. (2004) Neuron 43, 551–562 [DOI] [PubMed] [Google Scholar]
- 5.Speese S., Petrie M., Schuske K., Ailion M., Ann K., Iwasaki K., Jorgensen E. M., Martin T. F. (2007) J. Neurosci. 27, 6150–6162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Renden R., Berwin B., Davis W., Ann K., Chin C. T., Kreber R., Ganetzky B., Martin T. F., Broadie K. (2001) Neuron 31, 421–437 [DOI] [PubMed] [Google Scholar]
- 7.Liu Y., Schirra C., Stevens D. R., Matti U., Speidel D., Hof D., Bruns D., Brose N., Rettig J. (2008) J. Neurosci. 28, 5594–5601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jockusch W. J., Speidel D., Sigler A., Sørensen J. B., Varoqueaux F., Rhee J. S., Brose N. (2007) Cell 131, 796–808 [DOI] [PubMed] [Google Scholar]
- 9.Wojcik S. M., Brose N. (2007) Neuron 55, 11–24 [DOI] [PubMed] [Google Scholar]
- 10.Turner K. M., Burgoyne R. D., Morgan A. (1999) Trends Neurosci. 22, 459–464 [DOI] [PubMed] [Google Scholar]
- 11.Leenders A. G., Sheng Z. H. (2005) Pharmacol. Ther. 105, 69–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennett M. K., Miller K. G., Scheller R. H. (1993) J. Neurosci. 13, 1701–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirling H., Scheller R. H. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11945–11949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nielander H. B., Onofri F., Valtorta F., Schiavo G., Montecucco C., Greengard P., Benfenati F. (1995) J. Neurochem. 65, 1712–1720 [DOI] [PubMed] [Google Scholar]
- 15.Nagy G., Matti U., Nehring R. B., Binz T., Rettig J., Neher E., Sørensen J. B. (2002) J. Neurosci. 22, 9278–9286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagy G., Reim K., Matti U., Brose N., Binz T., Rettig J., Neher E., Sørensen J. B. (2004) Neuron 41, 417–429 [DOI] [PubMed] [Google Scholar]
- 17.Lonart G. (2002) Trends Neurosci. 25, 329–332 [DOI] [PubMed] [Google Scholar]
- 18.Klenchin V. A., Kowalchyk J. A., Martin T. F. (1998) Methods 16, 204–208 [DOI] [PubMed] [Google Scholar]
- 19.Grishanin R. N., Klenchin V. A., Loyet K. M., Kowalchyk J. A., Ann K., Martin T. F. (2002) J. Biol. Chem. 277, 22025–22034 [DOI] [PubMed] [Google Scholar]
- 20.Hosaka M., Hammer R. E., Südhof T. C. (1999) Neuron 24, 377–387 [DOI] [PubMed] [Google Scholar]
- 21.Lynch K. L., Martin T. F. (2007) J. Cell Sci. 120, 617–627 [DOI] [PubMed] [Google Scholar]
- 22.Lynch K. L., Gerona R. R., Kielar D. M., Martens S., McMahon H. T., Martin T. F. (2008) Mol. Biol. Cell 19, 5093–5103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Posewitz M. C., Tempst P. (1999) Anal. Chem. 71, 2883–2892 [DOI] [PubMed] [Google Scholar]
- 24.Liao P. C., Leykam J., Andrews P. C., Gage D. A., Allison J. (1994) Anal. Biochem. 219, 9–20 [DOI] [PubMed] [Google Scholar]
- 25.Loyet K. M., Ouyang W., Eaton D. L., Stults J. T. (2005) J. Proteome Res. 4, 400–409 [DOI] [PubMed] [Google Scholar]
- 26.Arnott D., Kishiyama A., Luis E. A., Ludlum S. G., Marsters J. C., Jr., Stults J. T. (2002) Mol. Cell. Proteomics 1, 148–156 [DOI] [PubMed] [Google Scholar]
- 27.Calvert R., Gratzer W. B. (1978) FEBS Lett. 86, 247–249 [DOI] [PubMed] [Google Scholar]
- 28.Blom N., Gammeltoft S., Brunak S. (1999) J. Mol. Biol. 294, 1351–1362 [DOI] [PubMed] [Google Scholar]
- 29.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., Bairoch A. (2003) Nucleic Acids Res. 31, 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishizaki T., Walent J. H., Kowalchyk J. A., Martin T. F. (1992) J. Biol. Chem. 267, 23972–23981 [PubMed] [Google Scholar]
- 31.Battistutta R., De Moliner E., Sarno S., Zanotti G., Pinna L. A. (2001) Protein Sci. 10, 2200–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarno S., Reddy H., Meggio F., Ruzzene M., Davies S. P., Donella-Deana A., Shugar D., Pinna L. A. (2001) FEBS Lett. 496, 44–48 [DOI] [PubMed] [Google Scholar]
- 33.Berwin B., Floor E., Martin T. F. J. (1998) Neuron 21, 137–145 [DOI] [PubMed] [Google Scholar]
- 34.Tandon A., Bannykh S., Kowalchyk J. A., Banerjee A., Martin T. F., Balch W. E. (1998) Neuron 21, 147–154 [DOI] [PubMed] [Google Scholar]
- 35.Brunk I., Blex C., Speidel D., Brose N, Ahnert-Hilger G. (2009) J. Biol. Chem. 284, 1050–1056 [DOI] [PubMed] [Google Scholar]
- 36.Waites C. L., Mehta A., Tan P. K., Thomas G., Edwards R. H., Krantz D. E. (2001) J. Cell Biol. 152, 1159–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blanquet P. R. (2000) Prog. Neurobiol. 60, 211–246 [DOI] [PubMed] [Google Scholar]
- 38.Litchfield D. W. (2003) Biochem. J. 369, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wassenberg J. J., Martin T. F. (2002) Ann. N.Y. Acad. Sci. 971, 201–209 [DOI] [PubMed] [Google Scholar]
- 40.Speidel D., Varoqueaux F., Enk C., Nojiri M., Grishanin R. N., Martin T. F., Hofmann K., Brose N., Reim K. (2003) J. Biol. Chem. 278, 52802–52809 [DOI] [PubMed] [Google Scholar]
- 41.Davletov B., Sontag J. M., Hata Y., Petrenko A. G., Fykse E. M., Jahn R., Südhof T. C. (1993) J. Biol. Chem. 268, 6816–6822 [PubMed] [Google Scholar]
- 42.Foletti D. L., Lin R., Finley M. A., Scheller R. H. (2000) J. Neurosci. 20, 4535–4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Risinger C., Bennett M. K. (1999) J. Neurochem. 72, 614–624 [DOI] [PubMed] [Google Scholar]
- 44.Charriaut-Marlangue C., Otani S., Creuzet C., Ben-Ari Y., Loeb J. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 10232–10236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chung H. J., Huang Y. H., Lau L. F., Huganir R. L. (2004) J. Neurosci. 24, 10248–10259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blanquet P. R. (1998) Neuroscience 86, 739–749 [DOI] [PubMed] [Google Scholar]
- 47.Cousin M. A., Robinson P. J. (2001) Trends Neurosci. 24, 659–665 [DOI] [PubMed] [Google Scholar]
- 48.Illies C., Gromada J., Fiume R., Leibiger B., Yu J., Juhl K., Yang S. N., Barma D. K., Falck J. R., Saiardi A., Barker C. J., Berggren P. O. (2007) Science 318, 1299–1302 [DOI] [PubMed] [Google Scholar]
- 49.Bhandari R., Saiardi A., Ahmadibeni Y., Snowman A. M., Resnick A. C., Kristiansen T. Z., Molina H., Pandey A., Werner J. K., Jr., Juluri K. R., Xu Y., Prestwich G. D., Parang K., Snyder S. H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15305–15310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hay J. C., Fisette P. L., Jenkins G. H., Fukami K., Takenawa T., Anderson R. A., Martin T. F. (1995) Nature 374, 173–177 [DOI] [PubMed] [Google Scholar]
- 51.James D. J., Khodthong C., Kowalchyk J. A., Martin T. F. (2008) J. Cell Biol. 182, 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Korolchuk V. I., Cozier G., Banting G. (2005) J. Biol. Chem. 280, 40796–40801 [DOI] [PubMed] [Google Scholar]