Abstract
Recently, expression profiling of breast carcinomas has revealed gene signatures that predict clinical outcome, and discerned prognostically relevant breast cancer subtypes. Measurement of the degree of genomic instability provides a very similar stratification of prognostic groups. We therefore hypothesized that these features are linked. We used gene expression profiling of 48 breast cancer specimens that profoundly differed in their degree of genomic instability and identified a set of 12 genes that defines the two groups. The biological and prognostic significance of this gene set was established through survival prediction in published datasets from patients with breast cancer. Of note, the gene expression signatures that define specific prognostic subtypes in other breast cancer datasets, such as luminal A and B, basal, normal-like, and ERBB2+, and prognostic signatures including MammaPrint® and Oncotype DX, predicted genomic instability in our samples. This remarkable congruence suggests a biological interdependence of poor-prognosis gene signatures, breast cancer subtypes, genomic instability, and clinical outcome.
INTRODUCTION
Breast carcinomas are among the most frequent tumors in women worldwide.1 The clinical course and disease free survival times are extremely heterogeneous. This is why there have been intensive efforts to introduce morphological, clinical and molecular markers for precise disease staging and prognostication over the past decades.2 These markers include tumor size, grade and stage, lymph node and hormone receptor status, expression of growth factor receptors, DNA content, S-phase fraction and proliferative activity, and, more recently, specific gene expression signatures of poor prognosis.3-6
Quantitative measurement of the nuclear DNA content was the first attempt to understand the genetic basis of the observed profound differences in prognostic profiles of breast cancer. Large retrospective and prospective studies showed that patients with tumors with large variability in the nuclear DNA content, i.e., aneuploidy, have a recurrence free survival rate that is about only half as long as the one for patients with a diploid distribution.7, 8 The strong association of nuclear DNA content with disease free survival is independent of lymph node status, and therefore reflects a tumor innate biological feature that directly influences outcome. More recently, this ploidy-based classification system has been refined to not only describe the status quo of nuclear DNA content, i.e., diploid or aneuploid, but to also appreciate the degree of genomic instability reflected as the variability of the DNA content in the tumor cell population.9, 10 Based on these data it appears that more than just the ploidy status alone, but rather an inherent level of genomic instability (which can be measured as a stemline scatter index) is associated with prognosis. Extending these observations to mechanistic conclusions, one could argue that the degree of plasticity of a tumor genome provides the means to acquire a genetic and genomic makeup that bestows the nimbleness required to successfully negotiate the challenges of continuing selective pressure, and to generate tumor cell populations that are best adapted to environmental changes, including therapeutic interventions.
Numerous groups have used parallel gene expression profiling to extract clinically relevant signatures from breast cancers with considerable success.3-6, 11, 12 These gene expression signatures also allowed identification of breast cancer subtypes, such as luminal A and B, basal, normal-like, and ERBB2+, which harbor prognostic significance as well.3, 11, 13, 14 These signatures reportedly carry prognostic information independent of lymph-node status, but showed association with tumor grade, e.g. in the report by van de Vijver and colleagues.4 The same applies when measurements of the nuclear DNA content and genomic instability were used for outcome prediction, i.e., a strong correlation to histopathologic grading, yet no correlation to axillary node status.7, 8, 15, 16 These similarities, along with the fact that the Kaplan-Meier curves for assessment of recurrence-free survival were essentially identical when using either gene expression signatures or genomic instability as nominal independent variables, provoked the assumption that the two are connected. In order to determine the nature of this connection, we formulated the following hypotheses: first, the degree of genomic instability finds its reflection in (or is determined by) an aneuploidy-specific gene expression signature; second, this signature is general in nature and therefore robust enough to allow prediction of the clinical course in independent datasets; and, third, breast cancer subtype specific gene expression signatures of poor prognosis allow classification of our genomically stable and unstable tumors, hence connecting the two classification systems in support of a biological relationship. These hypotheses were tested by, first, gene expression profiling of 48 primary breast carcinomas with defined patterns and degrees of genomic instability; second, through classification of published and clinically annotated breast cancer datasets using our gene expression signature of genomic instability; and, third, by classification of our datasets using other breast cancer prognostic genetic signatures, including MammaPrint® 4, 5 and Oncotype DX.12
MATERIALS AND METHODS
Patient Samples
48 patients with breast cancer were diagnosed and tumor samples collected at the Karolinska Institute and Hospital, Stockholm, Sweden, during 2000 and 2001 adhering to the guidelines of the local ethical review board. Clinical material was collected from surgically removed tumors, diagnosed on H&E-stained tissue sections at the Department of Oncology and Pathology, Cancer Center Karolinska, and graded according to Elston.17 The clinical data are summarized in Table 1A. After surgery clinical tissue was first used for touch preparation slides for quantitative measurement of the nuclear DNA content before it was snap frozen until further processing with TRIzol reagent (Invitrogen, Carlsbad, CA) for DNA and RNA extraction. In addition, paraffin-embedded specimens of the same tumors were used for histopathology and immunohistochemistry.
Table 1.
Summary of clinical and experimental parameters in 17 diploid genomic stable (dGS), 15 aneuploid genomic stable (aGS), and 16 aneuploid genomic unstable (aGU) breast carcinomas.
| Case | Ploidy | Age | Size (mm) | Elston Grade | Histology | Side | Lymphnode Metastasis | Cyclin A Positivity | Surgery | CGH | cDNA Array | Subtype |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D03 | dGS | 61 | ND | II | ductal | left | 0/0 | 0% | lumpectomy | Yes | Yes | Normal-like |
| D05 | dGS | 83 | 30 | II | ductal | right | 0/3 | 2% | mastectomy | Yes | Yes | Normal-like |
| D07 | dGS | 41 | 18 | II | mucin | left | 0/11 | 6% | lumpectomy | Yes | Yes | Normal-like |
| D08 | dGS | 51 | 12 | ND | lobular | right | 0/8 | 1% | mastectomy | Yes | Yes | Normal-like |
| D09 | dGS | 48 | 25×20 | II | lobular | left | 0/0 | 1% | mastectomy | Yes | Yes | Luminal A |
| D10 | dGS | 83 | 45 | II-III | ductal | left | 3/7 | 3% | mastectomy | Yes | No | |
| D11 | dGS | 60 | 12 | II | ductal | bilateral | 0/ND | 1% | lumpectomy | Yes | No | |
| D12 | dGS | 75 | 12 | I | tubular | right | 0/0 | 5% | lumpectomy | Yes | Yes | Luminal A |
| D13 | dGS | 86 | 10 | ND | lobular | left | 0/0 | 4% | recurrent | Yes | Yes | Luminal A |
| D14 | dGS | 86 | 26 | I | ductal | right | 4/6 | 3% | lumpectomy | Yes | Yes | Luminal A |
| D15 | dGS | 50 | ND | II | lobular | right | 3/9 | 2% | mastectomy | Yes | Yes | Normal-like |
| D16 | dGS | 54 | 70 | III | lobular | left | 1/12 | 2% | mastectomy | Yes | Yes | Luminal A |
| D18 | dGS | 48 | 14 | II | ductal | left | 0/7 | 4% | mastectomy | Yes | No | |
| D19 | dGS | 62 | 20 | I | ductal | right | 0/3 | 8% | lumpectomy | No | Yes | Luminal A |
| D21 | dGS | 79 | 22 | II | ductal | left | 4/16 | 4% | mastectomy | Yes | Yes | Luminal A |
| D23 | dGS | 34 | 10 | II | ductal | left | 0/0 | 4% | lumpectomy | No | Yes | Luminal A |
| D25 | dGS | 71 | 12 | I | lobular | ND | 0/0 | 1% | lumpectomy | Yes | Yes | ERRB2+ |
| NP01 | aGS | 55 | 60 | III | ductal | right | 9/9 | 2% | mastectomy | Yes | Yes | Luminal A |
| NP02 | aGS | 62 | 11 | I | ductal | right | 0/23 | 6% | mastectomy | Yes | Yes | Luminal A |
| NP06 | aGS | 72 | 15 | II | ductal | left | 0/5 | 1% | lumpectomy | Yes | Yes | Luminal A |
| NP08 | aGS | 58 | 14 | I | ductal | left | 0/0 | 3% | lumpectomy | Yes | Yes | Normal-like |
| NP10 | aGS | 71 | ND | II | Paget's | right | 0/0 | 4% | lumpectomy | No | Yes | Normal-like |
| NP11 | aGS | 81 | 26 | II | ductal | left | 1/7 | 2% | mastectomy | Yes | Yes | Luminal A |
| NP12 | aGS | 62 | 60+15 | I | lobular | right | 2/7 | 6% | mastectomy | Yes | Yes | Luminal A |
| NP13 | aGS | 75 | 13 | I | ductal | left | 0/0 | 0% | lumpectomy | Yes | Yes | Luminal A |
| NP14 | aGS | 54 | 12+9 | II | ductal | right | 0/0 | 1% | lumpectomy | Yes | Yes | ERRB2+ |
| NP16 | aGS | 49 | 54 | III | ductal/lobular | right | 0/0 | 0% | mastectomy | No | Yes | ERRB2+ |
| NP17 | aGS | 52 | 14 | II | ductal | right | 0/6 | 8% | mastectomy | Yes | Yes | Luminal A |
| NP18 | aGS | 63 | 30 | II | ductal | left | 0/0 | 4% | mastectomy | No | Yes | ERRB2+ |
| NP19 | aGS | 54 | 10 | II | ductal | right | 0/6 | 6% | lumpectomy | Yes | No | |
| NP20 | aGS | 43 | 20 | III | ductal | left | 0/6 | 6% | lumpectomy | Yes | Yes | Luminal A |
| NP22 | aGS | 88 | 20 | III | ductal | left | 0/0 | 6% | mastectomy | Yes | Yes | Luminal A |
| A01 | aGU | 54 | 20 | III | ductal | right | 0/5 | 20% | mastectomy | Yes | Yes | Basal |
| A02 | aGU | 62 | 35 | III | comedo | right | 0/35 | 15% | mastectomy | No | Yes | ERRB2+ |
| A03 | aGU | 43 | 35 | III | ductal | left | 0/0 | ND | mastectomy | No | Yes | ERRB2+ |
| A04 | aGU | 46 | 20 | III | ductal | left | 0/0 | 11% | mastectomy | Yes | Yes | Basal |
| A05 | aGU | 54 | 30 | III | medullar | ND | 14/15 | 30% | mastectomy | No | Yes | ERRB2+ |
| A06 | aGU | 74 | 40 | III | ductal | left | 0/17 | 10% | mastectomy | Yes | Yes | Basal |
| A07 | aGU | 55 | 40 | III | ductal | right | 0/0 | 1% | mastectomy | Yes | Yes | ERRB2+ |
| A10 | aGU | 71 | 30 | III | ductal | left | 0/0 | 54% | lumpectomy | Yes | Yes | Luminal A |
| A12 | aGU | 79 | 16 | III | ductal | left | 4/14 | 10% | mastectomy | No | Yes | Luminal A |
| A14 | aGU | 57 | 25 | III | ductal | left | 0/2 | 12% | mastectomy | Yes | Yes | ERRB2+ |
| A15 | aGU | 59 | 20 | III | ductal | left | 1/9 | 5% | mastectomy | No | Yes | ERRB2+ |
| A16 | aGU | 85 | 45 | III | comedo | right | 0/3 | 9% | mastectomy | Yes | Yes | ERRB2+ |
| A17 | aGU | 66 | 12 | III | lobular | left | 0/0 | 14% | lumpectomy | Yes | Yes | ERRB2+ |
| A18 | aGU | 62 | 12 | II | ductal | right | 0/10 | 23% | lumpectomy | Yes | Yes | Basal |
| A19 | aGU | 60 | 18+9 | III | ductal | ND | 0/ND | 62% | lumpectomy | Yes | Yes | Basal |
| A20 | aGU | 57 | 8 | III | metaplastic | right | 0/0 | 17% | lumpectomy | Yes | Yes | ERRB2+ |
ND, not determined. The column “subtype” refers to the classification established from our analyses, not to the actual clinical subtype.
Image Cytometry
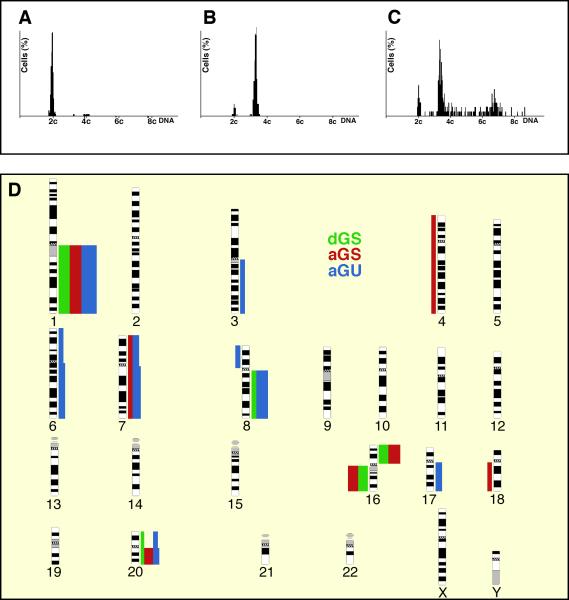
Image cytometry was performed on Feulgen-stained touch preparation slides. The staining procedure, internal standardization, and tumor cell selection were based on methods described previously.8 All DNA-values were expressed in relation to the corresponding staining controls which were given the value 2c, denoting the normal diploid DNA-content. The tumors were classified as belonging to three groups: (i) diploid cases with a distinct peak in the normal 2c region and no cells exceeding 5c, (ii) aneuploid cases with a main peak different from 2c and a stemline scatter index (SSI) below or equal 8.8, and (iii) aneuploid samples with a varying numbers of cells (>5%) exceeding 5c (SSI above 8.8). This novel classification system adheres to the parameters established by Kronenwett and colleagues,9 who defined the stemline scatter index (SSI) as a measurement of clonal heterogeneity in the tumor cell population. In our sample cohorts, seventeen tumors were classified as diploid, genomically stable (dGS), 15 tumors as aneuploid, yet genomic stable (aGS), and 16 tumors belonged to the aneuploid and genomic unstable (aGU) group of breast carcinomas. Examples of representative histograms for each group are provided in Figure 1A-C. The degree of genomic instability status was compared with grading using the Pearson's Chi-square test and with non-categorized parameters (age, tumor size, number of lymph nodes with metastases, cyclin A labeling index) using the Kruskall-Wallis test.
Figure 1.
Genomic instability in breast cancer: Examples of DNA histograms of diploid, genomically stable tumors (dGS) in A, aneuploid, yet genomically stable tumors (aGS) in B, and aneuploid and genomically unstable (aGU) tumors in C. Note the profound scattering of the ploidy stemline in C (for details of the ploidy classification see.9 D: Summary of genomic imbalances in 15 dGS (green), 12 aGS (red), and 11 aGU (blue) breast carcinomas analyzed by comparative genomic hybridization. Bars on the left side of the chromosome ideogram denote a loss of sequence in the tumor genome, while bars on the right side designate a gain. The width of the bars indicates the relative frequency of gains and losses observed.
Immunohistochemistry
All slides were deparaffinized with xylene, rehydrated and microwaved at 500W for 2×5 min in 10mM citrate buffer, pH 6.0. Intrinsic peroxidase activity was blocked with 3% hydrogen peroxide in methanol, followed by incubation with horse serum (1:20 dilution) in 0.1M PBS, pH 6.0. The levels of protein expression were revealed by overnight incubation with an antibody against cyclin A (Dilution 1:100, Novocastra, Newcastle upon Tyne, UK). The antibody was diluted in 1% (weight/volume) bovine serum albumin and visualized by standard avidin biotin-peroxidase complex technique (Vector Laboratories, Burlingame, CA). The cyclin A immunoreactivity was confined to the cell nuclei. In each specimen, the cyclin A labeling index (i.e., the percentage of stained tumor cells) was calculated.
Comparative Genomic Hybridization (CGH)
DNA was extracted from fresh frozen tissue using TRIzol. CGH was performed as described in detail elsewhere (http://riedlab.nci.nih.gov). Fluorescence intensity ratio plots were generated using Leica CW4000 Karyo V1.0 software (Leica Imaging Systems, Cambridge, UK). Interpretation of changes at 1pter, 16, 19, and 22 required careful examination because these loci are prone to artifacts due to the high proportion of repetitive sequences. CGH profiles of individual cases as well as the summary display of all cases can be found at http://www.ncbi.nlm.nih.gov/sky/skyweb.cgi.
Microarray analysis
Total RNA was extracted using TRIzol (Invitrogen) followed by Qiagen RNeasy column purification (Qiagen, Valencia, CA). Sample RNA and universal human reference RNA (Stratagene, La Jolla, CA) were then amplified for one round using RiboAmp RNA Amplification Kit (Arcturus, Mountain View, CA). All amplified RNA samples were hybridized against amplified reference RNA using a slightly modified protocol from Hedge and colleagues.18 Extraction and hybridization protocols used can be viewed in detail at http://www.riedlab.nci.nih.gov. In brief, 3 μg of amplified RNA was reverse transcribed using random primers and converted into cDNA using reverse transcriptase. After incorporation of aminoallyl-conjugated nucleotides, the RNA was indirectly labelled with Cy3 (tumor RNA) and Cy5 (reference RNA, Amersham, Piscataway, NJ). Each sample was hybridized against the reference RNA in a humid chamber (Arraylt™ Hybridization Cassette, TeleChem Intl., Sunnyvale, CA) for 16 hours at 42°C, washed, and scanned by the Axon GenePix 4000B Scanner (Axon Instruments, Union City, CA). In order to account for potential amplification bias, total RNA was hybridized following the same protocol for 11 samples (20 μg each). We used customized arrays obtained from the National Cancer Institute's microarray core facility. Arrays were used from one print batch and composed of 9,128 cDNAs denatured and immobilized on a poly-L-lysine-coated glass surface. The gene annotation file (GAL file) used (Hs-UniGEM2-v2px-32Bx18Cx18R.gal) can be found at the facilities website http://nciarray.nci.nih.gov. GenePix software version 4.0.1.17 was used to apply the GAL file through an interactive gridding process.19
Microarray quality assessment and data analysis (two-group class comparison)
After discarding arrays that did not pass our quality assessment criteria (Supplementary Methods, Section I), a total of 14 dGS samples, 14 aGS, and 16 aGU samples could be processed for further analysis. All values that did not meet the quality control criteria were treated as missing values as described in Supplementary Materials. Intensity ratios were calculated using the background corrected median intensities that were normalized with the locally weighted scatter plot smoother (LOWESS) algorithm for each print-tip group. The fraction of data points used in the local regression (f) was 0.1 and other parameters were adjusted as suggested by Cleveland.20 The value of f was determined using two self versus self experiments. All within-slide normalized ratios were log-transformed (natural base). A total of 7,657 genes were identified that did not show any missing values across all samples. Out of those 7,657 genes, differentially expressed genes were identified with pair-wise analysis: dGS against aGS, dGA against aGU, and aGS against aGU samples.
In order to produce a robust gene list, we used two methods and chose genes only when they appeared in both tests. First, we conducted the Wilcoxon rank-sum test with a permutation test so that if the p-value between the two groups was below 0.05, the values were randomly labeled into these two groups and the p-value was computed and repeated 10,000 times. All cases where the p-value using permuted labels was under 0.05 were summed and divided by the total number of permutations (10,000). This α-value denotes the probability that a gene had a smaller or equal significance by random permutation than the original significance as described earlier.21 Genes having α-value below 0.05 were considered to be differentially expressed. Second, we utilized a step-wise gene selection procedure.22, 23 The basic idea is to add genes one by one to a set of genes that discriminates two classes the best using Fisher's linear discriminant. The step-wise procedure was stopped if the weight (that marks the separation of the ratios between two classes) was less than 0.001. Finally, we took an intersection of the genes that were statistically significant using Wilcoxon test and also identified with the step-wise gene selection procedure.
Biological pathway analysis
We used Ingenuity Pathways Analysis (IPA) software (Ingenuity, Mountain View, CA) to assess the involvement of significantly differentially expressed genes in known pathways and networks. Differentially expressed genes as identified above were uploaded into the program and defined as focus genes if they were also part of the Pubmed-based IPA knowledge database. IPA was then used to determine groups of genes – involving focus genes and those known to interact with them - that together constitute networks. Such networks indicate how the genes of interest may influence each other above and beyond canonical pathways that are described in the Kyoto Encyclopedia of Genes and Genomes (KEGG, www.genome.jp/kegg). The IPA generated networks are listed in a certain order, with the top networks having a lower likelihood that the generation of the networks was serendipitous.
Identification of a gene signature (also by multiple-class comparison) that defines genomic instability and prognostic validation
The approach for the identification of a genomic instability signature and its prognostic validation is described below. A more detailed description is provided in Supplemental Methods, Section II.
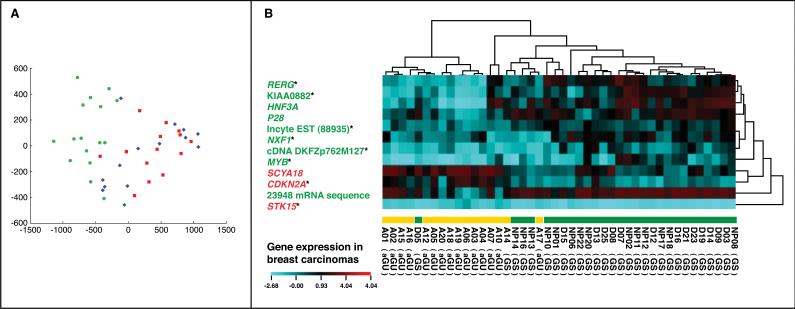
A supervised machine learning approach was applied in two sample settings to identify a genomic instability signature from the expression profiles of 7,657 genes in 44 primary breast cancer samples. In the first setting, genes were selected to discriminate genomically unstable (aGU) from genomically stable (aGS and dGS) tumors as binary classification. Random forests (the ValSelRF package)24 in R25 were used in the gene selection. In random forests, about one-third of the cases in the bootstrap sample are not used in growing the tree. These cases are called “out-of-bag” (OOB) cases and are used to evaluate the algorithm performance. A very important function of random forests is variable importance evaluation. Specifically, the variable evaluation process involves the following steps: 1) build a forest with 2,000 trees based on all 7,657 genes and obtain the importance measure of each gene variable; 2) based on the variable importance rank obtained in step 1, repeatedly remove 20% of the least important genes from the sample data; 3) in each iteration build a forest to get the out-of-bag (OOB) error estimate with the remaining genes; and 4) select the gene set with the smallest OOB error rate. The classification accuracy of the selected gene sets was evaluated with the Random Committee algorithm in WEKA26 during leave-one-out cross validation. Similar to Random Forests, Random Committee is an ensemble algorithm of classification trees. The first setting identified a 7-gene list. In the second setting, genes were selected to discriminate aGU, aGS, and dGS as multi-classification. Similarly, the random forests algorithm was used to identifiy a 70-gene list, which generated the smallest OOB error rate in the feature selection. Then, the Relief algorithm in WEKA26 was used to select the top 10 genes from this 70-gene set. This 10-gene list was able to further increase the classification accuracy of the 70-gene set. The accuracy of the gene sets in classifying aGU, aGS, and dGS was evaluated with the Naive Bayes algorithm in WEKA during leave-one-out cross-validation. Among the 7- and 10-gene sets, five genes were common. Therefore, these two gene lists were merged as a 12-gene genomic instability signature (Figure 2B). Eight signature genes (marked with asterisks in Figure 2B) were also selected in the previous differential gene expression analyses, providing an external validation to these results. This 12-gene signature was used to group the 44 breast carcinomas in hierarchical clustering analysis with CIMminer27 (Figure 2). The gene expression was aggregated based on Euclidean distance with average linkage. The distance of the samples was computed based on correlation and the cluster method was complete linkage.
Figure 2.
Principal Component Analysis (PCA) and Hierarchical Cluster Analysis of the gene expression data of the three classes of breast carcinomas. A. The PCA reflects pair-wise comparison of dGS (red dots) and aGU (green dots), which was then used to classify the third class (aGS, blue).
B. The cluster analysis shows aggregation into two groups separating genomically stable (dGS and aGS) from unstable tumors (aGU). The 12-gene genomic instability signature includes seven genes that were selected to classify genomically unstable (aGU) and stable (dGS and aGS) tumors, and 10 genes were selected to discriminate all three groups. There are five common genes among the 7- and 10-gene lists, namely, RERG, KIAA0882, cDNA DKFZp762M127, MYB, and STK15. The signature genes in red were over-expressed in genomically unstable breast carcinomas, whereas the genes in green were under-expressed in genomically unstable tumors. Genes marked with asterisks were also identified in the two-step differential expression analysis using Wilcoxon's tests, attesting to the robustness of the signature.
To evaluate the accuracy of this genomic instability signature in clinical outcome prediction, 496 tumor profiles in breast cancer4, 6, 11 were used as independent validation sets. The expression measurements of the signature genes were identified from the original microarray data with MatchMiner28 to predict disease-free survival and overall survival. The average expression centroids (profiles) of genomically stable patients and genomically unstable patients in our data were computed. Each patient in the validation cohorts was categorized into GS group or aGU group based on the correlation of this patient's gene expression profiles with the average expression profiles of the GS and aGU centroids in our data. In the external validation, patients are classified as GS (Genomically Stable) if the correlation of the gene expression with the average GS centroid is higher than that with the average aGU centroid. Similarly, they are classified as aGU (Genomically Unstable) if the correlation of the gene expression with the average aGU centroid is higher that with the average GS centroid. If there are multiple probes for the same annotated gene, the average of the gene expressions for all the probes is computed and used in the correlation analysis. The survival probabilities of the GS group and the aGU group were estimated by using Kaplan-Meier analyses with R. Statistical significance of the difference between the survival curves for different prognostic groups was assessed, using likelihood ratio tests and log-rank tests.
Classification of tumors to breast cancer intrinsic subtypes
Tumor samples in our data were classified into five breast cancer intrinsic subtypes (basal, ERBB2+, luminal A, luminal B, or normal-like) using the method from Chin et al.29 Specifically, the intrinsic gene list was downloaded from http://genome-www.stanford.edu/breast_cancer/robustness/data.shtml. The common genes in our data were identified by matching gene names. This common gene subset was used to compute the average expression profiles of each breast cancer subtype using 79 most tightly clustered samples from Sorlie et al.11 Pearson's correlation coefficient was determined between each tumor sample and each breast cancer subtype. Each tumor sample was classified to the subtype that has the highest correlation coefficient (p < 0.05). Detailed results are provided in Supplementary Table 5.
Classification of genomic instability by using Oncotype DX and MammaPrint
The signature genes in Oncotype DX12 and MammaPrint®4, 5 were identified in our data with MatchMiner.28 These two signatures were used to predict genomic instability in our data by using neural networks with WEKA, respectively. Leave-one-out cross validation was used to evaluate the predicted results.
Microarray data will be available online in a MIAME compliant format (http://www.riedlab.nci.nih.gov/).
RESULTS
The degree of genomic instability, as measured by the nuclear DNA content, directly impacts on a breast cancer patient's prognosis, independent from established parameters, such as lymph node status, and in addition to other clinical and histomorphological variables.7, 8 Patients with tumors that are genomically stable have a significantly better prognosis than those with unstable tumors.9, 10 We hypothesized that the profound differences between genomically stable and unstable breast carcinomas would reveal a measurable disparity in gene expression patterns as well. We have therefore analyzed 48 breast carcinomas using gene expression profiling on microarrays. In the present study, we selected 17 diploid tumors that were genomically stable (dGS), 15 tumors that were assessed as aneuploid, yet stable (aGS), and 16 carcinomas that were classified as aneuploid with substantial variability in the nuclear DNA content, and were hence assigned to the genomically unstable group (aGU); for details of the classification system see.9 Representative histograms of these tumor groups are displayed in Figure 1A-C, and the clinical data are summarized in Table 1. No significant differences were observed among the three groups regarding patients’ age, tumor size and number of lymph node metastases. However, a significant relationship existed between the degree of genomic instability and cyclin A labeling index (p < 0.0001 by Kruskal-Wallis test; data not shown). Moreover, tumors with high grade (Grade III) were significantly more frequently found in the aGU group compared with the dGS and aGS group (p < 0.0001 by Pearson's Chi-square test; data not shown). The higher degree of genomic instability in the aGU group was also reflected in an increase in chromosomal copy number changes as measured by comparative genomic hybridization. A detailed summary and comparison of chromosomal aberrations between the three groups is presented in Figure 1D. Chromosomal imbalances in the genomically stable tumors (dGS and aGS) were mostly restricted to gains of chromosome 1q and 16p, accompanied by losses on chromosome 16q, while aGU tumors showed more diverse changes, including frequent gain of the long arm of chromosome 17, the mapping position of the ERBB2 oncogene. This confirms earlier data from our group.30
Gene expression profiling using cDNA arrays
The similarity of the genomically stable groups (dGS and aGS), when contrasted to the tumors classified as genomically unstable (aGU), was further underlined by the results of our gene expression profiles. These similarity of the gene expression patterns of the two genomically stable groups, when compared to the genomically unstable group, is presented pictorially in Figure 2A as a Principal Component Analysis (PCA). The PCA was based on the genes summarized in supplemental Table 1A. Applying the Wilcoxon test with permutation test and the step-wise algorithm to identify differentially expressed genes for the pair-wise comparisons of the three groups we show that 38 genes were commonly differentially expressed for the comparisons aGU versus dGS and aGU versus aGS, whereas only two genes were commonly observed in the comparisons aGU versus aGS and aGS versus dGS, and three genes among aGS versus dGS and aGU versus dGS (Table 2). The gene lists that describe the exclusive differences between all groups are listed in supplemental Tables 1A-C.
Table 2.
Commonly differentially expressed genes for the comparisons of diploid genomically stable (dGS), aneuploid genomically stable (aGS), and aneuploid genomically unstable (aGU) breast carcinomas. The α-value denotes the probability that a gene had a smaller or equal significance by random permutation than the original significance as described earlier (Hyman et al., 2002). Genes having α-value below 0.05 were considered to be differentially expressed.
| Nr | Incyte PD | Gene name | ENTREZ Gene ID | Location | Gene function | Ratio aGU / dGS | Ratio aGU / aGS | α-value aGU / dGS | α-value aGU / aGS |
|---|---|---|---|---|---|---|---|---|---|
| Genes similar expressed for “aGU versus dGS” and “aGU versus aGS” | |||||||||
| 1 | 62144 | CCNE1 - cyclin E1 | 898 | 19q12 | Overexpression is known for many tumors and results in chromosome instability. | 2.23 | 2.34 | 0.0479 | 0.0407 |
| 2 | 520342 | EVL - Enah/Vasp-like | 51466 | 14q32.32 | Cytoskeletal regulator activity and cell surface receptor linked signal transduction. | 0.28 | 0.39 | 0.0478 | 0.0431 |
| 3 | 522991 | SUSD3 | 203328 | 9q22.32 | Sushi domain containing 3 | 0.27 | 0.35 | 0.0463 | 0.0405 |
| 4 | 606609 | VAV3 - vav 3 oncogene | 10451 | 1p13.3 | Function in signal transduction. Deregulation leads to marked cytoskeletal changes and cell division alterations. | 0.39 | 0.39 | 0.043 | 0.0459 |
| 5 | 644989 | RERG - Ras-like | 85004 | 12p13.1 | Estrogen-regulated, growth inhibitor | 0.32 | 0.26 | 0.0464 | 0.0482 |
| 6 | 690231 | CCL18 - chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated) | 6362 | 17q11.2 | Chemotactic activity for naive T cells, CD4+ and CD8+ T cells and nonactivated lymphocytes. It may play a role in humoral and cell-mediated immunity responses. | 4.25 | 6.34 | 0.0481 | 0.0439 |
| 7 | 1394835 | HOOK2 - hook homolog 2 | 29911 | 19p13.13 | Attachment to microtubules and binding to organelles. | 0.47 | 0.40 | 0.0464 | 0.0458 |
| 8 | 1453049 | SCNN1A | 6337 | 12p13 | Sodium channel, nonvoltage-gated 1 alpha | 0.25 | 0.20 | 0.0456 | 0.0402 |
| 9 | 1646030 | PTD008 | 51398 | 19p13.13 | PTD008 protein | 0.47 | 0.46 | 0.0424 | 0.0465 |
| 10 | 1662893 | C18orf1 | 753 | 18p11.2 | Chromosome 18 open reading frame 1 | 0.40 | 0.37 | 0.0462 | 0.0432 |
| 11 | 1672574 | PACE4 - proprotein convertase subtilisin/kexin type 6 | 5046 | 15q26 | One of its substrates is the transforming growth factor beta related protein. This gene plays a role in tumor progression. | 0.36 | 0.37 | 0.0495 | 0.0452 |
| 12 | 1698713 | ERBB3 - v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) | 2065 | 12q13 | Encodes a member of the epidermal growth factor receptor (EGFR) family. Function in cell proliferation or differentiation. Amplification of this gene and/or overexpression of its protein have been reported also in breast tumors. | 0.38 | 0.42 | 0.0457 | 0.0426 |
| 13 | 1711594 | FOXA1 - forkhead box A1 | 3169 | 14q12-q13 | HNF3alpha is amplified and overexpressed in esophageal and lung adenocarcinomas. | 0.16 | 0.14 | 0.043 | 0.0443 |
| 14 | 1734634 | NaN | NaN | 0.31 | 0.27 | 0.0491 | 0.0454 | ||
| 15 | 1749102 | INDO -indoleamine-pyrrole2,3dioxygenase | 3620 | 8p12-p11 | Antiproliferative effect on many tumor cells and inhibits intracellular pathogens. | 3.20 | 4.50 | 0.0464 | 0.0428 |
| 16 | 1755193 | FLJ20366 | 55638 | 8q23.2 | Hypothetical protein FLJ20366 | 0.32 | 0.30 | 0.0435 | 0.0476 |
| 17 | 1793853 | ALCAM | 214 | 3q13.1 | Activated leukocyte cell adhesion molecule | 0.38 | 0.37 | 0.0478 | 0.0439 |
| 18 | 1808121 | KIAA1324 | 57535 | 1p13.3-p13.2 | 0.31 | 0.30 | 0.0491 | 0.0478 | |
| 19 | 1844691 | ARMCX2 | 9823 | Xq21.33-q22.2 | Potential role in tumor suppression. | 0.40 | 0.41 | 0.0455 | 0.0419 |
| 20 | 1861614 | CREB3L4 | 148327 | 1q21.3 | cAMP response element-binding (CREB) proteins are transcription activators. | 0.41 | 0.42 | 0.0475 | 0.0463 |
| 21 | 1922038 | SYT17 | 51760 | 16p13.11 | Synaptotagmin XVII | 0.40 | 0.37 | 0.0463 | 0.0416 |
| 22 | 1998428 | AGR2 - anterior gradient 2 homolog (Xenopus laevis) | 10551 | 7p21.3 | Involvement in differentiation, associated with oestrogen receptor-positive breast tumours and interacts with metastasis gene C4.4a and dystroglycan. | 0.23 | 0.25 | 0.0437 | 0.0444 |
| 23 | 1998792 | DNALI1 | 7802 | 1p35.1 | Potential candidate for the immotile cilia syndrome (ICS). | 0.34 | 0.40 | 0.0448 | 0.0448 |
| 24 | 2026332 | NaN | NaN | 0.25 | 0.30 | 0.0462 | 0.0447 | ||
| 25 | 2106441 | ASAH1 | 427 | 8p22-p21.3 | Mutations are associated with a lysosomal storage disorder (Farber disease). | 0.33 | 0.32 | 0.0468 | 0.0452 |
| 26 | 2190664 | KIAA0882 | 23158 | 4q31.21 | Membrane-associated protein. | 0.22 | 0.26 | 0.0495 | 0.0463 |
| 27 | 2230088 | MGC18216 | 145815 | 15q26.3 | Hypothetical protein MGC18216. | 0.35 | 0.31 | 0.0463 | 0.0438 |
| 28 | 2242817 | TFF3 - trefoil factor 3 (intestinal) | 7033 | 21q22.3 | May protect the mucosa from insults, stabilize the mucus layer and affect healing of the epithelium. Expressed in goblet cells of the intestines and colon. | 0.34 | 0.17 | 0.048 | 0.0446 |
| 29 | 2366522 | TMEM101 | 84336 | 17q21.31 | Transmembrane protein 101 | 0.33 | 0.35 | 0.0478 | 0.0454 |
| 30 | 2555590 | MYB | 4602 | 6q22-q23 | V-myb myeloblastosis viral oncogene homolog | 0.27 | 0.20 | 0.0482 | 0.0418 |
| 31 | 2591494 | SELENBP1 - selenium binding protein 1 | 8991 | 1q21-q22 | Effects of selenium in preventing cancer and neurologic diseases may be mediated by selenium-binding proteins. | 0.41 | 0.34 | 0.0453 | 0.0451 |
| 32 | 2617968 | ShrmL - shroom | ShrmL | 4q21.22 | F-actin-binding protein | 0.51 | 0.44 | 0.0469 | 0.0479 |
| 33 | 2708596 | KIF13B | 23303 | 8p12 | Kinesin family member 13B | 0.33 | 0.42 | 0.0463 | 0.045 |
| 34 | 2740665 | NaN | NaN | 0.34 | 0.26 | 0.045 | 0.0431 | ||
| 35 | 2823476 | STC2 - stanniocalcin 2 | 8614 | 5q35.1 | Expression is induced by estrogen and altered in some breast cancers. | 0.34 | 0.35 | 0.0441 | 0.0451 |
| 36 | 3094261 | GREB1 - GREB1 protein | 9687 | 2p25.1 | Estrogen-responsive; plays important role in hormone-responsive tissues and cancer. | 0.53 | 0.33 | 0.0487 | 0.0457 |
| 37 | 3123244 | NaN | 15q22.32 | 0.17 | 0.21 | 0.0487 | 0.042 | ||
| 38 | 4256950 | NPHP1 - nephronophthisis 1 | 4867 | 2q13 | Signal transduction, cell-cell adhesion, actin cytoskeleton organization and biogenesis. | 0.37 | 0.22 | 0.0469 | 0.0455 |
| Genes similar expressed for “aGU versus dGS” and “aGS versus dGS” | |||||||||
| 1 | 1596916 | NOL4 | 8715 | 18q12 | Nucleolar protein 4 | 2.78 | 2.13 | 0.0458 | 0.0434 |
| 2 | 2060823 | S100P - S100 calcium binding protein P | 6286 | 4p16 | Regulation of cellular processes such as cell cycle progression and differentiation. | 2.36 | 2.77 | 0.0474 | 0.0479 |
| 3 | 2518249 | AFF3 - AF4/FMR2 family, member 3 | 3899 | 2q11.2-q12 | Tissue-restricted nuclear transcriptional activator preferentially expressed in lymphoid tissue. May function in lymphoid development and oncogenesis. | 0.21 | 0.43 | 0.046 | 0.044 |
| Genes similar expressed for “aGU versus aGS” and “aGS versus dGS” | |||||||||
| 1 | 2313368 | MS4A1 - membrane-spanning 4-domains, subfamily A, member 1 | 931 | 11q12 | Encodes a B-lymphocyte surface molecule with function in development and differentiation of B-cells into plasma cells. | 0.43 | 3.44 | 0.0429 | 0.0478 |
| 2 | 3970665 | NaN | NaN | 2.11 | 0.34 | 0.0469 | 0.0414 | ||
We were then interested in analyzing expression levels of genes that are potential candidates for being involved in the control of genetic stability. These genes include APC, BIRC5, BUB1, CDH1, FRS3, RB1CC1, SMC1A (structural maintenance of chromosome), ST7, AURKA, TP53, and the cyclins CCNA2, CCND1, CCND3, and CCNE1. Several of these genes have been previously studied by means of either quantitative RT-PCR or immunohistochemistry in a comparable set of genomically stable and unstable tumors.30, 31 Five of these genes were significantly differentially expressed when we compared the genomically stable (dGS and aGS) and unstable (aGU) tumors. We confirmed the increased expression of CCNA2 and CCNE1 in the unstable tumors, however, CCND1 was downregulated in this group. In addition, we could show that both BIRC5, which prevents apoptosis, and AURKA, a gene involved in centrosome duplication and genomic instability, were significantly higher expressed in the genomically unstable tumors (Supplemental Table 2).
Functional annotation of the genes that discern the genomically stable (dGS and aGS) from unstable (aGU) tumors using Ingenuity Pathway Analysis
Both, the CGH analyses and expression profiles suggest that those tumors that were classified as genetically unstable differed most from the genetically stable tumors, regardless of the actual ploidy status (i.e., the position of the stemline which is at 2c in the dGS, and different from 2c in the aGS group). We were therefore interested to functionally annotate the differentially expressed genes between the two groups (dGS and aGS versus aGU using Ingenuity Pathway Analysis (IPA) (Supplemental Table 3). For nomenclature of IPA please see Material and Methods section.
Thirty-eight genes were differentially expressed between the aGU and the dGS/aGS subtypes, of which 33 were present as focus genes. Four of these focus genes belonged to the highest ranked network. The top ranked networks are characterized by a low likelihood that the generation of the network was serendipitous. Genes of this network are involved in cancer development, cellular growth and proliferation and gene expression, whereas the second network contained one focus gene. The top functions in this network include cell-to cell signaling and interaction, and cellular assembly and organization.
Validation of the genomic-instability-dependent gene set for prediction of disease-free survival and overall survival in breast cancer
We were then interested in exploring to which extent the gene expression signature that defines genomic instability in the breast cancer samples analyzed here could be useful for prediction of disease outcome in previously published independent datasets. We therefore first explored the differences between all three groups. Using random forests, this classification generated a list of 70 genes. The top 10 genes of this set selected with Relief were then used to individually classify the three groups; this classification achieved an accuracy of 80%. Next, we used random forests to establish a gene list that discerns the genomically stable (dGS and aGS) tumors from the genomically unstable ones (aGU). In leave-one-out cross-validation, this 7-gene signature allowed classification of genomically stable versus unstable tumors with an accuracy of 93%. The 7-gene list and the 10-gene list showed an overlap of five genes. The resulting two largely concordant signatures from both approaches confirmed the relevance of the identified signature genes as descriptors of genomic instability. Combining the two gene lists therefore resulted in a 12-gene genomic instability signature list. The classification of the samples into two major groups (genomically stable and unstable) is shown as a Hierarchical Cluster analysis in Figure 2B. Eight of those signature genes were identified also in the previous two-step differential expression analyses (Wilcoxon test and step-wise algorithm, see above), indicating concordance despite different computational approaches. Among the 12-gene genomic instability signature, SCYA18, STK15, and CDKN2A were over expressed in genomically unstable breast carcinomas, whereas the remaining nine signature genes were under-expressed in genomically unstable tumors (p < 0.001, two-sided t-tests). This gene set was then used for predicting cancer outcomes in independent datasets.
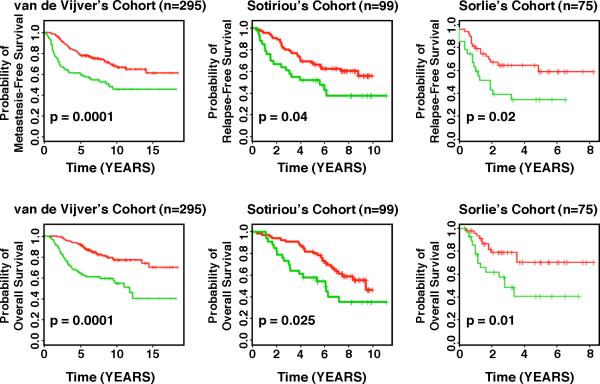
The breast cancer datasets that we used for validation purposes were included in the publications by Sorlie et al.,11 van de Vijver et al.,4 and Sotiriou and colleagues.6 These datasets comprise gene expression profiles from 469 patients with heterogeneous histology and different disease stages. The clinical parameters included tumor grade, tumor size, lymph node status, estrogen receptor status, progesterone receptor status, age, and histology. The clinical endpoints used for the validation of the classifiers included relapse-free survival, metastasis-free survival, disease-free survival (here a disease event refers to either breast cancer relapse or metastasis) and overall survival. Each patient in the three validation cohorts was classified as being more similar to either the genomically stable signature (groups dGS and aGS) or the aGU signature, based on the correlation of this patient's gene expression pattern with the average expression profiles of the genomically stable samples (referred to as GS) and aGU samples in our dataset. Kaplan-Meier analyses showed that the genomic instability-defined prognostic groups were associated with a distinct relapse-free survival and metastasis-free survival (p < 0.04, log-rank tests) in the patient cohorts from Sorlie et al.,11 van de Vijver et al.,4 and Sotiriou et al.6 Patients with the GS signature had longer relapse-free survival and metastasis-free survival than those with the aGU signature. Furthermore, the genomic-instability defined prognostic groups had remarkably different overall survival in Kaplan-Meier analyses (p < 0.025, log-rank tests), as shown in Figure 3, despite the fact that about 50% of patients died without having suffered from breast cancer recurrence.12
Figure 3.
Applying the 12-gene genomic instability signature for prediction of disease-free and overall survival in independent datasets using Kaplan-Meier analyses. The curves in red reflect the genomically stable signature, the curves in green the one representative of genomic instability. For all examples, we observed a statistically significant association of genomic instability with shorter disease-free and overall survival.
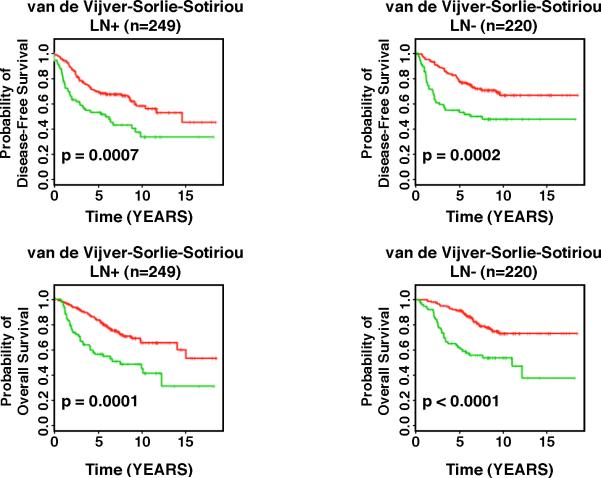
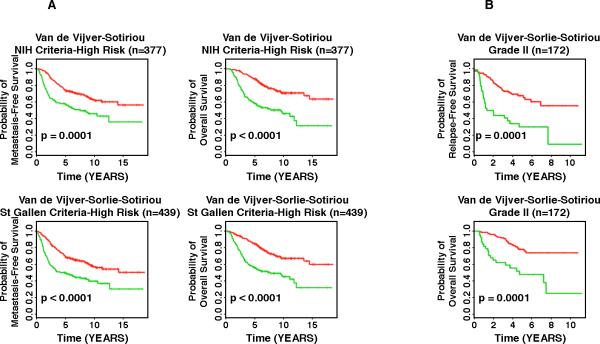
We then evaluated the association between genomic instability-defined risk groups and traditional prognostic factors of breast cancer, including lymph node status, tumor grade, the NIH consensus criteria,32 and the St. Gallen criteria,33 which are based on tumor size ≤ 1 cm (NIH low risk), and estrogen (positive) and/or progesterone (positive) receptor status, tumor size (≤ 2 cm), tumor grade (Grade I), and patient age (≥ 35 years) (St. Gallen low risk). To investigate whether the 12-gene signature is independent of lymph node status, the three external validation cohorts were combined, and lymph node-negative patients and node-positive patients were analyzed separately. In all studied lymph node-negative patients, the GS and the aGU groups had distinct disease-free survival (p < 0.0002, log-rank tests) and overall survival (p < 0.0001, log-rank tests) in Kaplan-Meier analyses. Similarly, in all lymph node-positive patients, the GS group and the aGU group had remarkably different disease-free survival (p < 0.0007, log-rank tests) and overall survival (p < 0.0001, log-rank tests). These results indicate that the 12-gene genomic instability signature is independent of lymph node status in breast cancer prognosis (Figure 4). We then investigated whether the 12-gene signature provides additional prognostic information within the high risk groups defined by the NIH criteria and the St. Gallen criteria. In van de Vijver's cohort,4 284 patients were defined as high risk according to the NIH criteria, and 273 patients were defined as high risk according to the St. Gallen criteria. In Sotiriou's cohort,6 93 patients were defined as NIH high-risk, and 91 patients were defined as St. Gallen high-risk. Since tumor size was not available in Sorlie's cohort,11 patients could not be classified according to the NIH criteria. All 75 patients in Sorlie's cohort11 were high-risk according to St. Gallen criteria. In high risk patients defined by the NIH criteria (n = 377), those with the GS signature had significantly better prognosis in terms of metastasis-free survival (p = 0.0001, log-rank tests) and overall survival (p < 0.0001, log-rank tests) than those with the aGU signature (Figure 5A). Similarly, in the high risk patients defined by the St. Gallen criteria (n = 439), patients with the GS signature had significantly better prognosis in terms of metastasis-free survival (p < 0.0001, log-rank tests) and overall survival (p <0.0001, log-rank tests) than those with the aGU signature (Figure 5A). Furthermore, the 12-gene genomic instability signature stratified Grade II breast cancers (n = 172) into subgroups with distinct relapse-free survival (p = 0.0001, log-rank tests) and overall survival (p = 0.0001, log-rank tests) in Sorlie's cohort and Sotiriou's cohort (Figure 5B). Together, these results demonstrate that the 12-gene genomic instability signature is independent of traditional clinicopathologic factors used for breast cancer prognostication.
Figure 4.
The 12-gene genomic instability signature quantified breast cancer outcomes in Kaplan-Meier analyses independent of lymph-node status in three different patient cohorts. The curves in red reflect the genomically stable signature, the curves in green the one representative of genomic instability. The signature of genomic instability was used to predict disease-free and overall survival in three combined datasets. We observed a statistically significant association with survival independent of lymph-node status.
Figure 5.
Comparison of the 12-gene signature of genomic instability with other predictors of high-risk. The curves in red reflect the genomically stable signature, the curves in green the one representative of genomic instability. A: the 12-gene signature allows further stratification of established high-risk signatures (the so called NIH and St. Gallen criteria). B: Refinement of relapse-free and overall survival in two independent datasets using the 12-gene signature of genomic instability. In Grade II tumors, application of the 12-gene signature allowed for improved prognostic classification.
Linking breast cancer subtype-specific gene expression signatures and signatures of poor prognosis with the genomic instability classification
In the recent past, several comprehensive gene expression based tumor profiling studies of large cohorts of breast cancer patients identified specific subtypes, i.e., luminal A and B, basal, ERBB2+, and normal-like.3, 11, 13, 14 These subtypes were reproduced in several independent studies, and are associated with distinct prognostic profiles. For instance, luminal A and normal-like are associated with longer disease-free survival, whereas patients with tumors classified as basal or ERBB2+ fare worse. We therefore speculated that the subtype-specific gene expression signatures and our instability based classification systems were linked, potentially pointing to overlapping biological mechanisms. This potential connection was explored by using subtype-specific gene expression signatures to classify our 48 samples. The results reveal a remarkable concordance of these independent classification systems. Of the 28 genomically stable tumors in our collection, 24 were assigned to subtypes luminal A (n = 18) or normal-like (n = 6). Only four tumors were assigned to the ERBB2+ group. In contrast, all but one genomically unstable tumor (n = 16) was assigned to either the ERBB2+ group (n = 10) or the basal group (n = 5), indicating pour prognosis (Table 3A). Of note, most of our genomically unstable tumors showed genomic amplification of chromosome arm 17q, the mapping position of the ERBB2 oncogene (Figure 1B).
Table 3.
The linkage between genomic instability and breast cancer signatures of poor prognosis subtypes. A. The concordance between genomic instability and breast cancer subtypes (luminal A and B, normal-like, ERBB2+, and basal). The tumors labeled dGS and aGS comprise the genomically stable groups, whereas those labeled aGU define the genomically unstable group. B. The prediction results of genomic instability by using Oncotype DX and MammaPrint® using neural networks in leave-one-out cross validation.
| A. | |||||
|---|---|---|---|---|---|
| Luminal A | Luminal B | ERRB2+ | Basal | Normal-like | |
| dGS | 9 | 0 | 1 | 0 | 4 |
| aGS | 9 | 0 | 3 | 0 | 2 |
| aGU | 1 | 0 | 10 | 5 | 0 |
| B. | |||
|---|---|---|---|
| Sensitivity (Genomically unstable) | Specificity (Genomically stable) | Overall Accuracy | |
| Oncotype DX | 13/16 (81%) | 27/28 (96%) | 91% |
| MammaPrint® | 13/16 (81%) | 24/28 (86%) | 84% |
We next explored whether other gene expression signatures for breast cancer prognosis would allow classification of genomic instability in our samples. Specifically, we used the 21-gene signature of the so-called Oncotype DX assay (consisting of 16 cancer-associated genes and five genes included for normalization purposes),12 and the 70-gene signature of the MammaPrint®4, 5 to predict genomic instability in the tumors in our series. Twelve of the 21 genes used in the Oncotype DX test were present on our platform, whereas 21 of the 70 genes employed by MammaPrint® could be utilized in our set. Using the Oncotype DX gene set, overall prediction accuracy (measured as correct classification of unstable tumors as unstable, and stable tumors as stable) was 91%, whereas the MammaPrint® set correctly classified 84% of cases. These results are proof of the linkage between genomic instability and poor prognosis in breast cancer (Table 3B).
In summary, our data show that, first, differences in the degree of genomic instability among tumor groups are reflected in the identified 12-gene signature. Second, this aneuploidy-specific gene expression signature can reliably predict outcome in published datasets, and, third, in turn, the gene expression signature of poor prognosis, independent of the specific platform, predicts the degree of genomic instability with significant (p < 0.001) accuracy. Therefore, the three hypotheses that we postulated appear to be valid ones.
DISCUSSION
The prognosis of breast cancer depends on the pattern and degree of genomic instability.7,8 Patients with breast carcinomas with a relatively stable genome have considerably longer disease free survival times compared to women with genomically unstable tumors.9, 10 The degree of genomic instability can be considered a tumor-biological feature that defines different subtypes. In general, this feature does not change over time, i.e., when malignancy advances. This is also reflected by the fact that the average tumor sizes of the dGS (22.5 mm), aGS (25.6 mm), and aGU (25.4 mm) samples were not significantly different from each other. Therefore, the degree of genomic instability is not just a measure of tumor size and time of in vivo propagation. In order to further elucidate the genetic basis of the strikingly different clinical course of genomically stable versus unstable tumors, we were interested in analyzing (i) whether we can identify aneuploidy specific global gene expression profiles, (ii) to which extent the genomic instability-associated gene expression patterns would allow the prediction of clinical outcomes in published breast cancer datasets and, (iii) lastly, we were curious as to whether the gene expression signatures that define several breast cancer subtypes and are associated with poor prognosis, namely those used in Oncotype DX and MammaPrint® would classify our samples. In order to address these questions, we used quantitative measurement of the nuclear DNA content, comparative genomic hybridization, and array-based gene expression profiling of 48 breast cancer samples. The relevance of the genomic instability-specific gene expression signature was then independently and comprehensively validated using published datasets. The 12-gene genomic instability signature predicted disease-free survival and overall survival in breast cancer patients (n = 469), independent of traditional prognostic factors.
The results of the CGH experiments revealed clearly different patterns of genomic imbalances in the groups with a low versus high degree of genomic instability, both in terms of the affected chromosomes, and in terms of the frequency of chromosomal gains and losses. The most common genomic gain was mapped to the long arm of chromosome 1. This is consistent with previous analysis from our own laboratory and with CGH results reported in the literature using chromosome and array CGH.14, 29, 30, 34-36 Additional frequent gains in the genomically stable tumors were mapped to chromosome arms 16p (gains) and 16q (losses). The aneuploid genomically unstable tumors showed additional gains on chromosome arm 8q and 17q, which include the known oncogenes MYC and ERBB2. Previous analyses using FISH probes for these oncogenes are consistent with this finding.37 These observations, and in particular the fact that tumors with gains of chromosome 1q and losses on 16q were predominantly found in the genomically stable group nicely supports findings by Chin and colleagues and Bergamaschi and colleagues, who could show that patients with tumors with the “1q/16q” signature have longer disease-free survival times compared to patients with more complex aberrations, such as those that belonged to the genomically unstable groups in our collection.14, 29 In our patient cohort, 47% of the dGS, 33% of the aGS and none of the aGU tumor samples showed this “1q/16q” pattern.
The conclusion that, based on the CGH analyses, the genomically stable tumors were more closely related, and different from the unstable group, was corroborated by the gene expression profiling data. There was a significant overlap of deregulated genes between the genomically unstable aGU group on one side, and the genomically stable dGS and aGS cases on the other. Therefore, two groups emerged from these analyses, i.e., tumors that were classified as genomically stable (dGS and aGS), and those that are not (aGU, see also Figure 1). Genes differentially regulated between these groups mapped predominantly to pathways that govern cancer development, cellular growth and proliferation and gene expression. In previous analyses we could show that two genes involved in cell cycle progression, CCNA2 and CCNE1, were expressed at higher levels in genomically unstable tumors, which translated into reduced disease free survival times.9, 31, 38 These data were generated using either quantitative RT-PCR or immunohistochemistry. Using array based gene expression profiling as described here, we could confirm these results in this new, extended dataset. The ratio of median expression values of CCNA2 and CCNE1 in aGU versus aGS and dGS tumors is 1.88 (p = 0.022) and 1.86 (p = 6.68e-5), respectively. The inhibitor of apoptosis, BIRC5, was also significantly higher expressed in the unstable tumors associated with poor prognosis, which is intuitive. It is tempting to speculate that unstable tumors express phenotypic features of chromosome segregation errors, one of which are centrosome amplifications. Indeed, we have shown recently that centrosome aberrations are significantly correlated with the degree of aneuploidy and in particular with the stemline scatter index.39 In addition to CCNE1, which is involved in the control of centrosome maturation and duplication,40 the serine/threonine kinase AURKA plays an integral role in the maintenance of centrosome integrity.41 Overexpression of AURKA in cell lines results in centrosome amplification, genomic instability and aneuploidy. Of note, the expression of AURKA was significantly increased in the genomically unstable tumors compared to the stable groups (median ration of 1.87, p = 1.08e-8). This suggests that this gene plays a major role in the development of aneuploidy in human breast cancer. Interestingly, members of the serine/threonine kinase family, including AURKA, were also identified as upregulated in chromosomally instable tumors by Carter and colleagues and so were members of the cyclin and kinesin families.42 Two other genes prominently overexpressed in the genomically unstable tumors include CDKN2A (p16), a cyclin-dependent kinase inhibitor that maps to the short arm of chromosome 9, and SCYA18 (CCL18), a gene involved in immune response. Interestingly, this gene maps to chromosome arm 17q, which is commonly gained in the unstable tumors. RERG, an estrogen regulated growth inhibitor is downregulated in the unstable tumors, and so are the transcriptional activator HNF3A (FOXA1), the ankyrin repeat protein p28 (PSMD10), the nuclear RNA export factor NXF1, and the oncogene MYB.
Having established distinct difference between the genomically stable and unstable tumors, including specific patterns of genomic imbalances and sets of differentially expressed genes, we were now eager to explore to which extent these differences would allow prediction of the clinical course in published independent datasets. Included were three datasets from patients with breast cancer (n = 469) for which different clinical endpoints were available.4, 6, 11 In three external validation sets, patients were classified as belonging to either the GS signature or to the aGU signature, based on the correlation with the average 12-gene GS and aGU centroids (i.e., profiles) in our data. There are strong correlation patterns with our GS and aGU centroids in the external validation sets (Supplementary Figures 4-6), rendering a robust classification of genomically stable and unstable breast cancers in the validation cohorts. Based on this direct mapping, our 12-gene signature reliably predicts disease-free survival and overall survival in breast cancer independently of traditional prognostic factors, including tumor grade, lymph node status, and the St. Gallen and NIH consensus criteria, suggesting inherent biological similarities between the parameters that define the classification systems.
The external validation sets used in this study consist of completely independent patient cohorts. The prognostic prediction based on the 12-gene genomic instability signature employed the “gold standard” of validation schemes, i.e., an independent training set and a validation in multiple, non-overlapping datasets. Fan et al.43 compared five breast cancer signatures, including Oncotype DX,12 MammaPrint®,4, 5 wound response predictor,44 intrinsic subtypes,3, 11, 13 and the “two-gene ratio”45 using the cohort from van de Vijver et al.4 This comparison represents an entirely independent test set only for Oncotype DX and the “two–gene ratio”, whereas the remaining three signatures used part of the samples from van de Vijver's cohort (n = 295) in model development. If the training samples were removed for testing these three signatures, the resulting test dataset would be greatly reduced to fewer than 147 samples and possibly as few as 72 samples.43 In this evaluation, all five signatures except the two-gene ratio allowed for prognostic categorization with respect to disease-free survival (log-rank p < 0.001) and overall survival (log-rank p < 0.001). Compared with these results in consideration of the bias toward MammaPrint®, intrinsic subtypes, and wound response predictor, our 12-gene genomic instability signature is as accurate as Oncotype DX and could potentially be more accurate than the other signatures in terms of predicting disease-free survival and overall survival in van de Vijver's cohort. Notably, our 12-gene genomic instability signature further stratified high risk patients defined by the St. Gallen criteria into distinct prognostic groups. These results suggest, upon further validation, that the signature presented in this study could extend the prognostic space of the current four concordant breast cancer signatures46 by linking gene expression patterns with tumour inherent genomic instability.
Several studies have generated evidence that primary breast cancer can be stratified into specific subtypes, namely luminal A and B, basal, normal-like, and ERBB+, based on gene expression profiles.3, 11, 13, 14 These subtypes are associated with a distinctly different prognosis, with luminal A and normal-like showing significantly prolonged disease free survival times, whereas the basal and ERBB2+ signatures indicate poor prognosis. It was therefore intriguing to speculate that there would be concordance between our genomically stable tumors and those characterized as luminal A and normal-like. In order to address this question, we used the subtype-specific gene expression signatures to classify our tumors. The congruence was striking: 24 of 28 of the genomically stable (i.e., good prognosis) tumors were identified as being either luminal A or normal-like, whereas 15 of 16 genomically unstable (i.e., poor prognosis) tumors belonged to either the basal or ERBB2+ groups. Gain of chromosome 17q, on which ERBB2 resides, was common in the genomically unstable tumors, hence supporting the ERBB2+ phenotype. This overlap points to a strong biological relationship of the groups, and suggests that tumor inherent genomic instability is a major determinant of prognosis. This of course supports the wealth of data showing that measurement of the nuclear DNA content by either image or flow cytometry contributes to disease prognostications in patients with breast cancer.47
Currently, two prognostic gene signatures are in clinical use for the management of patients with early stage primary breast cancer. Oncotype DX12 is applied for treatment stratification of node-negative, estrogen receptor positive, and tamoxifen treated breast cancer; and MammaPrint®4, 5 for node-negative breast cancer patients under age 61 with tumor size less than 5 cm (stages I and IIA). We were therefore eager to further explore whether the concordance between gene expression signatures that define specific breast cancer subtypes and genomic instability extents to the two poor prognosis classification systems. We explored this by investigating whether the 21-gene set of Oncotype DX and the 70-gene set of MammaPrint® can stratify our samples as being genomically stable or unstable. The accuracy with which that was possible, i.e., 84% for the MammaPrint® set and 91% for Oncotype DX corroborated our interpretation of an interrelationship of these different genetic predictors of disease outcome.
In summary, this study presents a systematic approach to explore whether there is linkage between a tumor-inherent degree of genomic instability, gene expression patterns, and clinical outcomes. The results establish a firm association of genomic instability and breast cancer subtypes. The identified 12-gene aneuploidy-specific signature is an independent predictor of clinical outcome in breast cancer patients, suggesting an essential and dominating role of genomic instability in cancer progression and recurrence. Clinically used gene signatures of poor prognosis, OncotypeDX and MammaPrint®, accurately predict the degree of genomic instability in our dataset, thereby linking the classification systems. Based on these results one could make the argument that disease prognostication based on gene expression signatures could be augmented by quantitative measurements of the nuclear DNA content in diagnostic samples, including those acquired by minimally invasive fine needle aspiration cytology.
Supplementary Material
ACKNOWLEDGEMENTS
The authors express their gratitude to Buddy Chen for editing the manuscript and illustrations, Joseph Cheng for IT support, Ulla Aspenblad and Birgitta Sundelin for excellent technical assistance with immunohistochemistry, image cytometry, and clinical data assessment, and Marian Grade for critical comments on the manuscript. This project was supported by the Swedish Cancer Society (Cancerfonden), the Cancer Society (Cancerföreningen), Stockholm, Sweden, the Biocentrum Helsinki, Finland and the Intramural Research Program of the NIH, National Cancer Institute. J.D. received a travel grant by the Medical University of Lübeck, Germany. N.L.G. is supported by NIH/NCRR P20 RR16440-03. All authors declare that they have no competing financial interest.
Footnotes
NOVELTY
Genomic instability, as reflected in heterogeneous nuclear DNA content, serves as one of the most powerful predictors of clinical outcome. We now elucidated the genetic basis of this heterogeneity using gene expression profiling and identified a 12-gene genomic instability signature that provides prognostic information independent of conventional predictive parameters. The striking overlap with recently established gene expression based classifiers suggests that the degree of genomic instability per se is the dominant breast cancer inherent prognostic trait.
REFERENCES
- 1.DeVita VT, Hellman S, Rosenberg SA. Principles and practice of oncology. ed. 7th. JB Lippincott; Philadelphia: 2005. Cancer. [Google Scholar]
- 2.Henry NL, Hayes DF. Uses and abuses of tumor markers in the diagnosis, monitoring, and treatment of primary and metastatic breast cancer. Oncologist. 2006;11(6):541–52. doi: 10.1634/theoncologist.11-6-541. [DOI] [PubMed] [Google Scholar]
- 3.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 5.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 6.Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, Martiat P, Fox SB, Harris AL, Liu ET. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A. 2003;100(18):10393–8. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Auer GU, Caspersson TO, Wallgren AS. DNA content and survival in mammary carcinomas. Anal. Quant. Cytol. 1980;2:161–64. [PubMed] [Google Scholar]
- 8.Auer GU, Eriksson E, Azavedo E, Caspersson T, Wallgren A. Prognostic significance of nuclear DNA content in mammary adenocarcinomas in humans. Cancer Res. 1984;44:394–96. [PubMed] [Google Scholar]
- 9.Kronenwett U, Huwendiek S, Ostring C, Portwood N, Roblick UJ, Pawitan Y, Alaiya A, Sennerstam R, Zetterberg A, Auer G. Improved grading of breast adenocarcinomas based on genomic instability. Cancer Res. 2004;64(3):904–9. doi: 10.1158/0008-5472.can-03-2451. [DOI] [PubMed] [Google Scholar]
- 10.Kronenwett U, Ploner A, Zetterberg A, Bergh J, Hall P, Auer G, Pawitan Y. Genomic instability and prognosis in breast carcinomas. Cancer Epidemiol Biomarkers Prev. 2006;15(9):1630–5. doi: 10.1158/1055-9965.EPI-06-0080. [DOI] [PubMed] [Google Scholar]
- 11.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100(14):8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 13.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 14.Bergamaschi A, Kim YH, Wang P, Sorlie T, Hernandez-Boussard T, Lonning PE, Tibshirani R, Borresen-Dale AL, Pollack JR. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45(11):1033–40. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- 15.Fallenius AG, Auer GU, Carstensen JM. Prognostic significance of DNA measurements in 409 consecutive breast cancer patients. Cancer. 1988;62(2):331–41. doi: 10.1002/1097-0142(19880715)62:2<331::aid-cncr2820620218>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 16.Fallenius AG, Franzen SA, Auer GU. Predictive value of nuclear DNA content in breast cancer in relation to clinical and morphologic factors. A retrospective study of 227 consecutive cases. Cancer. 1988;62(3):521–30. doi: 10.1002/1097-0142(19880801)62:3<521::aid-cncr2820620314>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 17.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19(5):403–10. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 18.Hegde P, Qi R, Abernathy K, Gay C, Dharap S, Gaspard R, Hughes JE, Snesrud E, Lee N, Quackenbush J. A concise guide to cDNA microarray analysis. BioTechniques. 2000;29(3):548–50. 52–4, 56. doi: 10.2144/00293bi01. passim. [DOI] [PubMed] [Google Scholar]
- 19.Korn EL, Habermann J, Upender M, Ried T, McShane LM. An objective method of comparing DNA microarray image analysis systems. BioTechniques. 2004;36(6):960–7. doi: 10.2144/04366BI01. [DOI] [PubMed] [Google Scholar]
- 20.Cleveland WS. Robust locally weighted regression and smoothing scatterplots. J Am Stat Assoc. 1979;74(368):829–36. [Google Scholar]
- 21.Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, Ringner M, Sauter G, Monni O, Elkahloun A, Kallioniemi OP, Kallioniemi A. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62(21):6240–5. [PubMed] [Google Scholar]
- 22.Xiong M, Li W, Zhao J, Jin L, Boerwinkle E. Feature (gene) selection in gene expression-based tumor classification. Mol Genet Metab. 2001;73(3):239–47. doi: 10.1006/mgme.2001.3193. [DOI] [PubMed] [Google Scholar]
- 23.Kauraniemi P, Hautaniemi S, Autio R, Astola J, Monni O, Elkahloun A, Kallioniemi A. Effects of Herceptin treatment on global gene expression patterns in HER2-amplified and nonamplified breast cancer cell lines. Oncogene. 2004;23(4):1010–3. doi: 10.1038/sj.onc.1207200. [DOI] [PubMed] [Google Scholar]
- 24.Diaz-Uriarte R, Alvarez de Andres S. Gene selection and classification of microarray data using random forest. BMC Bioinformatics. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rabe-Hesketh S, Evertitt BS. Handbook of Satistical Analyses using Stata. ed. Fourth Edition Chapman & Hall/CRC Press; Boca Raton, FL: 2006. [Google Scholar]
- 26.Witten IB, Knudsen EI. Why seeing is believing: merging auditory and visual worlds. Neuron. 2005;48(3):489–96. doi: 10.1016/j.neuron.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 27.Weinstein JN, Myers TG, O'Connor PM, Friend SH, Fornace AJ, Jr., Kohn KW, Fojo T, Bates SE, Rubinstein LV, Anderson NL, Buolamwini JK, van Osdol WW, et al. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275(5298):343–9. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- 28.Bussey KJ, Kane D, Sunshine M, Narasimhan S, Nishizuka S, Reinhold WC, Zeeberg B, Ajay W, Weinstein JN. MatchMiner: a tool for batch navigation among gene and gene product identifiers. Genome Biol. 2003;4(4):R27. doi: 10.1186/gb-2003-4-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, Chen F, Feiler H, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10(6):529–41. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 30.Ried T, Just KE, Holtgreve-Grez H, du Manoir S, Speicher MR, Schrock E, Latham C, Blegen H, Zetterberg A, Cremer T, Auer G. Comparative genomic hybridization of formalin-fixed, paraffin-embedded breast tumors reveals different patterns of chromosomal gains and losses in fibroadenomas and diploid and aneuploid carcinomas. Cancer Res. 1995;55(22):5415–23. [PubMed] [Google Scholar]
- 31.Blegen H, Ghadimi BM, Jauho A, Zetterberg A, Eriksson E, Auer G, Ried T. Genetic instability promotes the acquisition of chromosomal imbalances in T1b and T1c breast adenocarcinomas. Anal Cell Pathol. 2001;22(3):123–31. doi: 10.1155/2001/126030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eifel P, Axelson JA, Costa J, Crowley J, Curran WJ, Jr., Deshler A, Fulton S, Hendricks CB, Kemeny M, Kornblith AB, Louis TA, Markman M, et al. National Institutes of Health Consensus Development Conference Statement: adjuvant therapy for breast cancer, November 1−3, 2000. J Natl Cancer Inst. 2001;93(13):979–89. doi: 10.1093/jnci/93.13.979. [DOI] [PubMed] [Google Scholar]
- 33.Goldhirsch A, Glick JH, Gelber RD, Senn HJ. Meeting highlights: International Consensus Panel on the Treatment of Primary Breast Cancer. J Natl Cancer Inst. 1998;90(21):1601–8. doi: 10.1093/jnci/90.21.1601. [DOI] [PubMed] [Google Scholar]
- 34.Tirkkonen M, Tanner M, Karhu R, Kallioniemi A, Isola J, Kallioniemi OP. Molecular cytogenetics of primary breast cancer by CGH. Genes Chromosomes Cancer. 1998;21(3):177–84. [PubMed] [Google Scholar]
- 35.Fridlyand J, Snijders AM, Ylstra B, Li H, Olshen A, Segraves R, Dairkee S, Tokuyasu T, Ljung BM, Jain AN, McLennan J, Ziegler J, et al. Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer. 2006;6:96. doi: 10.1186/1471-2407-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heselmeyer-Haddad K, Chaudhri N, Stoltzfus P, Cheng JC, Wilber K, Morrison L, Auer G, Ried T. Detection of chromosomal aneuploidies and gene copy number changes in fine needle aspirates is a specific, sensitive, and objective genetic test for the diagnosis of breast cancer. Cancer Res. 2002;62(8):2365–9. [PubMed] [Google Scholar]
- 38.Blegen H, Will JS, Ghadimi BM, Nash HP, Zetterberg A, Auer G, Ried T. DNA amplifications and aneuploidy, high proliferative activity and impaired cell cycle control characterize breast carcinomas with poor prognosis. Anal Cell Pathol. 2003;25(3):103–14. doi: 10.1155/2003/491362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kronenwett U, Huwendiek S, Castro J, Ried T, Auer G. Characterisation of breast fine-needle aspiration biopsies by centrosome aberrations and genomic instability. Br J Cancer. 2005;92(2):389–95. doi: 10.1038/sj.bjc.6602246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nigg EA. Origins and consequences of centrosome aberrations in human cancers. Int J Cancer. 2006;119(12):2717–23. doi: 10.1002/ijc.22245. [DOI] [PubMed] [Google Scholar]
- 41.Katayama H, Brinkley WR, Sen S. The Aurora kinases: role in cell transformation and tumorigenesis. Cancer Metastasis Rev. 2003;22(4):451–64. doi: 10.1023/a:1023789416385. [DOI] [PubMed] [Google Scholar]
- 42.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38(9):1043–8. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 43.Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DS, Nobel AB, van't Veer LJ, Perou CM. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355(6):560–9. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 44.Chang HY, Nuyten DS, Sneddon JB, Hastie T, Tibshirani R, Sorlie T, Dai H, He YD, van't Veer LJ, Bartelink H, van de Rijn M, Brown PO, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A. 2005;102(10):3738–43. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma XJ, Hilsenbeck SG, Wang W, Ding L, Sgroi DC, Bender RA, Osborne CK, Allred DC, Erlander MG. The HOXB13:IL17BR expression index is a prognostic factor in early-stage breast cancer. J Clin Oncol. 2006;24(28):4611–9. doi: 10.1200/JCO.2006.06.6944. [DOI] [PubMed] [Google Scholar]
- 46.Massague J. Sorting out breast-cancer gene signatures. N Engl J Med. 2007;356(3):294–7. doi: 10.1056/NEJMe068292. [DOI] [PubMed] [Google Scholar]
- 47.Bagwell CB, Clark GM, Spyratos F, Chassevent A, Bendahl PO, Stal O, Killander D, Jourdan ML, Romain S, Hunsberger B, Wright S, Baldetorp B. DNA and cell cycle analysis as prognostic indicators in breast tumors revisited. Clin Lab Med. 2001;21(4):875–95, x. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.