Summary
Tumor cells increase the use of anabolic pathways to satisfy the metabolic requirements associated with a high growth rate. Transformed cells take up and metabolize nutrients such as glucose and glutamine at high levels that support anabolic growth. Oncogenic signaling through the PI3K/Akt and Myc pathways directly control glucose and glutamine uptake, respectively. In order to achieve elevated rates of nucleotide biosynthesis, neoplastic cells must divert carbon from PI3K/Akt-induced glycolytic flux into the non-oxidative branch of the pentose phosphate pathway to generate ribose 5-phosphate. This redirection of glucose catabolism appears to be regulated by cytoplasmic tyrosine kinases. Myc-induced glutamine metabolism also increases the abundance and activity of different rate-limiting enzymes that produce the molecular precursors required for de novo nucleotide synthesis. In this review, we will focus on recent progress in understanding of how glucose and glutamine metabolism is redirected by oncogenes in order to support de novo nucleotide biosynthesis during proliferation and how metabolic reprogramming can be potentially exploited in the development of new cancer therapies.
Introduction
Cancer is a disease with complex metabolic perturbations. Unlike normal cells which depend primarily on oxidative phosphorylation for ATP production, tumor cells rely preferentially on glycolysis for their energy needs even when oxygen is available [1]. The phenomenon of aerobic glycolysis, also known as the Warburg effect, is a common property of many types of cancer and the basis of the widespread clinical application of fluorodeoxyglucose positron-emission tomography (FdG PET) for identifying primary and metastatic tumors [2]. Concomitant with the metabolic switch from mitochondrial respiration to aerobic glycolysis is high lactate secretion from tumor cells, which can result in acid-mediated matrix degradation, invasiveness and metastasis [3]. The high rate of metabolism of glucose has been shown to lead to reprogramming of mitochondrial metabolism to support glucose-dependent phospholipid synthesis and to stimulate activity of TOR to redirect amino acid metabolism, stimulating tRNA charging and increasing protein synthesis [4,5].
It is also increasingly appreciated that transformed cells consume large amounts of glutamine which can also be used to support anabolic synthesis [6,7]. Glutamine metabolism provides cells with NADPH as well as a carbon source for production of non-essential amino acid and lipids [6]. Surprisingly, recent evidence suggests that the oncogenic activation of the PI3K/Akt and Myc pathways can directly affect cellular metabolism through effects on glucose and glutamine uptake and catabolism (Figure 1) [7-9]. Activation of the PI3K/Akt pathway induces neoplastic cells to take up excessive glucose and depend on high rate of aerobic glycolysis for continued growth and survival [8,10]. In these glucose addicted cancer cells, the high rate of glycolysis supports glucose-dependent lipid synthesis and non-essential amino acid production [11,12]. Overexpression of Myc induces transformed cells to take up glutamine in excess of their bioenergetic needs and increase the glutaminase flux [7,9]. Myc-transformed cells rely on glutamine anapleurosis to maintain the normal TCA cycle function and become addicted to glutamine, despite the availability of glucose [9].
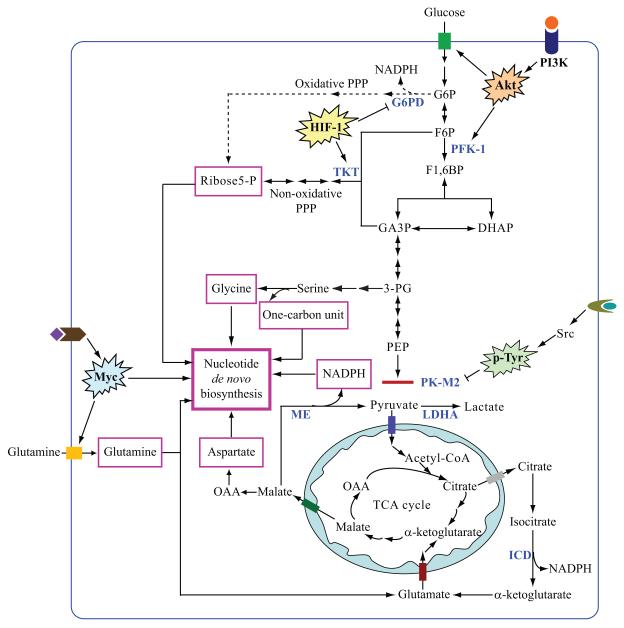
Figure 1.
Regulation of glycolysis, glutaminolysis and de novo nucleotide biosynthesis in tumor cells. Oncogenic activation of the PI3K/Akt and Myc pathways promotes glucose and glutamine uptake and catabolism. Tumor cells obtain precursors (in pink rectangles) including ribose 5-phosphate, glycine, glutamine, aspartate and NADPH for de novo biosynthesis from glucose and glutamine metabolism. Increased glucose uptake and PFK-1 activity by the PI3K/Akt pathway, inactivation of PK-M2 by p-Tyr signaling, as well as HIF-1-induced TKT activation allows glycolytic intermediates to enter the non-oxidative PPP for ribose 5-phosphate production in tumor cells. Hatched lines indicate that the oxidative arm of PPP is not the main pathway for ribose 5-phosphate or NADPH production in tumor cells. Glutamine is converted to lactate if mitochondrial malate is exported into the cytoplasm and decarboxylated to produce pyruvate by malic enzyme (ME). The glutaminolysis pathway and the reaction catalyzed by cytosolic isocitrate dehydrogenase (ICD) may serve as the major sources of NADPH for de novo nucleotide biosynthesis in tumor cells [9,52,53]. In addition to its role in providing tumor cells with NADPH, Myc also directly regulates several genes encoding key enzymes in the purine and pyrimidine biosynthetic pathway. Abbreviations: P, phosphate; G6P, glucose 6-phosphate; F6P, fructose 6-phosphate; F1,6BP, fructose 1,6-bisphosphate; GA3P, glyceraldehyde 3-phosphate; DHAP, dihydroxyacetone phosphate; 3-PG, 3-phosphoglycerate; PEP, phosphoenolpyruvate; OAA, oxaloacetate; G6PD, glucose 6-phosphate dehydrogenase; PFK-1, phosphofructokinase-1; TKT, transketolase; PK-M2, pyruvate kinase M2; LDHA, lactate dehydrogenase A; ME, malic enzyme; ICD, isocitrate dehydrogenase.
In order to divide and create two daughter cells, tumor cells must also utilize biosynthetic precursors derived from glycolytic and TCA cycle intermediates for nucleotide biosynthesis. For instance, carbon diverted from the glycolytic flux to the pentose phosphate pathway is utilized to generate ribose 5-phosphate for de novo nucleotide biosynthesis (Figure 1) [4]. The maximal proliferative capacity of a cell is limited by the abundance of its nucleotide pool as well as the level and activity of different rate-limiting enzymes of the nucleotide synthetic pathway. Tumor cells show an altered nucleotide metabolism compared with normal cells, as manifested by the larger size of the nucleotide pool, higher activity of the nucleotide anabolic pathway as well as lower activity of the nucleotide catabolic pathway [13]. Identification of the mechanism by which transformed cells achieve higher levels of nucleotide biosynthesis may offer possibilities for the design of selective therapies for human malignancy.
Many tumor cells generate ribose 5-phosphate through the non-oxidative arm of the pentose phosphate pathway for de novo nucleotide biosynthesis
Ribose 5-phosphate, the sugar component of nucleic acids, can be synthesized from glucose-6-phosphate by the oxidative branch of the pentose phosphate pathway (PPP) as well as from fructose-6-phosphate and glyceraldehyde-3-phosphate by the non-oxidative branch of the PPP (Figure 1). The oxidative branch of the PPP is catalyzed by glucose 6-phosphate dehydrogenase (G6PD) and 6-phosphate-gluconate dehydrogenase while the nonoxidative branch of the PPP is catalyzed by transketolase (TKT) and transaldolase. The oxidative PPP also generates NADPH which serves as the hydrogen donor in cellular reductive biosynthetic reactions and transforms glutathione to the reduced form to promote the scavenging of reactive oxygen species (ROS) [14].
In contrast to nontransformed cells which produce most of the ribose 5-phosphate for nucleotide biosynthesis through the oxidative arm of the PPP, the non-oxidative branch of PPP has been suggested to be the main source for ribose 5-phosphate synthesis in tumor cells [15-18]. Increased TKT and transaldolase activity has been observed in tumor cells [19,20]. Among the three members of the TKT gene family (TKT, TKTL1 and TKTL2), TKTL1 has been reported to be overexpressed in metastatic tumors and specific inhibition of TKTL1 mRNA is able to inhibit cell proliferation in several types of cancer cells [18,20-22].
All reactions in the non-oxidative branch of the PPP are reversible, which means that the relative level of the metabolic substrate and product determines the direction of the non-oxidative reactions of the PPP (Figure 1) [23]. Therefore, in order to direct glycolytic metabolites into PPP through the non-oxidative branch, tumor cells need to maintain high levels of fructose 6-phosphate and/or glyceraldehyde 3-phosphate. F1,6BP is the product of the phosphofructokinase-1 (PFK-1) reaction, the most important control point in glycolysis. Most tumors produce high levels of fructose 1,6-bisphosphate (F1,6BP) which breaks down into glyceraldehyde 3-phosphate by aldolase [24]. PFK-1 activity is significantly increased in cancer cell lines and primary tumor tissues [25,26]. Several oncoproteins including Myc and Ras have been demonstrated to activate PFK-1 in immortalized cells [27-29].
Besides PFK-1, another rate-limiting enzyme in glycolysis, pyruvate kinase M2 (PK-M2) also contributes to directing the carbon flux from glycolysis into PPP for biosynthetic pathways. Among the four isoenzymes of pyruvate kinase (type L, R, M1 and M2), PK-M2 is predominant in cells and tissues with high proliferative capacity such as tumor cells [24]. The M1 and M2 isoforms are encoded by the same gene and differ only in a 56-amino-acid region as a result of alternative splicing. These amino acid differences account for the specific allosteric activation of PK-M2 but not PK-M1 by F1,6BP [30]. Growth factor-dependent phosphotyrosine signaling inhibits PK-M2 activity by releasing F1,6BP from binding to PKM2 thus directs glycolysis intermediates towards biosynthetic pathways (Figure 1) [31,32]. In order to maintain high proliferation rate, this tyrosine kinase-dependent modulation of PK-M2 can upregulate the activity of the non-oxidative PPP pathway and assure a sufficient supply of glycolysis metabolites for nucleotide biosynthesis.
In addition to ribose 5-phosphate, serine synthesized from the glycolytic intermediate 3-phosphoglycerate is converted to glycine which is an important precursor for purine biosynthesis (Figure 1). As a consequence of its conversion to glycine, serine is also the donor of folate-linked one-carbon units which are required for nucleotide biosynthesis [33]. The activity of this serine and glycine biosynthetic pathway has been reported to be significantly upregulated in several tumor types [34].
HIF-1α increases the carbon flux into PPP via the non-oxidative arm
Another key regulator of tumor cell metabolism is the hypoxia-inducible factor 1 (HIF-1). Tumor cells select for increased HIF-1 activity through multiple pathways including increased HIF-1α transcription, translation, or protein stabilization. As a result, HIF-1α has been frequently found to be highly expressed in human cancers and HIF-1α stabilization induces the expression of a set of genes involved in glucose metabolism [35,36]. One important adaptation of HIF-1-mediated transcriptional regulation is the induction of anaerobic glycolysis, as well as suppression of mitochondrial pyruvate catabolism and oxygen consumption [37-41]. As a consequence of HIF-1 stimulation of glycolysis cancer cells exhibit reduced glucose flux through the oxidative arm of the PPP [42-44].
Our lab has recently reported that the non-hypoxic induction of HIF-1α regulates growth factor-stimulated glucose metabolism and allows cells to avoid producing harmful levels of mitochondrial ROS by secreting excess glycolytic pyruvate as lactate. This HIF-1α-dependent reprogramming of glucose metabolism promotes cell viability [41]. One mechanism by which HIF-1α suppresses ROS production is by reducing glucose flux through the TCA cycle [39]. In addition to stimulating anaerobic glycolysis, HIF-1α induces the expression of TKT and PK-M2. These effects appear to compensate for the suppression of the oxidative PPP by directly increasing glucose flux through the non-oxidative arm of the PPP to maintain ribose 5-phosphate synthesis (Zhao and Thompson, unpublished data).
Recent findings demonstrate that HIF-1 also activates the glycolytic regulatory bifunctional enzymes 6-phospho-2-kinase/fructose2, 6-biphosphatase (PFKFB) [37,38]. Among the four members of PFKFB, PFKFB3 is required for maintaining cellular level of phosphoribosyl pyrophosphate (PRPP) and de-novo nucleic acid synthesis in tumor cells because activation of PFKFB3 regulates the cellular level of F2,6BP, an allosteric activator of the glycolytic rate-liming enzyme PFK-1, which directs the carbon flow from glycolysis into PPP via the non-oxidative branch [45]. These findings suggest that inhibitors of the non-oxidative PPP may selectively inhibit nucleotide biosynthesis in tumors exhibiting constitutive HIF-1α activation.
The role of Myc in regulating nucleotide metabolism in tumor cells
The oncoprotein Myc is involved in regulating multiple growth-related processes [46-49]. Gene expression analysis has recently connected Myc with purine and pyrimidine synthesis and enzymes that direct glutamine uptake and catabolism [9,50,51]. Several genes encoding key enzymes in nucleotide biosynthesis including thymidylate synthase (TS), inosine monophosphate dehydrogenase 1 and 2 (IMPDH1 and 2) and phosphoribosyl pyrophosphate synthetase 2 (PRPS2), have been demonstrated to be direct Myc targets [50,51]. Myc-induced glutamine catabolism in addition provides the cell with an abundant supply of aspartate and amine groups with which to support nucleotide biosynthesis [9]. Myc depletion results in decreased cellular dNTP levels. Ectopic expression of TS, IMPDH2 and PRPS2 partially rescues the proliferative defect caused by Myc depletion in tumor cells [51].
In addition, Myc-induced glutaminolysis can provide cells with a robust way to produce the NAPDH required for de novo nucleotide biosynthesis [9]. This is especially important for tumor cells relying on the non-oxidative arm of the PPP for ribose 5-phosphate production. In these cells, the activity of G6PD is inhibited and the oxidative PPP cannot be used to produce NADPH to support macromolecular biosynthesis. Therefore, the ability of Myc to stimulate NADPH production through glutamine metabolism provides tumor cells with a compensatory mechanism to produce NADPH in order to support cell proliferation.
Conclusions
Transformed cells are metabolically reprogrammed to support cell growth and proliferation. It has long been known that tumor cells depend on de novo nucleotide synthesis to support increased RNA production and DNA replication. Furthermore, elevated nucleotide biosynthesis has been observed in many cancers. In this minireview, we review evidence that the ability to direct glucose and glutamine into de novo nucleotide synthesis depends on oncogenic activation of PI3K and Myc, respectively. Surprisingly though, the metabolic pathways for de novo nucleotide synthesis in cancer cells appear to be distinct from those used by quiescent cells. The intermediate steps in glucose and glutamine metabolism in cancer cells are affected by activation of cytoplasmic tyrosine kinases and HIF-1 dependent transcription. The effects of these oncogenes on the uptake and metabolism of glucose and glutamine is direct and results in the reprogramming of aerobic glycolysis and glutaminolysis. This review emphasizes how this anabolic transformation results in redirecting glucose and glutamine metabolites into de novo nucleotide biosynthesis. The preferential dependence of tumor cells on transketolase and transaldolase for ribose production, PK-M2 for controlling the level of glycolytic intermediates, and malic enzyme and isocitrate dehydrogenase for NADPH production [52,53], suggests that each of these enzymes may be potential targets for cancer therapy based on the tumor phenotype. Investigation of the possibilities is ongoing in a number of laboratories.
Acknowledgements
We thank members of the Thompson laboratory for useful discussion and critical reading of the manuscript. We apologize to authors whose work could not be cited due to space constraints. This work was supported in part by grants from the NCI.
Annotated references
* of special interest
** of outstanding interest
* Ref 6 (DeBerardinis et al): Using 13C NMR spectroscopy to study the metabolism of glioblastoma cells, the authors demonstrated that a high rate of glutamine uptake and catabolism provided transformed cells with precursors and NAPDH for biosynthetic processes.
** Ref 9 (Wise et al): The authors reported that Myc activated a transcriptional program which promotes glutamine uptake and catabolism in tumor cells, independent of the PI3K/Akt pathway. This Myc-dependent glutaminolysis resulted in the reprogramming of mitochondrial metabolism and addiction to glutamine for maintaining cellular viability.
** Ref 16 (Boros et al): The article reported that A-459 lung epithelial carcinoma cells generated ribose 5-phosphate through the non-oxidative arm of the pentose phosphate pathway for de novo nucleotide biosynthesis. Transforming Growth Factor β2 (TGFβ2) promoted the activity of this pathway.
* Ref 32 (Christofk et al): The authors provided evidence that phosphotyrosine signaling bound to PKM2 but not PKM1 and released F1,6BP from binding to PKM2, resulting in glycolytic intermediates entering biosynthetic pathways.
* Ref 41 (Lum et al): The authors reported that HIF-1α inhibited cell proliferation and promoted cell viability by reprogramming glucose metabolism. The article also indicated that the induction of HIF-1α allowed cells to convert pyruvate to lactate to avoid producing high level of ROS by mitochondria.
* Ref 45 (Chesney et al): The article reported that PFKFB3 was critical for maintaining cellular PRPP level for de novo nucleotide biosynthesis.
* Ref 50 (Liu et al): Using chromatin immunoprecipitation (ChIP) coupled with pair-end ditag sequencing analysis (ChIP-PET), the authors identified many genes encoding key enzymes in the nucleotide biosynthetic pathway as direct Myc targets.
* Ref 51 (Mannava et al): The authors demonstrated that Myc directly activated thymidylate synthase (TS), inosine monophosphate dehydrogenase 2 (IMPDH2) and phosphoribosyl pyrophosphate synthetase 2 (PRPS2) in human metastatic melanoma cell lines. By regulating these genes which encode enzymes critical for de novo nucleotide biosynthesis, Myc maintains cellular dNTP levels and promotes cell proliferation.
* Ref 52 (Parsons et al): The authors identified isocitrate dehydrogenase 1 (IDH1) as one of the most frequently mutated genes in glioblastoma multiforme. IDH1 is localized in the cytoplasm and peroxisomes and catalyzes the oxidative decarboxylation of isocitrate to α-ketoglutarate, resulting in NADPH production. This reaction may serve as an important cytosolic source of NADPH.
References
- 1.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Gambhir SS. Molecular imaging of cancer with positron emission tomography. Nat Rev Cancer. 2002;2:683–693. doi: 10.1038/nrc882. [DOI] [PubMed] [Google Scholar]
- 3.Gatenby RA, Gawlinski ET, Gmitro AF, Kaylor B, Gillies RJ. Acid-mediated tumor invasion: a multidisciplinary study. Cancer Res. 2006;66:5216–5223. doi: 10.1158/0008-5472.CAN-05-4193. [DOI] [PubMed] [Google Scholar]
- 4.Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 6.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 9.Wise DR, DeBerardinis RJ, A. M, M. S, X. Z, K. PH, I. N, E. D, M. Y, B. MS, et al. Myc Regulates a Transcriptional Program that Stimulates Mitochondrial Glutaminolysis and Leads to Glutamine Addiction. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.0810199105. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buzzai M, Bauer DE, Jones RG, Deberardinis RJ, Hatzivassiliou G, Elstrom RL, Thompson CB. The glucose dependence of Akt-transformed cells can be reversed by pharmacologic activation of fatty acid beta-oxidation. Oncogene. 2005;24:4165–4173. doi: 10.1038/sj.onc.1208622. [DOI] [PubMed] [Google Scholar]
- 11.Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–6322. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- 12.Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, Hingorani SR, Tuveson DA, Thompson CB. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Mandel P, Wintzerith M, Klein-Pete N, Mandel L. Comparative Investigation of the Free Nucleotides of an Ascitic Hepatoma and of Normal or Regenerating Liver. Nature. 1963;198:1000–1001. [Google Scholar]
- 14.Wood T. Physiological functions of the pentose phosphate pathway. Cell Biochem Funct. 1986;4:241–247. doi: 10.1002/cbf.290040403. [DOI] [PubMed] [Google Scholar]
- 15.Tian WN, Braunstein LD, Pang J, Stuhlmeier KM, Xi QC, Tian X, Stanton RC. Importance of glucose-6-phosphate dehydrogenase activity for cell growth. J Biol Chem. 1998;273:10609–10617. doi: 10.1074/jbc.273.17.10609. [DOI] [PubMed] [Google Scholar]
- 16.Boros LG, Torday JS, Lim S, Bassilian S, Cascante M, Lee WN. Transforming growth factor beta2 promotes glucose carbon incorporation into nucleic acid ribose through the nonoxidative pentose cycle in lung epithelial carcinoma cells. Cancer Res. 2000;60:1183–1185. [PubMed] [Google Scholar]
- 17.Cascante M, Centelles JJ, Veech RL, Lee WN, Boros LG. Role of thiamin (vitamin B-1) and transketolase in tumor cell proliferation. Nutr Cancer. 2000;36:150–154. doi: 10.1207/S15327914NC3602_2. [DOI] [PubMed] [Google Scholar]
- 18.Langbein S, Zerilli M, Hausen A Zur, Staiger W, Rensch-Boschert K, Lukan N, Popa J, Ternullo MP, Steidler A, Weiss C, et al. Expression of transketolase TKTL1 predicts colon and urothelial cancer patient survival: Warburg effect reinterpreted. Br J Cancer. 2006;94:578–585. doi: 10.1038/sj.bjc.6602962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinrich PC, Morris HP, Weber G. Behavior of transaldolase (EC 2.2.1.2) and transketolase (EC 2.2.1.1) Activities in normal, neoplastic, differentiating, and regenerating liver. Cancer Res. 1976;36:3189–3197. [PubMed] [Google Scholar]
- 20.Coy JF, Dressler D, Wilde J, Schubert P. Mutations in the transketolase-like gene TKTL1: clinical implications for neurodegenerative diseases, diabetes and cancer. Clin Lab. 2005;51:257–273. [PubMed] [Google Scholar]
- 21.Hu LH, Yang JH, Zhang DT, Zhang S, Wang L, Cai PC, Zheng JF, Huang JS. The TKTL1 gene influences total transketolase activity and cell proliferation in human colon cancer LoVo cells. Anticancer Drugs. 2007;18:427–433. doi: 10.1097/CAD.0b013e328013d99e. [DOI] [PubMed] [Google Scholar]
- 22.Zhang S, Yang JH, Guo CK, Cai PC. Gene silencing of TKTL1 by RNAi inhibits cell proliferation in human hepatoma cells. Cancer Lett. 2007;253:108–114. doi: 10.1016/j.canlet.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 23.Schenk G, Duggleby RG, Nixon PF. Properties and functions of the thiamin diphosphate dependent enzyme transketolase. Int J Biochem Cell Biol. 1998;30:1297–1318. doi: 10.1016/s1357-2725(98)00095-8. [DOI] [PubMed] [Google Scholar]
- 24.Mazurek S, Grimm H, Boschek CB, Vaupel P, Eigenbrodt E. Pyruvate kinase type M2: a crossroad in the tumor metabolome. Br J Nutr. 2002;87(Suppl 1):S23–29. [PubMed] [Google Scholar]
- 25.Hennipman A, Smits J, van Oirschot B, van Houwelingen JC, Rijksen G, Neyt JP, Van Unnik JA, Staal GE. Glycolytic enzymes in breast cancer, benign breast disease and normal breast tissue. Tumour Biol. 1987;8:251–263. doi: 10.1159/000217529. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez-Martinez C, Aragon JJ. Analysis of phosphofructokinase subunits and isozymes in ascites tumor cells and its original tissue, murine mammary gland. FEBS Lett. 1997;409:86–90. doi: 10.1016/s0014-5793(97)00496-1. [DOI] [PubMed] [Google Scholar]
- 27.Bosca L, Mojena M, Ghysdael J, Rousseau GG, Hue L. Expression of the v-src or v-fps oncogene increases fructose 2,6-bisphosphate in chick-embryo fibroblasts. Novel mechanism for the stimulation of glycolysis by retroviruses. Biochem J. 1986;236:595–599. doi: 10.1042/bj2360595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kole HK, Resnick RJ, Van Doren M, Racker E. Regulation of 6-phosphofructo-1-kinase activity in ras-transformed rat-1 fibroblasts. Arch Biochem Biophys. 1991;286:586–590. doi: 10.1016/0003-9861(91)90084-v. [DOI] [PubMed] [Google Scholar]
- 29.Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, Xu Y, Wonsey D, Lee LA, Dang CV. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- 30.Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44:9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- 31.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 32.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 33.de Koning TJ, Snell K, Duran M, Berger R, Poll-The BT, Surtees R. L-serine in disease and development. Biochem J. 2003;371:653–661. doi: 10.1042/BJ20021785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snell K, Natsumeda Y, Eble JN, Glover JL, Weber G. Enzymic imbalance in serine metabolism in human colon carcinoma and rat sarcoma. Br J Cancer. 1988;57:87–90. doi: 10.1038/bjc.1988.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pouyssegur J, Dayan F, Mazure NM. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 2006;441:437–443. doi: 10.1038/nature04871. [DOI] [PubMed] [Google Scholar]
- 36.Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minchenko A, Leshchinsky I, Opentanova I, Sang N, Srinivas V, Armstead V, Caro J. Hypoxia-inducible factor-1-mediated expression of the 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 (PFKFB3) gene. Its possible role in the Warburg effect. J Biol Chem. 2002;277:6183–6187. doi: 10.1074/jbc.M110978200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Obach M, Navarro-Sabate A, Caro J, Kong X, Duran J, Gomez M, Perales JC, Ventura F, Rosa JL, Bartrons R. 6-Phosphofructo-2-kinase (pfkfb3) gene promoter contains hypoxia-inducible factor-1 binding sites necessary for transactivation in response to hypoxia. J Biol Chem. 2004;279:53562–53570. doi: 10.1074/jbc.M406096200. [DOI] [PubMed] [Google Scholar]
- 39.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 41.Lum JJ, Bui T, Gruber M, Gordan JD, DeBerardinis RJ, Covello KL, Simon MC, Thompson CB. The transcription factor HIF-1alpha plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis. Genes Dev. 2007;21:1037–1049. doi: 10.1101/gad.1529107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gupte SA, Wolin MS. Hypoxia promotes relaxation of bovine coronary arteries through lowering cytosolic NADPH. Am J Physiol Heart Circ Physiol. 2006;290:H2228–2238. doi: 10.1152/ajpheart.00615.2005. [DOI] [PubMed] [Google Scholar]
- 43.Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006;25:4633–4646. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- 44.Tuttle SW, Maity A, Oprysko PR, Kachur AV, Ayene IS, Biaglow JE, Koch CJ. Detection of reactive oxygen species via endogenous oxidative pentose phosphate cycle activity in response to oxygen concentration: implications for the mechanism of HIF-1alpha stabilization under moderate hypoxia. J Biol Chem. 2007;282:36790–36796. doi: 10.1074/jbc.M700327200. [DOI] [PubMed] [Google Scholar]
- 45.Chesney J, Mitchell R, Benigni F, Bacher M, Spiegel L, Al-Abed Y, Han JH, Metz C, Bucala R. An inducible gene product for 6-phosphofructo-2-kinase with an AU-rich instability element: role in tumor cell glycolysis and the Warburg effect. Proc Natl Acad Sci U S A. 1999;96:3047–3052. doi: 10.1073/pnas.96.6.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boyd KE, Farnham PJ. Myc versus USF: discrimination at the cad gene is determined by core promoter elements. Mol Cell Biol. 1997;17:2529–2537. doi: 10.1128/mcb.17.5.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bello-Fernandez C, Packham G, Cleveland JL. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci U S A. 1993;90:7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, Dang CV. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nikiforov MA, Chandriani S, O’Connell B, Petrenko O, Kotenko I, Beavis A, Sedivy JM, Cole MD. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol Cell Biol. 2002;22:5793–5800. doi: 10.1128/MCB.22.16.5793-5800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu YC, Li F, Handler J, Huang CR, Xiang Y, Neretti N, Sedivy JM, Zeller KI, Dang CV. Global regulation of nucleotide biosynthetic genes by c-Myc. PLoS ONE. 2008;3:e2722. doi: 10.1371/journal.pone.0002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mannava S, Grachtchouk V, Wheeler LJ, Im M, Zhuang D, Slavina EG, Mathews CK, Shewach DS, Nikiforov MA. Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle. 2008;7:2392–2400. doi: 10.4161/cc.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ronnebaum SM, Ilkayeva O, Burgess SC, Joseph JW, Lu D, Stevens RD, Becker TC, Sherry AD, Newgard CB, Jensen MV. A pyruvate cycling pathway involving cytosolic NADP-dependent isocitrate dehydrogenase regulates glucose-stimulated insulin secretion. J Biol Chem. 2006;281:30593–30602. doi: 10.1074/jbc.M511908200. [DOI] [PubMed] [Google Scholar]