INTRODUCTION
Normal body weight is only maintained when there is a balance between energy taken in (food) and energy expended (metabolism and physical activity). To this end, individuals can conscientiously alter only their daily level of physical activity and their food intake. The Surgeon General recommends that individuals get a minimum of 30 min of moderate activity each day to maintain a healthy lifestyle. Still, approximately 23% of all adults surveyed by the U.S. Center for Disease Control and Prevention in 2006 performed no physical activity in the previous month (http://www.cdc.gov/BRFSS/). In societies like the United States with readily available food accompanied by sedentary lifestyles, obesity ensues. The latest U.S. data indicate that fewer than 38% of Americans have a body mass index of less than 24.9; thus, more than 60% of individuals in the United States are either obese or overweight (http://www.cdc.gov/BRFSS/).
Whereas environmental and social factors surely contribute to these trends, the latest edition of the Human Obesity Gene map (http://obesitygene.pbrc.edu/) lists 11 genes whose mutant forms directly contribute to single-gene mendelian inheritance of obesity. A total of 127 other genes are candidates for linkage to human obese phenotypes (13). There are also genes that could contribute to differences in either ability or motivation for physical activity, as identified using human and animal studies. The most recent update of the Human Gene Map for Performance and Health-Related Fitness Phenotypes lists 165 possible loci that could modulate physical activity, including 15 mitochondrial-encoded genes and 16 nuclear-encoded genes that have been directly linked to a human phenotype of exercise intolerance (12). Thus, evidence exists for the presence of genes controlling either the motivation or ability to exercise; individuals with polymorphisms that negatively affect these genes would be expected to show reduced physical activity and could display an overweight or obese phenotype.
We asked how many genes or linked loci were cross-listed on both the online Human Obesity Gene Map Database (13) (http://obesitygene.pbrc.edu/) search engine and on the Human Gene Map for Performance and Health-Related Fitness Phenotypes (12). Of the 165 loci on the Human Gene Map for Performance and Health-Related Fitness phenotypes, 66 loci (or ×40%) are colisted on the Human Obesity Gene Map Database (Table 1). Thus, more than one third of the genes or loci that affect physical activity also are implicated in body weight regulation.
TABLE 1.
Genes or loci found on both the Human Obesity Gene Map and the Human Gene Map for Performance and Health-Related Fitness Phenotypes, and available mouse models
| Chromosome | Genes or QTLs on Both Maps | Mouse Models Available | ||
|---|---|---|---|---|
| 1 | D101 | S100A1 | LEPR (db/db) | |
| LEPR | ATP1A2 | |||
| D1S1631 | ATP1B1 | |||
| APOA2 | AMPD1 | |||
| AGT | ||||
| 2 | D2S2952 | D2S1334 | GDF8 | |
| D2S1400 | GDF8 | |||
| 3 | PPARG | PPARG | ||
| 4 | PPARGC1A | UCP1 | PPARGC1a | |
| D3S1627 | UCP1 | |||
| 5 | D5S1725 | N53C1 | ADRB2 | |
| D5S1505 | ADRB2 | |||
| 6 | TNF | ESR1 | TNF | |
| 7 | IL6 | LEP | IL6 | |
| NPY | D7S495 | LEP (ob/ob) | ||
| PON1 | D7S2195 | NPY | ||
| PON2 | NOS3 | SERPINE1 | ||
| SERPINE1 | D7S3070 | |||
| CFTR | ||||
| 8 | LPL | ADRB3 | ADRB3 | |
| 9 | CNTFR | |||
| 10 | ADRA2A | GPR10 | ADRA2A | GPR10 |
| ADRB1 | ADRB1 | |||
| 11 | IGF2 | D11S2002 | UCP2 | |
| UCP2 | APOA1 | UCP3 | ||
| UCP3 | DRD2 | |||
| 12 | GNB3 | D10S1691 | ||
| VDR | IGF1 | |||
| 13 | D13S175 | |||
| 14 | D14S283 | |||
| 15 | CYP19A1 | D15S657 | CYP19A1 | |
| LIPC | LIPC | |||
| 17 | ACADVL | ACE | ACADVL | |
| 18 | D18S843 | MC4R | MC4R | |
| 19 | GYS1 | APOE | TGFB1 | |
| D19S254 | TGFB1 | |||
| 20 | D20S857 | |||
| 22 | COMT | PPARA | PPARA | |
Each gene or locus is listed by human chromosome number. Chromosomes 16 and 21 have no overlapping loci. Mouse models include naturally occurring mutants, transgenics, and targeted deletion (knockout) mutants. Models are only listed for genes that are on both maps and were identified using the Human Obesity Gene Map Database (13) and a textbook of models for biomedical research (6).
Unlike the map for performance and fitness, which focuses only on genes and loci identified through human studies, the Human Obesity Gene Map also lists 244 genes and 408 loci that affect body weight in transgenic or knockout mice (13). Mouse models of obesity and overweight can be divided into those models displaying juvenile-onset obesity and those models displaying adult-onset obesity. Interestingly, many transgenic and knockout mouse models have reduced energy expenditure through exercise. Some of these models are listed on Table 1, although many have not been specifically tested for relative exercise levels. Likewise, there are mouse models whose physical activity phenotypes are known, but the genes are not currently listed on the Human Gene Map for Performance and Health-Related Fitness phenotypes. Thus, it is likely that more genes will be added to this map once more human linkage and analysis studies analyzing those genes identified in mice are completed. We believe that one such gene that will be added in the future is nescient helix-loop-helix 2 (NHLH2, human; Nhlh2, mouse). NHLH2 is a basic helix-loop-helix transcription factor that controls body weight through control of physical activity levels. When Nhlh2 is deleted in mice (N2KO mice), adult-onset obesity ensues (3,7). Overeating does not cause obesity in N2KO mice; rather reduced spontaneous physical activity levels precede the overweight phenotype (3). Using new and published data from our laboratory and published articles from other laboratories, we provide evidence that the NHLH2 gene has the potential to influence either an individual's motivation or ability to perform physical activity.
THE Nhlh2/NHLH2 GENE AND GENETIC LINKAGE STUDIES
The murine Nhlh2 gene was originally cloned using its homology to the leukemia and red-blood-cell differentiation factor, SCL/tal-1 (5). The human gene was subsequently characterized and mapped to chromosome 1p11-1p12 (4). To determine whether any loci linked to performance and physical activity existed for the chromosomal region containing NHLH2, we used the Human Gene Map for Performance and Physical Activity Traits and compared it with the Human Genome Map Viewer (4). One locus marker, D1S189, on human chromosome 1 lies just approximately 300,000 base pair (bp) proximal to NHLH2 at 1p13 (Fig. 1). D1S189 marker is linked in 12 related individuals to a phenotype of autosomal-recessive exercise-induced polymorphic ventricular tachycardia (5), suggesting that the causative gene for this disorder is located near this region where the marker DNA is located. The relative proximity of NHLH2 to this marker region makes NHLH2 a putative candidate gene for this disorder. NHLH2 should be sequenced in these patients. Our laboratory has not studied the heart tissues or physiology from mice with a targeted deletion of the Nhlh2 gene, but mice with deletion of a related gene, Nhlh1, display a similar phenotype of stress-induced cardiac arrhythmias (6). Further analysis of NHLH2 in association with other heart disease using publicly available gene expression array data sets found that NHLH2 is elevated in 11 of 13 individuals with dilated cardiomyopathy (Gene Expression NCBI data sets, number GDS2206; [4]). In the original article, NHLH2 expression is listed as 1.3-fold higher in affected patients, with a significant difference between the affected and unaffected patients (1). Interestingly, the related bHLH transcription factor, NHLH1, shows a trend toward being reduced in these same samples (4). These two sets of data suggest that elevated NHLH2 expression could be either a cause or an effect of heart disease. Likewise, polymorphisms in the NHLH2 gene could be one cause of human exercise-induced performance phenotypes related to familial cardiac myopathy or tachycardia.
Figure 1.
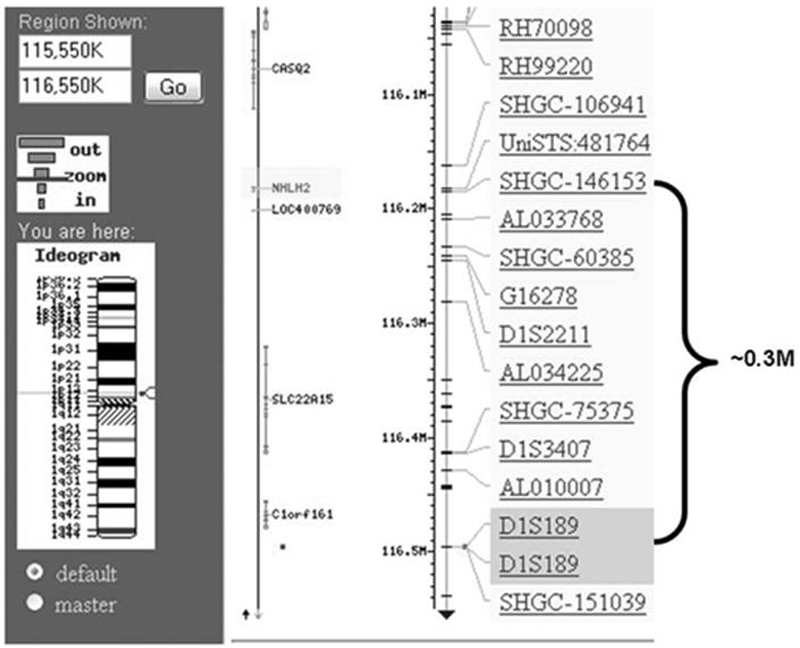
Screenshot from NCBI map viewer showing region of human chromosome 1 containing nescient helix-loop-helix 2 (NHLH2, human) and marker D1S189. National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/) map viewer was used to visualize the region of human chromosome 1 containing nescient helix-loop-helix 2 (NHLH2, human) and marker locus D1S189. A screenshot of the region was modified for use in this figure to highlight the two loci of interest and the distance between them.
Analysis of the more than 800 individuals listed in the NCBI single nucleotide polymorphism (SNP) database has identified no polymorphisms in the protein-coding region of the human NHLH2 gene. The protein-coding region of the gene is only 402 bp in length or just 6% of the total size of the messenger RNA (mRNA) for NHLH2. Within this region of chromosome 1, polymorphisms are predicted to be found every 400-600 bp of genomic DNA, so the expectation would be to find one polymorphism within that region. A total of five SNPs have been detected in the 3ôuntranslated region of the NHLH2 mRNA, which is approximately 1.5 kilobase in size. This is slightly higher than the estimated average of two-three polymorphisms predicted for this region. The 3ôuntranslated tail of NHLH2 mRNA is thought to mediate posttranscriptional control of NHLH2 through mRNA stability. One published article describes positive regulation of NHLH2 mRNA by the heterogeneous nuclear ribonucleoprotein, hnRNP-U (16), but the mechanism of this regulation and effects of any of these SNPs in human NHLH2 mRNA expression has not been further explored.
NHLH2 is implicated in the transcriptional control of another human gene, the melanocortin-4-receptor (MC4R). Functional polymorphisms in the MC4R gene are one of the most common causes of genetic obesity (4). MC4R is the receptor for the neuropeptide alpha melanocyte-stimulating hormone (α-MSH). Studies in animals have shown that the finite control of the levels for both α-MSH and MC4R are necessary for control of food intake, metabolism, and exercise (10,14). In two separate studies, polymorphisms in the MC4R gene have been linked to a lower exercise level in humans (2,11) In the first study, individuals with either a homozygous or heterozygous variation in the MC4R gene (MC4R-2745C/T or MC4R-2745T/T) were significantly more sedentary than individuals carrying the wild-type allele (MC4R-2745C/C) (11). In this study, physical activity levels were measured using a questionnaire assessing physical activity levels during the past year and a 3-d activity record kept during the study (11). In the second study, a QTL on chromosome 18q was linked to sedentary behavior, and 6 subjects in the study were found to harbor a mutation (MC4R-G55T) in MC4R gene (2). These data suggest that MC4R signaling may affect exercise behavior in humans, although the mechanism is not yet known. Recently, two unrelated human families with obesity were shown to carry a polymorphism in the promoter of MC4R within an E-box motif that binds human NHLH2 (15). These data suggest that NHLH2 can transcriptionally control levels of MC4R mRNA and, in conjunction with the studies linking MC4R to physical activity, ties NHLH2 activity to control of exercise in humans. Further analysis of NHLH2's transcriptional activity in the control of MC4R expression and the identification of other families with MC4R promoter mutations could allow for a comprehensive analysis of NHLH2 control of human exercise levels through MC4R regulation.
THE NHLH2 KNOCKOUT MICE AND PHYSICAL ACTIVITY
Exercise During Fed Conditions
In 1997, the adult-onset obese phenotype of mice containing a targeted deletion of Nhlh2 (N2KO mice) was described (5). Further characterization of this mouse model of adult-onset obesity showed that, although preobese adult N2KO mice have normal levels of food intake, spontaneous voluntary exercise was decreased by more than 50% in both male and female N2KO mice when given free access to running wheels. The mice showed no difference in the circadian pattern of running and no motor deficiencies in standard rotarod apparatus tests, indicating that lack of physical exercise was not caused by motor or balance problems (3). Careful study of that data shows that, although N2KO mice can get on and remain on the wheels, they show a reduced number of wheel turns compared with normal mice. Because of the nature of the collected data set, we were not able to discern whether N2KO mice run continuously but at a lower overall speed for the entire 20-min bin or if they get on and off the wheels, running in quick bursts of activity. This information could start to answer the question of whether N2KO mice have a reduced motivation for exercise or a reduced ability to engage in long bouts of exercise.
We then studied home cage ambulatory activity in the mutant animals in an attempt to elucidate the question of motivation versus ability. These data allowed us to analyze total daily regular activity via laser beam breaks in the home cage of individually housed animals. We identified a significant reduction in regular home-cage activity of both male and female N2KO mice (Fig. 2), and these differences were found in lower overall activity levels at each measured time point, especially during the lights-off period when normal mice showed maximal activity (data not shown). This home-cage activity pattern is similar to our published study using running wheels (3). These data suggest that N2KO mice show lower overall activity levels over long time periods, rather than fewer short bursts of activity. Analysis of the total activity each hour, which is a sum of 3 of the 20-min time points, underlines this point. Specifically, there is a 50% overall reduction in total hourly activity in N2KO mice compared with normal mice (P ≤ 0.001, data not shown). Thus, no matter which way activity levels are measured (20-min bins, hourly or daily) physical activity is reduced for N2KO mice. Although these additional data did not completely answer the question as to whether the motivation to start exercising or the ability to sustain exercise is affected, they do suggest that the maintenance of constant activity levels during any 24-h period is affected in the mutant animals. This phenotype is especially prominent during lights-off periods when normal mice have high activity levels but is also measurable during lights-on periods. Because reduced physical activity in N2KO mice occurs without the hyperphagia present in many obese mouse models (5), the N2KO mice may be a good model for humans who eat a healthy diet but have a sedentary lifestyle.
Figure 2.
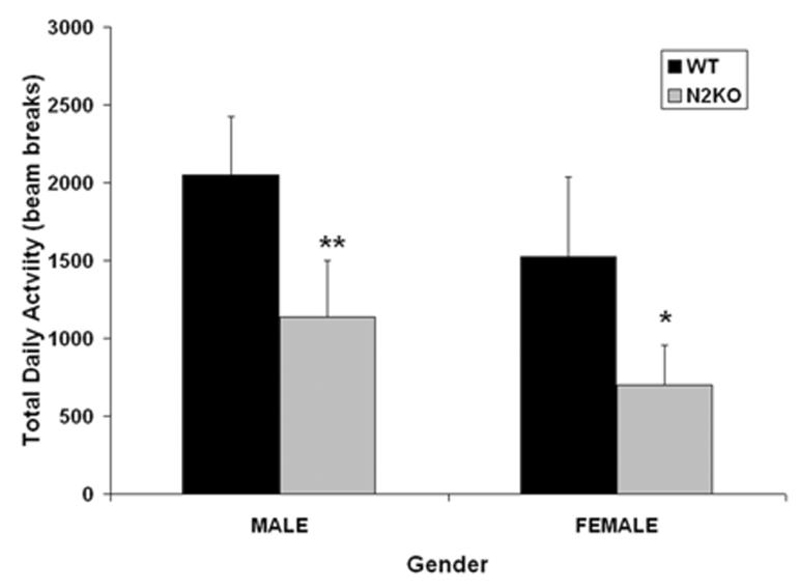
Home-cage activity levels for male and female N2KO mice. A laser beam-based home-cage activity system for mice (Columbus Instruments International, Columbus, OH) was used to assess general activity of the nescient helix-loop-helix 2 (Nhlh2)-deleted (N2KO) mice and normal (WT) age- and sex-matched mice. Data are based on the total average activity per 24-h period, measured over 3 d. Data are reported as the mean × SEM, n = 7 male WT, 7 male N2KO, 4 female WT, and 4 female N2KO. *P ≤ 0.05, **P ≤ 0.01
Exercise During Food Deprivation
In mammals, it is hypothesized that body weight is regulated by balancing energy (food) taken in, with energy expenditure in the form of metabolism and exercise. It is well known in humans and rodent models that calorie restriction alone will not result in sustained weight loss and that, after the restriction is lifted, weight gain occurs, sometimes in excess. One study in mice found that a 27% reduction in physical activity levels occurs during the calorie restriction period. Furthermore, the calorie-restricted group showed weight loss only initially and then continued to gain weight even while on a calorie-restricted diet (8). This is thought to occur through the compensatory reduction in energy expenditure by each individual. Although this is a longer study with calorie restriction and not total food deprivation, we sought to determine what would happen during total food deprivation in animals that already have reduced levels of physical activity. We modeled this condition using N2KO mice. As expected, during the ad libitum feeding conditions, N2KO mice showed a significant reduction in daily wheel running (Fig. 3A), as we had shown previously in the standard wheel running test (3). Both normal and N2KO mice were then subjected to a 24-h food deprivation period. As shown in Figure 3B, during food restriction, both normal and N2KO mice reduced their total wheel running activity by 20%-30%. This reduction occurred in the N2KO mice, although they were running less to begin with (compare normal (WT) vs N2KO levels; Fig. 3A). What is even more interesting is that the reduced activity levels of the mutant N2KO mice persist even when food is returned ad libitum for a 24-h period (Fig. 3B). Activity levels in normal mice returned to baseline with return of food. Surprisingly, there is no difference between the two genotypes in total food eaten after the food deprivation period, indicating that N2KO mice do not show the food deprivation- induced hyperphagia, which is also characteristic of many obese mice (Fig. 3C). Extending these data, we propose that short-term food restriction in N2KO mice could result in even higher body weight gain than animals that were never food deprived at all. In other words, whereas WT mice show a return to high exercise levels when food is returned, N2KO mice do not exercise at higher levels, although they are eating a similar level of food as WT mice. If translated to humans, individuals with polymorphisms in the NHLH2 gene or polymorphisms in the promoters of NHLH2-target genes could be at risk for higher weight gain after cessation of a dieting regimen if increased activity levels are not conscientiously maintained.
Figure 3.
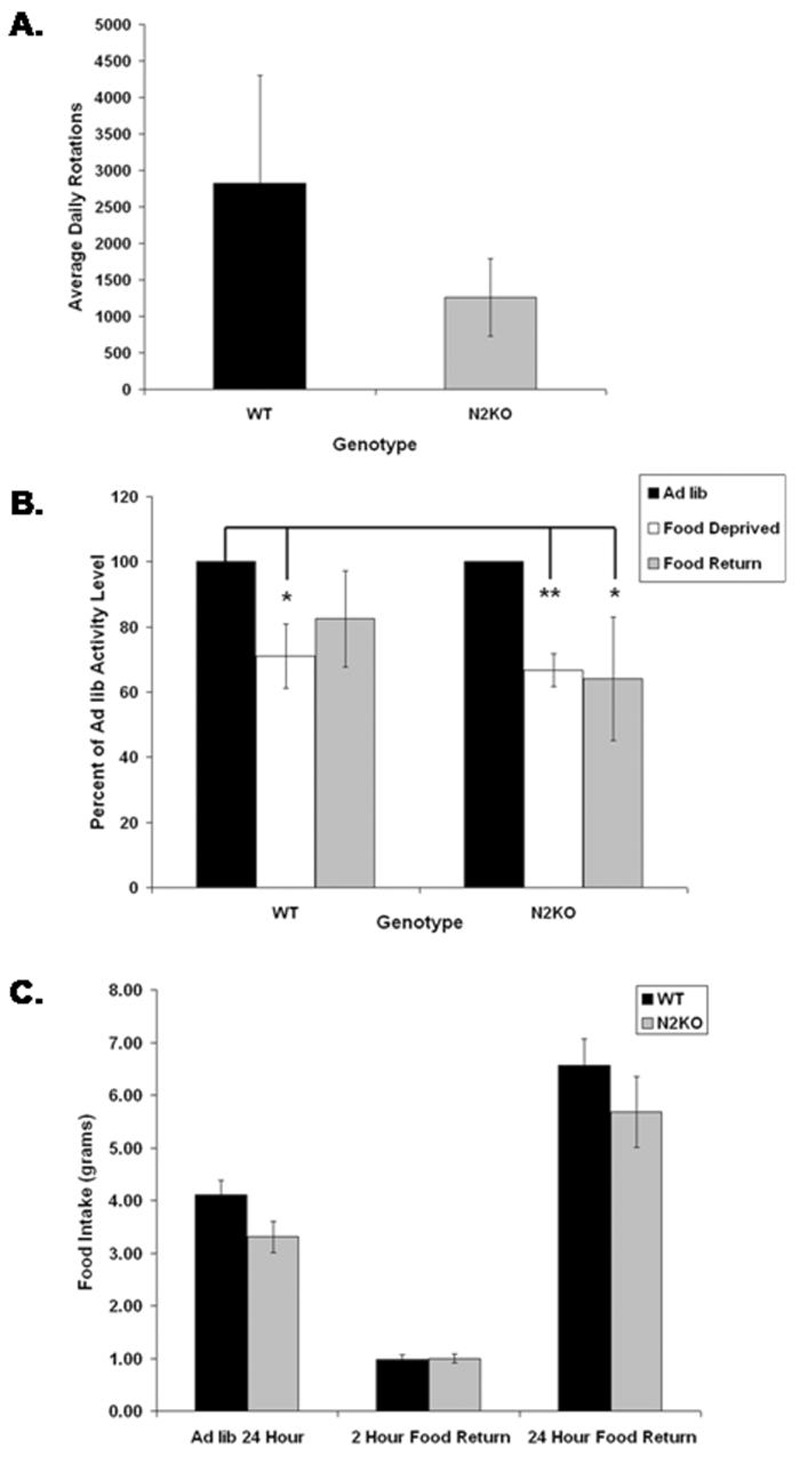
The effect of food deprivation on wheel running activity in N2KO mice. Mice were acclimated for 3 d to cages containing computerized running wheels (Columbus Instruments International). Wheel running was then measured for 24 h during ad libitum access to food. At 11 a.m. the following day, food was removed, and wheel running was accessed for 24 h of food deprivation. At 11 a.m. the following day, ad libitum food was returned to the animals, and wheel running was accessed for 24 h of food return. The data are reported as the percent of activity levels during ad libitum feeding for each genotype. Note that nescient helix-loop-helix 2 (Nhlh2)-deleted (N2KO) show a lower ad libitum activity level (2470 ± 175 turns per day, WT, compared with 1037 ± 141 turns per day, N2KO). N = 3 normal (WT) males, 4 N2KO males. *P ≤ 0.05, **P ≤ 0.01
EVIDENCE THAT NHLH2 COULD CONTROL EITHER THE MOTIVATION OR THE ABILITY TO EXERCISE
In this review, we present evidence that NHLH2 transcriptional activity may control some aspect of physical activity in humans, based on both human genetic evidence and phenotypic evidence from mice with a targeted deletion of the murine Nhlh2 gene. These data clearly demonstrate that functional inactivation of the Nhlh2 gene leads to decreased daily physical activity and that this is one of the contributing factors in adult-onset obesity observed in this mouse model. The genetic basis for body weight differences in humans can be linked to 11 single-gene mutations and to more than 600 other genes that could act together to influence various aspects of body weight control (13). Possible phenotypic influences are likely to include both the motivation and ability to perform moderate physical activity on a weekly basis. Although there are a growing number of genes with evidence for a genetic association to physical activity traits, more work in this area needs to be done. At this time, our data do not allow us to discriminate between whether N2KO mice are less motivated to exercise or whether physical problems affecting the muscle tissue of these animals contribute to the reduction in physical activity. Both of these possibilities will be discussed below.
Signals from the nervous system clearly participate in modulating motivational responses (5). One of the neuropeptides possibly involved in the motivation to continue sustained exercise is β-endorphin. β-Endorphin levels affect mood and pain perception and are increased after moderate exercise. We have previously shown that levels of the gene prohormone convertase I (PC1) are reduced by up to 60% in N2KO mice (5). PC1 activity is necessary for the initial step leading to the complete processing of the β-endorphin precursor molecule, pro-opiomelanocortin (POMC). Our original studies on PC1-mediated POMC processing in N2KO mice were focused on α-MSH processing (another POMC derivative). As such, these studies did not directly access β-endorphin levels, but reduced levels of PC1 in N2KO mice is likely to contribute to a decline in all POMC-derived neuropeptides in these animals. Simply put, it is possible that N2KO mice do not display the positive reinforcement in the form of endorphin secretion that would normally occur during exercise. This lack of positive reinforcement could lead to a lack of motivation for continued or subsequent exercise. This possibility has not been studied by our laboratory and has not been pursued in the β-endorphin knockout mice, which attain a body weight that is 10%- 15% higher than that of normal mice (14;15). These types of studies should be initialized to fully capture the consequences of lower pre- and post-β-endorphin levels on physical activity.
Physical activity also requires full muscle function without premature muscle fatigue. In addition to Nhlh2's well-documented expression in tissues of the central and peripheral nervous systems, anecdotal evidence indicates that NHLH2 also is expressed in human muscle tissue. We have already described studies of human heart mRNA expression profiles, which identify NHLH2 being up-regulated in individuals with dilated cardiomyopathy (1,9). Furthermore, online gene expression studies (www.genecard.org) in normal human tissues suggest that NHLH2 mRNA may be expressed in human skeletal muscle. Our laboratory has begun preliminary analyses of muscle function in N2KO mice, and our data also support a requirement for Nhlh2 to promote normal skeletal muscle appearance and function (data not shown). Should NHLH2 play a significant role in skeletal or heart muscle function, then individuals with inactivating or altered function mutations could have a diminished capacity for physical activity.
SUMMARY
This review has presented evidence to support the hypothesis that transcriptional activity by Nhlh2 promotes either the motivation or the ability to exercise. Loss of Nhlh2 through either directed mutation (N2KO mice) or spontaneous mutations (either in Nhlh2 or its target genes) leads to reduced physical activity and body weight gain. Significant additional work is needed to determine how Nhlh2 controls either the motivation or ability to engage in regular and high levels of physical activity (Fig. 4). Along these lines, hypothalamic β-endorphin levels should be measured, and if low, exercise studies should be conducted in the presence or absence of β-endorphin agonists to determine whether low β-endorphin contributes to reduced exercise. The possibility that a defective melanocortin signaling pathway may contribute to low levels of exercise in N2KO mice should be examined, including an in-depth analysis of the possible transcriptional control of MC4R by Nhlh2 and studies to replenish α-MSH levels in N2KO mice and examine resultant effects on exercise. Other studies to confirm NHLH2's expression in heart and skeletal muscle, to determine what conditions (exercise, leptin, food, etc.) change its expression level in these tissues, and to determine how functional inactivation of NHLH2 may affect the phenotype of muscle need to be conducted. Together, these studies will serve as a guide for future human research into NHLH2's role in human physical activity and body weight control.
Figure 4.
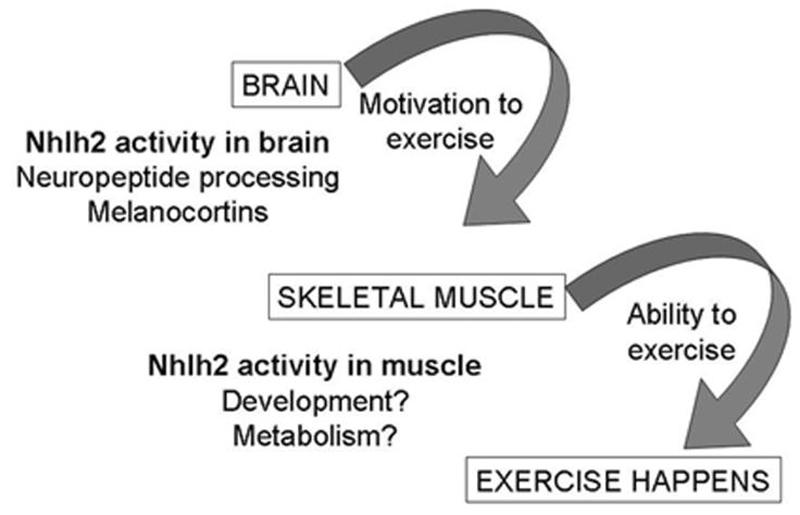
Diagram showing the possible role of nescient helix-loop-helix 2 (Nhlh2) in mediating brain and skeletal muscle function and exercise levels. Nescient helix-loop-helix 2 (Nhlh2) activity in the brain could control the motivation to exercise through either β-endorphin processing or melanocortin signaling, In skeletal muscle tissue, Nhlh2 could control the differentiation and/or regeneration of muscle tissue after exercise or muscle metabolism of glucose and fatty acid substrates.
ACKNOWLEDGMENT
The authors thank Mr. Franc-Eric Wiedmer, for careful reading of the manuscript, and Ms. Alison Bardwell, Mr. Brent Bowden, and Ms. Risa Pesapane, for excellent technical assistance.
This study was supported by a grant from the National Institute of Diabetes and Digestive Disorders No. DK-59903.
Footnotes
In mice, targeted deletion of the basic helix-loop-helix transcription factor, nescient helix-loop-helix 2 (Nhlh2), leads to adult-onset obesity and reduced physical activity. We propose the novel hypothesis that transcriptional activity by Nhlh2 (NHLH2 in humans) controls either the ability or the motivation for exercise.
References
- 1.Barth AS, Kuner R, Buness A, Ruschhaupt M, Merk S, Zwermann L, Kaab S, Kreuzer E, Steinbeck G, Mansmann U, Poustka A, Nabauer M, Sultmann H. Identification of a common gene expression signature in dilated cardiomyopathy across independent microarray studies. J. Am. Coll. Cardiol. 2006;48:1610–1617. doi: 10.1016/j.jacc.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 2.Cai G, Cole SA, Butte N, Bacino C, Diego V, Tan K, Goring HH, O'Rahilly S, Farooqi IS, Comuzzie AG. A quantitative trait locus on chromosome 18q for physical activity and dietary intake in Hispanic children. Obesity (Silver Spring) 2006;14:1596–1604. doi: 10.1038/oby.2006.184. [DOI] [PubMed] [Google Scholar]
- 3.Coyle CA, Jing E, Hosmer T, Powers JB, Wade G, Good DJ. Reduced voluntary activity precedes adult-onset obesity in Nhlh2 knockout mice. Physiol. Behav. 2002;77:387–402. doi: 10.1016/s0031-9384(02)00885-5. [DOI] [PubMed] [Google Scholar]
- 4.Farooqi S, O'Rahilly S. Genetics of obesity in humans. Endocr. Rev. 2006;27:710–718. doi: 10.1210/er.2006-0040. [DOI] [PubMed] [Google Scholar]
- 5.Fox DL, Vella KR, Good DJ. Energy balance pathways converging on the Nhlh2 transcription factor. Front Biosci. 2007;12:3983–3993. doi: 10.2741/2365. [DOI] [PubMed] [Google Scholar]
- 6.Good DJ. Obese mouse models. In: Conn PM, editor. Sourcebook of Models for Biomedical Research. Humana; Totowa, NJ: 2007. [Google Scholar]
- 7.Good DJ, Porter FD, Mahon KA, Parlow AF, Westphal H, Kirsch IR. Hypogonadism and obesity in mice with a targeted deletion of the Nhlh2 gene. Nat. Genet. 1997;15:397–401. doi: 10.1038/ng0497-397. [DOI] [PubMed] [Google Scholar]
- 8.Hambly C, Speakman JR. Contribution of different mechanisms to compensation for energy restriction in the mouse. Obes. Res. 2005;13:1548–1557. doi: 10.1038/oby.2005.190. [DOI] [PubMed] [Google Scholar]
- 9.Lahat H, Eldar M, Levy-Nissenbaum E, Bahan T, Friedman E, Khoury A, Lorber A, Kastner DL, Goldman B, Pras E. Autosomal recessive catecholamine- or exercise-induced polymorphic ventricular tachycardia: clinical features and assignment of the disease gene to chromosome 1p13-21. Circulation. 2001;103:2822–2827. doi: 10.1161/01.cir.103.23.2822. [DOI] [PubMed] [Google Scholar]
- 10.Lee M, Wardlaw SL. The central melanocortin system and the regulation of energy balance. Front Biosci. 2007;12:3994–4010. doi: 10.2741/2366. [DOI] [PubMed] [Google Scholar]
- 11.Loos RJ, Rankinen T, Tremblay A, Perusse L, Chagnon Y, Bouchard C. Melanocortin-4 receptor gene and physical activity in the Quebec Family Study. Int. J. Obes. (Lond) 2005;29:420–428. doi: 10.1038/sj.ijo.0802869. [DOI] [PubMed] [Google Scholar]
- 12.Rankinen T, Bray MS, Hagberg JM, Perusse L, Roth SM, Wolfarth B, Bouchard C. The human gene map for performance and health-related fitness phenotypes: the 2005 update. Med. Sci. Sports Exerc. 2006;38:1863–1888. doi: 10.1249/01.mss.0000233789.01164.4f. [DOI] [PubMed] [Google Scholar]
- 13.Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Perusse L, Bouchard C. The human obesity gene map: the 2005 update. Obesity (Silver Spring) 2006;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- 14.Seeley RJ, Drazen DL, Clegg DJ. The critical role of the melanocortin system in the control of energy balance. Annu. Rev. Nutr. 2004;24:133–149. doi: 10.1146/annurev.nutr.24.012003.132428. [DOI] [PubMed] [Google Scholar]
- 15.Valli-Jaakola K, Palvimo JJ, Lipsanen-Nyman M, Salomaa V, Peltonen L, Kontula K, Schalin-Jantti C. A two-base deletion -439delGC in the melanocortin-4 receptor promoter associated with early-onset obesity. Horm. Res. 2006;66:61–69. doi: 10.1159/000093469. [DOI] [PubMed] [Google Scholar]
- 16.Yugami M, Kabe Y, Yamaguchi Y, Wada T, Handa H. hnRNP-U enhances the expression of specific genes by stabilizing mRNA. FEBS Lett. 2007;581:1–7. doi: 10.1016/S0014-5793(07)01283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]


