Abstract
Background and Aims
Myrica rivas-martinezii is a critically endangered endemic of the laurel forest of the Canary Islands and co-occurs very close to M. faya. Some authors suggest that M. rivas-martinezii and M. faya are two morphs of the same species, so molecular markers were used to estimate the levels and structuring of genetic variation within and among natural populations in order to evaluate genetic relationships between these two congeners.
Methods
Six polymorphic microsatellite (simple sequence repeat, SSR) markers were used to determine the genetic diversity and the genetic relationship between both Myrica species.
Key Results
Most of the natural populations analysed were in Hardy–Weinberg equilibrium for both taxa. Analysis of molecular variance (AMOVA) for both species revealed that most of the genetic variability detected was contained within populations (92·48 and 85·91 % for M. faya and M. rivas-martinezii, respectively), which it is consistent with outcrossing and dioecious plants. Estimates of interpopulation genetic variation, calculated from FST and G′ST, were quite low in the two taxa, and these values did not increase substantially when M. rivas-martinezii and M. faya populations were compared. The UPGMA dendrogram based on Nei's genetic distance clustered the populations by their island origin, independently of taxon. In fact, the mixture of individuals of both taxa did not appreciably disrupt the intrapopulational genetic cohesion, and only 3·76 % variation existed between species.
Conclusions
All the results obtained using molecular markers indicate clearly that both taxa share the same genetic pool, and they are probably the same taxa. Considering that M. rivas-martinezii is classified as at risk of extinction, there should be a change of focus of the current management actions for the conservation of this putatively endangered Canarian endemic.
Key words: Canary Islands, conservation genetics, microsatellites, Myrica rivas-martinezii, Myrica faya, plant conservation
INTRODUCTION
Myrica rivas-martinezii A. Santos (Myricaceae) is a perennial, woody and dioecious tree. It is endangered, and endemic to the laurel forest of the Canary Islands. It was described for the first time in 1980 (Santos, 1980), and its range of distribution is restricted to only three islands: El Hierro, where the highest number of individuals is to be found, about 40, in an area of approx. 90 km2; La Gomera, with 12 isolated individuals, all in different and isolated geographic locations; and La Palma, where only two individuals (one male and one female) have been described, which are separated from each other by >20 km (Santos, 1983; Bañares et al., 2004). Due to the low number of individuals (<60 in total), M. rivas-martinezii has been classified as ‘critically endangered’ according to IUCN categories (VVAA, 2000). It was also catalogued as in danger of extinction by the Canarian Government (BOC, 2001) and by the European Habitat Directive (Beltrán et al., 1999).
Myrica rivas-martinezii co-occurs very close to M. faya Aiton (fayatree, firetree or firebush) which is quite abundant due to its colonizing capacity (Bañares et al., 2004). Myrica faya is native to the northern islands of Macaronesia (Azores, Madeira and the Canaries). Myrica faya, which is normally a dioecious plant (Gardner, 1985), has been recognized as one of the 12 most noxious plants alien to Hawaii, due to its ability to invade and colonize the Hawaiian environment rapidly and aggressively (Smith, 1985).
Both Myrica species are morphologically distinct, with M. rivas-martinezii having substantially smaller, more oval leaves, while the leaves of M. faya are larger, narrower and lanceolate (Santos, 1980).
However, the taxonomic range of M. rivas-martinezii in the Canary Islands has been questioned. Demographic studies conducted on M. rivas-martinezii showed no evidence of either asexual or sexual propagation (Bañares et al., 2004), and when ex situ germination tests were performed most of the viable offspring (90 %) showed the M. faya phenotype. Finally, no new offspring individuals of M. rivas-martinezii have been observed in the field, after >25 years of analysis. For that reason, some authors suggest that M. rivas-martinezii and M. faya are two morphs of the same species (M. Marrero, Teide National Park, Spain, pers. comm.).
Previous information regarding isozyme variation in both taxa of Myrica was available from Batista and Sosa (1998). They analysed six populations of both taxa with eight allozyme loci, and no genetic differences were detected among the populations of both congeners. However, the results were not conclusive, due to the high number of monomorphic loci detected. Similarly, Werner et al. (2007) did not find enough genetic differences between 40 samples of both taxa, using ISSR (inter simple sequence repeat), trnL intron sequences and the trnL–trnF intergenic spacer.
The general aims of this study are: (a) to use highly polymorphic molecular markers (microsatellites, i.e. simple sequence repeats, SSRs) in order to obtain essential information to enable exploration of the relationship between M. faya and M. rivas-martinezii in the Canary Islands (the hypothesis is that genetic differences between two taxa should be higher than the existing intrataxon differentiation); and (b) to use this molecular information as a tool for assessing the protection status of the endangered M. rivas-martinezii in order to formulate appropriate management and conservation strategies.
MATERIALS AND METHODS
Plant material
Forty-two plants of M. rivas-martinezii A. Santos were sampled from the three islands of the archipelago where it is present (El Hierro, La Gomera and La Palma). Also, 183 individuals of M. faya Aiton from eight localities were sampled in all the islands where M. rivas-martinezii occurs (Table 1).
Table 1.
Myrica rivas-martinezii and M. faya populations analysed in the Canary Islands
| Island | Population | Code | Species | n |
|---|---|---|---|---|
| La Gomera | Las Creces–Las Hayas | MFP1G | M. faya | 38 |
| Los Gallos–La Paisaita | MFP2G | M. faya | 35 | |
| La Cancela | MFCAG | M. faya | 25 | |
| Barranco de Garajonay | MFBGG | M. faya | 18 | |
| La Laguna Grande | MFLAG | M. faya | 16 | |
| La Gomera | MRG | M. rivas-martinezii | 10 | |
| Totals | M. rivas-martinezii | 10 | ||
| M. faya | 132 | |||
| El Hierro | El Fayal–La Caldereta | MFFCH | M. faya | 11 |
| Curva del Avión | MFCAH | M. faya | 27 | |
| El Fayal–La Caldereta | MRH | M. rivas-martinezii | 30 | |
| Totals | M. rivas-martinezii | 30 | ||
| M. faya | 38 | |||
| La Palma | Puntallana | MFP | M. faya | 13 |
| Garafía–Puntallana | MRP | M.rivas-martinezi | 2 | |
| Totals | M. rivas-martinezii | 2 | ||
| M. faya | 13 | |||
| Totals for all islands | M. rivas-martinezii | 42 | ||
| M. faya | 183 |
n = sample size.
DNA extraction and purification
DNA was extracted from silica-gel dried young leaves following the method of Dellaporta et al. (1983) modified by Corniquel and Mercier (1994). A 150 µL volume of total DNA samples was purified using the QIAquick PCR purification kit (Qiagen).
Microsatellite analysis and genotyping
Forward and reverse primers described by González-Pérez et al. (2008) were used to amplify six polymorphic microsatellite loci. PCR amplifications were carried out following the protocols in the aforementioned publication. Each 25 µL PCR contained approx. 20 ng of DNA, 10 pmol of each primer, 0·25 µL of bovine serum albumin (BSA; 0·4 %), as well as PCR Master Mix (Reddy-Mix, ABgene, Surrey, UK) that included 0·625 U of Taq DNA polymerase, 75 mm Tris–HCl, 20 mm (NH4)2SO4, 0·01 % Tween-20, 2·5 mm MgCl2 and 0·2 mm of each dNTP. Amplifications were carried out using the following thermal cycling conditions: 3 min denaturation at 95°C; 35 cycles of 30 denaturation at 95°C; 30 annealing at 55°C and 1·5 min elongation at 72°C, followed by 5 min elongation at 72°C.
The products were detected using an ABI 3100 Genetic Analyzer, and fragment sizes were determined using GENESCAN v. 2·02 and GENOTYPER v. 1·1 (Applied Biosystems, Inc.). Allele peak profiles were identified at each locus and a genotype was assigned to each individual.
Data analysis
Exact Hardy–Weinberg tests to measure the significance of deviations from the null hypothesis of random union of gametes were carried out on natural populations of M. faya and M. rivas-martinezii using Fisher's exact test, both for each pair of loci and within each population, using GENEPOP (Raymond and Rousset, 1995). In order to test if localities in which both species of Myrica were present form a panmictic unit, two exact tests were carried out by locality: one including individuals from both species and another excluding M. rivas-martinezii individuals from the analysis. For all tests, a sequential Bonferroni correction for multiple comparisons was applied (Rice, 1989).
To test genetic drift events in the natural populations, a bottleneck test was carried out using BOTTLENECK software (Piry et al., 1999), under the infinite allele model (IAM), the stepwise mutation model (SSM) and an intermediate two-phased model (TPM). Of the three statistical methods used by the BOTTLENECK software, the standardized differences test was not employed, because it requires at least 20 polymorphic loci to be reliable. The M. rivas-martinezii from La Palma population was not analysed because of its small sample size (n = 2). These tests are based on the fact that populations that have experienced a recent reduction in effective population size exhibit a more rapid reduction of allelic diversity than heterozygosity at polymorphic loci.
Due to the low number of M. rivas-martinezii individuals in each population, all the individuals from each island were considered together in the further analysis (Table 1).
Basic genetic diversity indices – mean number of alleles (A), the proportion of polymorphic loci at 95 % (P95), and the observed (Ho) and unbiased expected (He) heterozygosities (Nei, 1978) – were estimated using GENETIX version 4.02 (Belkhir et al., 2003).
In accordance with the hierarchical sampling design, allele frequency information was analysed using a nested analysis of variance [analysis of molecular variance (AMOVA) Excoffier et al. (2005)] to estimate the components of variance between and within species. The hierarchies were tested for ‘between species’, ‘among populations within species’ and ‘within populations’ for the entire data set, using ARLEQUIN software (Schneider et al., 2000). Subsequently, separate AMOVA models were analysed to test the distribution of genetic variance among and within populations of each species.
At individual level, the neighbor-joining tree, based on DAS, shared allele distance (Jin and Chakraborty, 1993) among individuals was estimated using POPULATION software (Langella, 2000). The resulting tree was visualized using MEGA 4 (Tamura et al., 2007).
To check the concordance between taxonomic status and genetic structure, all the genotypes were screened using a Bayesian admixture procedure implemented in STRUCTURE 2·21 (Pritchard et al., 2000), designed to identify the K (unknown) populations of origin of the sampled individuals and to assign the individuals simultaneously to the populations. The most likely value of K is assessed by comparing the likelihood of the data for different values of K. The model was assumed to be of population admixture and correlate allele frequencies. A series of independent runs was conducted for each value of K (the number of sub-populations) between 1 and 10. Analyses consisted of a 105 burn-in period replicated and a run length of 106 replicates. Populations or individuals were assigned to one cluster if their proportion of membership (qi) of that cluster was equal to or larger than an arbitrary threshold of 0·800.
Nei's genetic matrix distance (1972) between localities was calculated using GENALEX version 6.0 (Peakall and Smouse, 2006). The resulting tree was visualized using MEGA 4 (Tamura et al., 2007).
In order to clarify genetic differentiation among populations, pairwise divergences between localities were analysed using the FST (Wright, 1951), using GENALEX version 6·0 (Peakall and Smouse, 2006). Because the magnitude of FST (or GST) may depend on the variability of a locus, a standardized measure of differentiation, G′ST, which was developed by Hedrick (2005) to facilitate comparisons among loci that differ in variability, was also calculated. The standardized genetic differentiation measure (G′ST) was recoded using RecodeData version 0·1 (Merimans, 2006) and calculated with FSTAT version 1·2 (Goudet, 1995). In addition, correlation between both genetic differentiation measures was estimated using SPSS version 14·0 (SPSS Inc., Chicago, IL).
RESULTS
Levels of genetic variation
Six polymorphic microsatellite loci were investigated in a total of 225 individuals (183 of M. faya and 42 of M. rivas-martinezii). The basic indicators of genetic variability showed certain uniformity across the populations, with the exception of the M. rivas-martinezii population from La Palma, where the levels of genetic variability were very low due to the small number of plants (only two) analysed. The average number of alleles ranged from 2·33 to 7·17, while expected heterozygosites ranged from 0·437 to 0·715 (Table 2). Although at species level M. faya showed higher levels of genetic variability than M. rivas-martinezii, these differences were not significant.
Table 2.
Mean genetic diversity indices of M. rivas-martinezii and M. faya populations
| Species/Population | Island | n | A | P95 | Ho | He |
|---|---|---|---|---|---|---|
| M. rivas-martinezii | ||||||
| MRG | La Gomera | 10 | 5·7 | 1·000 | 0·668 | 0·669 |
| MRH | El Hierro | 30 | 4·7 | 1·000 | 0·419 | 0·475 |
| MRP | La Palma | 2 | 2·3 | 0·833 | 0·667 | 0·437 |
| Total | 42 | 6·5 | 1·000 | 0·488 | 0·560 | |
| M. faya | ||||||
| MFP1G | La Gomera | 38 | 7·2 | 1·000 | 0·585 | 0·636 |
| MFP2G | La Gomera | 35 | 6·3 | 1·000 | 0·571 | 0·671 |
| MFCAG | La Gomera | 25 | 5·7 | 1·000 | 0·496 | 0·565 |
| MFBGG | La Gomera | 18 | 6·2 | 1·000 | 0·718 | 0·715 |
| MFLAG | La Gomera | 16 | 5·5 | 1·000 | 0·691 | 0·657 |
| Total | 132 | 6·2 | 1·000 | 0·612 | 0·649 | |
| MFFCH | El Hierro | 11 | 4·5 | 1·000 | 0·545 | 0·597 |
| MFCAH | El Hierro | 27 | 5·7 | 1·000 | 0·515 | 0·609 |
| Total | 38 | 5·1 | 1·000 | 0·530 | 0·603 | |
| MFP | La Palma | 13 | 4·5 | 1·000 | 0·440 | 0·588 |
| Total | 183 | 9·3 | 1·000 | 0·570 | 0·668 |
A = average number of alleles, P95 = proportion of polymorphic loci, HO = observed heterozygosity, He = expected heterozygosity.
Most of the natural populations analysed were in Hardy–Weinberg equilibrium (data not shown) in both cases, i.e. when M. rivas-martinezii samples were included, and when they were excluded from analysis. With the exact test, 93 % (50 out of 54 possible tests) of comparisons were not significantly different from Hardy–Weinberg expectations after applying Bonferroni correction. Wilcoxon test performed under SSM was more appropriate to detect a bottleneck sign for Myrica microsatellites. Within M. rivas-martinezii only a slightly bottleneck signature (P < 0·05) in the population from El Hierro was recorded.
Genetic differentiation
AMOVA analyses (Table 3) revealed that for both species most of the genetic variability detected was contained within populations (92·48 and 85·91 % for M. faya and M. rivas-martinezii, respectively). This partitioned variation is consistent with an outcrossing and dioecious plant. Unexpectedly, this value was maintained when both taxa were artificially considered together (90·39 %, Table 3). In fact the mixture of individuals of both taxa did not disrupt appreciably the intrapopulational genetic cohesion, and only 3·76 % of the variation resided between species.
Table 3.
AMOVA at the three hierarchical levels considered from M. rivas-martinezii and M. faya
| Source of variation | d.f. | Percentage variation | F-statistics |
|---|---|---|---|
| M. rivas-martinezii vs. M. faya | |||
| Between species | 1 | 3·76 | FCT = 0·038ns |
| Among populations within specie | 9 | 5·74 | FSC = 0·060*** |
| Within populations | 439 | 90·39 | |
| Total | 449 | FST = 0·09*** | |
| M. faya | |||
| Among islands | 2 | 5·72 | FCT = 0·057*** |
| Among populations within islands | 5 | 1·80 | FSC = 0·019*** |
| Within populations | 358 | 92·48 | |
| Total | 365 | FST = 0·075*** | |
| M. rivas-martinezii | |||
| Among populations | 2 | 14·09 | |
| Within populations | 81 | 85·91 | |
| Total | 83 | FST = 0·141*** |
***P < 0·001; ns: non-significant.
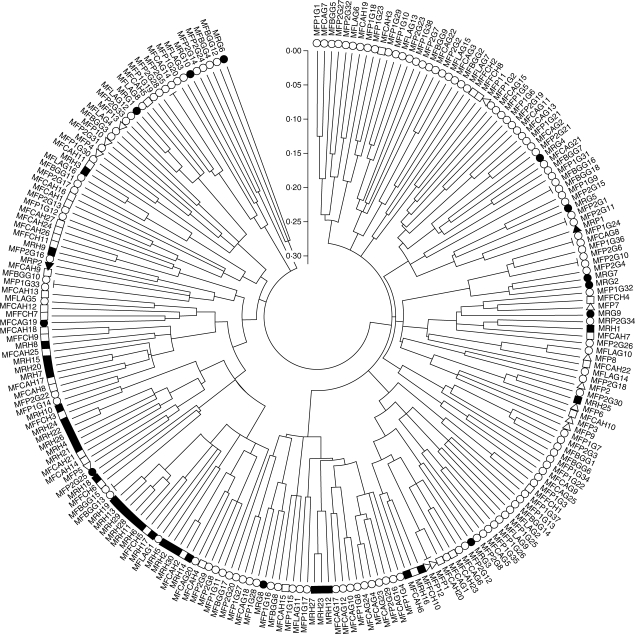
In accordance with these results, the neighbor-joining tree, based on DAS, shared allele distance (Jin and Chakraborty, 1993) showed no differences between M. rivas-martinezii and M. faya individuals (Fig. 1).
Fig. 1.
Neighbor-joining tree based on DAS, shared allele distance (Jin and Chakraborty, 1993) showing the relationship among individuals of M. rivas-martinezii and M. faya. Filled symbols, M. rivas-martinezii individuals; open symbols, M. faya individuals; circles, samples from Gomera island; squares, samples from El Hierro island; triangles, samples from La Palma island.
For the Bayesian analysis, using the total data set (225 individuals, 6 microsatellite loci, 11 localities) and K = 1–10, the probability of the data was maximum with K = 2, suggesting that the individuals analysed can be split into two distinct genetic clusters (Table 4). At K = 2, all the Myrica populations analysed were recognized as having descended from both inferred populations (cluster I and cluster II). All of them, except MRH assigned to cluster I, showed signs of admixture between two ‘genetic clusters’ inferred (Table 4). The proportion of membership of cluster I ranged from 0·238 (MFBGGqi) to 0·823 (MRHqi). There was only one population (MRH) that could be assigned to one cluster with a proportion of membership (qi) ≥0·800. There was a tendency for populations to be clustered according to their geographical origin. Thus, the populations from La Gomera showed a higher proportion of membership of cluster II than of cluster I, while populations from La Palma and El Hierro showed the inverse trend, i.e. a higher proportion of membership of cluster I than of cluster II. No grouping for taxonomic classification was observed (Table 4).
Table 4.
Bayesian clustering analysis of M. rivas-martinezii and M. faya individuals studied with the total sample set (225 individuals, 6 microsatellites loci, 11 sampled localities)
| Cluster |
||||
|---|---|---|---|---|
| Species | Island | Population | I | II |
| Myrica faya | La Gomera | MFP1G | 0·295 | 0·705 |
| Myrica faya | La Gomera | MFP2G | 0·428 | 0·572 |
| Myrica faya | La Gomera | MFCAG | 0·404 | 0·596 |
| Myrica faya | La Gomera | MFBGG | 0·238 | 0·762 |
| Myrica faya | La Gomera | MFLAG | 0·287 | 0·713 |
| Myrica faya | El Hierro | MFFCH | 0·544 | 0·456 |
| Myrica faya | El Hierro | MFCAH | 0·699 | 0·301 |
| Myrica faya | La Palma | MFP | 0·641 | 0·359 |
| Myrica rivas-martinezii | La Gomera | MRG | 0·382 | 0·618 |
| Myrica rivas-martinezii | El Hierro | MRH | 0·823 | 0·177 |
| Myrica rivas-martinezii | La Palma | MRP | 0·608 | 0·392 |
The table shows the proportion of membership (qi) of each pre-defined sampled population in each of two inferred clusters.
Estimates of interpopulation genetic variation, calculated from FST, are quite low in the two taxa, ranging from 0·010 to 0·109 (Table 5). In addition, the values of FST did not increase substantially when M. rivas-martinezii and M. faya populations were compared. In fact the lowest value of FST (0·010) was detected between two populations belonging to different taxa: the M. faya Los Gallos–La Paisaita population (MFP2G) and the M. rivas-martinezii population from La Gomera (MRG).
Table 5.
Genetic differentiation coefficient (FST) values (below the diagonal), and standardized gene differentiation G′ST (Hedrick, 2005) (above the diagonal) for all pairwise combinations of localities of M. rivas-martinezii and M. faya
|
M. faya |
M. rivas-martinezii |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Population | MFP1G | MFP2G | MFCAG | MFBGG | MFLAG | MFFCH | MFCAH | MFP | MRG | MRH | MRP |
| MFP1G | – | 0·045 | 0·013 | 0·011 | 0·069 | 0·054 | 0·219 | 0·218 | 0·026 | 0·277 | 0·203 |
| MFP2G | 0·019 | – | 0·058 | 0·006 | 0·037 | 0·049 | 0·095 | 0·076 | −0·056 | 0·229 | 0·023 |
| MFCAG | 0·015 | 0·028 | – | 0·058 | 0·123 | 0·061 | 0·213 | 0·163 | 0·065 | 0·229 | 0·135 |
| MFBGG | 0·014 | 0·013 | 0·031 | – | 0·012 | 0·079 | 0·176 | 0·149 | −0·012 | 0·291 | 0·088 |
| MFLAG | 0·026 | 0·021 | 0·045 | 0·016 | – | 0·048 | 0·133 | 0·146 | 0·083 | 0·287 | 0·172 |
| MFFCH | 0·030 | 0·029 | 0·039 | 0·037 | 0·031 | – | 0·031 | 0·122 | 0·061 | 0·067 | 0·053 |
| MFCAH | 0·063 | 0·033 | 0·076 | 0·051 | 0·045 | 0·027 | – | 0·051 | 0·095 | 0·108 | 0·053 |
| MFP | 0·062 | 0·029 | 0·057 | 0·050 | 0·053 | 0·054 | 0·034 | – | 0·080 | 0·200 | 0·066 |
| MRG | 0·025 | 0·010 | 0·038 | 0·020 | 0·038 | 0·042 | 0·043 | 0·042 | – | 0·241 | 0·047 |
| MRH | 0·087 | 0·072 | 0·083 | 0·088 | 0·091 | 0·037 | 0·040 | 0·079 | 0·084 | – | 0·080 |
| MRP | 0·109 | 0·075 | 0·107 | 0·089 | 0·102 | 0·088 | 0·081 | 0·096 | 0·085 | 0·093 | – |
The standardized genetic differentiation measure (G′ST), which corrects for differences in variability between loci, showed the same pattern as did FST (Table 5). Significant correlations were found between both genetic differentiation measures (r = 0·659; P < 0·01). For both measures, the lowest value (G′ST = −0·056) was found between populations MFP2G and MRG, and the highest (G′ST measure = 0·291) was between populations MRH and MFBGG.
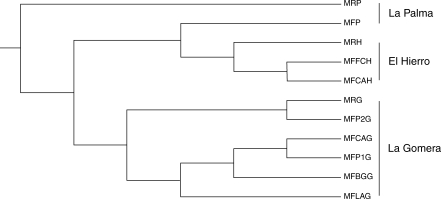
The UPGMA dendrogram based on Nei's genetic distance clustered the populations in two main groups (Fig. 2). The main cluster grouped all populations from La Gomera, independently of taxa. The second cluster included all populations from El Hierro and the population of M. faya from La Palma (MFP); only the population of M. rivas-martinezii from La Palma (MRP) was completely separated from the rest of the populations. However, it is important to remember that this population is formed by only two individuals, a fact which could explain the high differentiation of this population from the other congeric populations.
Fig. 2.
UPGMA dendrogram based on Nei (1972) genetic distance among the six localities of M. rivas-martinezii and M. faya analysed.
DISCUSSION
Genetic relationship between M. faya and M. rivas-martinezii
One of the main objectives of this research was to determine the genetic relationship between the only two congeneric species of Myrica from the Canary Islands: M. rivas-martinezii and M. faya. This is an important issue from a conservation standpoint due to the management policies for monitoring of endangered plant species which state that the species is the minimum unit for legal protection (IUCN, 2006). Numerous studies with a conservation and recovery focus have been carried out on M. rivas-martinezii since its discovery 25 years ago, but, in spite of this effort and input of resources, the number of individuals of this taxon remains the same. Under this scenario, one of the most important values of genetic data in addressing issues of plant conservation biology is in resolving taxonomic uncertainties (Frankham et al., 2002). In this respect, neutral markers are useful in estimating the relative evolutionary importance of genetic factors such as gene flow and genetic drift, and in clarifying the relationship between closely related taxa (Frankham et al., 2002), and microsatellites are especially suited to situations in which the taxa are genetically related (Steiner et al., 2006; Friar et al., 2007). Therefore, M. rivas-martinezii could be considered a paradigm for the application of molecular markers to a conservation programme of an endangered species. It is of clear conservation interest because its consideration as a species or subspecies of M. faya will determine, respectively, its inclusion in the lists of protected Canarian (and Spanish) endemics or its exclusion from this instrument of legal protection.
Although morphological differences between M. faya and M. rivas-martinezii are obvious, especially in the shape of their leaves, these differences are not likely to be a function of environmental or ecological variation between sites because they are found in different islands, and the habitat and conditions where M. rivas-martinezii grows are exactly the same as those for M. faya. In fact both taxa grow very close together, with no environmental or soil differences. In addition, all results obtained using molecular markers show clearly that both taxa share the same genetic pool. These results indicate that the threatening anthropic elements are acting on both taxa equally, and it is not likely therefore that the population size decrease of M. rivas-martinezii is exclusively due to human causes.
First, most natural populations were in Hardy–Weinberg equilibrium, although individuals of both species were included in the analysis, suggesting a very close relationship between both putative species. Therefore, values of FST are adequate to test the reproductive isolation between these two taxa (Wright, 1951), because this parameter reflects the role of gene flow as a force of genetic cohesion (Oliva-Tejera et al., 2006). The genetic history shown by microsatellites indicates that the geographic isolation, a consequence of its insular distribution, is probably the main factor that is acting in Myrica from the Canary Islands, and it is much more important than the putative genetic separation that could exist between M. faya and M. rivas-martinezii. Thus all populations from La Gomera, independently of taxonomy, showed a very high degree of similarity (Fig. 2).
Secondly, the present results coincide closely with those obtained previously for M. rivas-martinezii using RAPD (randomly amplified polymorphic DNA) markers (Batista et al., 2004). In the RAPD analysis, most individuals were found to be grouped by their island origin, plants from el Hierro being more closely related to those from La Palma, with plants from La Gomera in a separate cluster. These results stress that microsatellites and RAPD markers convey the same genetic information in this taxon, which could be used as another test of the validity of microsatellites results.
Thirdly, genetic distance and differentiation among M. rivas-martinezii and M. faya populations were lower than expected for populations of two differentiated congeneric taxa.
On the other hand, comparing genetic differentiation values (FST) obtained in each single species (M. rivas-martinezii FST = 0·141; M. faya FST = 0·075) with the estimates from the literature database (Morjan and Rieseberg, 2004), genetic differentiation coefficients were within the interquartile range of outcrossing plants, according to their description as primarily outbreeding species (Morjan and Rieseberg, 2004). In addition, both parameters showed that the level of differentiation was much lower within the species than between them (Table 5). Similarly, AMOVA showed that the percentage variation between species (3·76 %) was lower than the percentage variation attributed to differences between M. faya in different islands (5·72 %).
Friar et al. (2007), studying gene flow between two recently diverged species, Dubautia ciliolata and D. arborea, found higher average genetic differentiation values (FST = 0·260) than those detected in this research. In addition, both Dubautia species were effectively separated in principal component analysis, as well as in Bayesian cluster analysis.
Also, G′ST values detected within each species and among species (G′ST = 0·120) were much lower than those described for differentiation between hybridizing species of sunflowers (Helianthus annuus and Helianthus petiolaris, G′ST = 0·505; Yatabe et al., 2007).
Implications for conservation
Given its small population sizes, and its endemic character, with a very restricted geographic distribution, it was expected that M. rivas-martinezii populations would be genetically impoverished, as well as showing bottleneck signatures, especially in La Palma and La Gomera. However, the bottleneck detection test only shows slightly bottleneck signatures (P < 0·05) in M. rivas-martinezii populations from El Hierro, following Wilcoxon's test and under the SSM model (data not shown). Besides, genetic diversity detected in this endemic species is higher than those described for other endangered species (e.g. Borderea chouardii, Ho = 0·14; Segarra-Moragues et al., 2005) and other endemic species (e.g. Dubautia arborea, He = 0·221; and Dubautia ciliolata, He = 0·312; Friar et al., 2007). This result agrees with previous studies that show higher genetic diversity levels in M. rivas-martinezii using RAPD markers (Batista et al., 2004). In addition, higher genetic variability has been recorded in Canarian endemic flora than those distributed in other oceanic islands (Francisco-Ortega et al., 2000; González-Pérez et al., 2004). Francisco-Ortega et al. (2000) suggested that some Canarian endemics represent old lineages that took refuge in the Macaronesian region during glaciations and desertification in Europe and Northern Africa after the Miocene period. Under this ‘time’ hypothesis, the high level of genetic variation in this species could be accounted for by the progressive accumulation of mutation over time. On the whole, and in spite of the high genetic variability detected in both Myrica species, the FST index does not give underestimates of population divergence, since standardized G′ST also records a low genetic differentiation.
No significant differences were found between M. faya (He = 0·668) and the endemic M. rivas-martinezii (He = 0·560) at the genetic diversity level. With regard to genetic diversity values recorded in M. faya (He = 0·668), these were within those described in other tree and woody species (Pinus resinosa, He = 0·508; Boys et al., 2005; Juglans nigra, A = 23, He = 0·793; Victory et al., 2006; Quercus lobata, A = 13·6, He = 0·740; Dutech et al., 2005), but much lower than those detected in Myrica cerifera (A = 21; He = 0·833), another invasive species of the genus (Erickson et al., 2004).
Thus, if M. rivas-martinezii and M. faya constitute a single genetic pool of a widespread species this could explain the high genetic diversity values detected, as well as the low genetic distance and differentiation among populations of both putative species. All analyses carried out at different levels, i.e. the individual level (neighbor-joining and Bayesian cluster analysis), at the population level (Hardy–Weinberg exact test) and at the species/island level (genetic distance, genetic differentiation coefficient), support the view that there is no genetic differentiation between taxa.
It is true that in many island plant groups, extensive morphological and ecological divergence may occur with little or no divergence at allozyme loci (Crawford et al., 2006). However, as far as is known this decoupling has not been described for microsatellite markers, which are much more polymorphic, completely neutral and very suitable for the analysis of populations and closely related species (Oliveira et al., 2006).
In conclusion, it is considered that the individuals of M. rivas-martinezii are probably a morphotype of M. faya, the consequence of a morphological variation (Arafeh et al., 2002; Steiner et al., 2006). In addition, this could explain the high percentage of offspring (90 %) that showed the M. faya phenotype in the ex situ germination test carried out in M. rivas-martinezii. Therefore, the focus of current management actions for the conservation of this putatively endangered Canarian endemism should be revised.
ACKNOWLEDGEMENTS
The authors would like to thank Angel Fernández and Cito Chinea (Parque Nacional Garagonay), and Manuel Marrero and Eduardo Carqué (Parque Nacional Teide) for their assistance in collecting Myrica samples. English was revised by J. Sosa and A. Stephens. This research was supported by the Ministerio de Educación y Ciencia-Dirección General de Investigación (CGL2004-03839) of the Spanish Government. Financial support has gratefully been received from the MAEC-AECI Fellowship for the PhD studies of E.A.G.-G.
LITERATURE CITED
- Arafeh RMH, Sapir Y, Shmida A, Iraki N, Fragman O, Comes HP. Patterns of genetic and phenotypic variation in Iris haynei and I. atrofusca (Iris sect. Oncocyclus = the royal irises) along an ecogeographical gradient in Israel and the West Bank. Molecular Ecology. 2002;11:39–53. doi: 10.1046/j.0962-1083.2001.01417.x. [DOI] [PubMed] [Google Scholar]
- Bañares Á, Blanca G, Güemes J, Moreno JC, Ortiz S. Atlas y libro rojo de la flora vascular amenazada de España. Madrid: Dirección General de Conservación de la Naturaleza, Ministerio de Medio Ambiente; 2004. [Google Scholar]
- Batista FJ, Sosa PA. Estudio de la variabilidad genética de los géneros Echium, Ilex y Myrica por electroforesis isoenzimática. University of Las Palmas de Gran Canaria; 1998. Technical report. [Google Scholar]
- Batista F, Bouza N, González-Pérez MA, Sosa PA. High levels of genetic variation detected within and between populations of two endangered endemic species of the laurel forest from the Canary Islands, Myrica rivas-martinezii (Myricaceae) and Sideritis discolor (Lamiaceae) Australian Journal of Botany. 2004;52:471–480. [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4·04. Logiciel sous Windows pour la génétique des populations. Montpellier, France: Laboratoire Génome, Populations Interactions, Université de Montpellier II; 2003. [Google Scholar]
- Beltrán E, Wildpret W, León MC, García A, Reyes J. Libro rojo de la flora Canaria contenida en la Directiva-Hábitats Europea. La Laguna: Dirección General de Conservación de la Naturaleza, Ministerio de Medio Ambiente; 1999. [Google Scholar]
- BOC. Decreto 151, de 23 de julio, por el que se crea el catálogo de especies amenazadas de Canarias. Boletín Oficial de Canarias. 2001;97:11101–11111. [Google Scholar]
- Boys J, Cherry M, Dayanandan S. Microsatellite analysis reveals genetically distinct populations of red pine (Pinus resinosa, Pinaceae) American Journal of Botany. 2005;92:833–841. doi: 10.3732/ajb.92.5.833. [DOI] [PubMed] [Google Scholar]
- Corniquel B, Mercier I. Date palm (Phoenix dactylifera L.) cultivar identification by RFLP and RAPD. Plant Science. 1994;101:163–172. [Google Scholar]
- Crawford DJ, Archibald JK, Santos-Guerra A, Mont ME. Allozyme diversity within and divergence among species of Tolpis (Asteraceae-Lactuceae) in the Canary Islands: systematic, evolutionary and biogeographic implications. American Journal of Botany. 2006;93:656–664. doi: 10.3732/ajb.93.4.656. [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation, version II. Plant Molecular Biology Reporter. 1983;1:19–21. [Google Scholar]
- Dutech C, Sork VL, Irwin AJ, Smouse PE, Davis FW. Gene flow and fine-scale genetic structure in a wind-pollinated tree species, Quercus lobata (Fagaceaee) American Journal of Botany. 2005;92:252–261. doi: 10.3732/ajb.92.2.252. [DOI] [PubMed] [Google Scholar]
- Erickson DL, Hamrick JL, Kochert GD. Ecological determinants of genetic diversity in an expanding population of the shrub Myrica cerifera. Molecular Ecology. 2004;13:1655–1664. doi: 10.1111/j.1365-294X.2004.02139.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3·0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Francisco-Ortega J, Santos-Guerra A, Kim SC, Crawford DJ. Plant genetic diversity in the Canary Islands: a conservation perspective. American Journal of Botany. 2000;87:909–919. [PubMed] [Google Scholar]
- Frankham R, Ballou JD, Briscoe DA. Introduction to conservation genetics. Cambridge: Cambridge University Press; 2002. [Google Scholar]
- Friar EA, Crise-Sanders JM, McGlaughlin ME. Gene flow in Dubautia arborea and D. ciliolata: the roles of ecology and isolation by distance in maintaining species boundaries despite ongoing hybridization. Molecular Ecology. 2007;16:4028–4038. doi: 10.1111/j.1365-294X.2007.03423.x. [DOI] [PubMed] [Google Scholar]
- Gardner DE. Observations on some unusual flowering characteristics of Myrica faya. Newsletter of the Hawaiian Botanical Society. 1985;24:1417. [Google Scholar]
- González-Pérez MA, Caujapé-Castells J, Sosa PA. Allozyme variation and structure of the canarian endemic palm tree Phoenix canariensis (Arecaceae): implications for conservation. Heredity. 2004;93:307–315. doi: 10.1038/sj.hdy.6800507. [DOI] [PubMed] [Google Scholar]
- González-Pérez MA, Newton C, Sosa PA, Rivero E, González-González EA. Characterization of six microsatellite loci in the endangered endemic Myrica rivas-martinezii and the native M. faya (Myricaceae) Genetics and Molecular Biology. 2008 doi: 10.1590/S1415-47572009005000006. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. FSTAT (Version 1.2): a computer program to calculate F-statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- Hedrick PW. A standardized genetic differentiation measure. Evolution. 2005;59:1633–1638. [PubMed] [Google Scholar]
- IUCN. IUCN red list of threatened species. World Conservation Union. 2006. Available from http://www.iucnredlist.org .
- Jin L, Chakraborty R. Estimation of genetic distance and coefficient of gene diversity from single-probe multilocus DNA fingerprinting data. Molecular Biology and Evolution. 1993;11:120–127. doi: 10.1093/oxfordjournals.molbev.a040086. [DOI] [PubMed] [Google Scholar]
- Langella O. Populations (Logiciel de Genétique des Populations) 2000. CNRS. Available from http://www.legs.cnrs-gif.fr/ . Accessed 14 July 2008.
- Meirmans PG. Using the AMOVA framework to estimate a standardised genetic differentiation measure. Evolution. 2006;60:2399–2402. [PubMed] [Google Scholar]
- Morjan CL, Rieseberg LH. How species evolve collectively: implications of gene flow and selection for the spread of advantageous alleles. Molecular Ecology. 2004;13:1346–1356. doi: 10.1111/j.1365-294X.2004.02164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Genetic distance between populations. American Naturalist. 1972;106:283–292. [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva-Tejera F, Caujapé-Castells J, Navarro-Déniz J, Reyes-Betancort A, Scholz S, Baccarani-Rosas S, Cabrera-García N. Patterns of genetic divergence of three Canarian endemic Lotus (Fabaceae): implications for the conservation of the endangered. L. kunkelii. American Journal of Botany. 2006;93:1116–1124. doi: 10.3732/ajb.93.8.1116. [DOI] [PubMed] [Google Scholar]
- Oliveira EJ, Pádua JG, Zucchi MI, Vencovsky R, Vieira MLC. Origin, evolution and genome distribution of microsatellites. Genetics and Molecular Biology. 2006;29:294–307. [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piry S, Luikart G, Cornuet JM. BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. Journal of Heredity. 1999;90:502–503. [Google Scholar]
- Pritchard JK, Stephens M, Donnelli P. Inference of population structure from multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- Santos A. Contribución al conocimiento de la flora y vegetación de la Isla de Hierro (Islas Canarias) Fundación Juan March Serie Universitaria. 1980;114:1–50. [Google Scholar]
- Santos A. Vegetación y flora de La Palma. Santa Cruz de Tenerife, Spain: Interinsular Canaria; 1983. [Google Scholar]
- Schneider S, Kueffer JM, Roessli D, Excoffier L. Geneva: Genetics and Biometry Laboratory, University of Geneva; 2000. Arlequin ver. 2.000: A software for population genetic data analysis. [Google Scholar]
- Segarra-Moragues JG, Palop-Esteban M, González-Candelas F, Catalán P. On the verge of extinction: genetics of the critically endangered Iberian plant species, Borderea chouardii (Dioscoreaceae) and implications for conservation management. Molecular Ecology. 2005;14:969–982. doi: 10.1111/j.1365-294X.2005.02482.x. [DOI] [PubMed] [Google Scholar]
- Smith CW. Impact of alien plants in Hawaii's native biota. In: Stone SP, Scott JM, editors. Hawaii's terrestrial ecosystems: preservation and management. Honolulu: University of Hawaii's Cooperative National Park Studies Unit; 1985. pp. 180–250. [Google Scholar]
- Steiner FM, Schlick-Steiner BC, Konrad H, et al. No sympatric speciation here: multiple data sources show that the ant Myrmica microrubra is not a separate species but an alternate reproductive morph of Myrmica rubra. Journal of Evolutionary Biology. 2006;19:777–787. doi: 10.1111/j.1420-9101.2005.01053.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Victory E, Glaubitz J, Rhodes OE, Woeste K. Genetic homogeneity in Juglans nigra (Juglandaceae) at nuclear microsatellites. American Journal of Botany. 2006;93:118–126. [Google Scholar]
- VVAA. Lista roja de la flora vascular española (valoración según las categorías de la IUCN) Conservación Vegetal. 2000;6:11–38. [Google Scholar]
- Werner O, Ros RM, Fernández A. Caracterización genética de poblaciones de varias especies amenazadas en el Parque Nacional de Garajonay. Puerto de la Cruz, Spain: III Congreso de Biología de la Conservación de Plantas; 2007. [Google Scholar]
- Wright S. The genetical structure of populations. Annals of Eugenetics. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Yatabe Y, Kane NC, Scotti-Saintagne C, Rieseberg LH. Rampant gene exchange across a strong reproductive barrier between the annual sunflowers, Helianthus annuus and H. petiolaris. Genetics. 2007;175:1883–1993. doi: 10.1534/genetics.106.064469. [DOI] [PMC free article] [PubMed] [Google Scholar]