Abstract
Background and Aims
Bromeliads (Bromeliaceae) adapted to rock outcrops or ‘inselbergs’ in neotropical rain forests have been identified as suitable plant models for studying population divergence and speciation during continental plant radiations. Little is known about genetic relationships and variation in reproductive strategies within and among inselberg-adapted species, yet knowledge of these parameters is important for understanding divergence processes and for conservation planning.
Methods
Nuclear microsatellites were used to assess the role of clonal reproduction, estimate genetic diversity and explore genetic relationships and variation in reproductive strategies for a total of 15 populations of four closely related Alcantarea inselberg species in south-eastern Brazil: A. glaziouana, A. regina, A. geniculata and A. imperialis.
Key Results
Clonal propagation is frequent in coastal populations of A. glaziouana and A. regina, but absent in the high-altitude species A. geniculata and A. imperialis. Considerable variation in clonal diversity, gene diversity (He), allelic richness, and Wright's inbreeding coefficient (FIS) exists within and between species of Alcantarea. A Bayesian analysis of coastal inselberg species indicated pronounced genetic structure. A neighbor-joining analysis grouped populations of each species together with moderate bootstrap support, except for the high altitude species A. imperialis.
Conclusions
The coastal inselberg species A. glaziouana and A. regina tend to propagate asexually via vegetative clonal growth, and both reproductive strategies and breeding systems vary greatly between populations and species of Alcantarea. The microsatellite data indicate a history of hybridization and reticulation involving the high-altitude species A. geniculata and A. imperialis in areas of co-occurrence. The results highlight the need to understand similarities and differences in reproductive strategies both within and between related species for conservation planning and as a basis for understanding evolutionary processes in tropical radiations.
Key words: Alcantarea, Atlantic Rainforest, Bromeliaceae, clonality, gene flow, inselberg, microsatellites, reproductive strategy
INTRODUCTION
Integrating genetics into the ecological theory of adaptive radiation has become an important task for evolutionary biologists in recent years (Schluter, 2000), and research on rapidly radiating plant lineages has played a crucial role in working towards this goal (Givnish et al., 1997; Barrier et al., 2001; review by Seehausen, 2004). The proven usefulness of oceanic islands as model systems for studies of species' radiations (Darwin, 1859; MacArthur and Wilson, 1967; Emerson, 2002; Stuessy et al., 2006) provides concepts and tools that may also be applicable to evolutionary study of continental radiations (Porembski and Barthlott, 2000). Indeed, a growing number of studies explore plant species' evolution in archipelagos of ‘terrestrial islands’, e.g. in continental mountain ranges (Schönswetter et al., 2005; Hughes and Eastwood, 2006;) or other insular types of habitats (Givnish et al., 1997; DeChaine and Martin, 2005). So far, most population genetic or phylogeographic studies of this type have focused on inferring historical range expansions and contractions associated with paleoclimatic cycles (e.g. Dechaine and Martin, 2005; Schönswetter et al., 2005). Nevertheless, it is clear that terrestrial island archipelagos may also represent suitable models for studying ecological speciation during adaptive radiations, especially where populations are distributed across a range of different environments in an island-like manner (MacArthur and Wilson, 1967; Schluter, 2000).
Knowledge of natural variation in breeding systems and reproductive strategies within and between closely related species can help interpret the origin and maintenance of barriers to gene flow during speciation and species' radiations (Stebbins, 1957; Wendt et al., 2001, 2002; Coyne and Orr, 2004). The breeding system of a plant will determine how much variation is available to natural selection during bouts of ecological change and/or speciation (Seehausen, 2004). The breeding system will also have profound consequences for the build-up and maintenance of reproductive barriers during speciation (Stebbins, 1957), with obligate or facultative apomicts at one end of the continuum, and highly outcrossing, wind-pollinated species at the other. Two topics have aroused particularly great interest in the context of variation in breeding systems and reproductive strategies in recent years: the genetics of self-incompatibility (SI) in plants with mixed or outcrossing breeding systems, and the population dynamics of taxa with mixed sexual and asexual reproductive modes (Silvertown, 2008).
With respect to the population dynamics of mixed sexual/asexual taxa, new molecular and statistical tools to study clonal structure have led to a recent surge of studies on this topic (Arnaud-Haond et al., 2008). These studies have focused on the maintenance of genetic diversity in species with mixed sexual and asexual reproduction, the role of environmental effects and intraspecific competition in shaping clonal structure, and the role of asexuality in plant invasions (Chapman et al., 2004; Scheepens et al., 2007; recent reviews by Arnaud-Haond et al., 2008 and Silvertown, 2008). Despite the known importance of differences in breeding systems and reproductive strategies for population divergence and speciation alluded to above, few studies have addressed these topics for taxa that form part of well-described adaptive radiations (but see Whitkus et al., 2000).
Bromeliaceae is a well characterized example of an adaptive radiation in the neotropics (Benzing, 2000; Barfuss et al., 2005), comprising approx. 56 genera and 3000 species (Luther, 2004). Numerous adaptations and putative key innovations have been described for the family and have been the focus of much interest from botanists and evolutionary biologists, e.g. epiphytic and rupicolous growth habits (life on trees and bare rocks), the tank-habit (rosettes forming a tank-like structure able to hold large amounts of water), crassulacean acid metabolism (CAM), and several specialized strategies of nutrient uptake in nutrient-poor environments (Givnish et al., 1997; Benzing, 2000; Crayn et al., 2004). Systematic studies have been carried out on the family (e.g. Brown and Gilmartin, 1989; Givnish et al., 1997; Smith and Till, 1999) including a recent molecular phylogeny of Tillandsioideae (Barfuss et al., 2005), the subfamily that includes the species studied here. Population genetic studies are available only for a small number of bromeliads, e.g. for species of Tillandsia (Soltis et al., 1987; Gonzalez-Astorga et al., 2004), Aechmea (Murawski and Hamrick, 1990; Izquierdo and Pinero, 2000), Pitcairnia (Sarthou et al., 2001), Puya (Sgorbati et al., 2004) and Encholirium (Cavallari et al., 2006). Only one of these was focused on rock-adapted species, namely that on Pitcairnia geyskesii from rock outcrops in French Guiana (Sarthou et al., 2001).
Recently, we have begun to investigate within- and between-species patterns of genetic variability and gene flow in Vriesea and Alcantarea, two genera that have radiated into multiple species adapted to epiphytic life in continuous forest or rupicolous life on inselberg rock outcrops in the South American Atlantic Rainforest (Palma-Silva et al., 2007; Barbará et al., 2007). The 22 species of the genus Alcantarea are endemic to south-eastern Brazil, where they occur from 0–1900 m above see level on inselbergs of the Atlantic Rainforest or in Campo Rupestre (rocky field) vegetation of the Espinhaço mountain range (Versieux and Wanderley, 2007b). Alcantarea is phylogenetically and morphologicaly closely related to Vriesea (Grant, 1995; Barfuss et al., 2005) from which it is morphologically distinguished by its long spiralescent petals and seeds with both basal and apical comas (Grant, 1995). Ongoing research on Alcantarea has lead to the description of three new species (Versieux and Wanderley, 2007a, b) and has confirmed the identity of the four species included in the present study. Alcantarea regina is now thought to cover a wider range than previously suspected, as it was found to be synonymous with another Alcantarea species, A. edmundoi (Versieux and Wanderley, 2007c).
The inselbergs of south-eastern Brazil (from the German Insel = island and Berg = mountain) are ancient granitic rock outcrops embedded within a matrix of tropical forest (Safford and Martinelli, 2000) which exhibit much of the ‘insular’ nature alluded to at the beginning of this paper. Population genetic work on two high-altitude inselberg species in this group, Alcantarea imperialis and A. geniculata, found no evidence of isolation by distance, high levels of population differentiation, and extremely low amounts of historical gene flow (Nem < 1 for most pairs of populations), despite the predominantly outcrossing breeding systems of these animal-pollinated, wind-dispersed plants (Barbará et al., 2007). These findings are in agreement with the long-standing hypothesis that ‘inselbergs’ may resemble oceanic islands in their effects on patterns of variability and gene flow (Porembski and Barthlott, 2000).
Here, this earlier work is extended to two coastal inselberg species endemic to the same study area in the Brazilian Atlantic Rainforest, Alcantarea glaziouana and A. regina. To our knowledge, these two taxa have not previously been studied with any type of molecular genetic marker. Data are presented on patterns of genetic diversity and gene flow in coastal inselberg populations of these species, and we comprehensively analyse population and species relationships and compare variation in reproductive strategies across all four Alcantarea species studied to date, including the two high-altitude taxa studied previously, A. imperialis and A. geniculata. In particular, we ask the following questions. (1) How much variation in basic genetic parameters, such as clonal diversity, gene diversity (He), allelic richness and Wright's inbreeding coefficient (FIS), is there within and between these four Alcantarea inselberg species, and what are the mechanisms responsible for variation in reproductive strategies among populations and species? (2) Do populations of each species group together when analysed in a phylogenetic context, and what is the likely biological significance of departures from congruence between the genetic data and traditional taxonomic groupings? (3) How weak or strong are genetic structures in populations of the two coastal species, and what are the practical implications for conservation?
MATERIALS AND METHODS
Population sampling
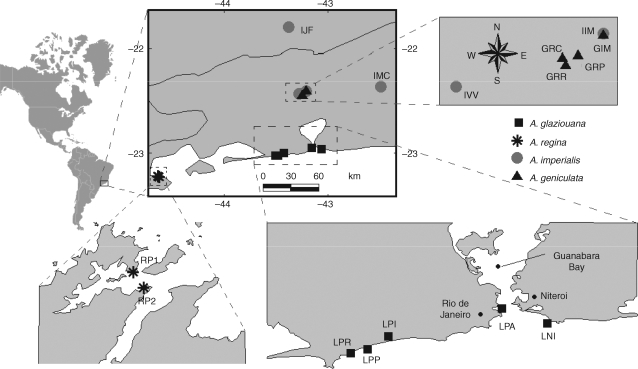
A total of 15 populations of Alcantarea glaziouana, A. regina, A. geniculata and A. imperialis were sampled on coastal (0–20 m elevation) and high-altitude (900–1300 m elevation) granitic inselbergs located in the Atlantic Rainforest of south-eastern Brazil (states of Rio de Janeiro and Minas Gerais; Fig. 1). Of these, data for seven coastal inselberg populations of A. glaziouana and A. regina are reported for the first time here. For A. glaziouana, the entire species range was sampled in this study. For A. regina, the only two previously described populations were included (Martinelli, 1994). Ongoing taxonomic revision indicates that the distribution range of A. regina is likely to be larger than previously assumed – another Alcantarea species has recently been found to be synonymous with A. regina (Versieux and Wanderley, 2007c). Nevertheless, the type specimen for A. regina is from exactly the same locality that was sampled in the present study (Parati, RJ, Brazil). Alcantarea imperialis and A. geniculata were sampled range-wide except for one locality per species, approx. 20 km west of population IJF in the case of A. imperialis and a few kilometres east of population GIM in the case of A. regina. The names, abbreviations and geographical co-ordinates of the sampled coastal inselberg populations are as follows: A. glaziouana: Glaziouana ‘Niterói’ or LNI (S22°58·617′, W43°2·825′), Glaziouana ‘Pão-de-Açúcar’ or LPA (S22°57·064′, W43°09·092′), Glaziouana ‘Pedra de Itaúna’ or LPI (S23°00·223′, W43°25·338′), Glaziouana ‘Pedra do Pontal’ or LPP (S23°2·105′, W43°28·247′) and ‘Glaziouana ‘Prainha’ or LPR (S23°2·376′, W043°30·258′); A. regina: Regina ‘Parati 1’ or RP1 (S23°13·369′, W44°37·616′) and Regina ‘Parati 2’ or RP2 (S23°14·071′, W44°37·208′) (Fig. 1). The geographic distances between populations of the narrow endemic A. glaziouana ranged from 3·5 to 47 km, whereas the two well-referenced coastal populations of A. regina were separated by 1·5 km. The sample sizes for all populations are given in Table 1. For each plant, leaf material for DNA extraction was collected and stored in silica gel.
Fig. 1.
Distribution map of Alcantarea inselberg populations sampled in the Atlantic Rainforest of Brazil. The present study is focused on the two coastal inselberg species seen at the bottom of the figure, A. glaziouana and A. regina. Data for the high-altitude inselberg species A. imperialis and A. geniculata were presented in detail elsewhere (Barbará et al., 2007) and are only used for comparative purposes here. For population abbreviations and details see Materials and Methods.
Table 1.
Characterization of microsatellite markers in ‘inselberg’ populations of Alcantarea glaziouana and A. regina, including marker source, repeat type, molecular size range in each species in basepairs (bp), and the following diversity and breeding system parameters calculated at the genet level (clones removed): number of alleles (A), expected (He) and observed (Ho) heterozygosity in each species, within-population inbreeding coefficient FIS and total-population inbreeding coefficient FIT
|
A. glaziouana |
A. regina |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Repeat type | Size range (bp) | A | He | Ho | FIS | FIT | Size range (bp) | A | He | Ho | FIS | FIT |
| Ai4·101 | di- | 186–189 | 3 | 0·364 | 0·257 | +0·229* | +0·312 | 184–188 | 3 | 0·633 | 0·705 | −0·149 | −0·080 |
| E192 | di- | 111–119 | 4 | 0·273 | 0·102 | +0·562**** | +0·644 | 111–123 | 4 | 0·244 | 0·227 | +0·040 | +0·096 |
| E62 | tri- | 106–166 | 13 | 0·820 | 0·419 | +0·362**** | +0·520 | 108 | 1 | – | – | – | – |
| Ai4·31 | di- | 185–191 | 2 | 0·019 | 0 | +1·00**** | +1·00 | 191–195 | 3 | 0·317 | 0·341 | −0·104 | −0·052 |
| CT52 | di- | 161–199 | 17 | 0·871 | 0·644 | +0·151*** | +0·290 | 165–189 | 10 | 0·887 | 0·727 | +0·118 | +0·237 |
| E6b2 | tri- | 131–143 | 3 | 0·364 | 0·308 | +0·060 | +0·181 | 131–143 | 4 | 0·715 | 0·682 | −0·193 | +0·209 |
| P2p192 | tri- | 194–205 | 4 | 0·548 | 0·381 | +0·147 | +0·343 | 199–214 | 5 | 0·758 | 0·682 | +0·007 | +0·180 |
| Pit83 | di- | 282–305 | 4 | 0·521 | 0·281 | +0·230* | +0·510 | 268–308 | 5 | 0·632 | 0·512 | −0·027 | +0·340 |
Microsatellite markers: 1isolated from A. imperialis in the Jodrell Laboratory at RBG Kew (Palma-Silva et al. 2007); 2isolated by Boneh et al. (2003); 3isolated by Sarthou et al. (2003).
Departures of within-population inbreeding coefficients (FIS) from HWE are indicated as follows: * P < 0·05, *** P < 0·005, **** P < 0·001.
Eight populations of the high-altitude inselberg species A. geniculata and A. imperialis described previously (Barbará et al., 2007) were included here to facilitate comparisons among high-altitude and coastal species (Fig. 1). Their names and abbreviations are as follows: Imperialis ‘Irmã Menor’ or IIM, Imperialis ‘Macaé-de-Cima’ or IMC, Imperialis ‘Juíz-de-Fora’ or IJF and Imperialis ‘Vale das Videiras’ or IVV; Geniculata ‘Irmã Menor’ or GIM, Geniculata ‘Ricardo's Clearing’ or GRC, Geniculata ‘Ricardo's Rock’ or GRR and Geniculata ‘Reserva Privada’ or GRP. In addition, two populations of Vriesea gigantea from the Atlantic Rainforest of Brazil (VG1, Florianópolis, Brazil; VG2, Santa Leopoldina, Brazil) were used for rooting the population tree of Alcantarea species.
Molecular markers and genotyping assays
The eight microsatellite markers used in this study (Table 2) were isolated from Alcantarea imperialis (Palma-Silva et al., 2007) or developed for the related bromeliad genera Tillandsia, Guzmania (Boneh et al., 2003) and Pitcairnia (Sarthou et al., 2003). Repeat types and molecular size ranges for all eight markers in the coastal inselberg species A. glaziouana and A. regina are given in Table 2. We chose to use the same set of markers also used for our recent study of the high-altitude species A. imperialis and A. geniculata (Barbará et al., 2007). This was done to facilitate direct comparisons among coastal and high-altitude populations and species and to be able to combine the datasets for construction of a phylogenetic tree of all populations.
Table 2.
Characterization of coastal ‘inselberg’ populations of Alcantarea glaziouana and A. regina with eight nuclear microsatellite markers, including sample size, genotypic diversity (G/N) and Simpson's index to document clonal diversity, and the following diversity and breeding system parameters calculated at the genet level: variance in allele size (Var), allelic richness, expected (He) and observed (Ho) heterozygosity, and within-population inbreeding coefficients FIS
| Species | Population | n | G/N | Simpson's D | Var | Allelic richness | He | Ho | FIS |
|---|---|---|---|---|---|---|---|---|---|
| A. glaziouana | LPR | 11 | 0·22 | 0·6 | 0·36 | 1·21 | 0·085 | 0·094 | −0·125 |
| LPP | 68 | 0·42 | 0·94 | 22·2 | 2·23 | 0·440 | 0·404 | +0·083* | |
| LPI | 29 | 0·81 | 0·98 | 9·52 | 1·66 | 0·221 | 0·178 | +0·197* | |
| LPA | 31 | 0·83 | 0·97 | 38·2 | 2·62 | 0·557 | 0·338 | +0·397*** | |
| LNI | 31 | 0·42 | 0·77 | 12·4 | 2·07 | 0·365 | 0·283 | +0·231* | |
| Overall: | 170 | 0·55 | 0·98 | 23·53 | 6·13 | 0·472 | 0·299 | – | |
| A. regina | RP1 | 26 | 1 | 1 | 26·2 | 3·514 | 0·505 | 0·505 | +0·001 |
| RP2 | 29 | 0·59 | 0·96 | 15·1 | 3·478 | 0·412 | 0·453 | −0·104 | |
| Overall: | 55 | 0·79 | 0·99 | 25·7 | 4·369 | 0·523 | 0·484 | – | |
| A. imperialis1 | Overall: | 124 | 1 | 1 | 27·96 | 6·250 | 0·615 | 0·362 | – |
| A. geniculata1 | Overall: | 84 | 1 | 1 | 23·84 | 5·250 | 0·429 | 0·357 | – |
1 Overall values for A. imperialis and A. geniculata from Barbará et al. (2007) are shown for comparative purposes.
Departures from Hardy–Weinberg equilibrium are indicated as follows: * P < 0·05, *** P < 0·005.
For molecular genotyping, total genomic DNA was extracted from silica-gel-dried leaves using a modified approach based on Doyle and Doyle (1987), and DNA was quantified using an Eppendorf BIO photometer. The eight nuclear microsatellites were amplified using polymerase chain reaction (PCR) as in Palma-Silva et al. (2007), making use of a standard touchdown cycling program with an annealing temperature (Ta) of 48°C and either FAM or JOE labelled forward primers, or a three-primer protocol including unlabelled M13-tagged forward and unlabelled/untagged reverse primers for each marker and a third ‘universal’ M13-primer labelled with one of the fluorescent dyes, FAM or JOE (Applied Biosystems, Foster City, CA, USA). Microsatellite genotypes were resolved on an ABI PRISM® 3100 Genetic Analyzer (Applied Biosystems), making use of the different fluorescent dyes for duplexing. Molecular sizes in base pairs were determined using the GENESCAN-500 ROX size standard (Applied Biosystems), and result files from the sequencers were analysed using GENESCAN and GENOTYPER software (Applied Biosystems).
Data analysis
Detection of clones and identification of genets for subsequent analysis
Because clonal reproduction had previously been suspected in coastal Alcantarea inselberg populations (Martinelli, 1994), we checked for the presence of identical genotypes in all populations of A. regina and A. glaziouana. Individuals with identical genotypes were identified using a simple neighbor-joining (NJ) tree in Phylip (Felsenstein, 2004) based on the proportion of shared alleles between individuals (Bowcock et al., 1994) and later confirmed using the GENCLONE software (Arnaud-Haond and Belkhir, 2007). In order to explore the power of our marker loci for detecting clones we calculated the probability of identity, PID (Waits et al., 2001), for each locus in A. regina and A. glaziouana using the GIMLET software (Valière, 2002) and the probability of clonal identity, Psex, using GENCLONE (Arnaud-Haond and Belkhir, 2007). In addition, all samples with identical genotypes were checked by eye for their geographic location using maps produced by GIS software and GPS co-ordinates of sampled specimens. This was done to check whether identical genotypes aggregated in patches as expected for vegetatively derived clonal copies of a genet. The detection of clones allowed us to focus all subsequent analyses at the genet level.
Genetic diversity of the sampled loci and populations
In order to characterize the microsatellite loci in coastal inselberg populations of A. glaziouana and A. regina, the number of alleles (A), expected heterozygosity (He), observed heterozygosity (Ho), within-population inbreeding coefficient (FIS) and total population inbreeding coefficient (FIT) were calculated at the genet level (clones removed) for each locus using the computer programs MSA (Dieringer and Schlötterer, 2003) and FSTAT (Goudet, 1995). In addition, within each population, each locus was tested for departure from Hardy–Weinberg equilibrium (HWE) using FSTAT. This was done to explore whether departures from HWE are due to locus-specific effects or rather due to systematic factors, i.e. deviations from random mating of the sampled genets. We note that the absence of null-alleles for these loci in Alcantarea spp. has been established previously (Barbará et al., 2007; Palma-Silva et al., 2007).
Subsequently, each coastal inselberg population was characterized using clonal diversity (G/N) and Simpson's diversity index calculated with Genclone (Arnaud-Haond and Belkhir, 2007) and the following diversity parameters at the genet level (clones removed): variance in allele size (Var), He and Ho calculated by MSA and allelic richness in FSTAT (Goudet, 1995). All of these genetic diversity parameters were corrected for sample size in MSA and FSTAT. Departures from HWE for each population at the genet level were identified using exact tests in GENEPOP (Raymond and Rousset, 1995).
Tests of basic assumptions for interpreting F-statistics in terms of historical gene flow
In order to determine if the sampled coastal inselberg populations are likely to meet the equilibrium conditions required for the interpretation of F-statistics in terms of historical gene flow, the possibility of founder effects due to recent colonization (bottlenecks) was tested using the ‘sign test’ and ‘Wilcoxon sign-rank’ test in the BOTTLENECK program (Piry et al., 1999). Both tests are able to detect recent reductions in effective population size due to genetic bottlenecks. The analyses were carried out both for the ‘infinite allele model’ (IAM) and for the ‘two-phased mutation model’ (TPM) recommended for microsatellites in the user manual. The detection of recent bottlenecks may indicate that Alcantarea inselberg populations are not in an equilibrium between gene flow and genetic drift, which would render the indirect estimation of gene flow via FST difficult (Whitlock, 1992). In many plant species, equilibrium between migration and drift can also be tested by studying correlations between genetic and geographic distance (‘isolation by distance’; Slatkin, 1993). This test is not suitable for the species studied here, since isolation by distance is not necessarily expected for species with island-like distribution patterns, including species on inselbergs (Barbará et al., 2007). The absence of isolation by distance in the coastal inselberg species A. glaziouana was established using non-parametric Mantel tests in FSTAT with 10 000 randomizations. This test could not be carried out for A. regina as only two populations were available.
Indirect analysis of gene flow via F-statistics and population phylogeny
Analysis of molecular variance (AMOVA) in GENALEX (Peakall and Smouse, 2006) was used to obtain F-statistics for microsatellites at different hierarchical levels using the genet-level data. We tested the hierarchies ‘among species’, ‘among populations within species’ and ‘within populations’ for the entire dataset. Subsequently, separate AMOVA models were analysed to test the distribution of genetic variance among and within populations of each species. The significance of each F-statistic was tested through 9999 permutations in GENALEX at the appropriate hierarchical level. In addition, FST was also estimated for pairs of coastal Alcantarea inselberg populations and gene flow (Nem) between pairs of populations was estimated from FST as (1/FST –1) × 0·25 under the assumption of migration-drift equilibrium.
In order to depict relationships between populations and species in a graphical way, a neighbor-joining (NJ) tree was constructed based on Nei's (1987) genetic distance. One thousand bootstrap replicates of the distance matrix were obtained in MSA, and NJ trees were generated and analysed in PHYLIP 3·6 (Felsenstein, 2004). To gain a more complete picture of genetic relationships among Alcantarea inselberg populations and species, eight previously described (Barbará et al., 2007) populations of the high-altitude species A. imperialis and A. geniculata were added to the dataset. In addition, two populations of Vriesea gigantea were included for comparative purposes.
Bayesian genetic structure analysis
Bayesian analysis in STRUCTURE v2 (Pritchard et al., 2000) was used to obtain insights into patterns of gene flow and population subdivision within coastal Alcantarea inselberg populations. Our aim was to determine the most likely number of populations (K) for each species, and to estimate admixture proportions (Q) for individuals of each population. A ‘burn-in’ period of 5 × 104 iterations and data collection for 106 iterations were identified as being appropriate based on the diagnostic tools available in STRUCTURE. The analyses were carried out under the admixture model for independent allele frequencies. All possible models from K = 1 to K = 10 were evaluated for A. glaziouana and all models from K = 1 to K = 7 for A. regina, based on the natural logarithm of their probability and on their variances. Individual admixture proportions (Q) for each sampled population in each genetic cluster found by STRUCTURE were recorded for the model that best explained the data. The STRUCTURE results were subsequently confirmed by analysis with BAPS (Corander et al., 2004) without the use of baseline information for population origin. In contrast to STRUCTURE, BAPS estimates K and Q separately, which may increase precision of parameter estimation in some situations. One hundred iterations, 200 reference individuals and 20 iterations for reference individuals were used in the admixture analysis, following the software manual.
RESULTS
Information content of microsatellite loci in coastal Alcantarea inselberg populations
All eight microsatellite loci were polymorphic in coastal Alcantarea inselberg populations, with up to 17 alleles per locus in A. glaziouana and up to ten alleles per locus in A. regina (Table 2). One locus, E6, was fixed for a single allele in A. regina but was polymorphic in A. glaziouana. The combined probability of identity (PID) for the eight microsatellite loci was 8·44 × 10−6 for A. regina and 2·32 × 10−5 for A. glaziouana, thus indicating a low probability that two unrelated individuals drawn from each of these two species' gene pools will share the same multi-locus genotype. Likewise, the probability of a particular genotype being present more than once as a result of sexual reproduction (as opposed to being part of the same genetic individual), Psex, was on average 2·10 × 10−3 for A. glaziouana and 6·0 × 10−4 for A. regina. Based on these values it was considered appropriate to use the microsatellite genotype data for the identification of clones for subsequent analysis.
Clonal reproduction, genetic diversity, and variation in reproductive strategies in coastal Alcantarea inselberg populations
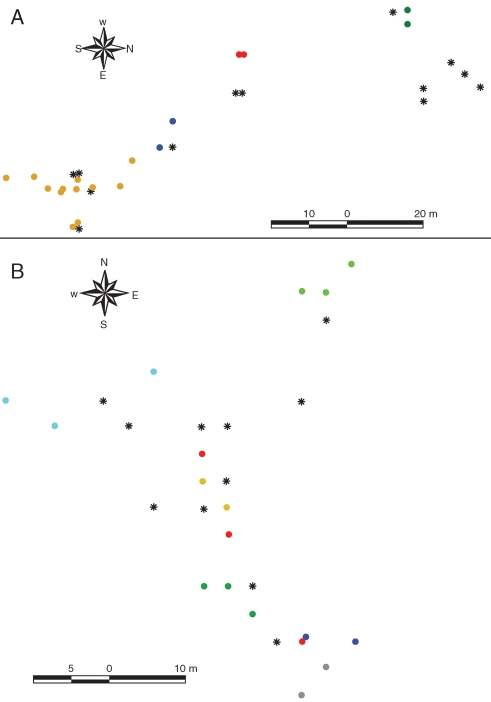
Inspection of microsatellite genotypes using individual genetic distances and GENCLONE revealed the presence of clones in all populations of A. glaziouana and in one population of A. regina, resulting in values for clonal diversity (G/N) and Simpson's diversity lower than 1 for these populations (Table 1). Individual genets comprised up to 12 ramets in A. glaziouana and up to three ramets in A. regina. Graphical inspection of geo-referenced, mapped individuals showed that ramets (clonal copies) were always spatially clustered, as expected if identical genotypes are due to vegetative clonal reproduction (Fig. 2). The largest distance observed between plants belonging to the same genet (= the same clone) was 9 m (rosettes of individual ramets typically have a diameter of up to 1 m). A detailed fine-scale spatial genetic structure analysis of all Alcantarea inselberg populations was presented previously (Barbará et al., 2008). Here, identification of clones allowed us to assess variation in reproductive strategies and to focus our analysis of genetic diversity and structure at the genet level.
Fig. 2.
Spatial distribution of clones (genets) in two coastal inselberg populations of Alcantarea: (A) population LNI of A. glaziouana; (B) population RP2 of A. regina. In each graph, colours indicate different clones as identified with eight nuclear microsatellite markers, and stars indicate plants that had unique genotypes for these markers. The two graphs exemplify the spatial clustering of plants belonging to the same clone in populations of these species, which supports the hypothesis of asexual reproduction via vegetative clonal growth rather than agamospermy.
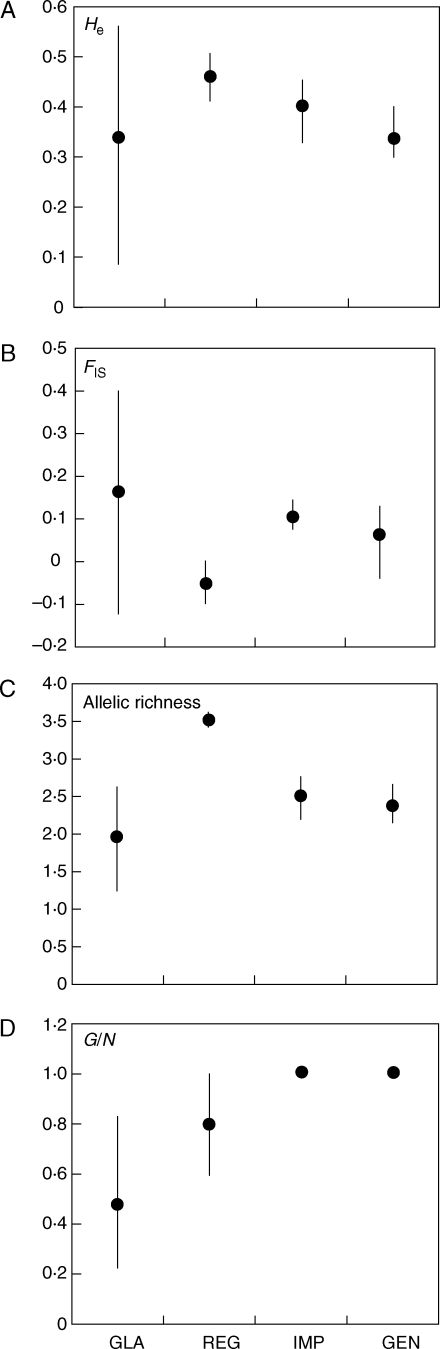
Genetic diversity evaluated over all loci at the genet level was generally higher in populations of A. regina than in A. glaziouana when estimated via the variance in allele size (Var), expected (He) or observed heterozygosity (Ho; Table 1), and only allelic richness was higher in A. glaziouana. The data facilitate a graphical comparison of genetic diversity (He, allelic richness, clonal diversity) and inbreeding coefficients (FIS) among populations of all four Alcantarea species studied to date, A. glaziouana, A. regina, A. geniculata and A. imperialis (Fig. 3), which allows us to discuss variation in breeding systems and reproductive strategies.
Fig. 3.
Comparative analysis of genetic diversity and breeding system parameters in four Alcantarea inselberg species, A. glaziouana, A. regina, A. imperialis and A. geniculata. (A) Expected heterozygosity (He); (B) within-population inbreeding coefficient (FIS); (C) allelic richness, (D) genotypic diversity (G/N). Data are the averages and ranges of genetic parameter estimates across populations of each species. The increased range of FIS, He, and allelic richness across populations of A. glaziouana, and the total absence of clonal structure in A. imperialis and A. geniculata, are clearly visible.
Tests for departure from Hardy-Weinberg equilibrium (HWE) at the genet level indicated significant heterozygote deficits (= significantly positive FIS) for most loci and populations in A. glaziouana, whereas in A. regina no significant departure from HWE was found (Tables 1 and 2). Total-population inbreeding coefficients FIT, included for completeness in Table 2, were positive for all loci in A. glaziouana and for most loci in A. regina, indicating species-level homozygote excess due to population subdivision.
Neither the sign test nor the Wilcoxon sign-rank test for recent population bottlenecks was significant for any of the populations, regardless of the mutation model used (data not shown), which allows us to interpret population differentiation (FST) in terms of inter-population gene flow. The Mantel correlation between geographic and genetic distance was not significant (data not shown), thus suggesting the absence of isolation by distance among inselberg populations of A. glaziouana, as previously observed for high-altitude inselberg species of this genus (Barbará et al., 2007).
Genetic divergence and relationships among inselberg populations and species of Alcantarea
Analysis of molecular variance (AMOVA) of coastal inselberg species attributed a significant proportion of the genetic variance (18 %) to the ‘among population within species’ level, and a similar and significant proportion to the ‘among species’ (16 %) level, whereas most of the variance resided within populations (67 %, Table 3; all P-values < 0·001). Separate AMOVA models for each species revealed that similar proportions of the genetic variance resided among populations in A. regina (20 %) and A. glaziouana (22 %) and a high and significant proportion was observed within populations of each species (80 % and 78 %, respectively; Table 3). Individual FST estimates between pairs of populations of A. glaziouana ranged from 0·116 to 0·315 (Table 4; all P-values < 0·001). The pairwise FST values translate into estimates of gene flow (Nem) ranging from 0·54 to 1·91 for coastal inselberg populations of A. glaziouana and Nem = 1·03 for the pair of coastal populations sampled for A. regina, respectively (Table 4).
Table 3.
Results of analysis of molecular variance (AMOVA) for the coastal inselberg species A. glaziouana and A. regina calculated at the genet level using three different hierarchical models: a three-level model including both species, and separate two-level models for each species
| Model | Partitioning | d.f. | SS | MS | Variation (%) | F-statistic |
|---|---|---|---|---|---|---|
| Three levels – two species | Among species | 1 | 70·719 | 70·719 | 16 | FRT = 0·158**** |
| Among populations within species | 5 | 96·580 | 19·316 | 18 | FSR = 0·210**** | |
| Within populations | 291 | 480·054 | 1·650 | 67 | FST = 0·334**** | |
| Two levels – A. glaziouana | Among populations | 4 | 75·395 | 18·849 | 22 | FST = 0·217**** |
| Within populations | 205 | 319·398 | 1·558 | 78 | ||
| Two levels – A. regina | Among populations | 1 | 21·185 | 21·185 | 20 | FST = 0·195**** |
| Within populations | 86 | 160·657 | 1·868 | 80 |
The significance of each F-statistic was tested through 9999 permutations at the appropriate hierarchical level. **** P < 0·001.
Table 4.
Genetic divergence (FST; below diagonal) and gene flow (Nem;above diagonal) for pairs of populations of the coastal inselberg species Alcantarea glaziouana, estimated at the genet level
| LPI | LPP | LNI | LPR | LPA | |
|---|---|---|---|---|---|
| LPI | – | 1·03 | 0·59 | 0·54 | 0·63 |
| LPP | 0·196 | – | 0·92 | 0·69 | 1·91 |
| LNI | 0·299 | 0·214 | – | 0·59 | 1·10 |
| LPR | 0·315 | 0·266 | 0·296 | – | 0·600 |
| LPA | 0·285 | 0·116 | 0·185 | 0·294 | – |
Gene flow was estimated from FST under the assumption of migration-drift equilibrium. Pairwise FST and Nem for the only pair of coastal populations sampled for A. regina were 0·195 and 1·03, respectively. All FST values were significant at the 0·001 level.
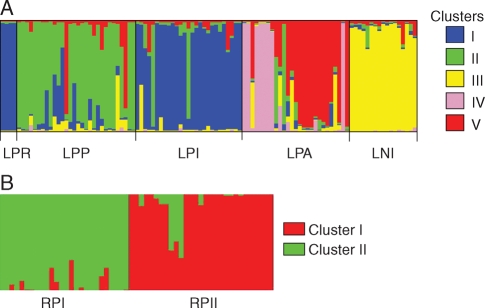
Bayesian genetic structure analysis confirmed clear population structure in both coastal species and provided additional insights into patterns of inter-population gene flow in A. glaziouana. Analysis in BAPS and STRUCTURE yielded comparable results, and the latter are presented here. A K = 5 population model was chosen to represent the data for A. glaziouana and a K = 2 model was chosen for A. regina, based on the pointers provided in the software manual (probabilities and variances of each model). For A. glaziouana, the Bayesian analysis indicated occasional gene exchange events beyond neighbouring populations (Fig. 4). Population LNI (Niteroi; yellow cluster) will be given particular attention in the discussion as it provides information on the potential of Guanabara bay, the bay that separates LNI from all other populations, to act as a barrier to gene flow (Fig. 1, bottom right).
Fig. 4.
Bayesian admixture proportions (Q) of individual plants of (A) A. glaziouana for a K = 5 population model, and (B) A. regina for a K = 2 population model, calculated at the genet level. The five and two ‘genetic clusters’ identified by STRUCTURE for A. glaziouana and A. regina, respectively, are indicated by different colors. For details of population abbreviations see Materials and Methods.
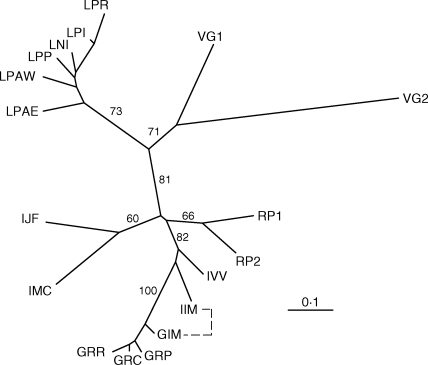
A phylogenetic tree of populations based on microsatellite genetic distances (Fig. 5) revealed relationships among inselberg populations of A. glaziouana, A. regina, A. geniculata and A. imperialis and two populations of Vriesea gigantea included for comparative purposes. Populations of all Alcantarea species except for A. imperialis were grouped together with moderate-to-high bootstrap support (100 for A. geniculata, 73 for A. glaziouana and 66 for A. regina, respectively). Populations of Alcantarea imperialis did not cluster together, with populations IIM and IVV being more closely related to A. geniculata than to their remaining conspecific populations IMC and IJF (Fig. 5). Populations IIM of A. imperialis and GIM of A. geniculata, connected by a dotted line in Fig. 5, co-occur on the same inselberg and were previously suspected to have experienced low levels of historical interspecific gene flow based on coalescent-based estimates of migration rates (Barbará et al., 2007).
Fig. 5.
Genetic distance-based neighbor-joining tree of populations of four Alcantarea inselberg species, A. glaziouana, A. regina, A. imperialis and A. geniculata, calculated at the genet level and including two populations of the closely related taxon Vriesea gigantea for comparative purposes. Bootstrap support values >60 % are shown for nodes of interest to the discussion of the data. Populations starting with V = Vriesea gigantea; starting with I = A. imperialis; starting with G = A. geniculata; starting with R = A. regina; starting with L = A. glaziouana. For details of population abbreviations see Materials and Methods. The two populations connected by a dashed line occur in sympatry on the same inselberg and were suspected to have experienced historical interspecific gene flow at the outset of the study (Barbará et al., 2007).
DISCUSSION
Variation in clonal diversity within and among Alcantarea inselberg species
The molecular marker-based estimates of clonal diversity, genetic variability and inbreeding provide a first glimpse at variation in reproductive strategies within and among four closely related Alcantarea bromeliad species endemic to ‘inselberg’ rock outcrops in the Atlantic Rainforest of Brazil. The results indicate great differences in clonal diversity between these four closely related inselberg species, with 100 % sexual reproduction in the high-altitude species A. imperialis and A. geniculata, resulting in clonal diversities equal to 1, and a considerable proportion of clonally derived rosettes in the two coastal species A. glaziouana and A. regina, resulting in clonal diversities lower than 1 (Figs 2 and 3D). As expected from the literature (see review by Ellstrand and Roose, 1987), increased levels of cloning, i.e. decreased clonal diversities, were not necessarily reflected by decreased genetic variability (Fig. 3, Table 1).
Other population genetic studies of bromeliads have also detected clonality, including studies on the rock-outcrop species Pitcairnia geyskesii (Sarthou et al., 2001), the terrestrial taxa Aechmea magdalenae (Murawski and Hamrick, 1990), Aechmea tuitensis (Izquierdo and Pinero, 2000) and Puya raimondii (Sgorbati et al., 2004), and the epiphyte Guzmania monostachia (Cascante-Martin, 2006). The degree of clonality detected by these studies varied greatly and was clearly highest in Puya raimondii, the ‘Queen of the Andes’ (Sgorbati et al., 2004). These authors reported a clonal diversity of only 0·09, based on a genetic analysis with amplified fragment length polymorphisms (AFLP). Indeed, studies reporting exclusive or predominant sexual reproduction in bromeliads, as exemplified by species of Encholirium (Cavallari et al., 2006), appear to be the exception rather than the rule. Clonal reproduction is thus an important reproductive strategy in bromeliads with different geographic distributions, habitats and life histories.
A limiting aspect of previous studies on clonality in bromeliads is that they used genetic markers and spatial sampling strategies that differed in their ability to detect clones. In addition, some studies deliberately avoided sampling clones as they were designed to address other questions (Soltis et al., 1987; Gonzalez-Astorga et al., 2004). Considering the increasing interest in population divergence, micro-evolution and speciation in bromeliads (Soltis et al., 1987; Murawski and Hamrick, 1990; Sarthou et al., 2001; Wendt et al., 2001, 2002; Barbará et al., 2007; Palma-Silva et al., 2007), more studies such as ours are needed that estimate clonal diversity and structure with highly informative molecular markers and that apply the same markers and sampling strategies to several closely related species in a comparative way.
It is of great botanical interest to know whether or not clonal reproduction in a given species may be due to agamospermy (apomixis; Richards, 2003). In bromeliads, controlled crossing experiments including experimental manipulations of a large number of flowers and plants suggested that agamospermy occurs in species of the genus Pitcairnia that live in the same type of habitat (inselberg rock outcrops) and in the same study area (south-eastern Brazil) as the species studied here. Agamospermy, in combination with high levels of selfing, may contribute to the maintenance of species-barriers in sympatric Pitcairnia species in the absence of temporal, geographic and post-pollination barriers in this group (Wendt et al., 2001, 2002). It is thus of relevance to test for the possibility of agamospermy in the inselberg Alcantarea species studied here, and our data allow us to do so.
In light-seeded, wind-dispersed bromeliads such as Alcantarea spp., agamospermy would be expected to result in the spread of clonally propagated seeds within populations (Richards, 2003). In contrast, asexual reproduction via vegetative clonal growth would result in spatial aggregation of clones. In the two coastal Alcantarea species studied here, ramets of the same clone were always spatially clustered, and neighbouring rosettes of the same genotype were never separated by more than 9 m (examples shown in Fig. 2). This is close to the maximum distance of 10 m observed between neighbouring vegetatively derived rosettes of Aechmea magdalene, a bromeliad with asexual propagation through vegetative clonal growth (Murawski and Hamrick, 1990). Thus, we tentatively conclude that asexual reproduction in coastal Alcantarea inselberg species is due to clonal growth rather than agamospermy. Crossing experiments of the type carried out by Wendt et al. (2001, 2002) would provide further information about the presence or absence of agamospermy in coastal Alcantarea species.
Variation in inbreeding coefficients within and among Alcantarea inselberg species
Estimates of clonal diversity and within-population inbreeding coefficients (FIS) estimated at the genet level (after the removal of clonal copies) differed between populations of Alcantarea species (Table 1, Fig. 3B). Alcantarea glaziouana was the species with the highest inbreeding coefficients and the lowest clonal diversities (Fig. 3), suggesting that positive FIS values may stem in part from reduced mate availability (i.e. biparental inbreeding) due to clonal growth (Honnay and Jacquemyn, 2008). Nevertheless, populations with a propensity to form clones were not always those with high inbreeding coefficients (Table 1), and thus we may assume that differences in FIS at the genet level capture other factors in addition to clonal propagation.
The coastal species A. glaziouana clearly displays the genetic signature of a primarily inbreeding species, with consistently positive inbreeding coefficients for all loci (Table 2) and most populations (Table 1), in contrast to the remaining three Alcantarea species studied (Fig. 2B). There also appears to be considerable variation in the strength of inbreeding among populations of A. glaziouana (Fig. 3B). Some – but certainly not all – of this intraspecific variation is due to population LPR for which FIS was neither significant nor positive (Table 1). This population is very small (almost all extant plants were sampled), and is thus subject to stochastic factors and offers low power for detecting inbreeding. Nevertheless, even if the ranges for FIS in Fig. 3 are corrected for the low inbreeding coefficient of population LPR, the range of inbreeding estimates for A. glaziouana is still greater than those of all other species studied (data not shown). Thus, there appears to be considerable variation in inbreeding levels not only between different Alcantarea species, but also within the coastal species A. glaziouana.
The evolution of breeding systems in plants with mixed mating has received great interest in recent years (Charlesworth and Charlesworth, 1987; Stephenson et al., 2000; Vogler and Kalisz, 2001). In bromeliads, the question has been raised as to whether evolution towards selfing may sometimes be driven by the need to avoid the fitness costs associated with hybridization between related species with incomplete pre- or postzygotic barriers (Wendt et al., 2001, 2002). If so, then an important question is to what extent differences in selfing rates are controlled by heritable factors versus ecological factors such as pollinator or resource limitation in particular localities (Wendt et al., 2002; Paggi et al., 2007). It is noteworthy in this context that two of the species studied here, A. imperialis and A. regina, have been the subject of controlled pollination experiments and observations of pollen tube growth (Martinelli, 1994). For A. imperialis, these experiments showed that ovule penetration was much higher after cross-pollination (range 52–75 %) than after self-pollination (range 0–29 %), suggesting partial self-incompatibility. Although no such difference in ovule penetration was observed for A. regina, marked protandry was observed in this species (Martinelli, 1994). Both observations are congruent with our population genetic data, since both A. imperialis and A. regina present the genetic signatures of near-random mating with low or non-significant inbreeding coefficients (Barbará et al., 2007; Fig. 3, Tables 1 and 2). No information is currently available on the timing of flowering or ovule penetration for the other two species studied, A. glaziouana and A. geniculata. Clearly, more work is needed to assess the role of variation in reproductive strategies and mating systems on the evolution and maintenance of reproductive barriers in these species.
Genetic relationships and gene flow among Alcantarea inselberg populations
Our study detected great genetic divergence and low levels of gene flow among inselberg populations of the coastal species A. glaziouana and A. regina, as also observed in our earlier analysis of the high-altitude species A. imperialis and A. geniculata (Barbará et al., 2007). This is reflected in a high and significant proportion of among-inselberg variation in our AMOVA analysis of coastal species (Table 3; 22 % and 20 % for A. glaziouana and A. regina, respectively). Indeed, gene flow (Nem) was lower than 1 for most pairs of populations of A. glaziouana and close to 1 migrant per generation for the pair of populations studied for A. regina (Table 4). In addition, departure from an isolation-by-distance model of migration was established for A. glaziouana, as shown previously for the two high-altitude species A. imperialis and A. geniculata. Thus, as alluded to in our earlier study on high-altitude species (Barbará et al., 2007), neotropical inselbergs indeed appear to resemble archipelagos of ‘terrestrial islands’ in terms of their effect on restriction of gene flow and structuring of genetic variability, as predicted by ecologists (Porembski and Barthlott, 2000). This opens the way to address the genetic underpinnings of adaptive radiation in bromeliads based on the well-developed conceptual framework for studying radiations on islands (MacArthur and Wilson, 1967; Emerson, 2002), in order to address the origin and evolution of bromeliad species adapted to the very different ecological conditions prevailing on coastal versus high-altitude inselbergs of south-eastern Brazil (Safford and Martinelli, 2000). In particular, the insular nature of these species facilitates the identification of independent ‘replicate’ populations for studies of population divergence and speciation. This has been discussed in more detail elsewhere (Barbará et al., 2007), and therefore the remainder of this discussion will focus on aspects that are specific to coastal inselberg species of Alcantarea, and on genetic relationships among populations of coastal and high-altitude species.
The Bayesian genetic analysis confirms pronounced genetic structure in coastal inselberg species, and it also reveals additional fine-scale spatial genetic patterns (Fig. 4). For instance, population subdivision becomes apparent in population LPA (clusters IV and V), whereas populations LPR and LPI are both dominated by genetic material from the same genetic cluster (cluster I; Fig. 4). The Bayesian analysis also suggests the possibility of occasional long-distance gene dispersal in the coastal species A. glaziouana, a species that is probably pollinated by bats (Martinelli, 1994). Perhaps most conspicuously, recent long-distance dispersal is indicated from population LNI of A. glaziouana, sampled on an outcrop near the city of Niteroi across the Guanabara bay from Rio de Janeiro (Fig. 1), into populations sampled to the west of the bay. A small number of immigrants with an approximately 50 % contribution of nuclear DNA from population LNI was detected in populations LPA, LPI and LPP of A. glaziouana. Assuming these intermediate genotypes are first-generation immigrants, they indicate recent long-distance dispersal (Fig. 4A, yellow bars). Considering the increasing interest in animal pollination syndromes in neotropical species in general and bat pollination in particular (I. Sazima et al., 1989; M. Sazima et al., 1999), it would be of great value to know whether long-distance dispersal across the bay was facilitated by dispersal of pollen or seeds. This question may be addressed by a comparison of genetic patterns for biparentally inherited nuclear and uniparentally inherited organellar markers (Ennos, 1994). Alternatively, and considering the low chance of detecting sufficient polymorphism in the plastid genomes of Alcantarea species (Barfuss et al., 2005; Barbará, 2008), the question may be answered by using nuclear markers only. In the absence of two-generation genotypic data, this can be achieved by an analysis of slope and shape components of the curve that describes the decrease of genetic relatedness between individual plants with geographic distance (Heuertz et al., 2003). Given sufficient replication, this type of analysis can reveal whether gene flow is more restricted by dispersal of pollen or seeds.
A NJ tree of populations allowed us to test genetic relationships between populations of coastal and high-altitude inselberg species of Alcantarea, including two populations of the related Vriesea gigantea that were included for comparative purposes (Fig. 5). Two trends are clearly visible from this analysis. Firstly, populations of A. geniculata, A. glaziouana and A. regina each group together with moderate bootstrap support. Thus, microsatellite data provide a picture of population relationships that is congruent with traditional taxonomic species' delimitation for these taxa (Martinelli, 1994). Secondly, populations of A. imperialis do not group together, as two populations from an area of sympatry with the closely related A. geniculata show a clear affinity with that species (Fig. 5). In general, bootstrap support values of branches that involve populations of A. imperialis tend to be low. Population IIM of A. imperialis co-occurs sympatrically with population GIM of A. geniculata on the same inselberg (Fig. 5, dashed line), and it has been shown previously that historical gene flow (Nem) between these two morphologically well-differentiated species on this outcrop is greater than that detected between most populations of A. imperialis (Barbará et al., 2007). Population IVV of A. imperialis, also affected by this affinity to A. geniculata, is from an outcrop just a few kilometres away (Fig. 1). This reticulate pattern (Fig. 5) suggests past hybridization between A. imperialis and A. geniculata. Our finding is in agreement with the hypothesis that recombination of genetic material from related species represents an important aspect of species' radiations (Seehausen, 2004).
The successful genotyping of populations of the closely related V. gigantea with the same set of nuclear microsatellites (Fig. 5) indicates a potential for microsatellites or other nuclear markers to resolve phylogenetic relationships among species of these two closely related genera. The long branches of the V. gigantea populations probably stem from the fact that they originate from different parts of the species' range – these populations also fall into different genetic clusters in a Bayesian analysis of the gene pool structure of V. gigantea (Palma-Silva, 2008). Note that V. gigantea has a mixed mating system with high levels of inbreeding due to selfing and/or biparental inbreeding (Paggi et al., 2007; Palma-Silva, 2008) and is placed next to the highly inbred A. glaziouana in the unrooted NJ tree (Fig. 5). This suggests a potential for the use of well-resolved molecular phylogenies to understand the evolution of mating systems in this group. Molecular systematic work with multiple plastid and nuclear DNA markers including all known species of Alcantarea and multiple species of Vriesea is currently in progress in our lab (L. M. Versieux et al., unpubl. res.).
Relevance for conservation and concluding remarks
The results not only provide information on population relationships and variation in reproductive strategies, they also have clear implications for conservation management. Firstly, the great and significant genetic differentiation between coastal inselberg populations of Alcantarea (Tables 3 and 4, Fig. 4) implies that efficient in situ or ex situ conservation strategies need to be based on a sufficient number of inselbergs to represent the gene pools of each species. Secondly, the strong genetic differentiation between inselberg populations opens the opportunity for the use of so-called genetic ‘assignment tests’ to match individual plants to particular populations (Cornuet et al., 1999; Pritchard et al., 2000). This type of test requires great among-population differences in allelic composition and frequencies, a precondition that is met by these species. This would allow conservation geneticists to test whether plant and seed material currently available in botanical gardens around the world is representative for the gene pool of each of these inselberg species, or whether more fieldwork is required to achieve adequate coverage of species' gene pools. Assignment tests would also allow a tracking of the population origin of plant material used in horticulture, e.g. to detect illegal extractions from natural populations. Thirdly, our results indicate that in situ conservation of coastal Alcantarea species must account for clonal reproduction – sampling plants at distances less than 10 m from each other would almost certainly result in clonal duplicates, thus decreasing the amount of genetic variability captured in ex situ collections.
In conclusion, reproductive strategies and breeding systems vary greatly both within and between Alcantarea inselberg species. Asexual reproduction via vegetative clonal growth occurs frequently in coastal inselberg populations whereas sexual reproduction prevails in high-altitude species. Great variation also exists for levels of inbreeding, but the relative contributions of genetic and ecological factors to this variation remain to be determined. Population relationships based on nuclear microsatellites, although supported only by moderate bootstrap support, are largely congruent with species' delimitation based on traditional taxonomy, except for indications of past hybridization and reticulation in areas where the two high-altitude species A. imperialis and A. geniculata overlap. The strong genetic structure observed for gene pools of coastal and high-altitude species provides an excellent basis for the application of genetic markers to conservation management in these ecologically and horticulturally important bromeliads. Our molecular marker study also provides a basis for future work on the genetic underpinnings of adaptive radiation in this group of Bromeliaceae.
ACKNOWLEDGEMENTS
We are thankful to Pedro Cavalcanti (‘Jardims de Altitude’ eco-tourism), Ricardo Alvarez, Norma Barbará, Daniela Zappi, João Silva, Márcia Botelho and Pedro Simões Lopes for support during field collections, the Brazilian IBAMA for processing of collection/export permits, Jeffrey Joseph for help in the lab, Simon Hiscock for helpful discussions, and Gecele Paggi and Myriam Heuertz for comments on the manuscript. We also thank Cecilia C. Faria of Parque Nacional da Serra dos Órgãos for providing access to digital vegetation maps and Walter Till for providing leaf samples for PCR amplification tests. Fieldwork was supported by the Kew Overseas Fieldwork Committee, the Jardim Botânico do Rio de Janeiro and British Airways. Thelma Barbará and Clarisse Palma-Silva received fellowship support from the Weston Foundation and Mellon Foundation, respectively.
LITERATURE CITED
- Arnaud-Haond S, Belkhir K. GENCLONE: a computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Molecular Ecology Notes. 2007;7:15–17. [Google Scholar]
- Arnaud-Haond S, Duarte CM, Alberto F, Serrao EA. Standardizing methods to address clonality in population studies. Molecular Ecology. 2008;16:5115–5139. doi: 10.1111/j.1365-294X.2007.03535.x. [DOI] [PubMed] [Google Scholar]
- Barbará T. Molecular population genetics of Alcantarea spp. adapted to inselbergs in the Atlantic rainforest of Brazil. UK: Open University; 2008. PhD thesis. [Google Scholar]
- Barbará T, Martinelli G, Fay MF, Mayo SJ, Lexer C. Population differentiation and species cohesion in two closely related plants adapted to neotropical high-altitude ‘inselbergs’, Alcantarea imperialis and Alcantarea geniculata (Bromeliaceae) Molecular Ecology. 2007;16:1981–1992. doi: 10.1111/j.1365-294X.2007.03272.x. [DOI] [PubMed] [Google Scholar]
- Barbará T, Lexer C, Martinelli G, Mayo S, Fay MF, Heuertz M. Within-population spatial genetic structure in four naturally fragmented species of a neotropical inselberg radiation, Alcantarea imperialis, A. geniculata, A. glaziouana and A. regina (Bromeliaceae) Heredity. 2008;101:285–296. doi: 10.1038/hdy.2008.65. [DOI] [PubMed] [Google Scholar]
- Barfuss MHJ, Samuel R, Till W, Stuessy TF. Phylogenetic relationships in subfamily Tillandsioideae (Bromeliaceae) based on DNA sequence data from seven plastid regions. American Journal of Botany. 2005;92:337–351. doi: 10.3732/ajb.92.2.337. [DOI] [PubMed] [Google Scholar]
- Barrier M, Robichaux RH, Purugganan MD. Accelerated regulatory gene evolution in an adaptive radiation. Proceedings of the National Academy of Sciences of the USA. 2001;98:10208–10213. doi: 10.1073/pnas.181257698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzing DH. Bromeliaceae: profile of an adaptive radiation. Cambridge: Cambridge University Press; 2000. [Google Scholar]
- Boneh L, Kuperus P, Van Tienderen PH. Microsatellites in the bromeliads Tillandsia fasciculata and Guzmania monostachya. Molecular Ecology Notes. 2003;3:302–303. [Google Scholar]
- Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL. High resolution of human evolutionary trees with polymorphic microsatellites. Nature. 1994;368:455–457. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- Brown GK, Gilmartin AJ. Chromosome numbers in Bromeliaceae. American Journal of Botany. 1989;76:657–665. [Google Scholar]
- Cascante-Martin AM. Establishment, reproduction and genetics of epiphytic bromeliad communities during premontane forest succession in Costa Rica. The Netherlands: University of Amsterdam; 2006. PhD thesis. [Google Scholar]
- Cavallari MM, Forzza RC, Veasey EA, Zucchi MI, Oliveira GCX. Genetic variation in three endangered species of Encholirium (Bromeliaceae) from Cadeia do Espinhaço, Brazil, detected using RAPD Markers. Biodiversity and Conservation. 2006;15:4357–4373. [Google Scholar]
- Chapman H, Robson B, Pearson ML. Population genetic structure of a colonising, triploid weed. Hieracium lepidulum. Heredity. 2004;92:182–188. doi: 10.1038/sj.hdy.6800392. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. Inbreeding depression and its evolutionary consequences. Annual Review in Ecology and Systematics. 1987;18:237–268. [Google Scholar]
- Corander J, Waldmann P, Marttinen P, Sillanpää MJ. BAPS 2: enhanced possibilities for the analysis of genetic population structure. Bioinformatics. 2004;20:2363–2369. doi: 10.1093/bioinformatics/bth250. [DOI] [PubMed] [Google Scholar]
- Cornuet J-M, Piry S, Luikart G, Estoup A, Solignac M. New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics. 1999;153:1989–2000. doi: 10.1093/genetics/153.4.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J, Orr H. Speciation. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- Crayn DM, Winter K, Smith JAC. Multiple origins of crassulacean acid metabolism and the epiphytic habit in the neotropical family Bromeliaceae. Proceedings of the National Academy of Sciences of the USA. 2004;101:3703–3708. doi: 10.1073/pnas.0400366101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeChaine EG, Martin AP. Marked genetic divergence among sky island populations of Sedum lanceolatum (Crassulaceae) in the Rocky Mountains. American Journal of Botany. 2005;92:477–486. doi: 10.3732/ajb.92.3.477. [DOI] [PubMed] [Google Scholar]
- Darwin CR. The origin of species by means of natural selection. London: John Murray; 1859. [Google Scholar]
- Dieringer D, Schlötterer C. Microsatellite Analyzer (MSA): a platform independent analysis tool for large microsatellite data sets. Molecular Ecology Notes. 2003;3:167–169. [Google Scholar]
- Doyle J, Doyle J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987;19:1–15. [Google Scholar]
- Ellstrand NC, Roose ML. Patterns of genotypic diversity in clonal plant species. American Journal of Botany. 1987;74:123–131. [Google Scholar]
- Emerson BC. Evolution on oceanic islands: molecular phylogenetic approaches to understanding pattern and process. Molecular Ecology. 2002;11:951–966. doi: 10.1046/j.1365-294x.2002.01507.x. [DOI] [PubMed] [Google Scholar]
- Ennos RA. Estimating the relative rates of pollen and seed migration among plant populations. Heredity. 1994;72:250–259. [Google Scholar]
- Felsenstein J. Seattle, WA, USA: Department of Genome Sciences and Department of Biology, University of Washington; 2004. PHYLIP (Phylogeny Inference Package), version 3.6. Available from the author. [Google Scholar]
- Givnish T, Sytsma K, Smith J, Hahn W, Benzing D, Burkhardt E. Molecular evolution and adaptive radiation in Brocchinia (Bromeliaceae: Pitcairnioideae) atop tepuis of the Guyana shield. In: Givnish T, Sytsma K, editors. Molecular evolution and adaptive radiation. Cambridge: Cambridge University Press; 1997. pp. 259–311. [Google Scholar]
- Gonzalez-Astorga JG, Cruz-Angon A, Flores-Palacios A, Vovides AP. Diversity and genetic structure of the Mexican endemic epiphyte Tillandsia achyrostachys E. Morr. ex Baker var. achyrostachys (Bromeliaceae) Annals of Botany. 2004;94:545–551. doi: 10.1093/aob/mch171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. FSTAT (Version 1.2): a computer program to calculate F-statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- Grant JR. Bromelienstudien. The resurrection of Alcantarea and Werauhia, a new genus. Tropische und subtropische Pflanzenwelt. 1995;91:1–57. [Google Scholar]
- Heuertz M, Vekemans X, Hausman JF, Palada M, Hardy OJ. Estimating seed vs. pollen dispersal from spatial genetic structure in the common ash. Molecular Ecology. 2003;12:2483–2495. doi: 10.1046/j.1365-294x.2003.01923.x. [DOI] [PubMed] [Google Scholar]
- Honnay O, Jacquemyn H. A meta-analysis of the relation between mating system, growth form and genotypic diversity in clonal plant species. Evolutionary Ecology. 2008;22:299–312. [Google Scholar]
- Hughes C, Eastwood R. Island radiation on a continental scale: exceptional rates of plant diversification after uplift of the Andes. Proceedings of the National Academy of Sciences of the USA. 2006;103:10334–10339. doi: 10.1073/pnas.0601928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo LY, Pinero D. High genetic diversity in the only known population of Aechmea tuitensis (Bromeliaceae) Australian Journal of Botany. 2000;48:645–650. [Google Scholar]
- Luther HE. An alphabetical list of bromeliad binomials. 9th edn. Orlando, FL: The Bromeliad Society International, Inc; 2004. [Google Scholar]
- MacArthur RH, Wilson EO. The theory of island biogeography. Princeton, NJ: Princeton University Press; 1967. [Google Scholar]
- Martinelli G. Reproductive biology of Bromeliaceae in the Atlantic rainforest of southeastern Brazil. Scotland: University of St. Andrews; 1994. PhD thesis. [Google Scholar]
- Murawski DA, Hamrick JL. Local genetic and clonal structure in the tropical terrestrial bromeliad, Aechmea magdalenae. American Journal of Botany. 1990;77:1201–1208. [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Paggi GM, Palma-Silva C, Silveira LCT, Kaltchuk-Santos E, Bodanese-Zanettini MH, Bered F. Fertility of Vriesea gigantea Gaud. (Bromeliaceae) in southern Brazil. American Journal of Botany. 2007;94:683–689. doi: 10.3732/ajb.94.4.683. [DOI] [PubMed] [Google Scholar]
- Palma-Silva C. Population genetic structure in Vriesea gigantea, a bromeliad species endemic to the Atlantic Rainforest of Brazil. Brazil: Federal University of Porto Alegre; 2008. PhD thesis. [Google Scholar]
- Palma-Silva C, Cavallari MM, Barbará T, Lexer C, Gimenes MA, et al. A set of polymorphic microsatellite loci for Vriesea gigantea and Alcantarea imperialis (Bromeliaceae) and cross-amplification in other bromeliad species. Molecular Ecology Notes. 2007;7:654–657. [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piry S, Luikart G, Cornuet JM. BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. Journal of Heredity. 1999;90:502–503. [Google Scholar]
- Porembski S, Barthlott W., editors. Inselbergs. Biotic diversity of isolated rock outcrops in tropical and temperate regions. Berlin: Springer-Verlag; 2000. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Richards AJ. Apomixis in flowering plants: an overview. Philosophical Transactions of the Royal Society of London, Series B. 2003;358:1085–1093. doi: 10.1098/rstb.2003.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safford HD, Martinelli G. Southeast Brazil. In: Porembski S, Barthlott W, editors. Inselbergs. Biotic diversity of isolated rock outcrops in tropical and temperate regions. Berlin: Springer; 2000. pp. 339–389. [Google Scholar]
- Sarthou C, Samadi S, Boisselier-Dubayle MC. Genetic structure of the saxicole Pitcairnia geyskesii (Bromeliaceae) on inselbergs in French Guiana. American Journal of Botany. 2001;88:861–868. [PubMed] [Google Scholar]
- Sarthou C, Boisselier-Dubayle MC, Lambourdiere J, Samadi S. Polymorphic microsatellites for the study of fragmented populations of Pitcairnia geyskesii L. B. Smith (Bromeliaceae), a specific saxicolous species of inselbergs in French Guiana. Molecular Ecology Notes. 2003;3:221–223. [Google Scholar]
- Sazima I, Vogel S, Sazima M. Bat pollination of Encholirium glaziovii, a terrestrial bromeliad. Plant Systematics and Evolution. 1989;168:167–179. [Google Scholar]
- Sazima M, Buzato S, Sazima I. Bat-pollinated flower assemblages and bat visitors at two Atlantic Forest Sites in Brazil. Annals of Botany. 1999;83:705–712. [Google Scholar]
- Scheepens JF, Veeneklaas RM, Van de Zande L, Bakker JP. Clonal structure of Elytrigia atherica along different successional stages of a salt marsh. Molecular Ecology. 2007;16:1115–1124. doi: 10.1111/j.1365-294X.2006.03213.x. [DOI] [PubMed] [Google Scholar]
- Schluter D. The ecology of adaptive radiation. Oxford, UK: Oxford University Press; 2000. [Google Scholar]
- Silvertown J. The evolutionary maintenance of sexual reproduction: evidence from the ecological distribution of asexual reproduction in clonal plants. International Journal of Plant Science. 2008;169:157–168. [Google Scholar]
- Schönswetter P, Stehlik I, Holderegger R, Tribsch A. Molecular evidence for glacial refugia of mountain plants in the European Alps. Molecular Ecology. 2005;14:3547–3555. doi: 10.1111/j.1365-294X.2005.02683.x. [DOI] [PubMed] [Google Scholar]
- Seehausen O. Hybridization and adaptive radiation. Trends in Ecology and Evolution. 2004;19:198–207. doi: 10.1016/j.tree.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Sgorbati S, Labra M, Grugni E, Barcaccia G, Galasso G, et al. A survey of genetic diversity and reproductive biology of Puya raimondii (Bromeliaceae), the endangered queen of the Andes. Plant Biology. 2004;6:222–230. doi: 10.1055/s-2004-817802. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Isolation by distance in equilibrium and nonequilibrium populations. Evolution. 1993;47:264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- Smith LB, Till W. Bromeliaceae. In: Kubitzki K, editor. The families and genera of vascular plants. Berlin: Springer; 1999. pp. 74–99. [Google Scholar]
- Soltis DE, Gilmartin AJ, Rieseberg L, Gardner S. Genetic variation in the epiphytes Tillandsia lonantha and Tillandsia recurvata (Bromeliaceae) American Journal of Botany. 1987;74:531–537. [Google Scholar]
- Stebbins GL. Self-fertilization and population variability in the higher plants. The American Naturalist. 1957;91:337–354. [Google Scholar]
- Stephenson AG, Good SV, Vogler DW. Interrelationships among inbreeding depression, plasticity in the self-incompatibility system, and the breeding system of Campanula rapunculoides L. (Campanulaceae) Annals of Botany. 2000;85:211–219. [Google Scholar]
- Stuessy TF, Jakubowsky G, Gomez RS, et al. Anagenetic evolution in island plants. Journal of Biogeography. 2006;33:1259–1265. [Google Scholar]
- Valière N. GIMLET: a computer program for analysing genetic individual identification data. Molecular Ecology Notes. 2002;2:377–379. [Google Scholar]
- Versieux LM, Wanderley MGL. A new species of Alcantarea (E. Morren ex Mez) Harms, Bromeliaceae. Hoehnea. 2007;34:409–413. [Google Scholar]
- Versieux LM, Wanderley MGL. Two new species of Alcantarea (Bromeliaceae, Tillandsioideae) from Brazil. Brittonia. 2007;59:57–64. [Google Scholar]
- Versieux LM, Wanderley MGL. Wanderley MGL, Shepherd GJ, Melhem TS, Giulietti AM, editors. Alcantarea (E. Morren ex Mez) Harms. Flora Fanerogâmica do Estado de São Paulo. (1st edn) 2007:59–62. São Paulo: Imprensa Oficial, 2007, v. 5. [Google Scholar]
- Vogler DW, Kalisz S. Sex among the flowers: the distribution of plant mating systems. Evolution. 2001;55:202–204. doi: 10.1111/j.0014-3820.2001.tb01285.x. [DOI] [PubMed] [Google Scholar]
- Waits LP, Luikart G, Taberlet P. Estimating the probability of identity among genotypes in natural populations: cautions and guidelines. Molecular Ecology. 2001;10:249–256. doi: 10.1046/j.1365-294x.2001.01185.x. [DOI] [PubMed] [Google Scholar]
- Wendt T, Canela MBF, De Faria APG, Rios RI. Reproductive biology and natural hybridization between two endemic species of Pitcairnia (Bromeliaceae) American Journal of Botany. 2001;88:1760–1767. [PubMed] [Google Scholar]
- Wendt T, Canela MBF, Klein DE, Rios RI. Selfing facilitates reproductive isolation among three sympatric species of Pitcairnia (Bromeliaceae) Plant Systematics and Evolution. 2002;232:201–212. [Google Scholar]
- Whitkus R, Doan H, Lowrey TK. Genetics of adaptive radiation in Hawaiian species of Tetramolopium (Asteraceae). III. Evolutionary genetics of sex expression. Heredity. 2000;85:37–42. doi: 10.1046/j.1365-2540.2000.00722.x. [DOI] [PubMed] [Google Scholar]
- Whitlock MC. Temporal fluctuations in demographic parameters and the genetic variance among populations. Evolution. 1992;46:608–615. doi: 10.1111/j.1558-5646.1992.tb02069.x. [DOI] [PubMed] [Google Scholar]