Abstract
Background and Aims
Tolerance of complete submergence is recognized in a small number of accessions of domesticated Asian rice (Oryza sativa) and can be conferred by the Sub1A-1 gene of the polygenic Submergence-1 (Sub1) locus. In all O. sativa varieties, the Sub1 locus encodes the ethylene-responsive factor (ERF) genes Sub1B and Sub1C. A third paralogous ERF gene, Sub1A, is limited to a subset of indica accessions. It is thought that O. sativa was domesticated from the gene pools of the wild perennial species O. rufipogon Griff. and/or the annual species O. nivara Sharma et Shastry. The aim of this study was to evaluate the orthologues of the Sub1 locus in the closest relatives of O. sativa to provide insight into the origin of the gene and allelic variation of the Sub1 locus.
Methods
Orthologues of the Sub1 genes were isolated from O. rufipogon and O. nivara by use of oligonucleotide primers corresponding to the most highly conserved regions of the Sub1 genes of domesticated rice. The phylogenetic relatedness of Sub1 genes of O. sativa and its wild relatives was evaluated.
Key Results and Conclusions
Both O. rufipogon and O. nivara possess two Sub1 gene orthologues with strong sequence identity to the Sub1B and Sub1C alleles of cultivated rice. The phylogeny of the Sub1 genes of the domesticated and wild rice suggests that Sub1A arose from duplication of Sub1B. Variation in Sub1B alleles is correlated with the absence or presence of Sub1A. Together, the results indicate that genetic variation at the Sub1 locus is due to gene duplication and divergence that have occurred both prior to and after rice domestication.
Key words: Oryza sativa, Oryza nivara, Oryza rufipogon, submergence tolerance, Sub1 genes, gene duplication
INTRODUCTION
Rice (Oryza sativa) is cultivated in flooded paddy fields since its culture requires a large amount of water. However, excessive rainfall and poor regulation of paddy water level can result in partial and complete inundation of aerial tissue, which compromises crop productivity. Indeed, most rice cultivars die within 2 weeks of complete submergence, causing serious loss of rice production in South and South-east Asia (Xu et al., 2006).
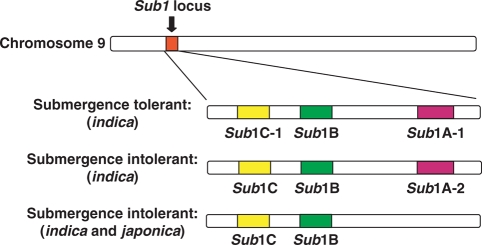
Rice varies dramatically in its response to flooding and submergence. Deep-water and most lowland rice accessions accelerate carbohydrate consumption and gibberellic acid-promoted elongation growth when submerged to escape the inundation (Fukao and Bailey-Serres, 2008). This response strategy is effective if floodwaters are shallow or rise gradually. Only a few exceptional rice cultivars, including Flood Resistant 13A (FR13A) can endure complete submergence for 2 weeks or longer (Setter and Laureles, 1996; Fukao et al., 2006; Xu et al., 2006). The submergence response in this as well as other rice accessions is regulated by the Submergence-1 (Sub1) locus, which encodes a variable cluster of up to three ethylene-responsive factor (ERF) genes: Sub1A, Sub1B and Sub1C (Fig. 1; Fukao et al., 2006; Xu et al., 2006). The Sub1 genes are members of group VII in the ERF gene family (Nakano et al., 2006) and are more closely related to one another than any other rice ERF genes (Gutterson and Reuber, 2004). The two ERF genes, Sub1B and Sub1C are present in all indica and japonica accessions examined to date, whereas Sub1A appears to be limited to a subset of indica accessions (Xu et al., 2006; S. Heuer, IRRI, Philippines, pers. comm.). Two alleles of Sub1A (Sub1A-1 and Sub1A-2) have been recognized, which are distinguished by a single amino acid substitution within the coding region. The transcripts of Sub1A-1 and Sub1A-2 are highly and poorly induced under submergence, respectively (T. Fukao, S. Heuer, D. Mackill and J. Bailey-Serres, unpubl. res.). Interestingly, only submergence-tolerant accessions possess the Sub1A-1 allele, whereas accessions that contain the less highly expressed Sub1A-2 are submergence-intolerant. Moreover, Sub1A is absent from all japonica and some indica accessions, all of which are intolerant to submergence. In support of Sub1A-1 as the key determinant of submergence tolerance, the over-expression of the allele in the intolerant japonica Liaogeng was confirmed to be sufficient to markedly enhance submergence tolerance (Xu et al., 2006).
Fig. 1.
Sub1 haplotypes of O. sativa. The Sub1 locus encodes two or three ethylene-responsive factors, Sub1A, Sub1B and Sub1C. Only submergence-tolerant accessions contain the Sub1A-1 allele at the locus, which confers submergence tolerance to rice (Fukao et al., 2006; Xu et al., 2006).
Oryza rufipogon and O. nivara as well as O. sativa belong to the A-genome group of the genus Oryza (Khush, 1997; Wing et al., 2005; Vaughan et al., 2008). Oryza rufipogon is a wild perennial species which adapts to persistently wet habitats, whereas O. nivara is an annual species which inhabits a seasonally dry environment (Li et al., 2006a; Vaughan et al., 2008). Oryza nivara is sometimes considered to be an ecotype or subspecies of O. rufipogon and these names are not differentiated when referring to wild rice (Londo et al., 2006; Kovach et al., 2007; Sweeney and McCouch, 2007). The history of rice domestication has long been controversial. Recent genome-scale analyses demonstrate that japonica and indica cultivars of domesticated rice (O. sativa) are closely related to different accessions of wild rice, suggesting that the two subspecies were domesticated from divergent wild populations (Lu et al., 2002; Cheng et al., 2003; Garris et al., 2005; Londo et al., 2006; Kovach et al., 2007; Sweeney and McCouch, 2007; Sang and Ge, 2007). However, allelic surveys of genes associated with shattering, pericarp colour and amylose content suggest that the key domestication genes originated only once (Yamanaka et al., 2004; Li et al., 2006b; Sweeney et al., 2006; Kovach et al., 2007; Lin et al., 2007; Sang and Ge, 2007; Vaughan et al., 2008). Based on these conflicting observations, single and multiple domestication hypotheses have been proposed. However, both hypotheses agree that O. sativa was domesticated from the gene pool of the wild-rice species, O. rufipogon and/or O. nivara.
Molecular genetic analyses have demonstrated that the polygenic Sub1 locus of O. sativa consists of a variable cluster of closely related ERF genes (Sub1A, -B, -C) and confirmed that ectopic expression of at least one allele of Sub1A is sufficient to confer submergence tolerance (Fukao et al., 2006; Xu et al., 2006). The polygenic nature of the Sub1 locus raises questions about the evolutionary timing of the gene duplication and source of the allelic variation at Sub1 that is associated with submergence tolerance. In this study, two orthologues of Sub1B and Sub1C of O. sativa were identified in both O. rufipogon and O. nivara. Phylogenetic analysis of Sub1 gene sequences in diverse accessions of domesticated rice revealed that alleles of Sub1B and Sub1C can be distinguished based on the absence of Sub1A and its allelic variation.
MATERIALS AND METHODS
Plant material and submergence treatment
Dehulled seeds of O. nivara (IRGC-101524 from the National Plant Germplasm System, USDA; originally collected in India) were sterilized in 70 % (v/v) ethanol for 10 min and in 2 % (v/v) sodium hypochlorite for 20 min. After rinsing thoroughly with sterilized deionized water, each seed was incubated in 3 mL of 1× Murashige and Skoog medium (pH 5·8) in a 16 mm × 15 cm test tube at 20 °C for 10 d (16 h light/8 h dark; light intensity, 50 µmol m−2 s−1). For the submergence treatment, the test tube was filled with 18 mL of sterilized distilled water, closed with a loose plastic cap, and incubated for 3 d at 25 °C in the light (50 µmol m−2 s−1). After treatment, aerial organs of submerged plants were immediately frozen in liquid nitrogen and stored at –80 °C until use.
Amplification of genomic DNA by polymerase chain reaction (PCR) and cloning
Genomic DNA of O. nivara (IRGC-80470; originally collected in India) and O. rufipogon (IRGC-80506) was kindly provided by Dr Tao Sang (Michigan State University). Genomic PCR was performed in a 50-μL reaction containing 100 ng genomic DNA, 5 µL 10× PCR buffer (Qiagen, CA, USA), 10 µL Q-solution (Qiagen), 1 µm primers, 0·3 mm deoxynucleotide triphosphates and 2·5 U Proofstart DNA polymerase (Qiagen). Primers used for amplification of Sub1 genes were: Sub1-forward (5′-GAVGAMTGGGAGGCCGCCTTCCRSGAGTTC-3′), and Sub1-reverse (5′-GTCGWAGSCGGCGAGGAGGCTGTCCATC-3′), where M = A or C, R = A or G, S = C or G, V = A or C or G, and W = A or T.
After denaturing the genomic DNA template at 95 °C for 5 min, PCR was performed with 30 cycles of denaturing at 95 °C for 45 s, annealing at 65 °C for 45 s, extension at 72 °C for 60 s, and final extension incubation at 72 °C for 15 min. Amplified DNA products were cloned into the pGEM-T vector (Invitrogen, CA, USA) and sequenced on both strands using standard procedures.
5′ and 3′ RACE
To isolate the full length of mRNA sequence, 5′- and 3′-RACE amplification was performed using the Gene Racer kit (Invitrogen) according to the manufacture's protocol. Total RNA was isolated from shoots of submerged O. nivara (IRGC-101524) plants using the RNeasy Plant Mini Kit (Qiagen). The GeneRacer RNA oligo containing 5′ adaptor was ligated to 5′ decapped mRNAs and the modified mRNAs were reverse transcribed using SuperScript III reverse transcriptase and the Gene Racer oligo dT containing 3′ adaptor. 5′- and 3′-RACE amplification was performed by PCR with gene-specific primers (Sub1-forward or Sub1-reverse) and Gene Racer primers supplied with the kit, as described for the amplification of genomic DNA by PCR. PCR products were gel purified, cloned into pCR4Blunt-TOPO vector (Invitrogen), and sequenced on both strands using standard procedures.
Semi-quantitative assessment of mRNA abundance by reverse transcriptase-polymerase chain reaction (RT-PCR)
Total RNA extraction from aerial tissue, cDNA synthesis and RT-PCR were performed exactly as described in Fukao et al. (2006). Sequences of primer pairs, annealing temperature and cycle number used for RT-PCR are given in Table S1 in Supplementary Information available online. The level of Actin1 mRNA was used as a loading control. The number of cycles for logarithmic amplification of RT-PCR was optimized by technical replications with a range of cycle numbers (i.e. 27, 30, 33 and 36 cycles) for each primer pair.
Sequence divergence and phylogenetic analyses
For direct comparison of Sub1 genes/alleles, the nucleotide and amino acid sequences were subjected to pairwise and multiple alignment analyses using EMBOSS (Labarga et al., 2007; http://www.ebi.ac.uk/emboss/align/) and ClustalW2 (Larkin et al., 2007; http://www.ebi.ac.uk/Tools/clustalw2/), respectively. The Sub1 orthologues of O. nivara and O. rufipogon used for this analysis are OnSub1B-1 (EU429442), OnSub1B-2 (EU429443), OnSub1C-1 (EU429445), OrSub1B-1 (EU429444) and OrSub1C-1 (EU429446). Alignments were adjusted manually upon visual inspection. For phylogenetic analyses, truncated nucleotide sequences were aligned with ERF2 (TIGR accession: LOC_Os01g21120), which belongs to the subgroup VII of the ERF gene family, as the outgroup sequence (Fig. S1 in Supplementary Information available online). Neighbor-joining analysis using uncorrected distances were calculated using PHYLIP, version 3·67 (Felsenstein, 1989). The reliability of the neighbor-joining output phylogeny was estimated using bootstrapping analysis with 100 replicates and one input order per replicate.
RESULTS
Sub1 orthologues of O. nivara and O. rufipogon
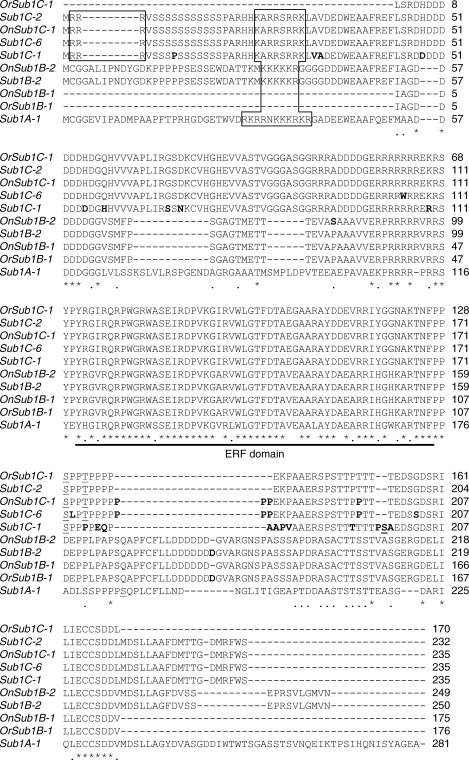
To isolate any orthologues of Sub1 genes from O. nivara and O. rufipogon by PCR, degenerate primers expected to bind to the two most highly conserved regions present in all rice Sub1 genes were designed based on the nucleotide sequence alignment of 16 alleles (two alleles of Sub1A, nine alleles of Sub1B and five alleles of Sub1C) (Fig. S2 in Supplementary Information available online). All Sub1 genes are classified into subgroup VII of the ERF transcription factor family (Gutterson and Reuber, 2004; Nakano et al., 2006). It was anticipated that the two degenerate primers would bind only to the Sub1 genes and not to subgroup VII or other ERF genes due to sequence dissimilarity in the primer binding region. The degenerate primers (Sub1-forward and Sub1-reverse) amplified a single band of the expected size (approx. 550 bp) from O. nivara and O. rufipogon genomic DNA (Fig. S3 in Supplementary Information available online]. Following gel purification and cloning of the PCR products, plasmid DNA was extracted from 25 independent clones and the nucleotide sequences of the inserts were determined. Sequence analysis revealed that both O. nivara and O. rufipogon encode two Sub1 orthologues which are more similar to Sub1B and Sub1C than the Sub1A gene of O. sativa (Fig. 2). The Sub1-like genes isolated from O. nivara (accession: IRGC-80470) and O. rufipogon (accession: IRGC-80506) using these degenerate primers were designated OnSub1B-1, OnSub1C-1, OrSub1B-1 and OrSub1C-1. To obtain the complete coding sequences of Sub1B and Sub1C genes of O. nivara, 5'- and 3'-RACE amplification was performed. Due to unavailability of seed from accession IRGC-80470, the O. nivara accession IRGC-101524 was used to obtain the full-length sequences of Sub1 cDNAs of this species. The Sub1B allele cloned from IRGC-101524 was designated OnSub1B-2 since a single non-conservative amino acid substitution distinguishes the allele from OnSub1B-1 of O. nivara accession (IRGC-80470; Fig. 2). The Sub1C sequence was identical in the region sequenced in both O. nivara accessions.
Fig. 2.
Amino acid alignment of representative Sub1A, Sub1B and Sub1C alleles of O. sativa and Sub1 orthologues of O. nivara and O. rufipogon. On, Oryza nivara; Or, Oryza rufipogon. Genes that do not have any prefix are from O. sativa. Sub1 orthologues isolated from O. nivara and O. rufipogon and representative Sub1A, Sub1B and Sub1C alleles were aligned using ClustalW2. Amino acid identity and similarity are indicated with an asterisk and dot, respectively. The highly conserved ERF domain responsible for DNA binding capability is underlined. An amino-terminal region of basic residues typical of a nuclear localization signal is indicated with a box. A bold residue represents amino acid variation between the Sub1B alleles or between Sub1C alleles. An underlined residue indicates a predicted mitogen-activated protein phosphorylation site. An amino acid alignment of all known Sub1 genes/alleles in O. sativa, O. nivara and O. rufipogon is available in Fig. S4 in Supplementary Information available online.
Nucleotide and amino acid sequences of the Sub1 orthologues isolated from O. nivara and O. rufipogon were compared with the reported sequences of Sub1B and Sub1C alleles of O. sativa (Fig. 2 and Table 1, and Fig. S4 in Supplementary Information available online; Xu et al., 2006). Direct sequence comparison revealed that the Sub1-like genes of wild rice are highly similar to alleles of Sub1B and Sub1C genes in O. sativa (approx. 99 % identity in nucleotide and amino acid sequences) (Table 1). The O. nivara Sub1 genes, OnSub1B-2 and OnSub1C-1 (complete coding sequences), are most closely related to Sub1B-2 and Sub1C-6 of domesticated rice, respectively. The two Sub1 genes of O. rufipogon, OrSub1B-1 and OrSub1C-1 (partial coding sequences) are most similar to Sub1B-2 and Sub1C-2, respectively. As previously shown, Sub1B-2 and Sub1C-2 are limited to japonica rice. Sub1C-6 was found in the indica accessions, Habiganj aman, IR24 and IRBB21, in combination with Sub1B-4, Sub1B-5 or Sub1B-8 (Table 2; Xu et al., 2006).
Table 1.
Percentage identity of Sub1 gene coding sequences in O. nivara, O. rufipogon and O. sativa
| GenBank accession | Most similar O. sativa allele | Nucleotide (% identity) | Amino acid (% identity) | ||
|---|---|---|---|---|---|
| O. nivara | OnSub1B-1* | EU429442 | Sub1B-2 | 99·4 (524/527) | 99·4 (175/176) |
| OnSub1B-2 | EU429443 | Sub1B-2 | 99·1 (746/753) | 99·2 (249/251) | |
| OnSub1C-1 | EU429445 | Sub1C-6 | 98·9 (700/708) | 98·7 (233/236) | |
| O. rufipogon | OrSub1B-1* | EU429444 | Sub1B-2 | 99·8 (526/527) | 100 (176/176) |
| OrSub1C-1* | EU429446 | Sub1C-2 | 100 (509/509) | 100 (170/170) |
* Partial coding sequences lacking 5′ (N-terminal) and 3′ (C-terminal) sequences.
Table 2.
Sub1 haplotypes of O. sativa, O. nivara and O. rufipogon
|
Sub1 haplotype |
||||||
|---|---|---|---|---|---|---|
| Species | Accession | Subspecies | Origin | Sub1A | Sub1B | Sub1C |
| O. sativa | FR13A | indica/aus | India | A-1 | B-1 | C-1 |
| Kurkaruppan | indica | Sri Lanka | A-1 | B-3 | C-1 | |
| Goda Heenati | indica | Sri Lanka | A-1 | B-6 | C-1 | |
| Teqing | indica | China | A-2 | B-1 | C-3 | |
| IR64 | indica | Philippines | A-2 | B-1 | C-3 | |
| 93–11 | indica | China | A-2 | B-1 | C-5 | |
| CO39 | indica | India | A-2 | B-7 | C-3 | |
| Habiganj aman | indica | Bangladesh | Absent | B-4 | C-6 | |
| IRBB21 | indica | Philippines | Absent | B-5 | C-6 | |
| IR24 | indica | Philippines | Absent | B-8 | C-6 | |
| Nipponbare | japonica | Japan | Absent | B-2 | C-2 | |
| Taipei309 | japonica | Taiwan | Absent | B-2 | C-2 | |
| Liaogeng | japonica | China | Absent | B-2 | C-2 | |
| M202 | japonica | USA | Absent | B-2 | C-2 | |
| Most similar O. sativa allele | ||||||
| O. nivara | IRGC-80470 | India | Absent | B-2 | C-6 | |
| IRGC-101524 | India | Absent | B-2 | C-6 | ||
| O. rufipogon | IRGC-80506 | India | Absent | B-2 | C-2 | |
Phylogenetic analysis of Sub1 genes in O. sativa, O. nivara and O. rufipogon
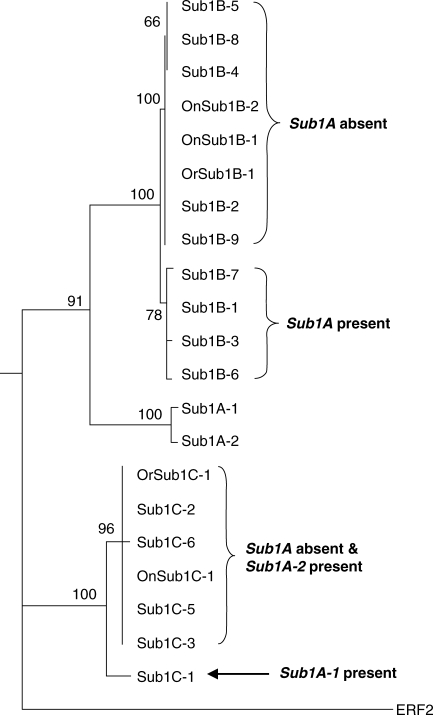
To estimate the evolutionary relatedness of Sub1 genes in O. sativa, O. nivara and O. rufipogon, a neighbor-joining method of phylogenetic analyses was used (Fig. 3). The three Sub1 genes, Sub1A, Sub1B and Sub1C, were separated into three distinct clades with significant bootstrap values supporting the phylogeny. The Sub1A gene, which is present only in some indica varieties, was more related to Sub1B than Sub1C. Interestingly, the O. sativa Sub1B alleles were resolved into two subgroups, which corresponded to the Sub1 locus haplotype. The subgroup with the Sub1B-1, -B-3, -B-6 and -B-7 alleles includes only rice accessions that encode Sub1A in addition to Sub1B and Sub1C (Table 2; Xu et al., 2006). By contrast, the O. sativa accessions with the Sub1B-2, -B-4, -B-5, -B-8 and -B-9 alleles do not possess the Sub1A gene. The Sub1B alleles of O. nivara and O. rufipogon co-clustered with the alleles of rice accessions that lack Sub1A, which is in accordance with the absence of the Sub1A gene in the wild-rice germplasms examined. The Sub1C alleles were resolved only in two subgroups. Unlike Sub1B, the Sub1C alleles were not distinguished by the absence and presence of Sub1A, due to high sequence similarity among alleles. Notably, the Sub1C-1 allele, which is limited to submergence-tolerant accessions (Sub1A-1 present), is distinct from the other known Sub1C alleles. The Sub1C-1 allele contains 16 non-synonymous and seven synonymous substitutions as compared with Sub1C-2, which is the most closely related allele. This amino acid sequence variation between these two alleles is concentrated in the region just carboxyl to the ERF binding domain and includes two potential phosphorylation sites (Fig. 2). Amino acid substitutions in this region are also observed amongst the other Sub1C alleles, particularly in the variable proline-rich region (Fig. S4 in Supplementary Information available online).
Fig. 3.
Phylogenetic tree based on nucleotide sequences of Sub1 gene alleles of O. sativa, O. nivara and O. rufipogon. A phylogenetic tree was generated by the neighbor-joining method using uncorrected distances in PHYLIP, version 3·67. ERF2 (TIGR accession: LOC_Os01g21120), another member of subgroup VII of the ERF gene family, was used as an outgroup. The central portion of the coding sequence was analysed (Fig. S1 in Supplementary Information available online). The length of each branch is proportional to sequence divergence. The numbers above branches indicate bootstrap values from 100 replicates.
Accumulation of transcripts encoding the Sub1 orthologues of O. nivara is promoted by submergence
Transcript levels of all three ERF genes of the Sub1 locus are elevated in response to submergence in aerial tissue of O. sativa (Fukao et al., 2006; Xu et al., 2006). To discern whether mRNA accumulation of the Sub1 orthologues is induced by submergence in O. nivara, RT-PCR analysis was carried out using gene-specific primers for OnSub1B and OnSub1C (Fig. 4). As reported previously for O. sativa (Fukao et al., 2006), 3 d of complete submergence increased the levels of OnSub1B and OnSub1C transcripts in aerial tissue of O. nivara. Transcript accumulation of orthologues of other submergence-inducible genes, such as α-amylase-3C (Amy3C) and alcohol dehydrogenase-2 (Adh2) were also examined in O. nivara using primers that are designed based on the genomic sequences of O. sativa (Fig. 4). The transcript levels of these two representative submergence inducible genes were also elevated in response to the stress.
Fig. 4.
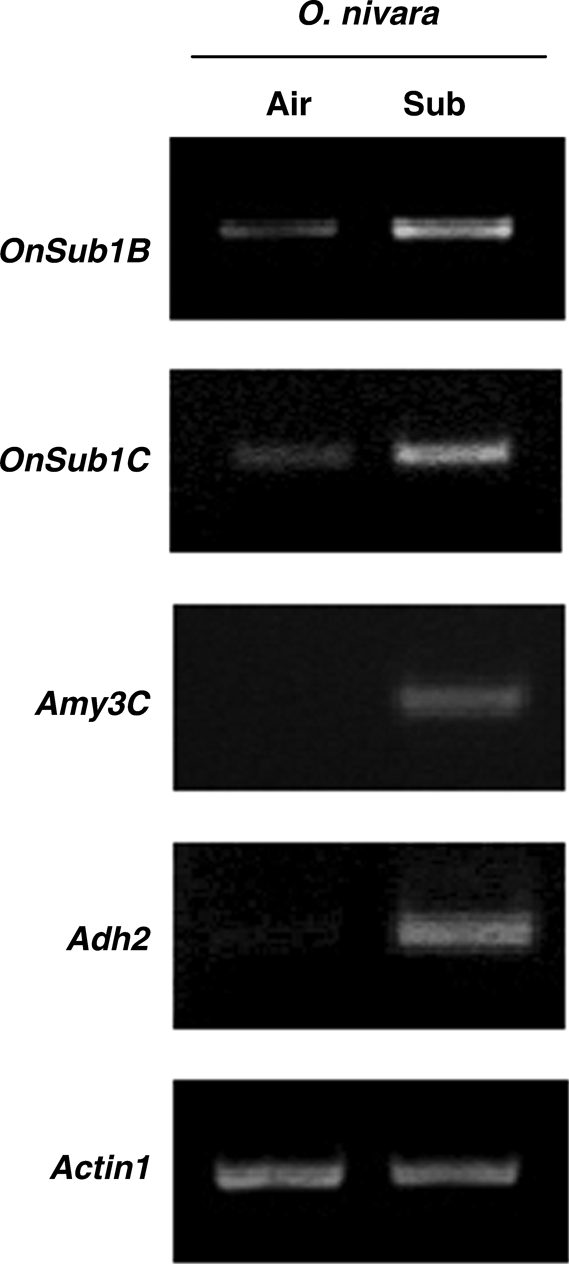
Accumulation of the Sub1 orthologue and representative gene mRNAs induced by submergence stress in O. nivara. Air, air-grown plants; Sub, submerged plants. Ten-day-old plants were completely submerged or aerobically grown in test tubes for 3 d. After treatment, total RNA was extracted from aerial tissue and analysed by RT-PCR. Gene-specific primers for OnSub1B and OnSub1C were designed based on the coding sequences of these genes in O. nivara. Primers for Amy3C, Adh2 and Actin1 were identical to those used for the transcript analysis in O. sativa (Fukao et al., 2006). The level of Actin1 mRNA was used as a loading control.
DISCUSSION
The Sub1 locus of O. sativa encodes a cluster of ERF proteins and is known to regulate responses to submergence. It is shown here that the two ERF genes that are orthologues of Sub1B and Sub1C are present in accessions of O. nivara and O. rufipogon, consistent with the detection of these genes in all O. sativa accessions examined to date (Xu et al., 2006; S. Heuer and D. Mackill, IRRI, Philippines, pers. comm.). The presence and submergence-induced expression of the Sub1B and Sub1C orthologues of O. nivara and O. rufipogon suggest that an orthologous Sub1 locus regulates submergence responses in these wild Oryza species. Direct sequence comparison revealed that Sub1B-2 and Sub1C-2 which are limited to japonica rice were most closely related to the Sub1B and Sub1C alleles in the O. rufipogon accession (Tables 1 and 2). By contrast, an O. nivara accession had a Sub1C allele that was most closely related to that of indica accessions. These observations are consistent with genome-scale analyses that show indica and japonica rice are more closely related to certain accessions of O. rufipogon and O. nivara than to each other (Lu et al., 2002; Cheng et al., 2003; Londo et al., 2006; Kovach et al., 2007; Sang and Ge, 2007; Sweeney and McCouch, 2007). Exceptionally, the O. nivara Sub1B allele isolated here was most related to Sub1B-2 which is restricted in japonica rice (Tables 1 and 2). Haplotype analysis of diverse domesticated and wild-rice accessions would provide further information to elucidate phylogenetic relatedness of polygenic Sub1 locus amongst O. sativa, O. nivara and O. rufipogon.
The three ERF genes of the Sub1 locus are most closely related to one another, as compared with the 139 other ERF-domain containing genes of rice (Gutterson and Reuber, 2004; Nakano et al., 2006; T. Fukao and J. Bailey-Serres, unpubl. res.), indicating that the polygenic locus arose via tandem duplication. The three SUB1 proteins are highly conserved within the ERF DNA binding domain, but are considerably divergent in the amino- and carboxyl-terminal regions (Fig. 2, and Fig. S4 in Supplementary Information available online). Although the data do not identify a founding member of this cluster of paralogues, the nucleotide and amino acid sequence analyses provide some intriguing insight into these genes. First, the phylogenetic analysis exposes diversification within the Sub1B and Sub1C clades that greatly exceeds that observed in the Sub1A clade, suggesting that the duplication that gave rise to the Sub1A gene is the most recent event (Fig. 3). This is also supported by the finding that the Sub1A gene is not present in all domestic and wild-rice accessions. In accordance with this, the subgroups of Sub1B alleles corresponded to accessions that either possess or lack the Sub1A gene (Fig. 3). Secondly, the phylogenetic analysis indicates that Sub1A is more closely related to Sub1B than Sub1C. This is consistent with the high amino acid sequence similarity at the N-terminus shared by Sub1A and Sub1B, but not Sub1C (Fig. 2 and Fig. S4 in Supplementary Information available online). This conserved N-terminal region resembles the MCGGAI(I/L) motif present in most known Group VII ERFs of diverse plant species (Nakano et al., 2006). Further support that Sub1A is more closely related to Sub1B than Sub1C is based on the site of the single intron in each of these genes. In Sub1A and Sub1B, the insertion is within the final portion of the coding sequence, whereas it is located at the 3'-untranslated region in Sub1C (Fig. S2 in Supplementary Information available online). Together, these results indicate that Sub1A arose by duplication of Sub1B.
Two Sub1A alleles have been recognized based on two single nucleotide polymorphisms within the coding region, one of which confers a non-conservative variation of a Ser (Sub1A-1) or Pro (Sub1A-2) (Xu et al., 2006). The Sub1A-2 allele is present in a subset of the indica accessions, all of which initiate an elongation growth response to submergence. The Sub1A-1 allele was identified in the submergence-tolerant accession FR13A collected in Orissa State of eastern India (Xu et al., 2006). FR13A is a member of the aus varietal group of domesticated rice. Aus varieties are distinguishable from indica by simple sequence repeat and single nucleotide polymorphism markers, but are considered a subgroup of indica (Garris et al., 2005; McNally et al., 2006). The presence of Sub1A-1 in FR13A and two indica cultivars from Sri Lanka (Goda Heenati and Kurkaruppan) is correlated with a submergence response that includes carbohydrate conservation and limited elongation growth (Fukao et al., 2006; Xu et al., 2006). The confirmation of Sub1A-1 or Sub1A-2 alleles in the genomes of a subset of indica and aus accessions indicates that Sub1A most likely arose from gene duplication at this locus prior to the divergence of the indica/aus lineages.
Submergence dramatically increases the transcript abundance of Sub1C as well as Sub1A in aerial tissue of submergence-tolerant and intolerant rice accessions (Fukao et al., 2006; Xu et al., 2006). Interestingly, the Sub1C-1 allele, which is limited to the submergence-tolerant accessions with Sub1A-1, is the most divergent of all of the Sub1C alleles (Fig. 3). The unusual Sub1C allele in submergence-tolerant rice could have arisen from positive nucleotide selection on this allele as a consequence of the function of Sub1A-1. In contrast to the observation for the Sub1C gene, more than one Sub1B allele was found in association with Sub1A-1 (Table 2) (Xu et al., 2006). Experimental analysis of the molecular function of the SUB1 proteins is required to understand better the observed diversity of the Sub1 genes. Of particular interest will be consideration of the variation in potential mitogen-activated protein kinase phosphorylation sites located immediately carboxyl-terminal to the ERF DNA binding domain (Fig. 2, and Fig. S4 in Supplementary Information available online; Xu et al., 2006).
Recent allelic survey of cloned domestication genes revealed that alleles which encode genes associated with shattering, pericarp colour and amylose content are common to all O. sativa accessions including indica and japonica (Yamanaka et al., 2004; Li et al., 2006b; Kovach et al., 2007; Sang and Ge, 2007; Sweeney et al., 2006). Unlike key domestication alleles, the submergence tolerance conferring Sub1A-1 allele is restricted to limited aus/indica accessions and is uncommon in modern cultivars. Interestingly, the occurrence of the submergence tolerance-conferring haplotypes of the Sub1 locus (Sub1A-1, Sub1B-1, -3, -6, Sub1C-1) in both aus (FR13A; Orissa, India) and indica (Goda Heenati and Kurkaruppan; Sri Lanka) most likely reflects human selection of the submergence tolerance trait in two geographic regions. An orally chronicled migration of followers of an outcast prince from the Orissa region to Sri Lanka circa 500 bc is supported by recent molecular studies of the frequency of subtype alleles encoding human leucocyte antigens (HLA-A*02) in Sri Lankan and eastern Indian populations (Malavige et al., 2007). Hence, it is feasible that rice with the submergence tolerance-conferring Sub1 haplotype was transported by immigrants and subsequently introgressed into local indica varieties grown in Sri Lanka. The Sub1 haplotypes that confer submergence tolerance has limited distribution amongst rice cultivars but may have been prized by rice farmers with submergence-prone paddies in Orissa and Sri Lanka.
SUPPLEMENTARY INFORMATION
Supplementary information is available online at www.aob.oxfordjournals.org and consists of the following. Figure S1: nucleotide sequence alignment of truncated Sub1 genes in O. sativa, O. nivara, and O. rufipogon. Figure S2: nucleotide sequence alignment of Sub1 genes in O. sativa. Figure S3: Sub1 orthologues amplified by genomic PCR in O. nivara and O. rufipogon. Figure S4: Amino acid alignment of Sub1 genes of O. sativa, O. nivara, and O. rufipogon.
ACKNOWLEDGEMENTS
We thank David Mackill, Sigrid Heuer and Angelika Mustroph for valuable comments and discussion. We also thank Tao Sang for providing genomic DNA samples of O. nivara and O. rufipogon. This work is supported by US Department of Agriculture (2006-35100-17288) and a USAID Linkage Grant from the International Rice Research Institute.
LITERATURE CITED
- Cheng CY, Motohashi R, Tsuchimoto S, Fukuta Y, Ohtsubo H, Ohtsubo E. Polyphyletic origin of cultivated rice based on the interspersion pattern of SINEs. Molecular Biology and Evolution. 2003;20:67–75. doi: 10.1093/molbev/msg004. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP – Phylogeny Inference Package (Version 3·2) Cladistics. 1989;5:164–166. [Google Scholar]
- Fukao T, Bailey-Serres J. Ethylene – a key regulator of submergence responses in rice. Plant Science. 2008;175:43–51. [Google Scholar]
- Fukao T, Xu K, Ronald PC, Bailey-Serres J. A variable cluster of ethylene responsive-like factors regulates metabolic and developmental acclimation responses to submergence in rice. The Plant Cell. 2006;18 doi: 10.1105/tpc.106.043000. 2021–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S. Genetic structure and diversity in Oryza sativa L. Genetics. 2005;169:1631–1638. doi: 10.1534/genetics.104.035642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutterson N, Reuber TL. Regulation of disease resistance pathways by AP2/ERF transcription factors. Current Opinion in Plant Biology. 2004;7:465–471. doi: 10.1016/j.pbi.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Khush GS. Origin, dispersal, cultivation and variation of rice. Plant Molecular Biology. 1997;35:25–34. [PubMed] [Google Scholar]
- Kovach MJ, Sweeney MT, McCouch SR. New insights into the history of rice domestication. Trends in Genetics. 2007;23:578–587. doi: 10.1016/j.tig.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Labarga A, Valentin F, Anderson M, Lopez R. Web services at the European Bioinformatics Institute. Nucleic Acids Research. 2007;35:6–11. doi: 10.1093/nar/gkm291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. ClustalW2 and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li C, Zhou A, Sang T. Genetic analysis of rice domestication syndrome with the wild annual species, Oryza nivara. New Phytologist. 2006;a 170:185–194. doi: 10.1111/j.1469-8137.2005.01647.x. [DOI] [PubMed] [Google Scholar]
- Li C, Zhou A, Sang T. Rice domestication by reducing shattering. Science. 2006;b 311 doi: 10.1126/science.1123604. 1936–1939. [DOI] [PubMed] [Google Scholar]
- Lin Z, Griffith ME, Li X, Zhu Z, Tan L, Fu Y, et al. Origin of seed shattering in rice (Oryza sativa L.) Planta. 2007;226:11–20. doi: 10.1007/s00425-006-0460-4. [DOI] [PubMed] [Google Scholar]
- Londo JP, Chiang Y-C, Hung K-H, Chiang T-Y, Schaal BA. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proceedings of the National Academy of Sciences of the USA. 2006;103:9578–9583. doi: 10.1073/pnas.0603152103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu BR, Zheng KL, Qian HR, Zhuang JY. Genetic differentiation of wild relatives of rice as assessed by RFLP analysis. Theoretical and Applied Genetics. 2002;106:101–106. doi: 10.1007/s00122-002-1013-2. [DOI] [PubMed] [Google Scholar]
- McNally KL, Bruskiewich R, Mackill D, Buell CR, Leach JE, Leung H. Sequencing multiple and diverse rice varieties: connecting whole-genome variation with phenotypes. Plant Physiology. 2006;141:26–31. doi: 10.1104/pp.106.077313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malavige GN, Rostron T, Seneviratne SL, Fernando S, Sivayogan S, Wijewickrama A, et al. HLA analysis of Sri Lankan Sinhalese predicts North Indian origin. International Journal of Immunogenetics. 2007;34:313–315. doi: 10.1111/j.1744-313X.2007.00698.x. [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiology. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang T, Ge S. The puzzle of rice domestication. Journal of Integrative Plant Biology. 2007;49:760–768. [Google Scholar]
- Setter TL, Laureles EV. The beneficial effect of reduced elongation growth on submergence tolerance of rice. Journal of Experimental Botany. 1996;47:1551–1559. [Google Scholar]
- Sweeney M, McCouch S. The complex history of the domestication of rice. Annals of Botany. 2007;100:951–957. doi: 10.1093/aob/mcm128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney MT, Thomson MJ, Pfeil BE, McCouch S. Caught red-handed: Rc encodes a basic helix–loop–helix protein conditioning red pericarp in rice. The Plant Cell. 2006;18:283–294. doi: 10.1105/tpc.105.038430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan DV, Lu B-R, Tomooka N. The evolving story of rice evolution. Plant Science. 2008;174:394–408. [Google Scholar]
- Wing RA, Ammiraju JSS, Luo M, Kim H-R, Yu Y, Kudrna D, et al. The Oryza map alignment project: the golden path to unlocking the genetic potential of wild rice species. Plant Molecular Biology. 2005;59:53–62. doi: 10.1007/s11103-004-6237-x. [DOI] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, et al. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature. 2006;442:705–708. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Nakamura I, Watanabe KN, Sato Y. Identification of SNPs in the waxy gene among glutinous rice cultivars and their evolutionary significance during the domestication process of rice. Theoretical and Applied Genetics. 2004;108:1200–1204. doi: 10.1007/s00122-003-1564-x. [DOI] [PubMed] [Google Scholar]