Abstract
Background and Aims
Submergence is a recurring problem in the rice-producing rainfed lowlands of south and south-east Asia. Developing rice cultivars with tolerance of submergence and with agronomic and quality traits acceptable to farmers is a feasible approach to address this problem. The objectives of this study were to (a) develop mega varieties with Sub1 introgression that are submergence tolerant, (b) assess the performance of Sub1 in different genetic backgrounds, (c) determine the roles of the Sub1A and Sub1C genes in conferring tolerance, and (d) assess the level of tolerance in F1 hybrids heterozygous for the Sub1A-1-tolerant allele.
Methods
Tolerant varieties were developed by marker-assisted backcrossing through two or three backcrosses, and their performance was evaluated to determine the effect of Sub1 in different genetic backgrounds. The roles of Sub1A and Sub1C in conferring the tolerant phenotype were further investigated using recombinants identified within the Sub1 gene cluster based on survival and gene expression data.
Key Results
All mega varieties with Sub1 introgression had a significantly higher survival rate than the original parents. An intolerant Sub1C allele combined with the tolerant Sub1A-1 allele did not significantly reduce the level of tolerance, and the Sub1C-1 expression appeared to be independent of the Sub1A allele; however, even when Sub1C-1 expression is completely turned off in the presence of Sub1A-2, plants remained intolerant. Survival rates and Sub1A expression were significantly lower in heterozygotes compared with the homozygous tolerant parent.
Conclusions
Sub1 provided a substantial enhancement in the level of tolerance of all the sensitive mega varieties. Sub1A is confirmed as the primary contributor to tolerance, while Sub1C alleles do not seem important. Lack of dominance of Sub1 suggests that the Sub1A-1 allele should be carried by both parents for developing tolerant rice hybrids.
Key words: Oryza sativa, Sub1, marker-assisted backcrossing, mega varieties, submergence tolerance, recombinant, hybrid, abiotic stress
INTRODUCTION
Submergence stress adversely affects poor farmers living on 15 million ha of rice-growing areas in the rainfed lowlands in south and south-east Asia. In some areas, farmers plant landraces that are moderately tolerant of submergence but have low yield. In areas where high-yielding but submergence-intolerant rice varieties have been cultivated, farmers suffer from crop losses caused by periodic flash floods during the monsoon season. Recently, the extent of submergence stress has increased due to extreme weather events such as unexpected heavy rains that have inundated wider areas across many regions in Asia. More sustainable and permanent solutions are needed to overcome this problem. One of the most promising solutions is to develop high-yielding varieties that are submergence tolerant and that are more likely to be rapidly adopted by the farmers in the target regions.
Submergence tolerance is controlled by a single major quantitative trait locus (QTL) on chromosome 9, along with a number of minor QTLs (Xu and Mackill, 1996; Nandi et al., 1997; Toojinda et al., 2003). All these studies have used the landrace FR13A, which is one of the most submergence-tolerant donor varieties. The major QTL, named Sub1, with a LOD score of 36 and an R2 value of 69 % (Xu and Mackill, 1996), provides tolerance to complete submergence for up to 2 weeks. The fine-mapping of Sub1 employed 2950 F2 segregating individuals and, although the region had a low recombination rate, Sub1 was delineated to a genomic region of approx. 0·06 cM (Xu et al., 2000). Sequencing the Sub1 region in an FR13A-derived line revealed the presence of three genes encoding putative ethylene responsive factors (ERF), Sub1A, Sub1B and Sub1C and Sub1A were subsequently identified as the major determinant of submergence tolerance (Xu et al., 2006). It was also observed that Sub1C alleles were associated with tolerance; however, it was not known if the tolerant Sub1C allele had any effect on the level of tolerance.
The cloning of the major QTL Sub1 has provided an excellent opportunity not only to gain a better understanding of the molecular mechanisms and unravel the pathways underlying submergence tolerance, but also to design gene-based or tightly linked markers for more precise genotyping. Previous studies have reported the development of submergence tolerant varieties by introducing the Sub1 locus using marker-aided selection (Siangliw et al., 2003; Toojinda et al., 2005). However, these efforts did not take advantage of the benefits of marker-aided selection in precisely and rapidly transferring the locus. More recently this gene has been successfully introgressed through marker-assisted backcrossing (MAB) into a popular high-yielding variety from India, Swarna, within a 2-year time frame (Neeraja et al., 2007). Swarna-Sub1, the first example of a submergence-tolerant mega variety, is currently being evaluated in submergence-prone areas of India and Bangladesh. Under non-submerged control conditions no significant differences in agronomic performance, grain yield and grain quality between Swarna and Swarna-Sub1 were observed indicating complete restoration of the Swarna background in Swarna-Sub1 (Sarkar et al., 2006; Neeraja et al., 2007), but Swarna-Sub1 showed a 2-fold or higher yield advantage over Swarna after submergence for 10 d or more during the vegetative stage (S. Singh et al., IRRI, The Philippines unpubl. res.). These results highlighted the opportunity to develop additional high-yielding varieties that are adapted to other regions or environments. In addition, hybrid rice has been viewed as a promising approach that can be used as a vehicle to boost rice production. In recent years, there has been increasing interest to grow hybrid rice in several countries, and the potential use of Sub1 in hybrid rice has great promise for flood-prone areas
Here, recent advances in converting additional mega varieties to submergence-tolerant varieties are reported and the role of Sub1 in different genetic backgrounds confirmed. The importance of the Sub1A and Sub1C genes for tolerance is further investigated, and the potential use of Sub1 in hybrid rice is assessed. Implications for the development of additional submergence-tolerant varieties with Sub1, and the development of varieties with a higher level of tolerance under longer durations of submergence are discussed.
MATERIALS AND METHODS
The procedures for developing submergence-tolerant mega varieties of rice (Oryza sativa L.), including molecular analyses and evaluation of submergence tolerance, followed those previously described for Swarna-Sub1 (Neeraja et al., 2007). Two breeding lines derived from FR13A, i.e. IR49830-7-1-2-2 and IR40931-33-1-3-2 (referred to as IR49830 and IR40931, respectively), were used as donors. The mega varieties Samba Mahsuri and CR1009 from India, IR64 from the Philippines (IRRI), Thadokkham 1 (TDK1) from Laos, and BR11 from Bangladesh were used as recipient parents. BR11 was used in an additional set of experiments to study MAB selection schemes and the development of BR11-Sub1 will therefore not be discussed in this paper aside from presentation of data on submergence tolerance. To transfer the tolerant Sub1 allele into the mega varieties, a MAB strategy was followed, with closely flanking markers used for recombinant selection to reduce the target introgression size and background markers used to select for recurrent parent alleles (Collard and Mackill, 2008). Population sizes ranged from 343 to 697 plants for the five MAB populations at the BC1 generation and 69 to 320 plants at the BC2 generation. The fully converted Sub1 lines were selected at the BC2F2 or BC3F2 generation (Table 1).
Table 1.
Development of new versions of mega-varieties with submergence tolerance
| Sub1 variety | Donor | Generation | IR designation | Fixed line designation | BC1F1 plants |
BC2 plants |
BC3 plants |
Total markers* | Sub1 region introgression size (Mb) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. plants | BG SSR | F1 | F2 | F1 | F2 | |||||||
| Swarna-Sub1 | IR49830 | BC2 | IR82809-237 | IR05F101 | 697 | 56 | 320 | 420 | 95 | 6·5–7·3 | ||
| BC3 | IR82810-407 | IR05F102 | 18 | 432 | 2·3–3·4 | |||||||
| S. Mahsuri-Sub1 | IR49830 | BC2 | IR84193-36 | IR07F101 | 392 | 50 | 320 | 130 | 63 | 6·5–9·2 | ||
| BC3 | IR84196-32 | IR07F287 | 14 | 48 | 0·8–2·1 | |||||||
| IR64-Sub1 | IR40931 | BC2 | IR84194-139 | IR07F102 | 440 | 48 | 188 | 162 | 68 | 6·5–7·8 | ||
| BC3 | IR84197-88 | IR07F292 | 86 | 690†/327‡ | 1·1–2·7 | |||||||
| TDK1-Sub1 | IR40931 | BC3 | IR85264-141 | IR07F289 | 347 | 45 | 69 | 55§ | 38 | 190 | 76 | 1·5–2·5 |
| CR1009-Sub1 | IR40931 | BC2 | IR85262-85 | IR07F291 | 343 | 54 | 199 | 193¶/127# | 72 | 2·7–6·4 | ||
*The total number of markers used across all generations for each population.
† No fixed double recombinant, but only heterozygous recombinant identified.
‡ BC3F3 plants.
§ Not continued due to remaining donor background introgressions.
¶ Proceeded to next generation due to remaining donor background introgressions.
# BC2F3 plants.
Submergence screening was performed in the greenhouse or in the field at the International Rice Research Institute (IRRI), Los Baños, Philippines. For submergence screenings, Sub1 lines along with parents and the susceptible check IR42 were planted into trays that were submerged when seedlings were 14–21 d old. When the susceptible check showed >50 % damage, usually after about 14 d of complete submergence, trays were de-submerged and the survival of plants was scored after 10–21 d recovery.
Gene-based and intragenic Sub1 DNA markers were developed based on DNA sequences published by Xu et al. (2006) and available in the NCBI database (http://www.ncbi.nlm.nih.gov/). Primer3 was used to design PCR primers (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3.cgi) and BLAST search was used to find regions of similarity and verify the specificity of the markers for the Sub1 target region. (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). Sequence alignments were done using CLUSTALW (http://align.genome.jp/). Four new markers were developed within the Sub1A gene, three within Sub1C, three between Sub1B and Sub1C, and one between Sub1A and Sub1B (Table 2).
Table 2.
Molecular markers developed in the Sub1 gene cluster
| Primer | Sequence | Tm (°C) | Position | CAPS*/indel†/mismatch | Acc‡ | Expected size (bp) | Method |
|---|---|---|---|---|---|---|---|
| IYT1F | TAGGGGCCCATGAGTACTTG | 60·0 | Sub1A promoter | BsaA1 | TQ§/93-11 | 109 and 554 | PCR |
| IYT1R | TCAGACAGCTAGCTCGCAAC | 59·5 | Sty1 | TQ§/93-11 | 173, 27 and 463 | ||
| Sty1 | 26D17¶ | 200 and 463 | |||||
| IYT3F | GTTGATAACCGGAGGAGACG | 59·6 | Sub1A promoter | NlaIII | TQ§/93-11 | 273, 32 and 107 | PCR |
| IYT3R | GTAACCCGACTGGTCTCAGG | 59·6 | NlaIII | 26D17¶ | 305 and 107 | ||
| AEXF | AGGCGGAGCTACGAGTACCA | 62·2 | Functional SNP for SUB1A | Mismatch primer | 26D17¶ | 231 | PCR |
| AEX1R | GCAGAGCGGCTGCGA | 62·4 | Specific for tolerance | ||||
| Sub1A203F | CTTCTTGCTCAACGACAACG | 59·6 | Exon of Sub1A | NA** | TQ§/93-11/26D17¶ | 203 | RT-PCR |
| Sub1A203R | AGGCTCCAGATGTCCATGTC | 60·1 | |||||
| ART3F | TCTGAACCGGATCATCATTG | 59·5 | Sub1C promoter | 7-bp deletion in NB/93–11 | NB#/93-11 | 330 | PCR |
| ART3R | AGTTTGTCTCCATTCGAAGTCA | 58·8 | 118k20¶ | 337 | |||
| ART5F | CAGGGAAAGAGATGGTGGA | 58·6 | Sub1C promoter | 15-bp insertion in NB/93–11 | NB#/93-11 | 217 | PCR |
| ART5R | TTGGCCCTAGGTTGTTTCAG | 60·1 | 118k20¶ | 202 | |||
| Sub1C173F | AACGCCAAGACCAACTTCC | 60·1 | Exon of Sub1C | Cac8I | NB#/93-11 | 108 and 56 | PCR, RT-PCR |
| Sub1C173R | AGGAGGCTGTCCATCAGGT | 59·7 | 118k20¶ | 126 and 47 | |||
| 9-bp deletion in NB/93-11 | NB#/93-11 | 164 | |||||
| 118k20¶ | 173 | ||||||
| Sub1AB1F | CATGTTCCATAGCCATCGACT | 60·0 | Between Sub1A and B | 2-bp deletion in 93-11 | 93-11 | 148 | PCR |
| Sub1AB1R | GAGCGAAGAGAGCTACCTGAA | 59·0 | BAC 101O9¶ | 150 | |||
| Sub1BC1F | CAATCGATGCGTGCTTCTT | 60·0 | Between Sub1B and C | 6-bp deletion in 93-11 | 93-11 | 168 | PCR |
| Sub1BC1R | CGCAACAAGGCAGAAAAATA | 59·0 | 118k20¶ | 174 | |||
| Sub1BC2F | AAAACAATGGTTCCATACGAGAC | 58·5 | Between Sub1B and C | 38-bp insertion in 93-11 | 93-11 | 231 | PCR |
| Sub1BC2R | GCCTATCAATGCGTGCTCTT | 60·4 | 118k20¶ | 239 | |||
| Sub1BC3F | CATGGGTAAAATTGCCATCC | 60·0 | Between Sub1B and C | 24-bp deletion in 93-11 | 93-11 | 193 | PCR |
| Sub1BC3R | GCTTGAGGGTGAGTGGAGAG | 60·0 | 118k20¶ | 217 |
*Cleaved amplified polymorphic sequences.
† Insertion–deletion.
‡ Accession.
§ Teqing.
¶ IR40931-26.
# Nipponbare.
**Not applied.
Segregating populations used for MAB were screened for plants with recombination within the Sub1 gene cluster. Recombinants were initially identified using two markers tightly flanking the Sub1 region and were subsequently confirmed by further genotyping using gene-based and intra-genic markers. Populations of 2240 BC1F2 plants for IR64/IR40931, and 2137 BC2F2 and 279 BC3F1 plants for BR11/IR40931 were screened with flanking markers to identify recombination events between Sub1A and Sub1C.
F1 plants heterozygous for Sub1 were derived from crosses between IR64 and IR64-Sub1. About 200 F1 plants were used for a submergence test along with the parents and some check varieties. Submergence tests and expression studies for both, recombinants and F1 materials were conducted as described by Xu et al. (2006).
To determine the level of submergence tolerance of Sub1A/Sub1C recombinant plants and F1 hybrids, a randomized complete block design (RCBD) was used in two independent trials for both experiments, with two or three replications used per trial. About 30 plants were used in each replication. In most cases, 14-d-old seedlings were submerged and de-submerged when the susceptible check showed a substantial level of damage (>50 %). DNA was extracted from young leaf tissues following a standard protocol (Zheng et al., 1995) and total RNA was isolated using TRIzol (Gibco). PCR and RT-PCR analyses were followed by standard protocols (Xu et al., 2006) with slight modifications as specified in Table 2. The sequences of Rubisco primer pair used as a positive control are 5′-CATTCACCGAGCAATGCATG-3′ and 5′-CTCCATTTGCAAGCTGATCG-3′ (De Leon, 2007) with melting temperatures (Tm) of 63·6 °C and 61·9 °C, respectively.
RESULTS
Development of submergence-tolerant mega varieties
Samba Mahsuri-Sub1, a tolerant version of the popular Indian cultivar Samba Mahsuri, was developed within two rounds of backcrossing and one generation of self-pollination following the same MAB procedure used to develop Swarna-Sub1 (Neeraja et al., 2007). Only a relatively small fragment at the tip of chromosome 9, where the QTL Sub1 is located, was introduced. In comparison to Swarna-Sub1, fewer plants were available in the BC1F1 and BC2F2 generations (697 for Swarna and 392 for Samba Mahsuri in BC1F1, and 420 and 130 in BC2F2, respectively; Table 1). Nonetheless, one plant with the desired genotype was successfully selected in the BC2F2 generation (plant #166-62-36, IR84193-36). In addition, another version of Samba Mahsuri-Sub1 with a smaller introgression of the Sub1 region, which was 0·8–2·1 Mb, was selected among 48 BC3F2 progenies derived from a double-side cross-over type of the BC3F1 plant.
The tolerant version of IR64 was developed following a similar approach. Among the 440 BC1F1 and 188 BC2F1 plants that were genotyped, plant #50-22 was selected (Table 1). Out of 162 BC2F2 progeny from the selected plant, plant # 50-22-139 (IR07F102) was selected based on its possession of the Sub1 locus and maximum recipient genome, with a similar size of Sub1 introgression as the first version of Swarna-Sub1 (IR05F101). To develop another version of IR64-Sub1 with a smaller size of Sub1 introgression, 690 BC3F2 plants derived from three selected BC3F1 plants were genotyped. However, no plants with the double cross-over event were identified. Nevertheless, several promising lines with smaller introgressions were identified. A total of 327 plants derived from recombinant plant #50-22-13-138 were genotyped and one plant with a relatively small introgression (1·2–2·7 Mb) was selected. This new version of IR64-Sub1 is plant #50-22-13-138-88 (IR07F292; Table 1).
Due to the small size of the TDK1/IR40931 population from the earlier backcrosses and segregation bias of several markers toward the tolerant parent (data not shown), there were no ideal Sub1 plants selected out of the BC2F2 population (Table 1). Screening of larger BC3F1 and BC3F2 populations subsequently facilitated the identification of several BC3F1 plants with a small Sub1 introgression. One of the selected BC3F1 plants, #130-38-34, was self-pollinated and among the 190 BC3F2 progenies, plant #130-38-34-141 (IR07F289) was identified with the desired small Sub1 introgression (1·5–2·6 Mb).
As was the case for TDK1, no ideal plant could be identified from the BC2F2 population for CR1009, a popular variety from India. Part of this might be due to the small BC1F1 population which was only about half the size of the one used for Swarna (Table 1). In addition, about 50 % of the plants identified in BC2F1 were self-pollinated which significantly reduced the size of the population. In the BC2F2 population the background introgression was therefore still high (12 background markers). Among the 193 BC2F3 progenies, plant #195-9-64 showed the least number of background markers (four) and was selected. Among the 127 of BC2F3 progenies of the selected plant, in the end, one best plant was identified as plant #195-9-64-85 (IR07F291) having an introgression size of 2·6–6·3 Mb. For this variety, no BC3 version was developed because the size of the introgression was considered reasonable and the photoperiod sensitivity of this variety had already delayed the development of CR1009-Sub1.
Development of Sub1-specific markers
In the early applications of MAB for developing submergence-tolerant varieties the diagnostic marker used was GnS2, a cleaved amplified polymorphic sequence (CAPS) marker that was used in combination with several markers flanking Sub1 (Neeraja et al., 2007). However, there were some cases where GnS2 did not distinguish between tolerant and intolerant varieties. For instance, the Sub1A PCR product of TDK1 was not restricted by AluI and the TDK1 allele therefore appeared to be the same as the tolerant allele of IR40931-33 (IR40931). To develop some alternative markers and to measure the introgressed region of the Sub1 locus more precisely, additional allele-specific and intragenic markers were developed (Table 2). A specific single nucleotide polymorphism (SNP) within the Sub1A coding region that causes an amino acid substitution (intolerant: CCG = proline; tolerant: TCG= serine) was targeted for marker design. Since no restriction enzyme sites were located at the SNP locus that could be used to develop a CAPS marker, a dominant sequence tag site marker was developed by designing a PCR primer with the SNP at the 3′ end, e.g. the AEX marker. This marker was specifically designed for the tolerant allele (IR40931). In addition, two CAPS markers were designed in the promoter region of Sub1A, namely IYT1 and IYT3 (Table 2).
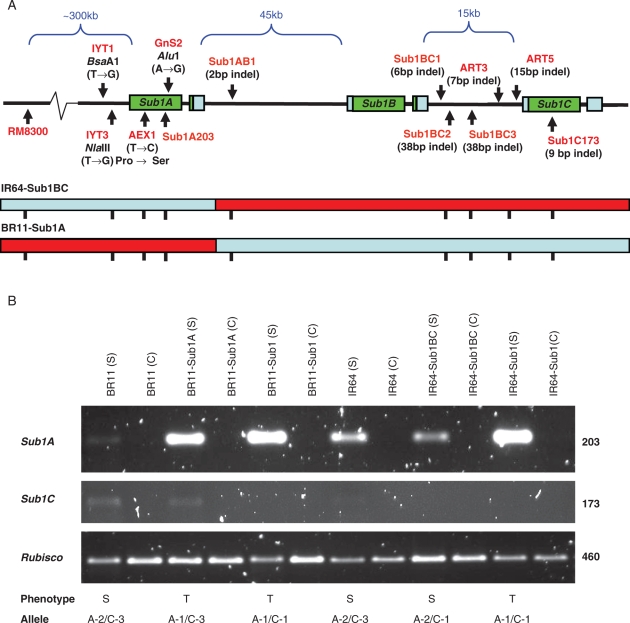
Sequencing of Sub1C revealed seven allelic groups and a unique phosphorylation site for the tolerant lines FR13A, Kurkaruppan, Goda Heenati, IR40931 and IR40981 (Xu et al, 2006). An insertion–deletion (indel) marker for Sub1C, SUB1C173, was designed and used in addition to the Sub1A markers. However, in some cases when there was no size polymorphism, this marker can also be used as a CAPS marker in order to differentiate the allele of Sub1C. Additionally two new indel markers were designed in the promoter region of Sub1C, namely ART3 and ART5 (Table 2). These indel markers could significantly reduce the cost of MAB by eliminating the need for the digestion step. In most cases, the indel markers ART5 or SUB1C173 are diagnostic and sufficient for foreground selection. GnS2 and other CAPS markers were used for rare cases where recombination occurred between ART5 or SUB1C173 (Sub1C) and the next Sub1A downstream marker (e.g. RM8300) (Fig. 1A, top panel) and for confirming introgression of the Sub1A-tolerant allele in the pre-selected material.
Fig. 1.
Graphical genotypes of the IR64-Sub1BC and BR11-Sub1A recombinant plants with their RT-PCR analysis. (A) Positions of the gene-based and intragenic markers within the Sub1 cluster. IR64-Sub1BC recombinant possess the intolerant Sub1A-2 allele from IR64 (blue bar), while the Sub1B-1 and Sub1C-1 alleles are derived from IR40931 (red bar). BR11-Sub1A has the Sub1A-1 allele from IR40931, while the Sub1B and Sub1C genes were derived from BR11. (B) RT-PCR analyses were performed with total RNA isolated from leaves of 13-d-old seedlings submerged (S) for 6 h and non-submerged controls (C), with their corresponding phenotypes (S = susceptible, T = tolerant) and allelic information.
In addition to the diagnostic markers developed within the Sub1 cluster, the development of microsatellite markers along the Sub1 region has been reported (Neeraja et al., 2007). So far RM8300 is one of the closest simple sequence repeat (SSR) markers downstream of Sub1A. This marker has been used as one of the recombinant markers in combination with different SSR markers located at about 1·5–2·5 Mb or 6–10 cM upstream of RM8300. Additionally, highly polymorphic SSR markers in the Sub1 region were identified from the International Rice Genome Sequencing Project (IRGSP; Matsumoto et al., 2005). Several of these markers are tightly linked and located upstream of the Sub1 locus, e.g. RM23835 (5·5 Mb), RM23865 (6·2 Mb) and RM23869 (6·3 Mb), and are very useful for limiting the size of the Sub1 introgression. The marker closest to Sub1 previously reported was RM23805 (4·5 Mb; fig. 3 in Neeraja et al., 2007).
Performance of mega varieties with submergence tolerance
The evaluation of the six converted mega varieties under submergence stress showed that all Sub1 varieties had significantly higher survival rates compared with the original recipient parents. Nevertheless, some differences in the level of tolerance among the Sub1 lines (Table 3) were observed. Based on preliminary data, IR64-Sub1, TDK1-Sub1 and CR1009-Sub1 showed the same high level of tolerance as Swarna-Sub1, whereas BR11-Sub1 was slightly less tolerant. Samba Mahsuri-Sub1 was the least tolerant among all the Sub1 lines, especially as seen in trials 2 and 4 (Table 3).
Table 3.
Submergence tolerance of the Sub1 mega varieties measured in different experiments at IRRI, Philippines
| Survival (%)* |
|||||||
|---|---|---|---|---|---|---|---|
| Variety | Sub1 present | Trial 1 | Trial 2 | Trial 3 | Trial 4 | Trial 5 | Trial 6 |
| IR42 | – | 11 | 0 | 0 | 0 | 0 | 21 |
| FR13A | + | 100 | |||||
| IR49830 | + | 87 | 89 | ||||
| IR40931 | + | 93 | 52 | 94 | 78 | ||
| Swarna | – | 34 | 0 | 24 | 4 | 37 | |
| Swarna Sub1 | + | 81 | 27 | 88 | 78 | 75 | |
| S. Mahsuri | – | 24 | 0 | 2 | 0 | 51 | |
| S. Mahsuri Sub1 | + | 56 | 2 | 50 | 21 | 83 | |
| IR64 | – | 20 | 0 | 27 | 30 | 28 | |
| IR64 Sub1 | + | 87 | 13 | 75 | 75 | 80 | |
| TDK1 | – | 0 | 4 | 3 | 0 | ||
| TDK1 Sub1 | + | 13 | 48 | 84 | 83 | ||
| CR1009 | – | 0 | 0 | ||||
| CR1009 Sub1 | + | 18 | 74 | ||||
| BR11 | – | 0 | 0 | 3 | |||
| BR11 Sub1 | + | 5 | 53 | 41 | 77 | ||
| l.s.d. (0·05) | 17 | 4 | 35 | 21 | 6 | 17 | |
*Trials 1–5 were conducted in a submergence tank (a concrete tank with a water depth of approx. 1·5 m) while test 6 was conducted in the field. RCBD was used with three, two, three, two, two and three replications in trials 1, 2, 3, 4, 5 and 6, respectively. Plants in trials 2–5 were submerged after they were 14 d old, while plants in trials 1 and 6 were submerged after they were 21 d old and were submerged until most of the susceptible control (IR42) were damaged. Survival was counted 21 d after desubmergence, except for trial 1 (10 d after).
Performance of Sub1A/Sub1C recombinants
The markers RM8300 and Sub1C173 or ART5 were used to identify recombinants within the Sub1 gene cluster. A second set of markers located between the two flanking markers was developed and subsequently used to further pinpoint the site of recombination (Table 2). After screening of 2240 BC1F2 plants derived from a IR64/IR40931 cross, one recombinant plant was identified and named IR64-Sub1BC. In this plant the tolerant Sub1B and Sub1C alleles were present but the Sub1A gene was still segregating. Additionally, a second recombinant named ‘BR11-Sub1A’ was identified that carried the tolerant Sub1A-1 allele but was still segregating for both Sub1B and Sub1C. This plant was identified after screening 2137 BC2F2 and 279 BC3F1 plants of the BR11/IR40931 population. Both types of recombinants were then allowed to self-pollinate and progenies were genotyped using gene-based and intragenic markers to select for homozygous recombinants (Fig. 1A).
The present submergence test data indicated that there were no significant differences in tolerance among the IR64-Sub1BC lines, some control lines taken from the same IR64/IR40931 population but having the entire Sub1 locus from IR64, and the original IR64. All lines showed an intolerant phenotype (Table 4). The present data also showed no significant differences in the level of tolerance among the BR11-Sub1A lines, lines selected from the same BR11/IR40931 population but having the entire Sub1 locus from the donor IR40931, and the tolerant check BR11-Sub1. In this case, all lines showed the tolerant phenotype.
Table 4.
Submergence tolerance screening of the recombinants in the Sub1 cluster
| Survival (%)* |
||||
|---|---|---|---|---|
| Variety | Alias/group | Sub1A and Sub1C alleles | Trial 1 | Trial 2 |
| IR42 | NA† | 0 | 0 | |
| IR64 | A-2/A-2, C-3/C3 | 0 | 18 | |
| IR64-14-2 | A-2/A-2, C-3/C3 | 2 | ||
| IR64-14-16 | A-2/A-2, C-3/C3 | 0 | ||
| IR64-14-22 | IR64-Sub1BC | A-2/A-2, C-1/C1 | 2 | |
| IR64-14-23-29 | IR64-Sub1BC | A-2/A-2, C-1/C1 | 2 | 10 |
| IR64-14-10 | IR64-Sub1BC | A-2/A-2, C-1/C1 | 6 | |
| IR84194-139 | IR64-Sub1 | A-1/A-1, C-1/C1 | 51 | 78 |
| BR11 | A-2/A-2, C-3/C-3‡ | 1 | 2 | |
| BR11-536-11 | BR11-Sub1A | A-1/A-1, C-3/C-3 | 49 | |
| BR11-536-15 | BR11-Sub1A | A-1/A-1, C-3/C-3 | 57 | |
| BR11-536-2 | A-2/A-2, C-1/C-3 | 36 | 60 | |
| BR11-536-25 | A-2/A-2, C-1/C-3 | 27 | ||
| BR11-536-1 | A-1/A-1, C-1/C1 | 35 | 66 | |
| BR11-536-7 | A-1/A-1, C-1/C1 | 38 | ||
| IR85260-148 | BR11-Sub1 | A-1/A-1, C-1/C1 | 23 | 55 |
| IR40931 | A-1/A-1, C-1/C1 | 82 | ||
| FR13A | A-1/A-1, C-1/C1 | 73 | ||
| l.s.d. (0·05) | 18 | 20 | ||
*Both trials were conducted in a submergence tank using RCBD with three and two replications for trials 1 and 2, respectively. Plants were submerged after they were 14 d old and desubmerged when most of IR42 were damaged. Scoring was done after 21 d recovery.
† Alleles were not determined.
‡ The Sub1C allele of BR11 was not determined by sequencing, but on the size of the Sub1C173 PCR amplicon after digestion with Cac8I. The size was identical to that of IR64 and the allele was therefore categorized as Sub1C-3.
RT-PCR gene expression analyses showed that under non-submerged conditions no Sub1A and Sub1C expression was detected in seedling leaves of the control and the recombinant plants (Fig. 1B). Under submerged conditions the tolerant Sub1A-1 allele showed much higher expression than the intolerant Sub1A-2 allele (e.g. when comparing Sub1A expression in IR64-Sub1 versus IR64), as was also seen in Xu et al. (2006). However, the IR64-Sub1BC recombinant showed much less Sub1A expression than IR64-Sub1 and slightly less when compared with IR64. The BR11-Sub1A recombinant showed slightly less expression than BR11-Sub1 but still much higher expression than was detected in BR11 (Fig. 1B). In contrast, Sub1C expression under submergence was undetectable in IR64-Sub1 and BR11-Sub1 plants, and was very low in IR64 and BR11. Sub1C appeared to have slightly lower expression in IR64 than in BR11, reflecting the inverse relationship between Sub1A and Sub1C expression that was originally suggested by Xu et al. (2006). Interestingly, the IR64-Sub1BC recombinant showed no detectable expression of Sub1C, while the BR11-Sub1A recombinant had slightly less Sub1C expression than BR11 itself.
Performance of the heterozygous Sub1A plants
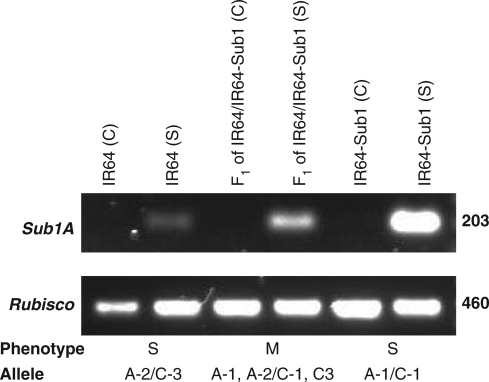
Submergence tolerance of F1 hybrids of IR64/IR64-Sub1 was compared with that of the parents. The heterozygous plants were significantly less tolerant than the plants homozygous for the tolerant allele (Table 5). In addition, the expression of the Sub1A allele in the heterozygotes was less than its expression in the homozygotes (Fig. 2). The results indicate that the Sub1 locus does not show dominance and tolerance is closely associated with the expression level and the dosage of the Sub1A alleles.
Table 5.
Submergence tolerance screening of heterozygotes for Sub1A-1 tolerance allele
| Survival (%)* |
||
|---|---|---|
| Variety | Trial 1 | Trial 2 |
| FR13A | 48 | |
| IR40931 | 86 | |
| IR184194 | 97 | 19 |
| F1 (IR64/IR184194) | 45 | 6 |
| F1 (IR64/IR184194) | 49 | |
| IR64 | 27 | 0 |
| IR42 | 0 | |
| l.s.d. (0·05) | 23 | 15 |
*Both trials were conducted in a submergence tank using RCBD with two and three replications for trials 1 and 2, respectively. In trial 1, 21-d-old seedlings were submerged and desubmerged after 14 d and survival was scored 10 d after desubmergence. In trial 2, 14-d-old seedlings were submerged and desubmerged when most of IR42 were damaged, and survival was scored after 21 d of recovery.
Fig. 2.
RT-PCR of Sub1A (top) with Rubisco as control (bottom) shown below each set of samples along with their corresponding phenotypes (S = susceptible, M = moderately tolerant, T = tolerant). RNA was extracted from leaf samples harvested from 21-d-old seedlings after submergence (S) for 6 h along with their respective unsubmerged controls (C).
DISCUSSION
Development of mega varieties with submergence tolerance through MAB
Submergence tolerance has been an important breeding objective for more than three decades (HilleRisLambers and Vergara, 1982; Mackill, 1986; Mohanty and Chaudhary, 1986). Breeders at IRRI made crosses between the tolerant donor FR13A and high-yielding varieties such as IR48 and IR36 in the mid- to late-1970s. Submergence tolerant lines with high yield potential were obtained in the early 1990s (Mackill et al., 1993). However, these tolerant prototypes were never widely adopted by farmers since they were inferior in grain quality or in other traits needed for local adaptation. On the other hand, mega varieties that possessed most traits desired by farmers but were submergence intolerant began to spread widely in both irrigated and rainfed lowland areas of south and south-east Asia (Mackill et al., 2006). To ensure the adoption of the final breeding product by the farmers, an approach of converting these mega varieties by MAB was undertaken. The objective was to convert these varieties to submergence-tolerant types while retaining the desirable traits of the original parents through a precise MAB approach (Collard and Mackill, 2008).
The development of six mega varieties with submergence tolerance was successfully completed through MAB within 3 years. In general, it was considered necessary to have about 500 or more BC1F1 plants to ensure that there would be sufficient plants to perform background selection following foreground and recombinant selection. Computer simulations estimating the genomic proportion of the recurrent parent in the backcross populations were in good agreement with the practical results of the Sub1 conversion of Swarna and Samba Mahsuri (Prigge et al., 2008). Due to a large number of monomorphic markers, the total numbers of markers used for background selection was <70 in some of the MAB populations in the present study. Nonetheless these markers were evenly spaced across the genome. Based on experience, one marker every 5 Mb or 20 cM, or a total of four to nine well-distributed markers per chromosome depending on the length of the chromosome is sufficient to monitor the donor introgressions. This result agrees with other MAB simulation studies (Hospital, 2003; Servin et al., 2004). Several empirical studies implied a similar marker distance with average donor segment lengths of about 34 cM in BC2F1 and 29 cM in BC3F1 (Xi et al., 2008), and 36 cM in BC2F2 (Septiningsih et al., 2003). This whole genome coverage will be easier to achieve with the availability of the approx. 18 000 SSR markers identified in the rice genome (McCouch et al., 2002; Matsumoto et al., 2005) in addition to a number of indel markers (Shen et al., 2004). Based on available sequence information, markers can also be developed to be able to identify the recombinant breakpoints and measure the introgression size more precisely. The sizes of the subsequent backcross and selfed populations can be adjusted accordingly depending on the number of the remaining background introgressions and the desired size of the target introgression. In the present Sub1 case study, the average size of introgression of the selected plants in BC2 and BC3 generations was 7 Mb (approx. 28 cM) and 1·5–2·5 Mb (approx. 6–10 cM), respectively. However, it is possible to identify plants with a very small introgression and a restored recipient background in the BC2F2 population by increasing the number of plants in BC1F1 and the following generations. This was the case with BR11-Sub1, which will be reported elsewhere (K. Iftekharuddaula et al., IRRI, The Philippines pers. comm.). The Sub1 lines with a limited introgression size at the target region were developed to prevent any negative linkage drag, which will be even more important when these lines are used as recipients of other QTLs in efforts to pyramid Sub1 with other traits.
Upon completion of the conversion of the mega varieties, evaluation of submergence tolerance showed that all Sub1 introgression lines had a significantly higher survival rate compared with their original parents (Table 3). This demonstrated that Sub1 confers tolerance in different genetic backgrounds. However, there were some differences in the level of tolerance among the Sub1 lines. Varieties that recovered faster after desubmergence tended to have better vegetative growth. This suggested that recovery is a very important component of submergence tolerance, and the underlying physiological mechanisms deserve further examination. In addition, the data suggested that there seemed to be a correlation between the amount of Sub1A expression in the original parents and the level of tolerance of their corresponding improved varieties. In this case, for example, the amount of Sub1A mRNA in IR64 was more than that of BR11 (Fig. 1B); correspondingly, the percentage survival rates of IR64 and IR64-Sub1 were higher than those of BR11 and BR11-Sub1 (Tables 3 and 4). However, whether this holds true for other varieties, and whether there are other genes that control the expression of Sub1A, requires further analysis. Evaluation of grain quality, yield performance and agronomically important traits of the first three Sub1 lines (Swarna-Sub1, Samba Mahsuri-Sub1 and IR64-Sub1) grown under control and stress conditions at IRRI was encouraging. The Sub1 lines were superior under stress and there was no significant difference under non-stress conditions (S. Singh et al., IRRI, The Philippines unpubl. res.). Preliminary field trials at different national agricultural research and extension systems (NARES) sites in India and Bangladesh confirmed these results (Sarkar et al., 2006). Evaluation of the six Sub1 mega varieties under shallow and submerged conditions at the IRRI farm as well as in other countries in flood-prone areas of south and south-east Asia is currently ongoing.
The MAB system developed will be useful in converting additional tolerant varieties, especially if they are derived from one of the mega varieties used in this study. IR64 is an example of a variety that has been used widely in breeding in south and south-east Asia, as well as in Africa, and many modern varieties in south-east Asia are derived from IR64. In cases where the recipient genome is closely related to the Sub1 donor, the MAB approach can be modified to facilitate identification of ideal plants as early as the BC1F2 generation and without using recombinant selection. PSB Rc82, PSB Rc18 (from the Philippines) and Ciherang (from Indonesia) are popular varieties that are closely related to IR64. An attempt is currently being made to develop Sub1 versions of these varieties with one backcross using IR64-Sub1 as a donor.
Performance of plants with recombination within the Sub1 gene cluster
The present RT-PCR analyses showed that Sub1A and Sub1C expression is undetectable under non-submerged conditions (Fig. 1B). Furthermore it was observed that the tolerant Sub1A-1 was associated with lower expression of the Sub1C-3 intolerant allele as indicated by the slightly lower expression in BR11-Sub1A1 as compared with that of BR11. These results are in agreement with data reported by Xu et al. (2006) showing reduced Sub1C-2 expression in response to the overexpression of Sub1A-1 in transgenic Liaogeng, a variety that does not naturally possess the Sub1A gene. However, suppression of Sub1C seems to be independent of the Sub1A allele since Sub1C-1 is neither expressed in IR64-Sub1BC plants with Sub1A-2 nor in IR64-Sub1 plants that carry the Sub1A-1 allele (Fig. 1B). In other words, the absence of Sub1C expression is observed in plants with high and low levels of Sub1A expression, suggesting that Sub1C expression is not solely regulated by Sub1A.
It was noted that limited expression of Sub1C was associated with tolerance (Xu et al., 2006). However, even though there was no expression of the Sub1C-1 allele, in the presence of Sub1A-2, as indicated in IR64-Sub1BC, the plants remained intolerant (Table 4). It was proposed that Sub1A represses shoot elongation and carbohydrate consumption under submergence stress due to reduced ethylene-mediated GA biosynthesis and Sub1C suppression (Fukao et al., 2006; Perata and Voesenek, 2007). According to these reports, suppression of Sub1C in tolerant plants is important to prevent the Sub1C mediated up-regulation of the α-amylase gene Ramy3D involved in starch degradation, thereby preserving carbohydrate reserves needed for recovery after de-submergence. At the same time, Sub1A-1 is important for down-regulation of Expansin A and the sucrose synthase Sus3. Based on the intolerant nature of the recombinant line IR64-Sub1BC, however, we speculate that most likely, the contribution of Sub1A mediated suppression of cell elongation and energy consumption is more substantial than the contribution of Sub1C-1 through its related downstream enzymes.
Interestingly, RT-PCR results also showed that under submerged conditions the Sub1C-1 allele was accompanied by slightly reduced expression of Sub1A-2 as observed for IR64-Sub1BC compared with that of IR64. In Sub1C-3 a similar trend was observed in both Sub1A alleles. Thus, when the Sub1C-3 allele was present, a more substantial decrease of Sub1A expression was observed in the allele Sub1A-2 (BR11) compared with that for the allele Sub1A-1 (BR11-Sub1A). We hypothesize that to some degree the Sub1C gene might have a feed-back restriction on Sub1A mRNA accumulation. This role can be more clearly seen when Sub1C-1 was combined with the Sub1A-2-intolerant allele. However, this early finding needs further investigation and confirmation. Nonetheless, since Sub1A with its related downstream genes most likely has the more prominent role in controlling cell growth and energy consumption as suggested, it overrides the role of Sub1C. And even in the situation when there was no detectable Sub1C-1 expression and this was accompanied by lower expression of the Sub1A-2-intolerant allele as in the case of IR64-Sub1BC, the plants remained intolerant. This suggested that the Sub1C-1 allele by itself does not provide a significant contribution to tolerance. Additionally, the present results also showed that even though the expression of Sub1A-1 was slightly reduced in the presence of the Sub1C-3-intolerant allele, it does not significantly reduce the level of tolerance (Table 4). This further confirms that Sub1A is the major determinant of submergence tolerance.
Performance of plants heterozygous for Sub1A allele
F1 progenies derived from crosses of IR64/IR64-Sub1 were significantly less tolerant compared with the tolerant parent IR64-Sub1. In addition, Sub1A expression in F1 plants was lower compared with IR64-Sub1 plants. It was also noted that BR11 breeding lines with a heterozygous Sub1 locus were less tolerant to submergence stress (K. Iftekharuddaula, IRRI, The Philippines pers. comm.). This preliminary study suggested that the tolerant allele of Sub1 should be in both parents in order to maintain the high level of tolerance in hybrids. However, the moderate tolerance of heterozygous hybrids may be beneficial under some circumstances where a combination of moderate submergence tolerance and moderate elongation ability may be desirable as is the case when stagnant longer-term partial flooding is anticipated before or after submergence.
Improving submergence tolerance beyond Sub1
Making further improvements in submergence tolerance will require identifying genes that confer a higher level of submergence tolerance and combining the Sub1 gene with other traits needed for adaptation to flooding. It is known that other QTLs in FR13A contribute towards its higher level of submergence tolerance (Nandi et al., 1997; Toojinda et al., 2003). In addition, several moderately tolerant varieties that do not possess the tolerant Sub1A-1 allele have been identified. F3 populations have been developed, and submergence screening is underway to determine if these have novel QTLs that could act additively with Sub1.
One potential drawback of the converted mega varieties is that they are not tolerant to conditions of prolonged partial flooding above 20 cm water depth. Such stagnant flooding or water logging is common in the rainfed lowlands of south and south-east Asia. Varieties like Swarna-Sub1 are quite short, and if water depth remains at or above the canopy level for longer than 2 weeks, the plants may not be able to elongate and continue growth, due to the Sub1-mediated suppression of elongation. This problem does not seem to occur in Sub1 varieties with taller plant stature, such as IR49830. In preliminary screening at IRRI using a water depth 20–60 cm for a 50-d duration, several rice accessions were identified as tolerant, based on the yield performance. While, the first three developed Sub1 lines were not tolerant of this condition, IRRI 119 (released as PSB Rc68 in the Philippines), which also possesses Sub1, survived well. This variety is taller compared with the first three converted Sub1 varieties. This observation suggests that Sub1 should be introgressed into taller plant types for areas where both stagnant water and submergence stress occur.
Varieties with tolerance to submergence during germination are needed especially in the regions where direct-seeding is a common practice. Some advancement has been made in the breeding of advanced lines conferring tolerance to submergence during germination using tolerant donors, such as Khaiyan, Khao Hlan Ohn and Ma-zhan (Red), in combinations with other beneficial traits, such as high-yielding and better adaptation, including varieties with the Sub1 gene (Ismail et al., 2009). Some promising lines having good grain quality and showing tolerance to both flooding during germination and submergence are now at the F6 generation (data not shown), and seed multiplication is ongoing. It has been possible to combine both traits in the same line, because they appear to be independent traits and are controlled by different mechanisms (Perata and Voesenek, 2007).
With the complexity of the problems associated with farming in marginal lands, it has been quite challenging to provide farmers with a packet of rice technologies that can be adopted under the erratic weather and severe environments caused by different kinds of abiotic stresses (Ismail et al., 2007). The dramatic effect of Sub1 in conferring submergence tolerance and recent progress in the identification of major QTLs conferring tolerance to some abiotic stresses, e.g. salinity, phosphorus deficiency and drought (Ismail et al., 2007) greatly enhances the prospects of combining this trait with tolerances to other important abiotic stresses.
In summary, it has been demonstrated that Sub1 confers submergence tolerance in different genetic backgrounds. Based on the recombinant genetic studies, Sub1A was confirmed as the major determinant of submergence tolerance, whereas alleles of the Sub1C gene did not significantly affect level of tolerance. The low level of Sub1A expression in heterozygous plants indicates that both parents used for hybrid rice production should carry the tolerant Sub1A-1 allele to reach the critical threshold of expression needed for tolerance. Lastly, significant progress has been achieved in developing improved varieties having a higher level of tolerance and wider adaptation by combining Sub1 with other traits. Some of our achievements have been very timely with the increasing vulnerability of rice farming to flash floods and other abiotic stresses currently being provoked by the recent trends in climate change.
ACKNOWLEDGEMENTS
We thank J. Mendoza, E. Suiton, G. Perez, R. Garcia, N. Ramos, K. Ifthekaruddaula, E. Ella and T. D. T. Minh for technical assistance, and Namrata Singh for designing the Sub1A203R primer. This work was partially supported by grants from the German Federal Ministry for Economic Cooperation and Development (03·7860·4-001·00) and the Japan Ministry of Foreign Affairs.
LITERATURE CITED
- Collard BC, Mackill DJ. Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philosophical Transactions of the Royal Society of London B Biological Sciences. 2008;363:557–572. doi: 10.1098/rstb.2007.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leon TB. Gene expression of selected candidate genes in near isogenic lines of rice Oryza sativa L.) during susceptible and resistance responses to rice tungro spherical virus (RTSV) The Philippines: University of the Philippines Los Baños; 2007. MS Thesis. [Google Scholar]
- Fukao T, Xu K, Ronald PC, Bailey-Serres J. A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation response to submergence in rice. The Plant Cell. 2006;18:2021–2034. doi: 10.1105/tpc.106.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HilleRisLambers D, Vergara BS. Summary results of an international collaboration on screening methods for flood tolerance. Proceedings of the 1981 International Deepwater Rice Workshop.; Los Baños: International Rice Research Institute; 1982. pp. 347–353. [Google Scholar]
- Hospital F. Marker-assisted breeding. In: Newbury HJ, editor. Plant molecular breeding. Oxford: Blackwell; 2003. pp. 30–59. [Google Scholar]
- Ismail AM, Heuer S, Thomson MJ, Wissuwa M. Genetic and genomic approaches to develop rice germplasm for problem soils. Plant Molecular Biology. 2007;65:547–570. doi: 10.1007/s11103-007-9215-2. [DOI] [PubMed] [Google Scholar]
- Ismail AM, Ella ES, Vergara GV, Mackill DJ. Mechanisms associated with tolerance to flooding during germination and early seedling growth in rice (Oryza sativa L.) Annals of Botany. 2009;103:197–209. doi: 10.1093/aob/mcn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M, et al. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.) DNA Research. 2002;9:199–207. doi: 10.1093/dnares/9.6.199. [DOI] [PubMed] [Google Scholar]
- Mackill DJ. Progress in rainfed lowland rice. Los Baños: International Rice Research Institute; 1986. Rainfed lowland rice improvement in South and Southeast Asia: results of a survey; pp. 115–144. [Google Scholar]
- Mackill DJ, Amante MM, Vergara BS, Sarkarung S. Improved semidwarf rice lines with tolerance to submergence of seedlings. Crop Science. 1993;33:749–753. [Google Scholar]
- Mackill DJ, Collard BCY, Neeraja CN, Rodriguez RM, Heuer S, Ismail AM. QTLs in rice breeding: examples for abiotic stresses. In: Brar DS, Mackill DJ, Hardy B, editors. Rice genetics 5: Proceedings of the International Rice Genetics Symposium. Manila: International Rice Research Institute; 2006. pp. 155–167. [Google Scholar]
- Matsumoto T, Wu JZ, Kanamori H, Katayose Y, Fujisawa M, Namiki N, et al. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Mohanty HK, Chaudhary RC. Progress in rainfed lowland rice. Los Baños: International Rice Research Institute; 1986. Breeding for submergence tolerance in rice in India; pp. 191–200. [Google Scholar]
- Nandi S, Subudhi PK, Senadhira D, Manigbas NL, Sen-Mandi S, Huang N. Mapping QTLs for submergence tolerance in rice by AFLP analysis and selective genotyping. Molecular and General Genetics. 1997;255:1–8. doi: 10.1007/s004380050468. [DOI] [PubMed] [Google Scholar]
- Neeraja C, Maghirang-Rodriguez R, Pamplona A, Heuer S, Collard B, Septiningsih E, et al. A marker-assisted backcross approach for developing submergence-tolerant rice cultivars. Theoretical and Applied Genetics. 2007;115:767–776. doi: 10.1007/s00122-007-0607-0. [DOI] [PubMed] [Google Scholar]
- Perata P, Voesenek LACJ. Submergence tolerance in rice requires Sub1A, an ethylene-response-factor-like gene. Trends in Plant Science. 2007;12:43–46. doi: 10.1016/j.tplants.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Prigge V, Maurer HP, Mackill DJ, Melchinger AE, Frisch M. Comparison of the observed with the simulated distributions of the parental genome contribution in two marker-assisted backcross programs in rice. Theoretical and Applied Genetics. 2008;116:739–744. doi: 10.1007/s00122-007-0707-x. [DOI] [PubMed] [Google Scholar]
- Sarkar RK, Reddy JN, Sharma SG, Ismail AM. Physiological basis of submergence tolerance in rice and implications for crop improvement. Current Science. 2006;91:899–906. [Google Scholar]
- Septiningsih EM, Prasetiyono J, Lubis E, Tai TH, Tjubaryat T, Moeljopawiro S, et al. Identification of quantitative trait loci for yield and yield components in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. Theory Applied Genetics. 2003;107:1419–1432. doi: 10.1007/s00122-003-1373-2. [DOI] [PubMed] [Google Scholar]
- Servin B, Martin OC, Mezard M, Hospital F. Toward a theory of marker-assisted gene pyramiding. Genetics. 2004;168:513–523. doi: 10.1534/genetics.103.023358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YJ, Jiang H, Jin JP, Zhang ZB, Xi B, He YY, et al. Development of genome-wide DNA polymorphism database for map-based cloning of rice genes. Plant Physiology. 2004;135:1198–1205. doi: 10.1104/pp.103.038463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siangliw M, Toojinda T, Tragoonrung S, Vanavichit A. Thai jasmine rice carrying QTLch9 (SubQTL) is submergence tolerant. Annals of Botany. 2003;91:255–261. doi: 10.1093/aob/mcf123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toojinda T, Siangliw M, Tragroonrung S, Vanavichit A. Molecular genetics of submergence tolerance in rice: QTL analysis of key traits. Annals of Botany. 2003;91:243–253. doi: 10.1093/aob/mcf072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toojinda T, Tragoonrung S, Vanavichit A, Siangliw JL, Pa-In N, Jantaboon J, et al. Molecular breeding for rainfed lowland rice in the Mekong region. Plant Production Science. 2005;8:330–333. [Google Scholar]
- Xi ZY, He FH, Zeng RZ, Zhang ZM, Ding XH, Li WT, et al. Characterization of donor genome contents of backcross progenies detected by SSR markers in rice. Euphytica. 2008;160:369–377. [Google Scholar]
- Xu K, Mackill DJ. A major locus for submergence tolerance mapped on rice chromosome 9. Molecular Breeding. 1996;2:219–224. [Google Scholar]
- Xu K, Xu X, Ronald PC, Mackill DJ. A high-resolution linkage map in the vicinity of the rice submergence tolerance locus Sub1. Molecular and General Genetics. 2000;263:681–689. doi: 10.1007/s004380051217. [DOI] [PubMed] [Google Scholar]
- Xu K, Xia X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, et al. Sub1A is an ethylene response factor-like gene that confers submergence tolerance to rice. Nature. 2006;442:705–708. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]
- Zheng K, Subudhi PK, Domingo J, Magpantay G, Huang N. Rapid DNA isolation for marker assisted selection in rice breeding. Rice Genetics Newsletter. 1995;12:255–258. [Google Scholar]