Abstract
Background and Aims
Large areas of the globe are becoming saline due to evapotranspiration and poor irrigation practices, and sustainability of agriculture is being seriously affected. Thiourea (TU) has been identified as an effective bioregulator imparting stress tolerance to crops. The molecular mechanisms involved in the TU-mediated response are considered in this study.
Methods
Differential display was performed in order to identify TU-modulated transcripts in Brassica juncea seeds exposed to various treatments (distilled water; 1 m NaCl; 1 m NaCl + 500 p.p.m. TU). The differential regulation of these transcripts was validated by quantitative real-time PCR.
Key Results
Thiourea treatment maintained the viability of seeds exposed to NaCl for 6 h. Expression analysis showed that the transcript level of alpha, beta, gamma, delta and epsilon subunits of mitochondrial ATPase (mtATPase) varied in seeds subjected to the different treatments for 1 h: expression level was significantly altered by 1 m NaCl relative to controls; however, in the NaCl + TU treatment it reverted back in an integrated manner. Similar results were obtained from time-kinetics studies of beta and delta subunits in roots of 8-d-old seedlings. These observations were also confirmed by the mtATPase activity from isolated mitochondria. The reversal in the expression and activity profile of mtATPase through the application of a bioregulator such as TU is a novel finding for any plant system.
Conclusions
The results suggest that TU treatment maintains the integrity and functioning of mitochondria in seeds as well as seedlings exposed to salinity. Thus, TU has the potential to be used as an effective bioregulator to impart salinity tolerance under field conditions, and might prove to be of high economic importance by opening new avenues for both basic and applied research.
Key words: Brassica juncea, differential display, mitochondrial ATPase, real-time PCR, salinity stress, thiourea
INTRODUCTION
The distinguishing characteristic of saline soils from the agricultural point of view is that they contain high concentration of neutral soluble salts. Saline soils can be defined as those that have an electrical conductivity of the saturated soil extract of more than 4 dS m−1 at 25 °C (Richards, 1954). Based on the FAO/UNESCO soil map of the world, the total area of saline soils is 397 million ha and that of sodic soils is 434 million ha, which are not necessarily arable but cover all salt-affected lands of the globe. Of the current 230 million ha of irrigated land, 45 million ha have salt-affected soils (19·5 %) and of the almost 1500 million ha of dry-land agriculture, 32 million have salt-affected soils (2·1 %), having varying degrees of salinity caused by human-induced processes (Oldeman et al., 1991). Salinity stress adversely affects the growth of several crop plants. It causes poor and patchy stands of crops, uneven and stunted growth and poor yields, with the extent of damage depending upon the degree of salinity. The primary effect of excess salinity is that it renders water less available to plants through adverse osmotic effects on the soil solution. In addition, excessive concentrations and absorption of individual ions may prove toxic to the plants and/or may retard absorption of other essential plant nutrients. In order to mitigate such adverse effects, an effective bioregulator molecule is required that can give enhanced tolerance under saline situations. In previous research, several field trials have shown that pre-treatment of mustard and wheat seeds with certain thiols such as thiourea (TU), dithiothreitol (DTT) and thioglycollic acid (TGA), or their foliar spray, increases yield under salt and drought conditions (Sahu and Singh, 1995; Burman et al., 2004; Sahu et al., 2005, 2006). Recently, it has also been shown that thiol treatment enhances the translocation of sucrose from source to sink in Brassica juncea (Srivastava et al., 2008). In order to establish whether TU acts as a bioregulatory molecule, we have attempted to study the molecular mechanism of its action. TU-responsive and salinity-dependent transcripts were identified from Brassica juncea seeds by differential display and their differential regulation was validated by quantitative real-time PCR. The data obtained showed that mitochondrial ATPases (FoF1-ATP synthase) play an important role in TU-induced salt tolerance in Brassica juncea.
Mitochondrial FoF1-ATP synthase is a multimeric enzyme, in which F1 is a peripheral component located on the face of the inner membrane carrying the catalytic sites for ATP synthesis/hydrolysis (Sane et al., 1996). The other component, Fo, is embedded in the hydrophobic region of the membrane so as to constitute a proton channel (Pedersen et al., 2000). Prokaryotes and eukaryotes have the same F1 structures, but a somewhat different Fo structure (Boyer, 1997; Jones et al., 1998; Kinosita et al., 1998; Mueller, 2000). The F1 structure consists of five subunits designated α, β, γ, δ and ε, in decreasing order of their molecular weights, having stoichiometries of α3β3γ1δ1ε1. Both of the two major subunits, α and β, contain a nucleotide binding site and play essential roles in the catalytic function of the F1 component. The amino acid sequences of these two major subunits are not only highly conserved during evolution but also related to each other. The other subunits (γ, δ and ε) are considered as minor subunits (Amzel and Pedersen, 1983).
The present study was aimed at investigating whether the effect of TU is mediated at the molecular level. The finding of an involvement of mitochondrial ATPase (mtATPase) is of interest for elucidating the molecular components of TU-mediated stress tolerance. This will also help us to decipher the regulation of mitochondrial ATP synthase activity and to understand the integrative function of each ATPsynthase subunit in response to environmental stresses.
MATERIALS AND METHODS
Plant material, stress induction and thiol treatment
Seeds of Brassica juncea (genetically homogeneous) were surface-sterilized with 30 % ethanol for 3 min and then washed thoroughly. Salinity stress was applied by soaking the seeds in 1 m NaCl. The optimum concentration of thiourea (TU) was standardized as 500 p.p.m. (6·5 µmol/mL), and to monitor its protective effects seeds were soaked in 1 m NaCl along with 500 p.p.m. TU. Seeds soaked in distilled water were considered as control treatments. For differential display, seeds were soaked in the respective solution for 1 h and then used for RNA isolation.
The differential germination response of seeds was assessed under the above treatments. Seeds were soaked for 6 h in the respective solutions and were then spread on a single Petri plate in separate lanes. Seeds were allowed to germinate in the presence of distilled water for 2 d under a 12-h light/dark cycle at 25 °C and 70 % humidity. A comparative germination percentage was followed under similar conditions for 2 d.
Temporal regulation of major (beta) and minor (delta) subunits was analysed in the roots of 8-d-old seedlings subjected independently to different treatments (distilled water; 1 m NaCl; 1 m NaCl + 500 p.p.m. TU). At time intervals of 15, 30 and 60 min, roots were harvested for expression profiling of beta and delta subunits.
In order to assess the change in mtATPase activity, four sets of seeds were allowed to germinate independently under a distilled water control for 24 h and were then exposed to either NaCl (0·7 m), NaCl (0·7 m) + TU (500 p.p.m.), or TU (500 p.p.m.), with one set of seeds being maintained in distilled water as a control. After 12 h of stress treatment, 4 g of seeds from each set was taken for mitochondria isolation and determination of ATPase activity.
RNA isolation and DNase treatment
RNA extraction was done using TRI reagent (Sigma, T 9424), as per the manufacturer's instructions. A message clean kit (GenHunter Corporation, Nashville, TN) was used for the removal of genomic DNA contamination. The integrity and quantity of RNA was checked by electrophoresis of total RNA (1 µg) on 1·2 % denaturing agarose gel (Sambrook et al., 1989; Vincze and Bowra, 2005). An optical density ratio (260/280) of >1·95 and the presence of two intact 28S and 18S rRNA bands were used as quality control parameters for the RNA to be used for differential display studies.
Production of differential transcripts
DNA-free total RNA (0·2 µg) was reverse transcribed using the oligo-dT primer. The cDNA obtained was subjected to differential display using an RNA image kit (GenHunter Corporation, Nashville, TN) as per the protocol and recommendations, with some adjustment and modifications (Minglin et al., 2005; Casimiro et al., 2006); details of the primers used for differential display are given in Table 1. During amplification, radioactive labelling was achieved by adding α32P-dATP (procured from BRIT-CCMB, India). After amplification, the products were run on a 6 % denaturing polyacrylamide gel. After a complete run, the gel was dried on a Whatman 3MM filter paper and an autoradiogram was developed. Once differentially induced bands had been identified from the autoradiogram, the hyperfilm was aligned with the dried gel and bands of interest were cut from the gel. The DNA from the dried gel was eluted by boiling it with 100 µL of water at 94 °C for 20 min. To prevent the selection of any false-positive bands all the amplifications were conducted three times and only those bands were selected that showed a similar pattern in all three independent experiments. One parallel DNA control was also maintained for every treatment along with the corresponding test reaction. The band detected in the test lane that was similar to the corresponding band present in the DNA control was not considered for further analysis.
Table 1.
Details of the anchored and arbitrary primers used for differential display
| Properties | Anchored primer | Arbitrary primer |
|---|---|---|
| Sequence | 5′-AAGCTTTTTTTTTTTG-3′ | 5′-AAGCTTCGACTGT-3′ |
| Length | 16 nt | 13 nt |
| Tm | 38·0 | 38·0 |
| GC % | 18·8 % | 46·2 % |
| ΔG | −29·7 kCal mol−1 | −23·3 kCal mol−1 |
| Molecular weight | 4920·2 g mol−1 | 4011·6 g mol−1 |
| Loop formation | None | None |
Re-amplification and cloning of differential transcripts
The DNA eluted from differential bands was subjected to re-amplification using the same set of the anchored and arbitrary primers (Liang et al., 1994). A portion of the re-amplified DNA was run on a 6 % polyacrylamide gel to make sure that amplification had taken place. This gel was used to quantify the amount of DNA and its size. After quantification, each of the differential transcripts was cloned in pTZ57R/T (MBI, Fermentas), as per the manufacturer's instructions.
Sequencing and data analysis
Differential products were sequenced using an automated capillary sequencer. All the sequences obtained in this study are currently available in dbEST database (Boguski et al., 1993) under the accession numbers. Sequence alignment and homology searches were performed with the BLAST N search algorithm (Altschul et al., 1990) in the GenBank NR database.
Design of primers and optimization of concentration
All the primers used for the SyBr green real-time RT-PCR were obtained from the Arabidopsis thaliana RT-PCR primer pair database (Han and Kim, 2006; see Table 2 for details of the primers used). Primers were obtained from Metabion International (www.metabion.com/). Specificity of all the primers was confirmed by sequence analysis of RT-PCR amplicons derived from Brassica juncea.
Table 2.
Gene-specific primers used for real-time PCR
| Subunits for mtATPase | Genopp ID* | Expected amplicon size (bp) |
|---|---|---|
| Alpha | At2g07698·1f97r535 | 349 |
| Beta | At5g08680·1f24r380 | 340 |
| Gamma | At2g33040·1f258r362 | 142 |
| Delta | At5g13450·1f100r5 | 149 |
| Epsilon | At1g51650·1f148r20 | 143 |
| Reference Gene | ||
| Actin† | At3g12110·1f348r361 | 155 |
* The Genopp ID can be used to locate details of the primer from the AtRTPrimer database (http://pombe.kaist.ac.kr/blan/genoPP.pl).
† Actin is used as a reference gene, allowing the gene expression values to be normalized.
Primer optimal concentrations of target and reference genes were determined with serial dilutions of cDNA obtained from 10 µg of RNA isolated from Brassica juncea seeds soaked for 1 h. For all the genes analysed, 12·5 pmoles of the primer were used in 25 µL of PCR reaction mix.
Real-time quantitative RT-PCR
DNA-free intact RNA (10 µg) was taken and then subjected to cDNA synthesis using a Stratagene high fidelity 1st-strand cDNA synthesis kit, as per the manufacturer's instructions (www.stratagene.com/). To minimize the potential effects of synthesis efficiency during the RT reaction, three separate cDNA synthesises were performed and pooled for each RNA preparation. The cDNA were then stored at –20 °C until being used for real-time PCR. Two-step RT-PCR was chosen in order to avoid the problem of primer dimer formation, which is associated with one-step RT-PCR (Brownie et al., 1997). The oligo-dT primer was used for cDNA synthesis so that the same cDNA pool could be used for analysis of all the genes. Real-time quantitative RT-PCR was carried out using a Corbett rotor gene 3000 (Corbett Life Science, www.corbettlifescience.com/; Ali-Benali et al., 2005; Bustin et al., 2005; Nolan et al., 2006). Arabidopsis thaliana actin gene was amplified in parallel with the target gene, allowing for gene-expression normalization and providing quantification. Before using it as a reference gene (Radoni et al., 2004), it was verified that its level remained unchanged under all the given treatments. The PCR efficiency of the reference and target genes was also checked and found to be approximately equal in a range of 1·96–1·99 (Tichopad et al., 2003). Detection of real-time RT-PCR products was done using SYBR Green 2x Master Mix kit (S 4320, Sigma), following the manufacturer's instructions. The quantity of cDNA used as a template for PCR was 2·5 µg (the equivalent of 500 ng of total RNA). The PCR cycling conditions comprised of an initial cycle at 50 °C for 2 min followed by 1 cycle at 95 °C for 10 min and 40 cycles each comprising of 95 °C for 50 s, 57 °C for 50 s and 72 °C for 40 s. For each sample, reactions were set up in triplicate to ensure the reproducibility of the results.
Data analysis
At the end of each PCR run, a melting curve was generated and analysed with the dissociation curve software built into the Corbett rotor gene 3000 (Corbett Life Science, www.corbettlifescience.com). The melt curve obtained depends on the GC/AT ratio and the overall length of the amplicon. This analysis allowed products to be distinguished from one another and can identify primer dimers or other erroneous dsDNA (Ririe et al., 1997). A relative expression ratio plot was generated using the software REST-MCS (Pfaffl et al., 2002). Significant differences in the expression profile were tested by one-way ANOVA.
Isolation of mitochondria
Isolation of mitochondria was performed using the percoll density gradient method as described by Hajek et al. (2004) with some modifications in the steps of percoll removal: the pellet was washed twice with wash solution (0·3 m sorbitol, 1 mm DTT and 20 mm HEPES-NaOH, pH 7·5) at 25 000 g for 20 min. The final mitochondria pellet was suspended in the minimum volume of suspension medium (same as wash solution plus BSA at 0·2 mg mL−1). Succinate dehydrogenase activity was used as a marker for detecting the purity of the mitochondrial fraction (Gopalkrishnan and Rao, 2006).
Measurement of ATPase activity
Activity of mtATPase was measured by the method described by Gallagher and Leonard (1982) by estimating the phosphate (Pi) produced after the reaction (Gopalkrishnan and Rao, 2006). Significant differences in the specific activity between different treatments were tested by one-way ANOVA (P < 0·01, n = 6).
RESULTS
Differential germination response of seeds in response to thiourea
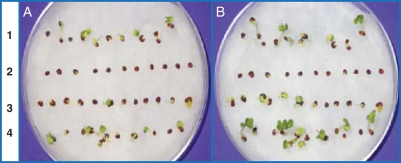
The differential germination responses of seeds under the various treatments are shown in Fig. 1. After Day 1, seeds soaked in distilled water and TU showed 100 % germination and were characterized by a fully developed radical and cotyledons (Fig. 1A). In contrast, the germination of seeds soaked in NaCl was only 18 %. Seeds soaked in NaCl + TU showed intermediate behavior; germination percentage was 100 % but the growth of seeds was severely reduced. After Day 2, the growth pattern of TU-soaked seeds was better than any other treatments (Fig. 1B). In seeds soaked in NaCl growth was completely inhibited, while in the NaCl + TU treatment the growth pattern was almost comparable to that of the control.
Fig. 1.
Differential germination responses of Brassica juncea seeds under different treatments. Lanes marked as 1, 2, 3 and 4 represent seeds soaked in distilled water, 1 m NaCl, 1 m NaCl + 500 p.p.m. TU, and 500 p.p.m. TU, respectively for 6 h and then allowed to germinate under normal conditions: (A) after 1 d, and (B) after 2 d.
Identification of salinity-dependent thiourea-modulated genes
Out of the total cDNA bands obtained (around 150), eight were selected on the basis of their differential regulation with respect to TU as well as salinity stress. These bands showed either up- or down-regulation in the presence of salinity, but they matched towards the level of distilled water when TU was provided along with the NaCl treatment. Out of the eight, three bands could not be re-amplify due to insufficient DNA, and two more gave small-sized, poorly defined bands upon re-amplification; thus, for these five bands we could not proceed with further analysis. However, the remaining three cDNAs, which gave well-defined, single bands, were cloned into the pTZ57R/T vector. The size of the cloned cDNA fragments ranged from 150–350 bp. A detailed description of all three cDNA clones is given in Table 3.
Table 3.
Details of the differential display data
| Clone | Accession number | Size of PCR fragment (bp) | Source | Nature of expression |
|---|---|---|---|---|
| DD1 (AJR001) | EE111313 | 290 | Seed | Present in all the treatments except NaCl |
| DD2 (AJR002) | EE111314 | 137 | Seed | Present in all the treatments except NaCl |
| DD3 (AJR003) | EE191543 | 337 | Seed | Absent in all the treatments, but present in NaCl |
Sequencing and identification of cDNA clones
The results of the NCBI BLAST are summarized in Table 4. Out of three cDNA clones, two were associated with the different subunit composition of the mtATPase. This provides evidence for the involvement of mtATPase in the protective effect of TU under salinity stress.
Table 4.
Sequence analysis details of the differential clones
| Description of the sequence showing maximum homology with the query sequence |
|||||
|---|---|---|---|---|---|
| Clone | Accession number | E-Value | % identity | Score | Description |
| DD1 (EE111313) | NM_126747·2 | 2e–135 | 98 | 489 | Arabidopsis thaliana ATP synthase alpha chain, mitochondrial, putative (AT2G07698) mRNA, complete cds |
| DD2 (EE111314) | EE464322·1 | 1e–43 | 92 | 183 | Brassica napus seed coat BNSCS2CT, Brassica napus cDNA |
| DD3 (EE191543) | AJ271468·1 | 3e–45 | 100 | 190 | Arabidopsis thaliana mRNA for mitochondrial F1 ATP synthase beta subunit (p_beta gene) |
Sequence alignment and homology searches were performed with the BLAST N search algorithm (Altschul et al., 1990) in the GenBank NR database.
Expression profiling of mtATPase subunits in Brassica juncea seeds
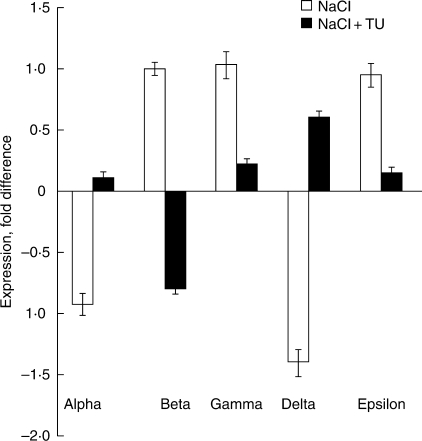
Treatment of seeds with NaCl for 1 h led to a 0·931- and 1·409-fold down-regulation of the alpha and delta subunits, respectively, as compared to the control (Fig. 2). But under similar conditions there was up-regulation of 0·999-, 1·027- and 0·944-fold for the beta, gamma and epsilon subunits, respectively. When TU was provided along with NaCl, changes in the expression profile were seen: with the exception of beta, all other subunits were up-regulated by 0·103-, 0·221-, 0·597- and 0·141-fold for alpha, gamma, delta and epsilon, respectively. The beta subunit was down-regulated 0·808-fold as compared to the control (Fig. 2).
Fig. 2.
Integrated expression profile of the various mtATPase subunits in treated seeds. The relative expression level is shown for the various subunits in seeds treated with NaCl or NaCl + TU for 1 h. The baseline expression (i.e. 0 on y-axis) represents the expression level observed in distilled-water controls. Values represent the means of three technical and two biological replicates (± s.d.). Differences in the mean values were found to be statistically significant at P < 0·01 (one-way ANOVA).
Temporal regulation of major (beta) and minor (delta) subunits of mtATPase in roots of 8-d-old seedlings
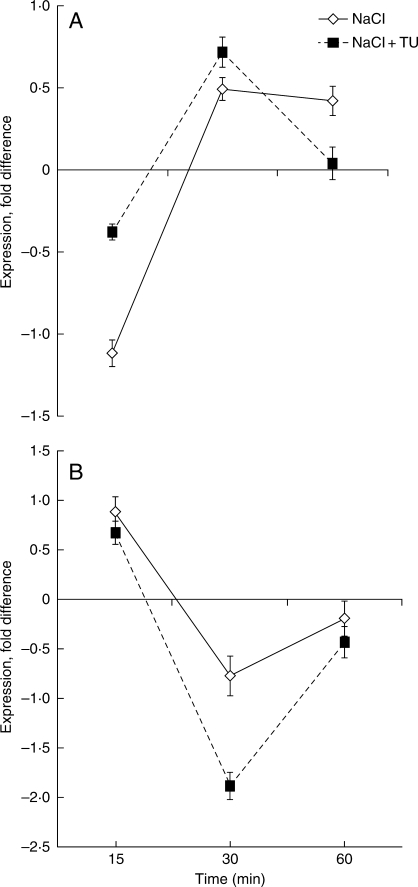
Temporal regulation of major and minor subunits was analysed in the roots of 8-d-old seedlings subjected independently to different treatments (distilled water; 1 m NaCl; 1 m NaCl + 500 p.p.m. TU). At time intervals of 15, 30 and 60 min, roots were harvested for expression profiling of beta and delta subunits. At 15 min, the beta subunit showed down-regulation in both the NaCl and NaCl + TU treatments; however, in NaCl the extent of down-regulation was more (−1·11-fold) as compared to NaCl + TU (−0·38-fold). After 15 min in the NaCl treatment there was a gradual increase in the expression that continued until 30 min and stabilized by 60 min, when the level of the beta subunit was 0·419-fold higher as compared to the control. In contrast, in the NaCl + TU treatment expression of the beta subunit increased after 15 min until 30 min and then it decreased up to 60 min; at 60 min the expression profile in the NaCl + TU treatment was similar to that of the control (Fig. 3A). Unlike the beta subunit, the delta subunit exhibited an opposite temporal regulation; at 15 min an up-regulation was seen at levels of 0·87- and 0·67-fold for the NaCl and NaCl + TU treatments, respectively. The expression decreased with time until 30 min and then an increasing trend was seen that continued up to 60 min (Fig. 3B). At both 30 and 60 min the extent of down-regulation appeared more in NaCl + TU than in NaCl alone.
Fig. 3.
Temporal regulation of (A) beta and (B) delta subunits of mtATPase in roots of 8-d-old seedlings. Expression levels were monitored in roots exposed to NaCl or NaCl + TU treatments for different time periods and are presented as relative to the values in distilled-water controls (i.e. 0 on y-axis). Values represent the mean of three technical and two biological replicates (± s.d.). Differences in the mean values were found to be statistically significant at P < 0·01 (one-way ANOVA).
Effect of thiourea on mtATPase activity
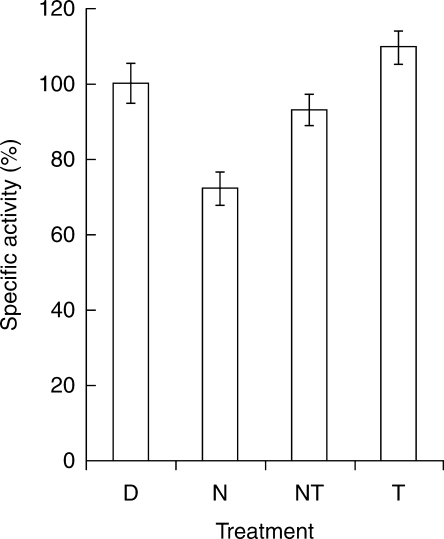
The activity of mtATPase was decreased by 28 % in response to NaCl as compared to the control. However, if TU was provided together with NaCl there was approximately 75 % reversal in the enzyme activity, which was comparable with that of the distilled-water control (Fig. 4).
Fig. 4.
Differential activity of mtATPase in 36-h-old seeds. Activity was monitored in 36-h-old seeds given different treatments after 24 h of germination. The data is represented in terms of percentage specific activity with reference to control in distilled water, D. Other treatments: N, NaCl; NT, NaCl + TU; T, TU. The values represent the mean of six technical and three biological replicates (± s.d.). Differences in the mean values were found to be statistically significant at P < 0·01 (one-way ANOVA).
DISCUSSION
Seeds of higher plants are known to be desiccation-tolerant and their mitochondria exhibit unique properties (Macherel et al., 2007). Respiration starts after a few minutes of imbibition and this affects the process of germination in several ways. For example, during imbibition the energy production by the mitochondria helps the seed to cope with the desiccation. This is dependent upon the most simple and robust mechanism of ATP production by ATP synthase. Another important role of mitochondria is to generate the hypoxic environment that is essential for proper seed germination. Hence, a decrease in the integrity and performance of the mitochondria during the early stages of germination can hamper the above-mentioned processes; thus, stress conditions lead to a decrease in germination and, ultimately, crop yield. Hence the application of any external stimuli or effector molecule that can help in maintaining mitochondrial functions may also maintain the viability of the seeds under stress. We have conducted field trials using various thiols, such as dithiothreitol (DTT), thiourea (TU) and thioglycollic acid (TGA), and found increased yields of mustard and wheat crops; application of TU was found to be the most cost effective and eco-friendly of these thiols (Srivastava et al., unpubl. res.). In the experiments desribed in this current study, the molecular mechanisms of TU action were investigated using Brassica juncea. Brassica was chosen instead of wheat because the whole-genome sequence of Arabidopsis thaliana is available, which has been helpful for conducting expression analysis in Brassica juncea (Girke et al., 2000).
Application of NaCl + TU was found to be effective in maintaining the functions of seed mitochondria under salinity stress. The primary evidence in support of this was derived from the seed viability assay. When seeds were exposed to 1 m NaCl for 6 h, they exhibited a drastic reduction in their germination ability and were not able to sprout even after then being subject to normal conditions (Fig. 1, lane 2). However, in 1 m NaCl + 500 p.p.m. TU, the germination ability of the seeds was retained to about control levels (Fig. 1, lane 3). Seeds soaked in 500 p.p.m. TU germinated normally, which showed that TU itself has no negative effect on the germination process. Differential display of seeds exposed to the various treatments for 1 h was performed in order to identify differentially induced transcripts. This technique was chosen because of its high sensitivity and its capability for simultaneous comparison of multiple RNA samples (Liang and Pardee, 1992; Landrum, 2006). Unlike the seed germination assay, the soaking time was kept to only 1 h because of our interest in identifying the regulatory genes associated with the TU treatment. Previous work with a mustard crop in the field had shown a salinity level of approx. 0·4 m, and thus a higher experimental NaCl concentration of 1 m was chosen in order ensure that a signalling effect would be observed (Sanders, 2000). Out of a total of 150 transcripts obtained, eight were found to be differentially expressed; among these eight, we could identify two as various subunits of mtATPase. Given that mtATPase has some role in TU-induced salt tolerance, expression of the five subunits (alpha, beta, gamma, delta and epsilon) was monitored using RT-PCR.
As the integrity and functioning of mitochondria is very important, the modulated expression of its various subunits points towards an important regulatory mechanism, which is somehow being controlled through the application TU. The alpha subunit of mtATPase has a critical function with respect to mitochondrial bioenergetics, and its turnover might be involved in the control of mitochondrial activity during germination (Rajjou et al., 2004). In the present study, it was found that it is down-regulated in seeds treated with NaCl for 1 h (Fig. 2). This could be because of increased generation of reactive oxygen species or because of changes in the Na+/K+ ratio associated with Na+ toxicity (Arora et al., 2002; Sairam and Tyagi, 2004). Stress caused by exposure to hydrogen peroxide has been shown to damage subunits of the ATP synthase of mitochondria (Sweetlove et al., 2002), and such an effect during the early stages of germination might have disturbed mitochondrial homeostasis, ultimately resulting in the poor seed germination. In contrast, in the NaCl + TU treatment the level of the alpha subunit was close to that of the control, which suggests a positive role of TU in maintaining mitochondrial function under salinity stress (Fig. 2). This was also reflected in terms of an increased germination percentage and a better growth pattern in the NaCl + TU treatment (Fig. 1, lane 3). Since TU is a thiol compound, it presumably changes the redox state of the system in such a way that the harmful effects of salinity stress are neutralized (Buchanan et al., 1994). A TU-mediated up-regulation of the alpha subunit under salinity stress has not been reported before, and thus appears to be a novel response. Some kind of dual role for this subunit has been suggested in animals system, with it being assigned the status of a heat-shock protein (Luis et al., 1990). It seems possible that the alpha subunit in plants may also perform a dual function; thus, its induction would lead not only to the restoration of mitochondrial function but also impart stress tolerance to the plant. In a recent study, Zhang et al. (2006) identified a salt-responsive gene from rice and found it to be a subunit of ATPsynthase. Its over-expression in tobacco made the transgenic plants more tolerant towards salinity stress.
The responses of the mtATPase subunits show considerable variation under different environmental stress and across species (Hamilton et al., 2001). For instance, when Arabidopsis thaliana cells are exposed to oxidative stress, the expression levels of both the alpha and beta subunits were significantly decreased (Sweetlove et al., 2002), whereas in case of aluminum stress ATPase activity increased significantly whilst the level of the alpha subunit remained the same (Hamilton et al., 2001). In the present study, NaCl treatment for 1 h to the seeds of Brassica juncea lead to the up-regulation of the beta subunit, up-regulation was not seen when TU was supplied along with the NaCl treatment (Fig. 2). In case of the gamma, delta and epsilon subunits also, it was found that the presence of TU along with NaCl reversed the expression profile seen under salinity stress (Fig. 2). In Oryza sativa, NaCl stress leads to an increase in the expression of the delta subunit (Zhang et al., 2006); interestingly, in Brassica juncea we found that this subunit is down-regulated under salinity stress. Such an integrated regulation in the transcript level of these subunits in response to TU treatment is likely to impose significant flux restrictions on the tricarboxylic acid cycle and electron transport, thereby limiting the synthesis of ATP (Sweetlove et al., 2002). This probably constitutes one of the mechanisms by which TU treatment reduces the extent of salinity-induced oxidative stress.
After evaluating the effect of TU on seed mitochondria, its effect were also monitored on mitochondria from other plant parts. Roots were chosen for this purpose as they are the first organs to come in direct contact with the stress; the integrity and functioning of root mitochondria need to be maintained in order to reduce damage under environmental stresses. The levels of the beta (Fig. 3A) and delta (Fig. 3B) subunits were monitored in roots of 8-d-old seedlings exposed to NaCl and NaCl + TU treatments for time intervals of 15, 30 and 60 min and were compared with controls. The beta and delta subunits were selected for this purpose as they showed opposite regulation in the NaCl and NaCl + TU treatments in seed mitochondria. After 60 min, similar profiles of the beta and delta subunits were obtained in roots treated with NaCl and NaCl + TU (Fig. 3A, B) as those observed in seeds (Fig. 2), but their levels were different. This is probably due to the fact that seed mitochondria are much more tolerant than those present in any other plant part (Macherel et al., 2007). The time-kinetics showed that in the NaCl + TU treatment the level of the beta subunit was very close to that of the control at 60 min, but at the same time in the NaCl treatment it was up-regulated, presumably due to minimization of stress-induced damage. In contrast to the beta subunit, the temporal regulation of the delta subunit showed an opposite trend; its expression was higher at 60 min in NaCl compared to that in NaCl + TU. Similar results were observed in rice under salt stress (Zhang et al., 2006). However, this is in contrast with the seeds, where the NaCl + TU treatment up-regulated the level of the delta subunit (Fig. 2). The expression analysis of the beta and delta subunits in roots and the results observed in seeds indicate that the regulation in the expression profile of the individual mtATPase subunits is an integrated and complex mechanism that is being modulated by a number of factors. It appears that TU affects this mechanism probably through redox regulation, which is supported by its chemical nature (Mittler, 2002). The other possibility is that as it is a compound containing –SH, it can also participate in ROS-mediated signalling and hence modulate some effectors that can neutralize the effect of stress (Pfannschmidt et al., 2001).
The activity of mtATPase was also assayed to see how it is regulated by the change in the expression profile of the various subunits. For this experiment, a salinity stress of only 0·7 m NaCl was used so that the viability of seed could be maintained for a longer duration. The results showed that mtATPase activity decreased in NaCl; however, it was at almost the same level as the control in the NaCl + TU treatment (Fig. 4). The relatively increased ATP production in the NaCl + TU treatment might support the energy-demanding processes associated with salt tolerance. Such an observation that TU changes the expression profile of the various subunits of mtATPase subunits in an integrated manner (Fig. 2) with an overall consequence of an increase in mtATPase activity (Fig. 4) indicates that the activity of mtATPase may possibly be regulated at the post-translational level as well (Hamilton et al., 2001).
In conclusion, the proper functioning of the mitochondria has to be maintained for proper germination to be possible under stress; salinity stress hampers the efficiency of components within the mitochondria (in the form of the various subunits of mtATPase), which leads to poor germination and a disrupted growth pattern. The presence of TU together with the NaCl treatment reverses the effect produced by salinity stress and thus maintains the effectiveness of the mitochondria, which is ultimately reflected in the form of proper germination and growth pattern under the stress. This is one of the mechanisms by which TU maintains mitochondrial homeostasis; however, further studies in this direction are needed to gain more information about the mechanisms of TU action. Although the full mechanism of TU action is not clear at present, its application as a bioregulatory technology certainly has the potential to provide an easy and applicable solution to ensure sustainable agriculture in salt-affected lands.
ACKNOWLEDGEMENTS
The authors wish to thank Dr S.K Apte, Associate Director (B) Biomedical Group and Head, Molecular Biology Division for providing the facilities for doing the differential display. We would also like to thank Mr S. Jay Kumar, Radiation Biology and Health Science Division, for his constant help while doing the real-time PCR.
LITERATURE CITED
- Ali-Benali MA, Alary R, Joudrier P, Gautier MF. Comparative expression of five lea genes during wheat seed development and in response to abiotic stresses by real-time quantitative RT-PCR. Biochimica et Biophysica Acta. 2005;1730:56–65. doi: 10.1016/j.bbaexp.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amzel LM, Pedersen PL. Proton ATPases: structure and mechanism. Annual Review of Biochemistry. 1983;52:801–824. doi: 10.1146/annurev.bi.52.070183.004101. [DOI] [PubMed] [Google Scholar]
- Arora A, Sairam RK, Srivastava GC. Oxidative stress and antioxidative system in plants. Current Science. 2002;82:1227–1238. [Google Scholar]
- Boguski MS, Lowe TMJ, Tolstoshev CM. dbEST – database for ‘expressed sequence tags. Nature Genetics. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- Boyer PD. The ATP synthase-a splendid molecular machine. Annual Review of Biochemistry. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- Brownie J, Shawcross S, Theaker J, et al. The elimination of primer-dimer accumulation in PCR. Nucleic Acid Research. 1997;25:3235–3241. doi: 10.1093/nar/25.16.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan BB, Schurmann P, Decottignies P, Lozano RM. Thioredoxin: a multifunctional regulatory protein with a bright future in technology and medicine. Archieves of Biochemistry and Biophysics. 1994;314:257–260. doi: 10.1006/abbi.1994.1439. [DOI] [PubMed] [Google Scholar]
- Burman U, Garg BK, Kathju S. Interactive effects of thiourea and phosphorus on clusterbean under water stress. Biologia Plantarum. 2004;48:61–65. [Google Scholar]
- Bustin SA, Benes V, Nolan T, Pfaffl MW. Quantitative real-time RT-PCR – a perspective. Journal of Molecular Endocrinology. 2005;34:597–601. doi: 10.1677/jme.1.01755. [DOI] [PubMed] [Google Scholar]
- Casimiro S, Tenreiro R, Monteiro AA. Identification of pathogenesis-related ESTs in the crucifer downy mildew oomycete Hyaloperonospora parasitica by high-throughput differential display analysis of distinct phenotypic interactions with Brassica oleracea. Journal of Microbiological Methods. 2006;66:466–478. doi: 10.1016/j.mimet.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Gallagher SR, Leonard RT. Effect of vanadate, molybdate and azide on membrane-associated ATPase and soluble phosphatase activities of corn roots. Plant Physiology. 1982;70:1335–1340. doi: 10.1104/pp.70.5.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke T, Todd J, Ruuska S, White J, Benning C, Ohlrogge J. Microarray analysis of developing Arabidopsis seeds. Plant Physiology. 2000;124:1570–1581. doi: 10.1104/pp.124.4.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalkrishnan A, Rao MV. Amelioration by vitamin A upon arsenic induced metabolic and neurotoxic effects. Journal of Health Science. 2006;52:568–577. [Google Scholar]
- Hajek T, Honys D, Capkova V. New method of plant mitochondria isolation and sub-fractionation for proteomic analyses. Plant Science. 2004;167:389–395. [Google Scholar]
- Hamilton CA, Good AG, Taylor GJ. Induction of vacuolar ATPase and mitochondrial ATP synthase by aluminum in an aluminum-resistant cultivar of wheat. Plant Physiology. 2001;125:2068–2077. doi: 10.1104/pp.125.4.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Kim D. AtRTPrimer: database for Arabidopsis genome-wide homogeneous and specific RT-PCR primer-pairs. BMC Bioinformatics. 2006;7:179–188. doi: 10.1186/1471-2105-7-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PC, Jiang W, Fillingame RH. Arrangement of the multicopy H + -translocating subunit c in the membrane sector of the Escherichia coli F1F0-ATP synthase. Journal of Biological Chemistry. 1998;273:17178–17185. doi: 10.1074/jbc.273.27.17178. [DOI] [PubMed] [Google Scholar]
- Kinosita K, Jr, Yasuda R, No Ji H, Ishiwata S, Yoshida M. F1-ATPase: a rotary motor made of a single molecule. Cell. 1998;93:21–24. doi: 10.1016/s0092-8674(00)81142-3. [DOI] [PubMed] [Google Scholar]
- Landrum CP. Differential display polymerase chain reaction Nature Methods. 2006;3:325–326. [Google Scholar]
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–970. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Liang P, Zhu W, Zhang X, et al. Differential display using one-base anchored oligo-dT primers. Nucleic Acid Reseasrch. 1994;22:5763–5764. doi: 10.1093/nar/22.25.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis AM, Alconada A, Cuezva JM. The alpha regulatory subunit of the mitochondrial F1-ATPase complex is a heat-shock protein. Journal of Biological Chemistry. 1990;265:7713–7716. [PubMed] [Google Scholar]
- Macherel D, Benamar A, Avelange-Macherel MH, Tolleter D. Function and stress tolerance of seed mitochondria. Physiologia Plantarum. 2007;129:233–241. [Google Scholar]
- Minglin L, Yuxiu Z, Tuanyao C. Identification of the genes up regulated in response to Cd exposure in Brassica Juncea. Gene. 2005;363:151–158. doi: 10.1016/j.gene.2005.07.037. [DOI] [PubMed] [Google Scholar]
- Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Science. 2002;9:405–410. doi: 10.1016/s1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- Mueller DM. Partial assembly of the yeast mitochondrial ATP synthase. Journal of Bioenergetics and Biomembranes. 2000;32:391–400. doi: 10.1023/a:1005532104617. [DOI] [PubMed] [Google Scholar]
- Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nature Protocols. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- Oldeman LR, Hakkeling RTA, Sombroek WG. World map of the status of human-induced soil degradation: an explanatory note. Wageningen, The Netherlands: International Soil Information and Reference Centre; 1991. [Google Scholar]
- Pedersen PL, Ko YH, Hong S. ATP synthase in the year 2000; evolving views about the structures of these remarkable enzyme complexes. Journal of Bioenergetics and Biomembranes. 2000;32:325–332. doi: 10.1023/a:1005594800983. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acid Research. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T, Allen JF, Oelmüller R. Principles of redox control in photosynthesis gene expression. Physiologia Plantarum. 2001;112:1–9. [Google Scholar]
- Radoni A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochemical and Biophysical Research Communications. 2004;313:856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- Rajjou L, Gallardo K, Debeaujon I, Vandekerckhove J, Job C, Job D. The effect of α-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiology. 2004;134:1598–1613. doi: 10.1104/pp.103.036293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards LA. Diagnosis and improvement of saline and alkali soils. Science. 1954;120:800. [Google Scholar]
- Ririe KM, Rasmussen RP, Wittwer CT. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Analytical Biochemistry. 1997;245:154–160. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- Sahu MP, Singh D. Role of thiourea in improving productivity of wheat (Triticum aestivum L.) Journal of Plant Growth and Regulation. 1995;14:169–173. [Google Scholar]
- Sahu MP, Kumawat SM, D'souza SF, Ramaswamy NK, Singh G. Sulphydryl bioregulator technology for increasing mustard production. Research Bulletin RAU-BARC. 2005:1–52. [Google Scholar]
- Sahu MP, Kumawat SM, Ramaswamy NK, D'souza SF. Sulphydryl bioregulator technology for increasing wheat productivity. Research bulletin RAU-BARC. 2006:1–56. [Google Scholar]
- Sairam RK, Tyagi A. Physiology and molecular biology of salinity tolerance in plants. Current Science. 2004;86:407–421. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanders D. Plant biology: the salty tale of Arabidopsis. Current Biology. 2000;10:486–488. doi: 10.1016/s0960-9822(00)00554-6. [DOI] [PubMed] [Google Scholar]
- Sane AP, Sane VA, Sane PV. Purification and characterization of the mitochondrial F1 ATPase from Sorghum. Phytochemistry. 1996;43:561–564. [Google Scholar]
- Srivastava AK, Nathawat NS, Ramaswamy NK, et al. Evidence for thiol-induced enhanced in situ translocation of 14C-sucrose from source to sink in Brassica juncea. Environmental and Experimental Botany. 2008;64:250–255. [Google Scholar]
- Sweetlove LJ, Heazlewood JL, Herald V, et al. The impact of oxidative stress on Arabidopsis mitochondria. The Plant Journal. 2002;32:891–904. doi: 10.1046/j.1365-313x.2002.01474.x. [DOI] [PubMed] [Google Scholar]
- Tichopad A, Dilger M, Schwarz G, Pfaffl MW. Standardized determination of real-time PCR efficiency from a single reaction set-up. Nucleic Acid Research. 2003;31:122–128. doi: 10.1093/nar/gng122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincze E, Bowra S. Northerns revisited: a protocol that eliminates formaldehyde from the gel while enhancing resolution and sensitivity. Analytical Biochemistry. 2005;342:356–357. doi: 10.1016/j.ab.2005.02.026. [DOI] [PubMed] [Google Scholar]
- Zhang X, Takano T, Liu S. Identification of a mitochondrial ATP synthase small subunit gene (RMtATP6) expressed in response to salts and osmotic stresses in rice (Oryza sativa L.) Journal of Experimental Botany. 2006;57:193–200. doi: 10.1093/jxb/erj025. [DOI] [PubMed] [Google Scholar]