Abstract
Background and Aims
There is an extensive literature on the diversity of karyotypes found in genera within Liliaceae, but there has been no attempt to analyse these data within a robust phylogenetic framework. In part this has been due to a lack of consensus on which genera comprise Liliaceae and the relationships between them. Recently, however, this changed with the proposal for a relatively broad circumscription of Liliaceae comprising 15 genera and an improved understanding of the evolutionary relationships between them. Thus there is now the opportunity to examine patterns and trends in chromosome evolution across the family as a whole.
Methods
Based on an extensive literature survey, karyo-morphometric features for 217 species belonging to all genera in Liliaceae sensu the APG (Angiosperm Phylogeny Group) were obtained. Included in the data set were basic chromosome number, ploidy, chromosome total haploid length (THL) and 13 different measures of karyotype asymmetry. In addition, genome size estimates for all species studied were inferred from THLs using a power regression model constructed from the data set. Trends in karyotype evolution were analysed by superimposing the karyological data onto a phylogenetic framework for Liliaceae.
Key Results and Conclusions
Combining the large amount of data enabled mean karyotypes to be produced, highlighting marked differences in karyotype structure between the 15 genera. Further differences were noted when various parameters for analysing karyotype asymmetry were assessed. By examining the effects of increasing genome size on karyotype asymmetry, it was shown that in many but not all (e.g. Fritillaria and all of Tulipeae) species, the additional DNA was added preferentially to the long arms of the shorter chromosomes rather than being distributed across the whole karyotype. This unequal pattern of DNA addition is novel, contrasting with the equal and proportional patterns of DNA increase previously reported. Overall, the large-scale analyses of karyotype features within a well-supported phylogenetic framework enabled the most likely patterns of chromosome evolution in Liliaceae to be reconstructed, highlighting diverse modes of karyotype evolution, even within this comparatively small monocot family.
Key words: C-value, chromosomes, genome size, karyotype asymmetry, karyotype evolution, Liliaceae, Liliales, polyploidy
INTRODUCTION
Comparative karyotype analysis of related species has traditionally been used to describe patterns and directions of chromosomal evolution within a group and to infer the evolutionary role that such karyotype changes may have played (e.g. Stebbins, 1971; Stace, 1978; Gonzalez-Aguilera and Fernandez-Peralta, 1984; Watanabe et al., 1995; De Melo Nationiel et al., 1997; Vanzela et al., 1997; Das et al., 1999; Shan et al., 2003). Whereas earlier studies were, to some extent, flawed by the lack of a rigorous phylogenetic framework in which to analyse the data, more recently this problem has been addressed and there are now an increasing number of papers which have analysed karyological data using a well-supported phylogenetic tree. These include the works of Cox et al. (1998) on slipper orchids (Orchidaceae), Watanabe et al. (1999) on Brachyscome (Asteraceae), Venora and collaborators (Venora et al., 2000; Frediani et al., 2005; Caputo et al., 2006; Ruffini Castiglione et al., 2007) on Vicia (Fabaceae), Leitch and colleagues on Nicotiana (Solanaceae) (Lim et al., 2000, 2004, 2006, 2007), Lysak and collaborators on Brassicaceae (Lysak et al., 2005, 2006, 2007; Lysak and Lexer, 2006; Lysak and Mandakova, 2007) and Pires et al. (2006) on Asparagales.
The karyotype is the phenotypic aspect of the chromosome complement as seen at mitotic metaphase. A description of the karyotype typically includes the chromosome number [the product of two variables, basic number (x) and ploidy, resulting in chromosome number (2n)], the absolute and/or relative length of chromosomes (reflecting genome size), the position of primary and secondary constrictions (Levan et al., 1964), the distribution of material with different staining properties (Stebbins, 1971; Stace, 2000; Levin, 2002; Lysak and Lexer, 2006) and the degree of symmetry. A symmetrical karyotype is characterized by mainly metacentric and sub-metacentric chromosomes of approximately equal size. Changes to an asymmetric karyotype can arise by shifts in centromere position towards the telomere (intrachromosomal) and/or by the addition or deletion of chromatin from some but not all chromosomes, leading to differences in size between the largest and smallest chromosomes (interchromosomal). These processes are not usually correlated, though there are some reports that they are in some groups (Stebbins, 1971).
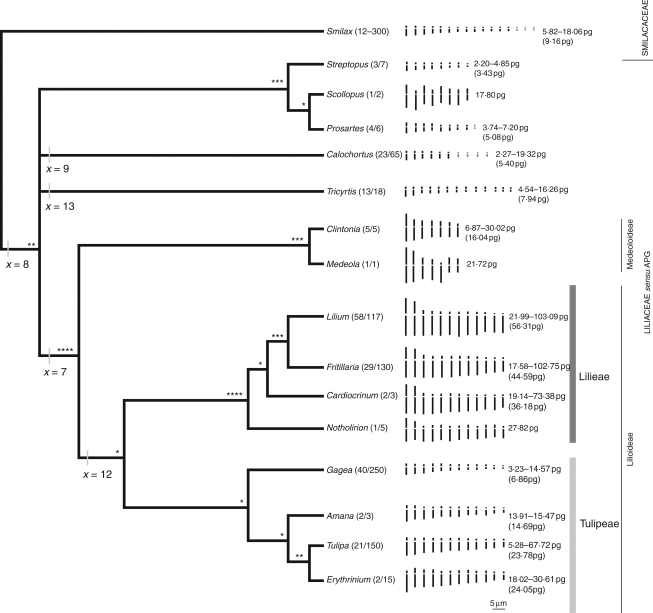
The chromosomes of Liliaceae have long attracted attention, given their diversity in size, number and structure (e.g. Sato, 1943; Cave, 1970; Sen, 1975; Tamura, 1995). However, attempts to infer patterns of chromosome evolution by comparative karyotype analysis have been hampered by the lack of a rigorous phylogenetic framework together with a lack of agreement over the genera comprising Liliaceae. Examples of some of these contrasting circumscriptions are outlined in Table 1. There is now a general consensus of the genera comprising subfamily Lilioideae [Amana, Cardiocrinum, Erythronium, Fritillaria, Gagea (including Lloydia), Lilium (including Nomocharis), Notholirion and Tulipa] and the phylogenetic relationships between them (Fay and Chase, 2000; Patterson and Givnish, 2002; Fay et al., 2006). Similarly, genera comprising subfamily Medeoloideae (Clintonia and Medeola) and their relationships are well supported (Fay et al., 2006). However, the placement of Tricyrtis, Calochortus and the clade comprising Prosartes, Scoliopus and Streptopus (Streptopeae) has been more problematic (Patterson and Givnish, 2002; Rønsted et al., 2005; Fay et al., 2006). Nevertheless, by taking a broad circumscription of Liliaceae, as used by Chase et al. (2000), Fay and Chase (2000) and the Angiosperm Phylogeny Group (APG II, 2003), the opportunity to examine the wealth of cytogenetic data available for the 15 genera comprising Liliaceae sensu APG II (2003) within a phylogenetic context now exists. Figure 1 summarizes the current understanding of these phylogenetic relationships.
Table 1.
Taxonomic treatment of genera currently included in Liliaceae which have appeared in the last 30 years
|
Dahlgren et al. (1985) |
Takhtajan (1997) |
Chase et al. (1995), Tamura (1995, 1998a, b) |
Patterson and Givnish (2002) |
Fay and Chase (2000), APG II (2003) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genus | Tribe | Family | Order | Tribe | Family | Order | Tribe | Subfamily | Family | Order | Subfamily | Family | Order | Family | Order |
| Streptopus | Uvularieae | Uvulariaceae | Liliales | Streptopeae | Uvulariaceae | Colchicales | Tricyrtideae | – | Calochortaceae | Liliales | ‘Streptopoideae’ | Calochortaceae | Liliales | Liliaceae | Liliales |
| Scoliopus | – | Trilliaceae | Dioscoreales | – | Scoliopaceae | Colchicales | Tricyrtideae | – | Calochortaceae | Liliales | ‘Streptopoideae’ | Calochortaceae | Liliales | Liliaceae | Liliales |
| Prosartes | Uvularieae | Uvulariaceae (in Disporum) | Liliales | Streptopeae | Uvulariaceae | Colchicales | Tricyrtideae | – | Calochortaceae | Liliales | ‘Streptopoideae’ | Calochortaceae | Liliales | Liliaceae | Liliales |
| Calochortus | – | Calochortaceae | Liliales | – | Calochortaceae | Colchicales | Calochorteae | – | Calochortaceae | Liliales | ‘Calochortoideae’ | Calochortaceae | Liliales | Liliaceae | Liliales |
| Tricyrtis | Tricyrtideae | Uvulariaceae | Liliales | – | Tricyrtidaceae | Colchicales | Tricyrtideae | – | Calochortaceae | Liliales | ‘Calochortoideae’ | Calochortaceae | Liliales | Liliaceae | Liliales |
| Medeola | – | Trilliaceae | Dioscoreales | – | Medeolaceae | Liliales | – | Medeoloideae | Liliaceae | Liliales | Medeoloideae | Liliaceae | Liliales | Liliaceae | Liliales |
| Clintonia | Uvularieae | Uvulariaceae | Liliales | Streptopeae | Uvulariaceae | Colchicales | – | Medeoloideae | Liliaceae | Liliales | Medeoloideae | Liliaceae | Liliales | Liliaceae | Liliales |
| Cardiocrinum | – | Liliaceae | Liliales | Lilieae | Liliaceae | Liliales | Lilieae | Lilioideae | Liliaceae | Liliales | Lilioideae | Liliaceae | Liliales | Liliaceae | Liliales |
| Notholirion | – | Liliaceae | Liliales | Lilieae | Liliaceae | Liliales | Lilieae | Lilioideae | Liliaceae | Liliales | Lilioideae | Liliaceae | Liliales | Liliaceae | Liliales |
| Nomocharis* | – | Liliaceae | Liliales | Lilieae | Liliaceae | Liliales | Lilieae | Lilioideae | Liliaceae | Liliales | Lilioideae | Liliaceae | Liliales | Liliaceae | Liliales |
| Fritillaria | – | Liliaceae | Liliales | Lilieae | Liliaceae | Liliales | Lilieae | Lilioideae | Liliaceae | Liliales | Lilioideae | Liliaceae | Liliales | Liliaceae | Liliales |
| Lilium | – | Liliaceae | Liliales | Lilieae | Liliaceae | Liliales | Lilieae | Lilioideae | Liliaceae | Liliales | Lilioideae | Liliaceae | Liliales | Liliaceae | Liliales |
| Gagea | – | Liliaceae | Liliales | Lloydieae | Liliaceae | Liliales | Tulipeae | Lilioideae | Liliaceae | Liliales | Lilioideae | Liliaceae | Liliales | Liliaceae | Liliales |
| Lloydia† | – | Liliaceae | Liliales | Lloydieae | Liliaceae | Liliales | Tulipeae | Lilioideae | Liliaceae | Liliales | Lilioideae | Liliaceae | Liliales | Liliaceae | Liliales |
| Amana‡ | – | Liliaceae (in Tulipa) | Liliales | Tulipeae | Liliaceae | Liliales | Tulipeae | Lilioideae | Liliaceae | Liliales | Lilioideae | Liliaceae | Liliales | Liliaceae | Liliales |
| Tulipa | – | Liliaceae | Liliales | Tulipeae | Liliaceae | Liliales | Tulipeae | Lilioideae | Liliaceae | Liliales | Lilioideae | Liliaceae | Liliales | Liliaceae | Liliales |
| Erythronium | – | Liliaceae | Liliales | Tulipeae | Liliaceae | Liliales | Tulipeae | Lilioideae | Liliaceae | Liliales | Lilioideae | Liliaceae | Liliales | Liliaceae | Liliales |
*Nomocharis has been shown to be embedded within Lilium (Rønsted et al., 2005).
†Lloydia has been shown to be embedded within Gagea (Peterson et al., 2004; Peruzzi et al., 2008a, b; Peterson et al., 2008; M. Zarrei et al., unpubl. res.).
‡The restoration of Amana, which was formerly included in Tulipa, was based on the work by Tan et al. (2005).
Fig. 1.
Mean haploid idiograms and range of inferred 1Cx values (means in parentheses) for the genera of Liliaceae and Smilacaceae superimposed onto a phylogenetic framework. Inferred basic chromosome numbers and the strength of the clades (* = supported by one molecular marker only; ** = supported by two molecular markers; *** = supported by three molecular markers; **** = supported by four or more molecular markers) are given. Sub-divisions of Liliaceae are indicated on the right of the figure. For each genus, the number in parenthes corresponds to the number of species studied karyologically followed by the total number of species considered to comprise the genus. For genera in which the basic chromosome number is variable, chromosomes which are not found in all species are indicated in grey.
Despite phylogenetic issues noted above, some previous karyotype analyses have been informative. For example, Tamura (1995, 1998a, b) highlighted differences in karyotype structure between the small and variable number of chromosomes in genera such as Calochortus, Prosartes, Scoliopus, Streptopus and Tricyrtis, and the larger and more stable chromosome numbers found in subfamily Lilioideae (e.g. Lilium, Fritillaria, Tulipa). Tamura (1995. 1998a, b) also noted clear differences in karyotype structure between genera in subfamily Medeoloideae (x = 7) and Lilioideae (x = 12) and the distinct karyotype shared by all representatives of tribe Lilieae, comprising two longer metacentric chromosome pairs and ten shorter telocentric pairs.
More recently, research in Liliaceae sensu APG II (2003) revealed considerable variability in genome size, broadly paralleling chromosome heterogeneity (Leitch et al., 2007). Large genomes (mean 1C = 35·9 pg) characterized subfamilies Lilioideae and Medeloloideae, whereas smaller genomes (mean 1C = 5·6 pg) characterized the remaining genera (Leitch et al., 2007). In their analysis, genome size data were compiled from the Plant DNA C-values database (Bennett and Leitch, 2005b) and Zonneveld et al. (2005), together with new genome size estimates for 23 species of Liliaceae to give a total data set of 78 species. Despite containing some of the largest genomes so far reported in the angiosperms, a phylogenetic analysis revealed that the ancestral genome size reconstructed for the family was just 1C = 6·67 pg, similar to that reported for Smilax in Smilacaceae (sister to Liliaceae) (1C = 5·0 pg).
Given recent advances in understanding phylogenetic relationships within Liliaceae noted above, this paper aims to summarize quantitatively the large amount of karyotype data available in the literature for all genera of Liliaceae (sensu APG II, 2003) and analyse them within a phylogenetic framework to assess possible modes and directions of karyotype evolution.
MATERIALS AND METHODS
Source of karyotype data
An extensive literature survey was conducted to collate available karyotype data for Liliaceae and Smilaacceae. The index to plant chromosome numbers (eds, P. Goldblatt and D. E. Johnson, Missouri Botanical Garden, St Louis) was consulted (hard copies up to 1979 and then electronically at http://mobot.mobot.org/W3T/Search/ipcn.html) together with other chromosome atlases (e.g. Darlington and Wylie, 1955; Fedorov, 1969) and internet resources (e.g. Kew Bibliographic Databases, ISI Web of Science, Google Scholar).
In total, karyotype data were collected for 405 accessions of Liliaceae (386) and Smilacaceae (19). Data were available for 217 species, including representatives of all genera comprising these two families. Only literature with published idiograms, and/or karyotype measurements and/or good metaphase plates with magnification or scale bar indicated were considered. When 2n mitotic metaphase plates were measured, the graphic method proposed by Plummer et al. (2003) was used to match homologous chromosomes. This involves plotting the relative length of each chromosome [= (length of individual chromosome/total length of all chromosomes)/100] against its arm ratio, in order to pair chromosomes.
Karyotype analysis
A data matrix of 53 karyotype features was assembled including basic chromosome number (x), ploidy, chromosome number (2n) and chromosome total haploid length (THL). For analysis of karyotype asymmetry, all known methods of assessing this character were used (Table 2). A review of each method and a summary of how each asymmetry index was calculated is given in Paszko (2006). For the present analyses, two coefficients of variation (CVs) were found to be particularly informative measures of asymmetry. The CVCI index evaluates differences in centromere position for each chromosome in the karyotype and provides a measure of intrachromosomal asymmetry. In contrast, the CVCL gives a measure of interchromosomal asymmetry as it reflects how variable the chromosome sizes are in the karyotype. In both cases, the larger the value the greater the asymmetry in the karyotype.
Table 2.
Measures of karyotype asymmetry used in the present work
| Name of asymmetry index | Abbreviation (if applicable) | Reference |
|---|---|---|
| Stebbins asymmetry index | StebA–C | Stebbins (1971) |
| Stebbins asymmetry index | Steb1–4 | Stebbins (1971) |
| Intrachromosomal asymmetry index | A1 | Romero Zarco (1986) |
| Interchromosomal asymmetry index | A2 | Romero Zarco (1986) |
| Percentage karyotype asymmetry index | AsK% | Arano (1963) |
| Total form % | TF% | Huziwara (1962) |
| Index of chromosome size resemblance | Rec | Greilhuber and Speta (1976) |
| Index of karyotype symmetry | Syi | Greilhuber and Speta (1976) |
| Karyotype dispersion index | DI | Lavania and Srivastava (1992) |
| Degree of karyotype asymmetry | A | Watanabe et al. (1999) |
| Coefficient of variation (CV) of the centromeric index (centromeric index is the ratio of the length of the short arm to that of the total chromosome length) | CVCI | Paszko (2006) |
| Coefficient of variation (CV) of chromosome lengths | CVCL | Paszko (2006) |
| Asymmetry index (CVCI × CVCL) ÷ 100 | AI | Paszko (2006) |
For each genus, a mean haploid idiogram was built as follows. Using karyoytype data for each species, chromosome pairs were arranged into decreasing lengths, then the absolute lengths of the long and short arms of each chromosome pair were taken. The data were then pooled to produce a mean length of each chromosome pair for a genus (e.g. the data for the longest chromomosome pair of all Lilium species were pooled to give the mean value for Lilium chromosome pair I). It is noted that since the study was based almost exclusively on measurements of Feulgen-stained chromosomes, there was no possibility of identifying and analysing homoeologous chromosomes between taxa. Instead the ‘mean karyotype’ of each genus refers only to the shape of the haploid karyotype.
Genome size estimates inferred from the THL
According to Levin (2002, and literature cited therein), the correlation between THL and 1C values typically exceeds r = 0·85 within species and between species in related genera, and THL has thus been considered a good proxy for genome size. In the present study, THLs were measured for all species analysed. To transform THL into an inferred 1Cx (= monoploid genome size, which represents the DNA amount in the basic haploid chromosome complement and is calculated by dividing the 2C DNA amount by the ploidy level; Greilhuber et al., 2005) estimate for each species, first a predictive regression model was built by plotting the mean THL for each genus (Table 3, based on data in Supplementary Data 1, available online) against the mean 1Cx genome size for each genus using data taken from the Plant DNA C-values database (Bennett and Leitch, 2005b), Leitch et al. (2007) and Zonneveld et al. (2005) (Table 3, Fig. 2A). Using these data, 11 common linear regression models were tested. All were significant at the 0·01 level. The highest r2 value (0·92) was obtained with a Cubic curve equation, but this was rejected because it altered the extreme values too much. Among the remaining models, a linear curve (r2 = 0·86) was excluded as it gave negative 1Cx estimates for small THL values. From six other equally well fitting curves (r2 = 0·85–0·86), the power regression model was selected as it gave the most reasonable outputs for both small and large THL values (Fig. 2A).
Table 3.
Mean monoploid genome sizes (1Cx) using data taken from the literature (see Materials and Methods) and mean total haploid length (THL) used to build the predictive regression analysis shown in Fig. 2A
| Taxon | Mean 1Cx (pg) | No. of taxa analysed | Mean THL (µm) | No. of taxa analysed |
|---|---|---|---|---|
| Genera | ||||
| Amana | 21·5 | 1 | 67·75 | 2 |
| Calochortus | 5·4 | 1 | 28·66 | 23 |
| Cardiocrinum | 38·6 | 1 | 130·62 | 2 |
| Clintonia | 9·5 | 1 | 71·83 | 5 |
| Erythronium | 31·8 | 4 | 97·67 | 2 |
| Fritillaria | 46·7 | 23 | 153·1 | 29 |
| Gagea | 6·6 | 2 | 38·52 | 40 |
| Lilium | 39·6 | 10 | 174·38 | 58 |
| Medeola | 14·2 | 1 | 90·89 | 1 |
| Notholirion | 35·3 | 1 | 109·4 | 1 |
| Prosartes | 3·4 | 1 | 30·36 | 4 |
| Scoliopus | 9·2 | 1 | 78·27 | 1 |
| Streptopus | 3·3 | 1 | 22·63 | 3 |
| Tricyrtis | 4·5 | 2 | 39·22 | 13 |
| Tulipa | 22·5 | 26 | 93·14 | 21 |
| Tribe | ||||
| Lilieae | 44·3 | 34 | 136·95 | 130 |
| Streptopeae | 5·3 | 3 | 28·92 | 8 |
| Tulipeae | 22·8 | 31 | 92·37 | 25 |
| Subfamily | ||||
| Lilioideae | 33·7 | 66 | 133·91 | 155 |
| Medeoloideae | 11·9 | 2 | 73·19 | 6 |
| Family | ||||
| Liliaceae | 32·1 | 70 | 114·2 | 205 |
| Smilacaceae | 5·0 | 1 | 45·11 | 12 |
Fig. 2.
(A) Scatter plot of the power regression model from chromosome total haploid length (THL; x-axis) to 1Cx (y-axis), with the related regression formula and significance parameters. Original data are taken from Table 3. (B) Comparison between mean 1Cx estimates inferred from the regression analysis of THL (circles) and 1Cx values estimated using either using flow cytometry or Feulgen microdenstometry (squares); see Materials and Methods. Data are arranged by increasing 1Cx values inferred from THLs.
Correlations
For continuous quantitative data, Pearson's correlation coefficient was used, whereas for mixed scalar and ordinal data Spearman's rho coefficient was calculated. Correlations were considered as weak (up to 0·3), average (up to 0·5), good (up to 0·80) and high (>0·80). Only correlations significant at the 1 % level or higher are discussed.
Phylogenetic framework and inferred basic chromosome numbers
The phylogenetic framework for Liliaceae used in this study (Fig. 1) was derived mainly from the work of Patterson and Givnish (2002) and Fay et al. (2006), together with additional data from the phylogenetic analysis of Tan et al. (2005, restoration of Amana), Rønsted et al. (2005, sinking of Nomocharis), Peterson et al. (2004, 2008), Peruzzi et al. (2008a, b) and M. Zarrei et al. (unpubl. res.) concerning the sinking of Lloydia in Gagea. Smilacaceae is the sister family to Liliaceae (Patterson and Givnish, 2002; Fay et al., 2006). All nodes for which bootstrap support and branch length did not fit the requirements for the 0·95 binomial confidence interval (Zander, 2004) were collapsed. The resulting phylogenetic tree is almost identical to the one used by Leitch et al. (2007) in their genome size study (except for the relationships between Amana, Erythronium and Tulipa).
The ancestral basic chromosome number for Liliaceae was inferred to be x = 8 based on the frequency of 2n = 32 counts in Smilax (Smilacaceae) and cytological data suggesting the genus to be palaeopolyploid (Vijayavalli and Mathew, 1987). This is also considered to be the ancestral basic number for Streptopus, Scoliopus and Prosartes given the frequency of species in these genera based on x = 8. An ancestral basic chromosome number of x = 9 was adopted for the highly variable Calochortus (x ranging from 6 to 10) based on the parsimony analysis of Patterson and Givnish (2004), and x = 13 for the chromosomally stable genus Tricyrtis as suggested by Nakamura (1968). An ancestral basic number of the Medeoloideae + Lilioideae clade is more equivocal, although basic numbers of x = 7 for Medeloideae and x = 12 for Lilioideae are well supported by previous karyotype studies together with the suggestion that their karyotypes may be related by Robertsonian rearrangements (e.g. Tamura, 1998b).
RESULTS
Inferring genome size from THLs
In general there was a broad agreement between the mean inferred 1Cx value for each genus and values from genome size studies, although there were some discrepancies in Lilium and Fritillaria. Nevertheless this approach was considered robust enough to infer 1Cx values from THL measurements for all accessions studied, and these are given in Supplementary Data 1, available online.
Variabilities in inferred 1Cx values for each genus are illustrated by the boxplots shown in Fig. 3 which are superimposed onto the consensus phylogenetic framework of Liliaceae (from Fig. 1). This figure highlights the contrast between the large and wide range of genome sizes in subfamily Lilioideae, especially in tribe Lilieae, compared with the smaller and narrow range of genome sizes encountered in the remaining genera. The only exceptions to this were found in Gagea and Scoliopus. The mean genome size for Gagea (inferred mean 1Cx = 6·86 pg) was less than half the next smallest genome size in the tribe (for Amana with an inferred mean 1Cx of 14·69 pg). Scoliopus stood out because the genome size for this genus (inferred 1Cx = 17·80 pg) was more than three times larger than that for Prosartes (inferred mean 1Cx = 5·08 pg) and Streptopus (inferred mean 1Cx = 3·43 pg) in the same clade.
Fig. 3.
Boxplots illustrating the variability of 1Cx genome size, as inferred from the regression model given in Fig. 2A. The outlined central box depicts the middle 50 % of the data extending from the upper to lower quartile; the horizontal bar is at the median. The ends of the vertical lines (‘whiskers’) indicate the minimum and maximum data values, unless outliers are present, in which case the whiskers extend to a maximum of 1·5 times the interquartile range. Circles indicate outliers, unless extreme outliers are present, in which case the circles extend to a maximum of three times the interquartile range and the extreme outliers are indicated as asterisks. Taxa are ordered by phylogenetic grouping (according to the phylogenetic tree on the bottom of the graph, taken from Fig. 1).
Constructing mean haploid idiograms for each genus
Mean haploid idiograms for all genera superimposed onto the consensus phylogenetic framework of Liliaceae are shown in Fig. 1 using data taken from Table 4. The different sizes of the karyotypes largely reflect the patterns already noted from the boxplots in Fig. 3, with the relatively small chromosomes of Gagea compared with other genera in subfamily Lilioideae and the larger chromosomes in Scoliopus compared with Calochortus, Prosartes, Streptopus and Tricyrtis standing out as exceptions.
Table 4.
Mean chromosome features (± s.d.) for genera studied
| I | II | III | IV | V | VI | VII | VIII | IX | X | XI | XII | XIII | XIV | XV | XVI | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Smilax (N = 19) | 4·31 ± 1·55 | 3·75 ± 1·21 | 3·18 ± 0·94 | 2·80 ± 0·91 | 2·43 ± 0·89 | 2·28 ± 0·65 | 2·13 ± 0·67 | 2·01 ± 0·70 | 1·76 ± 0·57 | 1·60 ± 0·49 | 1·57 ± 0·57 | 1·42 ± 0·51 | 1·33 ± 0·38 | 1·20 ± 0·39 | 1·14 ± 0·44 | 0·84 ± 0·36 |
| 1·11 ± 1·42 | 0·80 ± 0·56 | 0·85 ± 0·34 | 0·89 ± 0·34 | 0·89 ± 0·31 | 0·81 ± 0·41 | 0·75 ± 0·34 | 0·64 ± 0·29 | 0·71 ± 0·35 | 0·69 ± 0·29 | 0·58 ± 0·22 | 0·61 ± 0·23 | 0·60 ± 0·27 | 0·54 ± 0·24 | 0·49 ± 0·18 | 0·42 ± 0·26 | |
| Streptopus (N = 9) | 3·42 ± 0·68 | 2·69 ± 0·70 | 2·07 ± 0·48 | 1·84 ± 0·52 | 1·66 ± 0·36 | 1·39 ± 0·44 | 1·27 ± 0·40 | 0·83 ± 0·43 | ||||||||
| 1·38 ± 0·80 | 1·00 ± 0·28 | 0·94 ± 0·31 | 0·95 ± 0·16 | 0·90 ± 0·26 | 0·87 ± 0·21 | 0·87 ± 0·31 | 0·56 ± 0·39 | |||||||||
| Scoliopus (N = 1) | 7·69 | 9·23 | 5·58 | 6·92 | 4·81 | 5·96 | 6·35 | 3·46 | ||||||||
| 5·58 | 2·12 | 4·81 | 2·50 | 4·42 | 3·08 | 2·69 | 3·08 | |||||||||
| Prosartes (N = 5) | 3·32 ± 0·80 | 2·87 ± 0·52 | 2·59 ± 0·42 | 1·95 ± 0·31 | 2·50 ± 0·86 | 2·15 ± 0·81 | 1·61 ± 0·74 | 1·25 ± 0·34 | 0·75 ± 0·51 | |||||||
| 2·32 ± 0·50 | 2·02 ± 0·44 | 1·66 ± 0·42 | 1·58 ± 0·36 | 0·99 ± 0·28 | 0·79 ± 0·32 | 0·72 ± 0·39 | 0·91 ± 0·18 | 0·38 ± 0·11 | ||||||||
| Calochortus (N = 25) | 3·93 ± 1·68 | 3·33 ± 1·47 | 2·98 ± 1·17 | 2·65 ± 1·30 | 2·59 ± 1·22 | 2·64 ± 1·35 | 2·00 ± 1·13 | 1·38 ± 1·15 | 1·11 ± 1·22 | 0·77 ± 0·88 | ||||||
| 1·21 ± 0·92 | 1·34 ± 0·82 | 1·14 ± 0·90 | 1·04 ± 0·56 | 0·84 ± 0·52 | 0·59 ± 0·33 | 0·67 ± 0·38 | 0·40 ± 0·40 | 0·37 ± 0·33 | 0·33 ± 0·71 | |||||||
| Tricyrtis (N = 34) | 4·52 ± 1·33 | 3·86 ± 1·16 | 2·51 ± 0·92 | 2·17 ± 0·79 | 2·06 ± 0·64 | 1·85 ± 0·60 | 1·78 ± 0·51 | 1·64 ± 0·44 | 1·62 ± 0·59 | 1·61 ± 0·60 | 1·51 ± 0·57 | 1·42 ± 0·51 | 1·08 ± 0·29 | |||
| 1·05 ± 0·32 | 0·97 ± 0·31 | 1·24 ± 0·43 | 1·30 ± 0·36 | 1·15 ± 0·34 | 1·18 ± 0·32 | 1·11 ± 0·38 | 1·12 ± 0·43 | 1·05 ± 0·29 | 0·96 ± 0·26 | 0·92 ± 0·23 | 0·82 ± 0·22 | 0·79 ± 0·32 | ||||
| Clintonia (N = 13) | 8·03 ± 1·58 | 7·66 ± 1·48 | 6·70 ± 1·46 | 6·08 ± 1·50 | 5·63 ± 0·81 | 5·59 ± 1·43 | 5·78 ± 1·82 | |||||||||
| 7·50 ± 1·66 | 4·28 ± 1·38 | 3·68 ± 1·08 | 3·46 ± 0·85 | 3·07 ± 1·06 | 2·53 ± 0·84 | 1·90 ± 0·74 | ||||||||||
| Medeola (N = 1) | 10·07 | 9·64 | 7·88 | 8·76 | 10·07 | 4·38 | 4·38 | |||||||||
| 10·07 | 7·99 | 4·38 | 3·29 | 1·75 | 4·38 | 3·83 | ||||||||||
| Lilium (N = 124) | 13·00 ± 3·49 | 12·87 ± 3·63 | 12·94 ± 2·82 | 12·70 ± 2·78 | 13·05 ± 3·43 | 12·67 ± 3·61 | 13·36 ± 4·47 | 12·87 ± 4·21 | 12·95 ± 4·58 | 10·98 ± 3·45 | 11·10 ± 3·69 | 11·20 ± 4·50 | ||||
| 9·91 ± 2·83 | 7·92 ± 2·29 | 2·59 ± 1·34 | 2·20 ± 1·08 | 1·92 ± 0·85 | 1·74 ± 0·78 | 1·72 ± 0·67 | 1·53 ± 0·71 | 1·37 ± 0·72 | 1·24 ± 0·69 | 0·96 ± 0·54 | 0·79 ± 0·54 | |||||
| Fritillaria (N = 61) | 11·10 ± 3·57 | 10·68 ± 3·57 | 12·19 ± 3·88 | 11·90 ± 3·60 | 11·31 ± 3·54 | 10·85 ± 3·81 | 10·34 ± 3·61 | 10·05 ± 3·34 | 9·53 ± 3·02 | 8·38 ± 2·92 | 8·10 ± 2·88 | 7·32 ± 2·83 | ||||
| 7·67 ± 3·18 | 6·29 ± 2·74 | 2·79 ± 2·94 | 2·27 ± 2·58 | 2·20 ± 2·38 | 1·85 ± 1·04 | 1·89 ± 0·99 | 1·70 ± 0·78 | 1·68 ± 0·99 | 1·44 ± 0·84 | 1·25 ± 0·73 | 1·30 ± 1·02 | |||||
| Cardiocrinum (N = 8) | 9·67 ± 3·85 | 9·69 ± 3·41 | 10·23 ± 3·45 | 10·07 ± 2·94 | 9·55 ± 3·23 | 8·80 ± 2·76 | 8·95 ± 4·12 | 8·67 ± 4·14 | 8·11 ± 4·28 | 7·53 ± 2·61 | 7·80 ± 3·36 | 6·48 ± 3·42 | ||||
| 7·15 ± 3·02 | 6·00 ± 3·27 | 1·82 ± 1·41 | 1·27 ± 0·77 | 1·33 ± 0·64 | 1·69 ± 0·83 | 1·33 ± 0·50 | 1·19 ± 0·53 | 1·13 ± 0·43 | 0·81 ± 0·28 | 0·60 ± 0·24 | 0·81 ± 0·39 | |||||
| Notholirion (N = 1) | 8·00 | 7·10 | 8·50 | 8·00 | 7·10 | 8·00 | 7·10 | 6·50 | 6·50 | 6·00 | 5·50 | 6·10 | ||||
| 6·00 | 4·20 | 1·10 | 1·40 | 2·00 | 1·20 | 1·30 | 1·90 | 1·20 | 1·60 | 2·00 | 1·10 | |||||
| Gagea (N = 56) | 4·49 ± 1·31 | 3·98 ± 1·28 | 3·39 ± 1·17 | 2·59 ± 0·94 | 2·15 ± 0·69 | 1·99 ± 0·69 | 1·78 ± 0·52 | 1·64 ± 0·56 | 1·53 ± 0·48 | 1·37 ± 0·40 | 1·35 ± 0·44 | 1·17 ± 0·36 | ||||
| 1·06 ± 0·47 | 0·97 ± 0·41 | 0·9 5 ± 0·41 | 1·14 ± 0·38 | 1·11 ± 0·42 | 0·94 ± 0·31 | 0·90 ± 0·35 | 0·85 ± 0·26 | 0·81 ± 0·30 | 0·82 ± 0·32 | 0·67 ± 0·26 | 0·63 ± 0·22 | |||||
| Amana (N = 2) | 6·89 ± 0·08 | 5·77 ± 1·12 | 5·91 ± 0·49 | 5·04 ± 0·53 | 4·40 ± 0·41 | 4·26 ± 1·24 | 4·22 ± 0·44 | 3·61 ± 0·86 | 3·22 ± 1·33 | 3·33 ± 0·60 | 2·42 ± 0·65 | 1·94 ± 0·00 | ||||
| 1·89 ± 0·64 | 2·14 ± 1·07 | 1·42 ± 0·67 | 1·25 ± 0·01 | 1·70 ± 0·01 | 1·53 ± 0·40 | 1·07 ± 0·04 | 1·44 ± 0·18 | 1·48 ± 0·67 | 0·90 ± 0·17 | 1·25 ± 0·12 | 0·78 ± 0·04 | |||||
| Tulipa (N= 43) | 8·71 ± 4·94 | 8·20 ± 4·67 | 7·35 ± 4·37 | 7·04 ± 3·92 | 6·66 ± 3·82 | 6·04 ± 3·51 | 5·69 ± 3·29 | 5·39 ± 3·07 | 5·16 ± 3·02 | 4·78 ± 2·74 | 4·34 ± 2·40 | 3·81 ± 2·29 | ||||
| 2·15 ± 1·16 | 1·80 ± 1·08 | 1·92 ± 1·05 | 1·79 ± 1·09 | 1·70 ± 0·83 | 1·70 ± 0·94 | 1·54 ± 0·89 | 1·63 ± 0·98 | 1·49 ± 0·83 | 1·47 ± 0·79 | 1·45 ± 0·79 | 1·35 ± 0·72 | |||||
| Erythronium (N = 3) | 10·71 ± 2·17 | 8·00 ± 3·60 | 8·00 ± 2·13 | 7·44 ± 2·03 | 7·11 ± 1·34 | 6·61 ± 1·57 | 5·96 ± 1·46 | 5·23 ± 0·27 | 4·59 ± 0·53 | 4·45 ± 0·70 | 3·79 ± 1·11 | 3·54 ± 1·02 | ||||
| 1·60 ± 0·41 | 2·91 ± 0·67 | 2·07 ± 0·52 | 2·02 ± 0·83 | 1·69 ± 0·71 | 1·66 ± 0·39 | 1·93 ± 0·23 | 1·94 ± 0·52 | 1·65 ± 0·43 | 1·50 ± 0·53 | 1·65 ± 0·41 | 1·63 ± 0·39 | |||||
| Total: 405 |
Chromosome pairs are named with roman numerals, according to their decreasing length. For each chromosome pair the absolute lengths in micrometres of the long (top) and short (bottom) arm are given. Genera are arranged in phylogenetic order according to Fig. 1. The number of accessions studied is given following the genus name.
Karyotype asymmetry
Results for the 13 different measures of karyotype asymmetry are given in Supplementary Data 1, available online. Pearson's correlation coefficients between each of the different measures varied from weak to strong, negative to positive and insignificant to significant (Supplementary Data 2, available online).
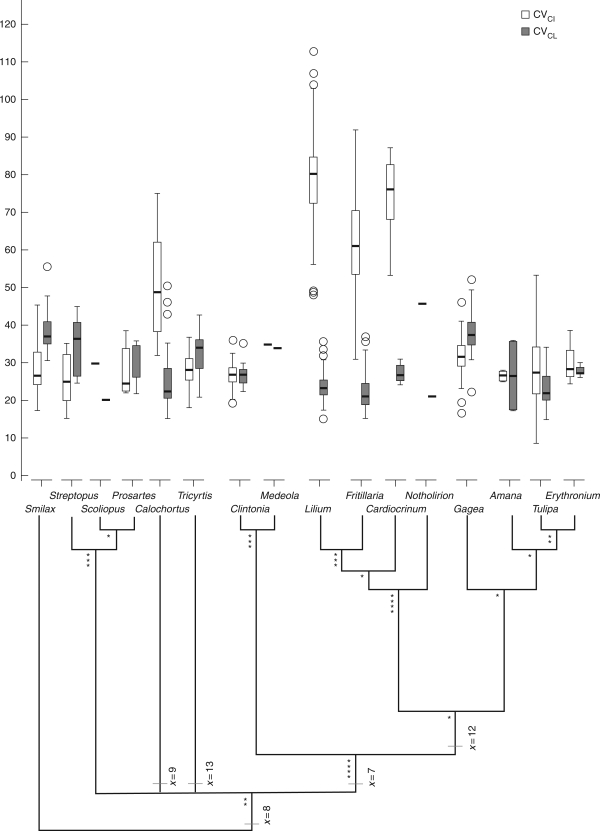
Two of these measures, CVCL and CVCI, which represent the degree of heterogeneity within a chromosome complement in terms of chromosome length (interchromosomal variation) and centromere position (intrachromosomal variation), respectively, were analysed in greater detail. Boxplots showing the range of values obtained for these two parameters for each genus are given in Fig. 4, arranged phylogenetically. Figure 5 summarizes differences between genera in these two measures of karyotype asymmetry by plotting the CVCI and CVCL values for each species.
Fig. 4.
Clustered boxplots illustrating the variability of both the coefficient of variation (CV) of the centromeric index (CVCI) and chromosome length (CVCL; see Fig. 3 for more explanations concerning the boxplots.) Taxa are ordered by phylogenetic grouping (according to the phylogenetic tree on the bottom of the graph, taken from Fig. 1).
Fig. 5.
Analysis of karyotype asymmetry in Liliaceae and Smilacaceae. Coefficients of variation (CV) of the centromeric index (CVCI) and chromosome length (CVCL) were plotted for each species studied. Increasing karyotype asymmetry is reflected in increasing values of CVCI and/or CVCL; (A) genera in Lilieae, (B) genera in Tulipeae, (C) the remaining genera in Liliaceae and Smilax in Smilacaceae.
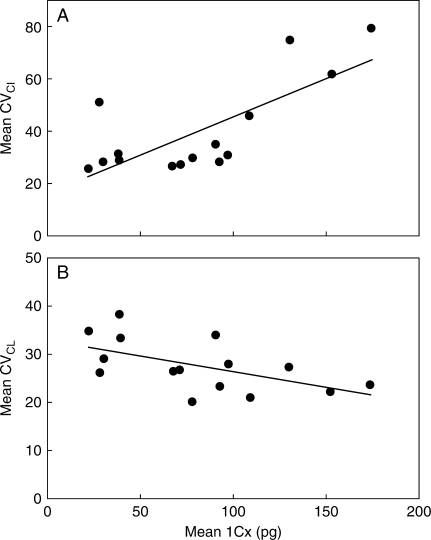
Karyotype asymmetry and genome size
In general, there was a good positive correlation between 1Cx and CVCI across Liliaceae (r = 0·65, Fig. 6A; Supplementary Data 2, available online), indicating that increases in genome size were generally accompanied by increasing karyotype asymmetry through increasing variability in centromere position. This correlation was also found for various subgroups within Liliaceae including Tricyrtis (r = 0·56), Lilioideae (r = 0·59), Lilieae (r = 0·23) and Lilium (r = 0·32). However, the correlation was negative for Fritillaria (within Lilieae, r = −0·33) and for all Tulipeae (r = −0·37).
Fig. 6.
Relationship between genome size (1Cx) and karyotype asymmetry. Mean 1Cx values inferred for each genus (taken from Table 3) were plotted against the mean values for the coefficient of variation (CV) of (A) the centromeric index (CVCI) and (B) the chromosome length (CVCL) for each genus.
The relationship between 1Cx and CVCL in Liliaceae was generally negative (r = −0·49, Fig. 6B). This correlation was also found for many sub-groups analysed alone, including Tricyrtis (r = −0·36), Lilioideae (r = −0·46) and Tulipeae (r = −0·46). The least negative Pearson's coefficient values were found in Lilium (r = −0·22) and Lilieae (r = −0·17).
Trends in karyotype asymmetry and genome size were also analysed for individual chromosomes. This was done by plotting, for each chromosome in the karyotype, its R (arm ratio) value against the inferred 1Cx for the accession (data not shown). [The R value is a measure of chromosomal asymmetry calculated by dividing the length of the long arm by the length of the short arm for each chromosome; see Levan et al. (1964). A metacentric chromosome has an R value of 1 whereas higher R values are associated with increasingly acrocentric chromosomes.] This approach revealed that the larger, often metacentric (i.e. chromosome pairs I and II) and smaller sub-metacentric to telocentric chromosomes (chromosome pairs IV and higher) undergo different patterns of karyotype evolution. While there was a strong positive correlation with smaller chromosomes from IV to XII (r = 0·92–0·98), indicating that the additional DNA is associated with increasing intrachromosomal asymmetry, chromosome pairs I–III had a low negative correlation with genome size (r values ranging from −0·14 to −0·18), implying that as genome size increases these chromosomes become more symmetric. This pattern was observed for all genera except Fritillaria and genera in Tulipeae.
When Smilacaceae were analysed alone, there were no relevant differences in the relationships between 1Cx and R values for the larger and shorter chromosomes. In all cases, the correlations were positive and high (r = 0·70–0·98) or not significant (depending on the chromosome).
Polyploidy
Overall the percentage of polyploids in Liliaceae was low (16 % for the whole data set). Many genera were exclusively diploid (Cardiocrinum, Erythronium, Medeola, Notholirion, Prosartes, Scoliopus and Tricyrtis) and only three genera had 50 % or more polyploids (Amana, Clintonia and Gagea) (Fig. 7). Plotting the relationship between the occurrence of polyploids and mean inferred 1Cx genome size for each genus revealed that most polyploids (94 %) occurred in species with 1Cx genome sizes <25 pg (Fig. 7); above this DNA amount, the number of polyploid species was small (Fig. 8). In all genera, the species with the highest inferred 1Cx values were always diploid (Fig. 8).
Fig. 7.
The relationship between the inferred mean 1Cx genome size and percentage of polyploids (based on data in Supplementary Data 1, available online) for the 15 genera of Liliaceae. The dotted line corresponds to a mean 1Cx value of 25 pg.
Fig. 8.
Scatter plots of the 386 accessions of Liliaceae and of eight selected genera (including Smilax from Smilacaceae), arranged by increasing 1Cx values (x-axis). The left-hand y-axis represents inferred 1Cx values (squares), and the right-hand y-axis represents ploidy levels (circles). The percentage of diploids is reported for all genera.
DISCUSSION
The reliability of inferring genome size from THLs
Whereas previous studies have highlighted the potential for using chromosome data as a proxy for genome size (e.g. Narayan and Rees, 1976; Raina and Rees, 1983; Ceccarelli et al., 1995), it has also been shown that chromosomes of a species can vary considerably in size without any change in DNA amount depending on the method of chromosome preparation as well as the phosphate concentration of the growth medium or type of root analysed (Bennett and Rees, 1967, 1969; Bennett, 1970). Given this, it was necessary to check if the karyotype data available for Liliaceae could be used to infer genome sizes. Using the approaches outlined, a new predictive regression model for inferring genome size from the THL of chromosome complements was produced specifically for Liliaceae and the closely allied Smilacaceae. In general there was broad agreement between ‘real’ genome size values obtained using ‘best practice’ genome size estimation techniques (flow cytometry and Feulgen microdensitometry) and those inferred from THL measurements (Fig. 2B). The largest differences occurred in Lilium. One explanation is that Lilium is a large genus (approx. 100 species) and, while there are karyological data for 58 species, genome size data are only available for ten species. It is possible that the smaller mean 1Cx values from genome size data are due to the small number of species analysed. From the data given in Supplementary Data 1 (available online), Lilium species with the largest THLs have not had their genome sizes measured using best practice techniques. Whilst it is recognized that accurate genome size estimates will never be obtained from THLs, given the wealth of karyotype information available compared with the amount of genome size information, it was felt that the current approach was appropriate for providing some insights into patterns of genome evolution across Liliaceae.
Patterns of genome size evolution across Liliaceae
Superimposing inferred genome size data onto a phylogenetic tree of Liliaceae revealed similar patterns to those predicted from chromosome data by Tamura (1995, 1998a, b) and observed by Leitch et al. (2007), most notably the striking differences between the genome size profiles of (a) genera in subfamily Lilioideae and Medeoloideae and (b) the remaining genera (Calochortus, Tricyrtis, Prosartes, Scoliopus and Streptopus) (Figs 1 and 3). However, the considerably larger data set available in the present work (217 species compared with 78 species in Leitch et al., 2007) highlighted two exceptions to this general pattern: (1) the relatively large genomes in Scoliopus (inferred 1Cx = 17·2 pg) compared with Prosartes (mean inferred 1Cx = 5·1 pg), Streptopus (inferred mean 1Cx = 3·4 pg), Calochortus (inferred mean 1Cx = 5·4 pg) and Tricyrtis (inferred mean 1Cx = 7·9 pg); and (2) the small Gagea genomes (inferred mean 1Cx = 6·9 pg) compared with the remaining genera in subfamily Lilioideae (inferred mean 1Cx range 14·7–56·3 pg).
Overall, although statistical analysis of genome size evolution across the family has revealed it to be punctuated with a significantly large shift and subsequent radiation in genome size at the base of Lilioideae (Leitch et al., 2007), more localized genome size changes in the branches leading to Gagea and Scoliopus have also taken place. For Scoliopus the larger genomes most probably represent the result of localized genome size increases, whereas for Gagea genome downsizing would appear to be the more parsimonious explanation. However, the possibility that its small genome size represents the ancestral state with all other genera in the clade having experienced an increase cannot be ruled out, particularly in light of recent data suggesting that the predominant direction of genome size evolution in angiosperms is upwards (Hawkins et al., 2009). The nature of the evolutionary forces driving these changes remains unknown, although analysis suggests that genome size evolution in Liliaceae is passive rather than adaptive (Leitch et al., 2007).
In general, there is a positive correlation between genome size and the percentage of repetitive DNA for both tandem and dispersed repeats (Levin, 2002; Gregory, 2005). As for the effects that repetitive non-coding DNA has on the organism, there are broadly two different viewpoints. One of them interprets non-coding DNA as ‘junk’ DNA, whereas the other interprets repetitive DNA as adaptive in various aspects of plant structure and function, i.e. a larger genome is correlated with larger cell volumes and longer cell cycles (reviewed in Gregory, 2001; Bennett and Leitch, 2005a; Leitch and Bennett, 2007). The latter view seems to be the best supported, at least for Liliaceae, since genome size is related to morphological, ecological and/or adaptive changes (see Patterson and Givnish, 2002, for fuller discussion), although, as noted above, the analysis by Leitch et al. (2007) suggested that large genomes evolved passively.
Karyotype structure in Liliaceae
Figure 1, which gives a mean haploid karyotype for each genus, and data in Supplementary Data 1 (online) highlight the considerable diversity in karyotype structure in Liliaceae, already noted in previous studies (e.g. Tamura, 1995, 1998a, b). There is considerable variation in chromosome number (x = 6–13), ploidy (2x–6x; and up to 11x in Gagea; Peruzzi, 2008, and references cited therein), chromosome size (1·00–37·73 µm) and karyotype asymmetry. By superimposing these data onto the phylogenetic framework, patterns and trends in karyotype evolution can be seen.
Karyotype asymmetry
Significant differences in karyotype asymmetry are apparent within Liliaceae based on the various measures of karyotype asymmetry analysed. Most of the correlations found among different karyotype asymmetry measures (Supplementary Data 2, available online) agreed with data reported by Paszko (2006) for Calamagrostis (Poaceae), and overall the present study supports her conclusions concerning the general validity of the different measures of karyotype asymmetry tested. However, from the present data, the asymmetry index (AI) which Paszko proposed as a new measure of asymmetry, aiming to capture the degree of asymmetry in a single value, is strongly and positively correlated with CVCI (r = 0·85, a measure of asymmetry based on variation in centromere position between chromosomes in the karyotype) but not significantly correlated with CVCL (r = 0·08, a measure of asymmetry based on variation in chromosome lengths between chromosomes in the karyotype). For this reason, it was felt that the AI value proposed by Paszko does not adequately reflect all aspects of karyotype asymmetry in Liliaceae. Instead it seems more meaningful to retain these two separate parameters which reflect two different aspects of karyotype asymmetry, variation in chromosome length (CVCL) and centromere position (CVCI).
Boxplots summarizing the values of these two parameters for each genus (Fig. 4) highlight the wide range and large values of CVCI in Calochortus and tribe Lilieae compared with other genera. This indicates that their karyotypes are considerably more asymmetric in terms of variation in the position of the centromeres than those of other genera of Liliaceae. In contrast, there was less variation in the range of values obtained for CVCL between and within different genera.
Plotting CVCL and CVCI values for each genus highlights further differences in karyotype asymmetry between different genera (Fig. 5). For example, the asymmetric karyotypes that characterize species in Lilieae are due more to variation in the position of the centromere (large CVCI values) than to differences between the chromosome sizes in the karyotype (i.e. lower values of CVCL; Fig. 5A). Karyotypes in Tulipeae are generally less asymmetric than in Lilieae as most have a narrower range of values for both CVCL and CVCI (Figs 4 and 5B), as also found in Medeoloideae (Figs 4 and 5C). An exception to this pattern is the relatively high CVCL values in Gagea within Tulipeae. For the remaining genera in Liliaceae, increasing karyotype asymmetry arises through increasing variation in chromosome lengths (wide range of CVCL values) rather than shifts in centromere position (narrow range of CVCI; Fig. 5C), a pattern also observed in Smilax in Smilacaceae. There were no species with large CVCL and CVCI values, indicating that there are no karyotypes that are asymmetric in terms of both large differences in chromosome length and centromere position in Liliaceae. The only comparable study using CVCL and CVCI to describe karyotype asymmetry was that of Paszko (2006). However, this study was restricted to just eight species of Calamagrostis (Poaceae) and the data were not viewed within a phylogenetic framework. To what extent the patterns observed here are specific to Liliaceae or represent more general patterns of karyotype asymmetry evolution is currently unknown.
Karyotype asymmetry and genome size
Overall, an analysis of the relationship between 1Cx and CVCI showed these two parameters were positively correlated (Fig. 6A), indicating that genome size increases are generally accompanied by increasing intrachromosomal asymmetry through increased variability in centromere position within the karyotype (r = 0·65). Such a pattern suggests that the additional DNA is not being added uniformly across the karyotype. This correlation was also found for various subgroups within Liliaceae including Tricyrtis (r = 0·56), Lilioideae (r = 0·59), Lilieae (r = 0·23) and Lilium (r = 0·32). However, for Fritillaria (in Lilieae) and the whole of Tulipeae, the correlation was negative (r = −0·33 for Fritillaria and −0·37 for Tulipeae), suggesting that the distribution of the additional DNA followed a different pattern from that noted for the rest of the family.
The relationship between 1Cx and CVCL was generally negative (Fig. 6B), indicating that an increase in genome size is accompanied by decreasing size differences between chromosomes of the karyotype (i.e. a more symmetrical karyotype). This correlation was also found for some sub-groups analysed alone: Tricyrtis, Lilioideae and Tulipeae.
The analysis of changes in individual chromosome asymmetry with genome size provided some insights into how the additional DNA is distributed across the karyotype. Whereas the large metacentric chromosomes (e.g. chromosome pairs I and II) became more symmetrical with increasing genome size, the smaller sub-metacentric and telocentric chromosomes (i.e. chromosome pairs IV and above) became more asymmetrical. Such a pattern implies that the additional DNA is added mainly to the long arms of the smaller chromosomes rather than being distributed evenly between all chromosome arms of the karyotype complement. As far as we are aware, this type of chromosome evolution pattern has not been noted before.
To date, two main patterns for the addition of DNA in a chromosome complement have been described (reviewed by Levin, 2002). For ‘proportional increase’, the amount of DNA added to each chromosome arm is proportional to its length. This pattern does not result in a change in karyotype asymmetry with increasing DNA amount and thus the values for CVCI and CVCL remain unchanged as the genome size changes. This pattern has been observed in several genera including Aloe and Gasteria (Xanthorrhoeaceae) (Brandham and Doherty, 1998) and in some species of Oxalis (Oxalidaceae) (De Azkue and Martinez, 1988). For ‘equal increase’, the same amount of DNA is added to each chromosome arm regardless of its size. This will result in an increase in karyotype symmetry, and hence a decrease in CVCI and CVCL with increasing genome size. Examples of genera showing this pattern include Vigna (Fabaceae) (Parida et al., 1990), Vicia (Fabaceae) (Raina and Rees, 1983), Papaver (Papaveraceae) (Srivastava and Lavania, 1991) and Hypochaeris (Asteraceae) (Cerbah et al., 1998a, b).
In many genera in Liliaceae, however, a third pattern is apparent. Here there is an ‘unequal increase’, i.e. the amount of DNA added varies between longer and shorter chromosome arms unequally, leading to an overall increase in karyotype asymmetry with genome size. This pattern gives rise to the observed increase in CVCI but a slight decrease in CVCL with increasing genome size.
Such a pattern may, in part, be influenced by an upper limit on chromosome size for a particular karyotype, as reported for both monocots and eudicots (Schubert and Oud, 1997; Hudakova et al., 2002). Nevertheless, other factors such as the involvement of Robertsonian translocations will also play a role as exceptions to the unequal increase pattern are found in Fritillaria (with some of the largest chromosomes of Liliaceae) and Tulipeae. In these groups, the increase in genome size is accompanied by a general decrease in asymmetry (decreasing CVCI) and is typical of the equal increase pattern noted above.
Of course there are other types of chromosome change which can contribute to asymmetry variation between species and genera (reviewed by Schubert, 2007) including pericentric inversions and/or differential translocations of DNA between smaller and larger chromosomes (if there is no change in chromosome number) and Robertsonian fissions and fusions (if accompanied by changes in chromosome number); such karyotype changes have also been reported in some species of Liliaceae. Indeed the low, but significant, negative correlation between CVCI and CVCL probably in part reflects the occurrence of Robertsonian translocations which are found in several genera including Cardiocrinum, Fritillaria, Streptopus and Tricyrtis (Darlington, 1973; Zhongyan et al., 1989; Tamura, 1995; Zhang and Gu, 2005).
Ploidy, genome size and karyotype asymmetry
The average negative correlation between the percentage of polyploid species and genome size found in almost all investigated taxa (Fig. 8) agrees with many other published studies for other groups of angiosperms (e.g. D'Ovidio and Marchi, 1990; Levin, 2002) and across angiosperms as a whole (Leitch and Bennett, 2004). The occurrence of polyploidy was also found to be related to genome size (Fig. 7). Thus genera with mean 1Cx values up to 25 pg showed a wide range of polyploidy from 0 to 80 % depending on the genus, whereas the occurrence of polyploidy in genera with mean 1Cx values above 25 pg was low (0·8 % in Lilium and 3 % in Fritillaria) or non-existent (in Notholirion and Cardiocrinum). These results agree broadly with those previously reported by Grif (2000) based on an analysis at family level, namely that the percentage of polyploids in a family decreased with increasing mean genome size for the family. It is noted that out of the 11 monocot families analysed by Grif, Liliaceae had the largest mean genome size (1C = approx. 20 pg) and the lowest percentage of polyploids (20 %).
Trends and patterns of karyotype evolution in Liliaceae
According to the present study (and also Leitch et al., 2007, concerning genome size evolution), ancestral karyotypes in Liliaceae are likely to have had small genomes (1Cx = approx. 6 pg), low CVCI values, relatively high CVCL values (similar to Smilax) and x = 8. From this evolutionary starting point, different trends in karyotype evolution at the generic level are apparent.
Streptopus, Scoliopus and Prosartes
In the small clade comprising Streptopus, Scoliopus and Prosartes (15 species in total), although there was some variation in chromosome number (in Prosartes; 2n = 12, 16 and 18) and ploidy (Streptopus; up to 6x based on x = 8), the karyotypes are largely similar to those in the outgroup Smilacaceae in terms of structure (i.e. range of CVCL and CVCI) and genome size, with the notable exception of the >2-fold increase in genome size in Scoliopus. Given that only one species of Scoliopus (out of two recognized) was analysed, the mode of DNA addition is difficult to determine.
Tricyrtis
In Tricyrtis the karyotype has maintained similar karyotypic features to those in Smilacaceae, Streptopus and Prosartes (small and narrow range of genome sizes and a karyotype that is not strongly asymmetric). The main difference in Tricyrtis was the increase in the basic chromosome number from x = 8 to x = 13 which may have arisen via an ancient palaeopolyploid event followed by loss of DNA. The predominant chromosome number for Tricyrtis is 2n = 26 although counts of 2n = 24 have been reported in T. hirta, possibly linked to Robertsonian translocations (Takahashi, 1991).
Calochortus
Karyotypically, Calochortus (comprising approx. 65 species) is more variable than the genera mentioned above, with considerable variation in basic chromosome number (x = 6, 7, 8, 9 and 10), genome size (inferred 1Cx values range 8·5-fold) and asymmetry (especially CVCI, Fig. 5C) compared with the presumed ancestral state in, for example, Streptopus and Smilax. The mechanisms involved in generating this karyotype diversity are currently unknown, although a role for translocations, fusions and fissions has been proposed (Beal, 1939). (For a more detailed discussion of chromosome diversity and evolution in the genus see Patterson and Givnish, 2004.)
Lilioideae + Medeoloideae
The Lilioideae + Medeoloideae clade is the most diverse of Liliaceae. This clade has experienced large increases in genome size (2- to 25-fold), and the two types of karyotype asymmetry (i.e. intra- and interchromosomal, reflected in the values for CVCI and CVCL, respectively) have contrasting influences on karyotype evolution, depending on the taxonomic group concerned.
Subfamily Medeoloideae
The most plesiomorphic karyotype in the Lilioideae + Medeoloideae clade is probably found in Medeoloideae which, apart from an increase in genome size (2− to 3-fold) and a reduction in the basic chromosome number from x = 8 to x = 7, has maintained low karyotype asymmetry features with a karyotype consisting of seven large, more or less metacentric chromosome pairs. Of the five species of Clintonia, only one is diploid (C. udenesis, 2n = 2x = 14). The remaining four are tetraploid, making this the genus with the highest percentage of polyploids in Liliaceae. Medeola is monotypic (M. virginiana is the sole species) and has 2n = 14 chromosomes.
Subfamily Lilioideae
In this subfamily the basic chromosome number increased to x = 12. It has been suggested that this arose through multiple reciprocal centric fissions (Tamura, 1995) probably involving the five smaller chromosome pairs of a Medeoleae-like ancestor or, less probably, through polyploidy. The majority of species in this subfamily have retained a base number of x = 12, with just a few exceptions in some Fritillaria (e.g. F. montana and F. ruthenica with 2n = 18) and Gagea species. However, whereas polyploidy is fairly common in Tulipeae (40·7 %, according to the present data set), it is rare in Lilieae (1·54 %).
Tribe Lilieae
In this tribe the karyotype evolved towards a further increase of intrachromosomal asymmetry (up to 3-fold increase in CVCI, Fig. 5A) and genome size (up to 7-fold in some Fritillaria and Lilium species), mostly through unequal addition of DNA to the long arms of the ten smaller telocentric pairs (III–XII; with Fritillaria being an exception). As a consequence of this unequal addition of DNA, this mode of karyotype evolution was accompanied by a small reduction in interchromosomal asymmetry (0·5− to 0·7-fold, Fig. 5A compared with 5C).
Tribe Tulipeae
In contrast to the sister tribe Lilieae, the main mechanism of chromosome evolution in Tulipeae was shown to be an equal change in the amount of DNA for each chromosome arm, regardless of its length, although there was some evidence of preferential addition or subtraction of DNA from the smaller chromosomes. Consequently, the two largest chromosome pairs are telocentric/sub-telocentric rather than typically metacentric (as in Lilieae), and these changes are accompanied by variations in interchromosomal asymmetry (i.e. CVCL, Fig. 5B). Within this framework, two distinct pathways of karyotype evolution are apparent: (1) a massive reduction in genome size (up to 0·3- to 0·5-fold) in Gagea accompanied by an increase in interchromosomal asymmetry (CVCL up to 2- to 3-fold), leaving the intrachromosomal asymmetry (CVCI) almost unchanged; and (2) an increase in genome size in Amana, Erythronium and Tulipa through the addition of equal amounts of DNA to each chromosome regardless of length, leading to a reduction in both their inter- and intrachromosomal asymmetries (i.e. CVCL and CVCI) with respect to Gagea (Fig. 5B).
CONCLUSIONS
In summary, through this combined approach of collating all available information (karyomorphometric data, genome size, phylogeny) for species of Liliaceae it has been possible to reconstruct the most likely patterns of chromosome evolution in this family. Such an approach has highlighted the diverse modes of karyotype evolution to be found even within this comparatively small angiosperm family.
SUPPLEMENTARY DATA
Supplementary Data are available at Annals of Botany online and consist of (1) karyotype features in the studied literature accessions of Liliaceae and Smilacaceae; and (2) Pearson's correlation coefficients among all published modalities of karyotype asymmetry measurements and genome size.
ACKNOWLEDGEMENTS
The authors wish to thank Professor F. Garbari, Professor R. Cremonini (University of Pisa) and Professor C. G. Vosa (Oxford University) for discussions about the work, comments and suggestions, and two anonymous reviewers who greatly improved an earlier version of the manuscript. Financial funding (EX60%) from the University of Pisa is gratefully acknowledged.
LITERATURE CITED
- APG II. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants. Botanical Journal of the Linnean Society. 2003;141:399–436. [Google Scholar]
- Arano H. Cytological studies in subfamily Carduoideae (Compositae) of Japan. IX. The karyotype analysis and phylogenetic considerations on Pertya and Ainsliaea. Botanical Magazine (Tokyo) 1963;76:32–39. [Google Scholar]
- Beal JM. Cytological studies in relation to the classification of the genus Calochortus. Botanical Gazette. 1939;100:528–547. [Google Scholar]
- Bennett MD. Natural variation in nuclear characters of meristems in Vicia faba. Chromosoma. 1970;29:317–335. doi: 10.1007/BF00325946. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ. Genome size evolution in plants. In: Gregory TR, editor. The evolution of the genome. a. San Diego: Elsevier; 2005. pp. 89–162. [Google Scholar]
- Bennett MD, Leitch IJ. Plant DNA C-values database. 2005. (release 4.0, October 2005). http://www.kew.org/genomesize/homepage . [DOI] [PubMed]
- Bennett MD, Rees H. Natural and induced changes in chromosome size and mass in meristems. Nature. 1967;b 215:93–94. doi: 10.1038/215093a0. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Rees H. Induced and developmental variation in chromosomes of meristematic cells. Chromosoma. 1969;27:226–244. [Google Scholar]
- Brandham PE, Doherty MJ. Genome size variation in the Aloaceae, an angiosperm family displaying karyotypic orthoselection. Annals of Botany. 1998;82:67–73. [Google Scholar]
- Caputo P, Frediani M, Venora G, Ravalli C, Ambrosio M, Cremonini R. Nuclear DNA contents, rDNAs, and karyotype evolution in subgenus Vicia: III. The heterogeneous section Hypechusa. Protoplasma. 2006;228:167–177. doi: 10.1007/s00709-006-0158-2. [DOI] [PubMed] [Google Scholar]
- Cave MS. Chromosomes of the California Liliaceae. University of California Publications in Botany. 1970;57:1–58. [Google Scholar]
- Ceccarelli M, Minelli S, Maggini F, Cionini PG. Genome size variation in Vicia faba. Heredity. 1995;74:180–187. [Google Scholar]
- Cerbah MJ, Coulaud J, Siljak-Yakovlev S. rDNA organization and evolutionary relationships in the genus Hypochaeris (Asteraceae) Journal of Heredity. 1998;a 89:312–318. [Google Scholar]
- Cerbah MJ, Souza-Chies T, Joubier MF, Lejeune B, Siljak-Yakovlev S. Molecular phylogeny of the genus Hypochaeris using internal transcribed spacers of nuclear rDNA: inference for chromosomal evolution. Molecular Biology and Evolution. 1998;b 15:345–354. doi: 10.1093/oxfordjournals.molbev.a025931. [DOI] [PubMed] [Google Scholar]
- Chase MW, Duvall MR, Hillis HG, et al. Molecular phylogenetics of Lilianae. In: Rudall PJ, Cribb PJ, Cutler DF, Humphries C, editors. Monocotyledons: systematics and evolution. Kew: Royal Botanic Gardens; 1995. pp. 109–137. [Google Scholar]
- Chase MW, Soltis DE, Soltis PS, et al. Higher-level systematics of the monocotyledons: an assessment of current knowledge and a new classification. In: Wilson KL, Morrison DA, editors. Monocots: systematics and evolution. Melbourne: CSIRO; 2000. pp. 3–16. [Google Scholar]
- Cox AV, Abdelnour GJ, Bennett MD, Leitch IJ. Genome size and karyotype evolution in the slipper orchids (Cypripedioideae: Orchidaceae) American Journal of Botany. 1998;85:681–687. [PubMed] [Google Scholar]
- Dahlgren RMT, Clifford HT, Yeo PF. The families of the monocotyledons: structure, evolution and taxonomy. Berlin: Springer-Verlag; 1985. [Google Scholar]
- Darlington CD. Chromosome botany and the origins of cultivated plants. London: George Allen & Urwin; 1973. [Google Scholar]
- Darlington CD, Wylie AP. Chromosome atlas of flowering plants. London: George Allen & Unwin Ltd; 1955. [Google Scholar]
- Das AB, Mohanty S, Marrs RH, Das P. Somatic chromosome number and karyotype diversity in fifteen species of Mammillaria of the family Cactaceae. Cytobios. 1999;97:141–151. [Google Scholar]
- De Azkue D, Martinez A. DNA content and chromosome evolution in the shrubby Oxalis. Genome. 1988;30:52–57. [Google Scholar]
- De Melo Nationiel F, Guerra M, Benko-Iseppon AM, De Menezes NL. Cytogenetics and cytotaxonomy of Velloziaceae. Plant Systematics and Evolution. 1997;204:257–273. [Google Scholar]
- D'Ovidio R, Marchi P. DNA content, karyotype structure analysis and karyotype symmetry in Ranunculus L. (Ranunculaceae). Italian species belonging to sections Flammula (Webb) Benson and Micranthus (Ovcz.) Nyarady. Caryologia. 1990;43:99–115. [Google Scholar]
- Fay MF, Chase MW. Modern concepts of Liliaceae, with a focus on the relationships of Fritillaria. Curtis's Botanical Magazine. 2000;17:146–149. [Google Scholar]
- Fay MF, Chase MW, Rønsted N, et al. Phylogenetics of Liliales: summarized evidence from combined analyses of five plastid and one mitochondrial loci. In: Columbus JT, Friar EA, Porter JM, Prince LM, Simpson MG, editors. Monocots:comparative biology and evolution (excluding Poales). Claremont, CA: Rancho Santa Ana Botanic Garden; 2006. pp. 559–565. [Google Scholar]
- Fedorov A. Chromosome numbers of flowering plants. Leningrad: Nauka Publishers; 1969. [Google Scholar]
- Frediani M, Caputo P, Venora G, Ravalli C, Ambrosio M, Cremonini R. Nuclear DNA contents, rDNAs and karyotype evolution in subgenus Vicia: II. Section Peregrinae. Protoplasma. 2005;226:181–190. doi: 10.1007/s00709-005-0114-6. [DOI] [PubMed] [Google Scholar]
- González-Aguilera JJ, Fernández-Peralta AM. Phylogenetic relationships in the family Resedaceae. Genetica. 1984;64:185–198. [Google Scholar]
- Gregory TR. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biological Reviews. 2001;76:65–101. [Google Scholar]
- Gregory TR. Synergy between sequence and size in large-scale genomics. Nature Reviews Genetics. 2005;6:699–708. doi: 10.1038/nrg1674. [DOI] [PubMed] [Google Scholar]
- Greilhuber J, Doležel J, Lysak MA, Bennett MD. The origin, evolution and proposed stabilization of the terms ‘genome size’ and ‘C-value’ to describe nuclear DNA contents. Annals of Botany. 2005;95:255–260. doi: 10.1093/aob/mci019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greilhuber J, Speta F. C-banded karyotypes in the Scilla hohenackeri group, S. persica and Puschkinia (Liliaceae) Plant Systematics and Evolution. 1976;126:149–188. [Google Scholar]
- Grif VG. Some aspects of plant karyology and karyosystematics. International Review of Cytology. 2000;196:131–175. doi: 10.1016/s0074-7696(00)96004-2. [DOI] [PubMed] [Google Scholar]
- Hawkins JS, Grover CE, Wendel JF. Repeated big bangs and the expanding universe: directionality in plant genome size evolution. Plant Science. 2009 in press. [Google Scholar]
- Hudakova S, Kunzel G, Endo TR, Schubert I. Barley chromosome arms longer than half of the spindle axis interfere with nuclear divisions. Cytogenetic and Genome Research. 2002;98:101–107. doi: 10.1159/000068530. [DOI] [PubMed] [Google Scholar]
- Huziwara Y. Karyotype analysis in some genera of Compositae. VIII. Further studies on the chromosomes of Aster. American Journal of Botany. 1962;49:116–119. [Google Scholar]
- Lavania UC, Srivastava S. A simple parameter of dispersion index that serves as a adjunct to karyotype asymmetry. Journal of Biosciences. 1992;17:179–182. [Google Scholar]
- Leitch IJ, Bennett MD. Genome downsizing in polyploid plants. Biological Journal of the Linnean Society. 2004;82:651–663. [Google Scholar]
- Leitch IJ, Bennett MD. Genome size and its uses: the impact of flow cytometry. In: Doležel J, Greilhuber J, Suda J, editors. Flow cytometry with plant cells. Wiley-VCH Verlag GmbH & Co; 2007. pp. 153–176. [Google Scholar]
- Leitch IJ, Beaulieu JM, Cheung K, Hanson L, Lysak M, Fay MF. Punctuated genome size evolution in Liliaceae. Journal of Evolutionary Biology. 2007;20:2296–2308. doi: 10.1111/j.1420-9101.2007.01416.x. [DOI] [PubMed] [Google Scholar]
- Levan A, Fredga K, Sandberg AA. Nomenclature for centromeric position on chromosomes. Hereditas. 1964;52:201–220. [Google Scholar]
- Levin DA. The role of chromosome change in plant evolution. New York: Oxford University Press; 2002. [Google Scholar]
- Lim KY, Matyasek R, Lichtenstein CP, Leitch AR. Molecular cytogenetic analyses and phylogenetic studies in the Nicotiana section Tomentosae. Chromosoma. 2000;109:245–258. doi: 10.1007/s004120000074. [DOI] [PubMed] [Google Scholar]
- Lim KY, Matyasek R, Kovarik A, Leitch AR. Genome evolution in allotetraploid Nicotiana. Biological Journal of the Linnean Society. 2004;82:599–606. [Google Scholar]
- Lim KY, Kovarik A, Matyasek R, et al. Comparative genomics and repetitive sequence divergence in the species of diploid Nicotiana section Alatae. The Plant Journal. 2006;48:907–919. doi: 10.1111/j.1365-313X.2006.02930.x. [DOI] [PubMed] [Google Scholar]
- Lim KY, Kovarik A, Matyasek R, et al. Sequence of events leading to near-complete genome turnover in allopolyploid Nicotiana within five million years. New Phytologist. 2007;175:756–763. doi: 10.1111/j.1469-8137.2007.02121.x. [DOI] [PubMed] [Google Scholar]
- Lysak MA, Lexer C. Towards the era of comparative evolutionary genomics in Brassicaceae. Plant Systematics and Evolution. 2006;259:175–198. [Google Scholar]
- Lysak MA, Mandakova T. Unexpected paleopolyploid evolution in Stenopetalum – a genus with the lowest chromosome numbers in the Brassicaceae. Chromosome Research. 2007;15:11–12. [Google Scholar]
- Lysak MA, Koch MA, Pecinka A, Schubert I. Chromosome triplication found across the tribe Brassiceae. Genome Research. 2005;15:516–525. doi: 10.1101/gr.3531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proceedings of the National Academy of Sciences; USA. 2006. pp. 5224–5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak MA, Cheung K, Kitschke M, Bures P. Ancestral chromosomal blocks are triplicated in Brassiceae species with varying chromosome number and genome size. Plant Physiology. 2007;145:402–410. doi: 10.1104/pp.107.104380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T. Cytological studies in family Liliaceae of Japan. 3. Karyotype analysis in genus Tricyrtis. Botanical Magazine (Tokyo) 1968;81:590–599. [Google Scholar]
- Narayan RKJ, Rees H. Nuclear DNA variation in Lathyrus. Chromosoma. 1976;54:141–154. [Google Scholar]
- Parida A, Raina SN, Narayan RKJ. Quantitative DNA variation between and within chromosome complements of Vigna species (Fabaceae) Genetica. 1990;82:125–133. [Google Scholar]
- Paszko B. A critical review and a new proposal of karyotype asymmetry indices. Plant Systematics and Evolution. 2006;258:39–48. [Google Scholar]
- Patterson TB, Givnish TJ. Phylogeny, concerted convergence, and phylogenetic niche conservatism in the core Liliales: insights from rbcL and ndhF sequence data. Evolution. 2002;56:233–252. doi: 10.1111/j.0014-3820.2002.tb01334.x. [DOI] [PubMed] [Google Scholar]
- Patterson TB, Givnish TJ. Geographic coehesion, chromosomal evolution, parallel adaptive radiations, and consequent floral adaptations in Calochortus (Calochortaceae): evidence from a cpDNA phylogeny. New Phytologist. 2004;161:253–264. [Google Scholar]
- Peruzzi L. Contribution to the cytotaxonomical knowledge of the genus Gagea Salisb. (Liliaceae). III. New karyological data from the C Mediterranean area. Caryologia. 2008;61:92–106. [Google Scholar]
- Peruzzi L, Peterson A, Tison J-M, Peterson J. Phylogenetic relationships of Gagea Salisb. (Liliaceae) in Italy, inferred from molecular and morphological data matrices. Plant Systematics and Evolution. 2008;a in press. doi: 10.1007/s00606-008-0081-4. [Google Scholar]
- Peruzzi L, Tison JM, Peterson A, Peterson J. On the phylogenetic position and taxonomic value of Gagea trinervia (Viv.) Greuter and the whole Gagea sect. Anthericoides A. Terracc. (Liliaceae) Taxon. 2008;b 57:1201–1214. [Google Scholar]
- Peterson A, John H, Koch E, Peterson J. A molecular phylogeny of the genus Gagea (Liliaceae) in Germany inferred from non-coding chloroplast and nuclear DNA sequences. Plant Systematics and Evolution. 2004;245:145–162. [Google Scholar]
- Peterson A, Levichev IG, Peterson J. Systematics of Gagea and Lloydia (Liliaceae) and infrageneric classification of Gagea based on molecular and morphological data. Molecular Phylogenetics and Evolution. 2008;46:446–465. doi: 10.1016/j.ympev.2007.11.016. [DOI] [PubMed] [Google Scholar]
- Pires JC, Maureira IJ, Givnish TJ, et al. Phylogeny, genome size, and chromosome evolution of Asparagales. Aliso. 2006;22:287–304. [Google Scholar]
- Plummer JA, Shan FC, Galwey N, Yan GJ. New methods for comparison of chromosomes within and between species. Caryologia. 2003;561:227–231. [Google Scholar]
- Raina SN, Rees H. DNA variation between and within chromosome complements of Vicia species. Heredity. 1983;51:335–346. [Google Scholar]
- Romero Zarco C. A new method for estimating karyotype asymmetry. Taxon. 1986;35:526–530. [Google Scholar]
- Rønsted N, Law S, Thornton H, Fay MF, Chase MW. Molecular phylogenetic evidence for the monophyly of Fritillaria and Lilium (Liliaceae; Liliales) and the infrageneric classification of Fritillaria. Molecular Phylogenetics and Evolution. 2005;35:509–527. doi: 10.1016/j.ympev.2004.12.023. [DOI] [PubMed] [Google Scholar]
- Ruffini Castiglione M, Frediani M, Gelati MT, et al. Cytological and molecular characterization of Vicia esdraeolensis Warb. & Eigr: a rare taxon. Protoplasma. 2007;231:151–156. doi: 10.1007/s00709-007-0256-9. [DOI] [PubMed] [Google Scholar]
- Sato D. Karyotype alteration and phylogeny in Liliaceae and allied families. Japanese Journal of Botany. 1943;12:57–161. [Google Scholar]
- Schubert I. Chromosome evolution. Current Opinion in Plant Biology. 2007;10:109–115. doi: 10.1016/j.pbi.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Schubert I, Oud JL. There is an upper limit of chromosome size for normal development of an organism. Cell. 1997;88:515–520. doi: 10.1016/s0092-8674(00)81891-7. [DOI] [PubMed] [Google Scholar]
- Sen S. Cytotaxonomy of Liliales. Feddes Repertorium. 1975;86:255–305. [Google Scholar]
- Shan F, Yan G, Plummer JA. Karyotype evolution in the genus Boronia (Rutaceae) Botanical Journal of the Linnean Society. 2003;142:309–320. [Google Scholar]
- Srivastava S, Lavania UC. Evolutionary DNA variation in Papaver. Genome. 1991;34:763–768. [Google Scholar]
- Stace HM. Cytoevolution in the genus Calotis R. Br. (Compositae: Astereae) Australian Journal of Botany. 1978;26:287–307. [Google Scholar]
- Stebbins GL. Chromosomal evolution in higher plants. London: Edward Arnold; 1971. [Google Scholar]
- Takahashi H. Karyotype variations of Tricyrtis hirta Hook. (Liliaceae) Acta Phytotaxonomica et Geobotanica. 1991;42:113–124. [Google Scholar]
- Takhtajan A. Diversity and classification of flowering plants. New York: Columbia University Press; 1997. [Google Scholar]
- Tamura MN. A karyological review of the orders Asparagales and Liliales (Monocotyledonae) Feddes Repertorium. 1995;106:83–111. [Google Scholar]
- Tamura MN. Calochortaceae. In: Kubitzki K, editor. The families and genera of vascular plants. III. Flowering plants – monocotyledons, Lilianae (except Orchidaceae) a. Berlin: Springer-Verlag; 1998. pp. 164–172. [Google Scholar]
- Tamura MN. Liliaceae. In: Kubitzki K, editor. The families and genera of vascular plants. III. Flowering plants – monocotyledons, Lilianae (except Orchidaceae) b. Berlin: Springer-Verlag; 1998. pp. 343–353. [Google Scholar]
- Tan D-Y, Zhang Z, Li X-R, Hong D-Y. Restoration of the genus Amana Honda (Liliaceae) based on a cladistic analysis of morphological characters. Acta Phytotaxonomica Sinica. 2005;43:262–270. [Google Scholar]
- Vanzela ALL, Ruas PM, Marin-Morales MA. Karyotype studies of some species of Dalechampia Plum. (Euphorbiaceae) Botanical Journal of the Linnean Society. 1997;125:25–33. [Google Scholar]
- Venora G, Blangiforti S, Frediani M, et al. Nuclear DNA contents, rDNAs, chromatin organization, and karyotype evolution in Vicia sect. Faba. Protoplasma. 2000;213:118–125. [Google Scholar]
- Vijayavalli B, Mathew PM. Cytology of five species of Smilax from South India. The Nucleus. 1987;30:57–60. [Google Scholar]
- Watanabe K, King RM, Yahara T, et al. Chromosomal cytology and evolution in Eupatorieae (Asteraceae) Annals of the Missouri Botanical Garden. 1995;82:581–592. [Google Scholar]
- Watanabe K, Yahara T, Denda T, Kosuge K. Chromosomal evolution in the genus Brachyscome (Asteraceae, Astereae): statistical tests regarding correlation between changes in karyotype and habit using phylogenetic information. Journal of Plant Research. 1999;112:145–161. [Google Scholar]
- Zander RH. Minimal values for reliability of bootstrap and jacknife proportions, decay index, and Bayesian posterior probability. Phyloinformatics. 2004;2:1–13. [Google Scholar]
- Zhang T, Gu Z-J. A new basic chromosome number of x = 7 for the genus Streptopus (Liliaceae) Acta Phytotaxonomica Sinica. 2005;43:533–538. [Google Scholar]
- Zhongyan S, Rujuan L, Shouyi Y. Studies on karyotypes of two species in Fritillaria (Liliaceae) In: Deyuan H, editor. Plant chromosome research 1987. Hiroshima: Nishiki Print Co., Ltd; 1989. pp. 323–326. [Google Scholar]
- Zonneveld BJM, Leitch IJ, Bennett MD. First nuclear DNA amounts in more than 300 angiosperms. Annals of Botany. 2005;96:229–244. doi: 10.1093/aob/mci170. [DOI] [PMC free article] [PubMed] [Google Scholar]