Abstract
Background
Photosynthetic electron transport is performed by a chain of redox components that are electrochemically connected in series. Its efficiency depends on the balanced action of the photosystems and on the interaction with the dark reaction. Plants are sessile and cannot escape from environmental conditions such as fluctuating illumination, limitation of CO2 fixation by low temperatures, salinity, or low nutrient or water availability, which disturb the homeostasis of the photosynthetic process. Photosynthetic organisms, therefore, have developed various molecular acclimation mechanisms that maintain or restore photosynthetic efficiency under adverse conditions and counteract abiotic stresses. Recent studies indicate that redox signals from photosynthetic electron transport and reactive oxygen species (ROS) or ROS-scavenging molecules play a central role in the regulation of acclimation and stress responses.
Scope
The underlying signalling network of photosynthetic redox control is largely unknown, but it is already apparent that gene regulation by redox signals is of major importance for plants. Signalling cascades controlling the expression of chloroplast and nuclear genes have been identified and dissection of the different pathways is advancing. Because of the direction of information flow, photosynthetic redox signals can be defined as a distinct class of retrograde signals in addition to signals from organellar gene expression or pigment biosynthesis. They represent a vital signal of mature chloroplasts that report their present functional state to the nucleus. Here we describe possible problems in the elucidation of redox signalling networks and discuss some aspects of plant cell biology that are important for developing suitable experimental approaches.
Conclusions
The photosynthetic function of chloroplasts represents an important sensor that integrates various abiotic changes in the environment into corresponding molecular signals, which, in turn, regulate cellular activities to counterbalance the environmental changes or stresses.
Key words: Photosynthesis, redox signals, gene expression, regulatory network, retrograde signalling, cross-talk, plastids, higher plants
INTRODUCTION
Photosynthesis is the principle energy fixing process that sustains life on earth. It harvests sunlight to drive electron transport from the low-potential electron donor water to the high-potential electron end-acceptor NADP+. During this reaction, energy (ATP) and reduction (NADPH) equivalents are generated that are used in subsequent steps of carbon dioxide fixation and reduction in the Calvin–Benson cycle, the dark reaction. Here ADP and NADP+ are regenerated to be used again in the light reaction. Thus, the light-driven redox chemistry of the light reactions and the temperature-dependent enzymatic reactions of the dark reaction are tightly coupled. This coupling renders photosynthesis and its efficiency highly dependent on the prevailing environmental conditions. Changes in various abiotic factors such as the intensity and quality of the incident light (directly affecting the light reaction), the temperature and the nutrient and water availability (directly or indirectly affecting the dark reaction) have an immediate impact on photosynthetic efficiency and ultimately on plant yield. Changes in photosynthetic electron transport, however, have an effect on the components involved in it: the balance between reduced and oxidized forms is changed. Photosynthetic organisms including cyanobacteria, algae and higher plants have therefore evolved regulatory responses that acclimate the photosynthetic process to the prevailing environment and optimize photosynthetic electron transport (Aro and Andersson, 2001; Blankenship, 2002; Walters, 2005). So far as is currently known, most (if not all) acclimation responses are triggered by those changes in the redox state of photosynthetic components that are the target for the counteracting effect of the acclimation response. By this principle a feedback control is established, in which photosynthesis serves as a sensor that signals distinct changes in the environment and that controls respective acclimation responses (Arnon, 1982).
Acclimation responses to imbalances in excitation energy distribution between photosystem II (PSII) and photosystem I (PSI) are well studied. Since the two photosystems work electrochemically in series, any illumination situation in which one PS is favoured over the other results in either reduction or oxidation of the intersystem electron transport chain. Such imbalances are counteracted by two different mechanisms: state transitions and adjustment of photosystem stoichiometry. State transitions represent a short-term response that occurs in the order of minutes. It re-distributes excitation energy between the photosystems by variation of their relative antennae cross-sections (Allen and Forsberg, 2001; Haldrup et al., 2001; Wollman, 2001). This is achieved by lateral movement of parts of the light-harvesting complex of PSII (LHCII). Upon reduction of the plastoquinone (PQ) pool, which carries the electrons from PSII to the cytochrome b6f (Cytb6f) complex, a redox-sensitive kinase is activated that specifically phosphorylates the mobile LHCII, resulting in its attachment to PSI, the so-called state II. Under PQ oxidizing conditions the kinase is inactive, LHCII is or becomes dephosphorylated (presumably by constitutively active phosphatases) and is relocated to PSII (state I). The mediation of the PQ redox signal toward the kinase is not understood yet; however, it involves the action of the PQ oxidation (QO) site at the cytb6f complex (Vener et al., 1997; Zito et al., 1999). Recently kinases in the alga Chlamydomonas rheinhardtii (Depege et al., 2003) and the higher plant Arabidopsis thaliana (Bellafiore et al., 2005) were identified that appear to be essential for these processes. They were called Stt7 and STN7, respectively.
Photosystem stoichiometry adjustment is a long-term response (LTR) that requires hours to days. It re-directs the excitation imbalances by changing the relative amounts of the two photosystems (Anderson et al., 1995; Melis et al., 1996; Murakami et al., 1997). In contrast to state transitions that represent a purely post-translational acclimation mechanism, adjustment in PS stoichiometry depends on targeted changes in the expression of photosynthesis genes both in the chloroplast and the nucleus (Pfannschmidt et al., 1999, 2001). Interestingly, this acclimation response is also regulated by the redox state of the PQ pool. Most species investigated so far exhibit opposing expression changes in the reaction-centre genes of PSI and PSII. As a general model, it appears that upon reduction of the PQ pool expression of PSI genes is favoured while upon its oxidation expression of PSII genes is favoured. The molecular details may vary from species to species, but one general picture emerges: the redox state of the PQ pool indicates which photosystem is rate-limiting and initiates appropriate counterbalancing changes in gene expression (Pfannschmidt, 2003). Recent analyses have demonstrated that this also involves the action of the STN7 kinase (Bonardi et al., 2005). This points to a functional coupling of state transitions and photosystem stoichiometry adjustment, which has been proposed earlier (Allen and Pfannschmidt, 2000; Rintamaki et al., 2000).
The two responses described above are just an example of how photosynthesis can control acclimation via redox signals. Both typically occur under low-light conditions, e.g. in dense plant populations where the light intensity is low and the light spectrum is shifted towards the far-red wavelength range. Under different conditions resulting in high or excess excitation pressure other acclimation responses are activated, such as non-photochemical quenching, the D1 repair cycle or various stress-response programmes. These responses are also controlled via redox signals from photosynthesis involving the PQ redox state and signals from the PSI acceptor side, but also involve responses controlled by reactive oxygen species (ROS) such as hydrogen peroxide or singlet oxygen. This review does not aim to give a comprehensive overview on the existing literature and can not describe all redox-regulated mechanisms in detail. The interested reader, therefore, is referred to several excellent reviews focused on these topics (Noctor and Foyer, 1998; Niyogi, 2000; Apel and Hirt, 2004; Mittler et al., 2004; Baier and Dietz, 2005; Fey et al., 2005a; Mullineaux and Rausch, 2005). Here, we address central questions and experimental problems regarding the investigation of all these mechanisms. We explain typical limitations of various experimental set-ups and describe examples of how to circumvent at least some of these problems.
IDENTIFICATION OF NATURE AND ORIGIN OF PHOTOSYNTHETIC REDOX SIGNALS
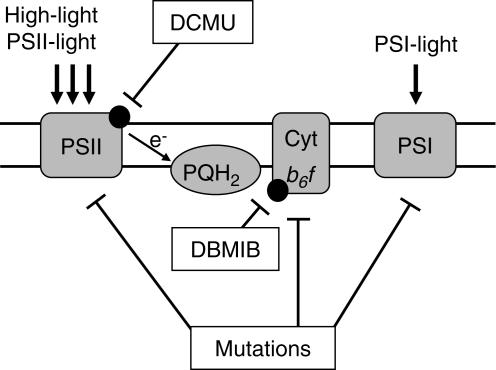
For investigation of redox signals that control gene expression three general approaches have been applied using (1) inhibitors of photosynthetic electron transport, (2) diverse light regimes with effects on electron transport, and (3) mutants with genetic defects in the photosynthetic apparatus (Fig. 1; for reviews see Durnford and Falkowski, 1997; Rodermel, 2001; Pfannschmidt, 2003). Each of these approaches has its specific advantages but also its limitations, which are now briefly presented and discussed.
Fig. 1.
Principle approaches to identify the nature and origin of photosynthetic redox signals. The photosynthetic electron transport chain is depicted schematically. Site-specific inhibitors act at different sites of the chain. Various light regimes can be used to manipulate the electron flow through the chain [PSII-light and high-light conditions lead to preferential or excess excitation of PSII and reduction of plastoquinone (PQH2); PSI-light leads to presumed excitation of PSI and oxidation of the PQ pool]. Mutations may interrupt or affect the electron flow and generate redox signals (reduction of components before the genetic defect and oxidation after it). Combinations of these approaches can help to define the nature and origin of a redox signal. Negative effects are indicated by bars.
The most commonly used electron transport inhibitors are 3-(3′,4′-dichlorophenyl) 1, 1′-dimethyl urea (DCMU) and 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB), which exhibit specific affinities to the QB binding site of the D1 protein of PSII and the plastoquinol oxidizing site at the Cytb6f complex, respectively (Trebst, 1980). Application of DCMU, therefore, blocks the electron transport at PSII and oxidizes all following components of the transport chain, while treatment with DBMIB results in a block at the Cytb6f complex that reduces the components before it and oxidizes those following it. An antagonistic effect of these inhibitors on the expression of a gene thus indicates that the redox state of the plastoquinone pool is the decisive determinant for its regulation. Equal effects of both inhibitors point to consecutive components of the Cytb6f complex as regulators, for instance on stromal components reduced by PSI such as thioredoxins. These inhibitors thus represent useful tools in identifying the origin of the redox signal. Nevertheless, a number of pitfalls exist that should be avoided when working with it. DCMU is a very stable compound that easily penetrates tissues and closes the reaction centres completely for several days (usually the plant dies if not supplemented with sugar). When sprayed on Arabidopsis leaves its effect can be easily monitored by illumination with UV-light, since this light will be dissipated by the PSII antenna as visible red fluorescence generating a high chlorophyll fluorescence phenotype. This can be used to test the evenness of spraying (T. Pfannschmidt et al, pers. obs.). In contrast, DBMIB is unstable in tissues and looses its effect within hours (Pfannschmidt et al., 2001). It is, therefore, desirable to repeat its application during the course of an experiment when working with plants. When working with photosynthetic organisms that can be cultured in liquid media such as algae or cyanobacteria, however, the inhibitor can be applied into the media, resulting in a permanent supply of the inhibitor (Alfonso et al., 2000; Kovacs et al., 2000). In Chlamydomonas reinhardtii it has been reported that stable inhibition at the QO site could be achieved by the plastoquinone analog stigmatellin, while comparable effects with DBMIB could only be observed when the compound was added in light but not in the dark (Finazzi et al., 2001). The concentrations applied are another crucial point for a successful experiment. Concentrations that completely block the electron transport, in general, should be avoided in gene expression studies since they cause a number of side effects. In addition, applied in high concentrations DBMIB loses its specificity and binds also to the QB binding site, exacerbating the interpretation of results. Furthermore, a complete block of electron transport destroys the ability of photosynthesis for regulation, since a block prevents dynamic physiological changes of electron flow. Redox signals, however, are generated by perturbation in the redox balance of a compound that requires a permanent flow of electrons, but not by arresting it in a completely reduced or oxidized state. It is, therefore, useful to perform a careful titration of the concentration to be used for the final experiment of interest in order to determine the physiologically relevant range of regulation. In our experience, 30–50 % inhibition of electron flow was found to be sufficient to induce a significant response at the level of gene expression (Pfannschmidt et al., 1999, 2001). Since it is difficult to determine the effective concentration within the tissue, the optimal concentration for this effect must be determined experimentally. This can be done by measuring Chl fluorescence before and after application of the inhibitor using a pulse amplitude-modulated fluorometer, which demonstrates the direct effect on the photosynthetic electron flow. It should be noted that – depending on leaf structure (thickness of cuticule and mesophyll) – the necessary concentration may vary between species (T. Pfannschmidt et al, pers. obs.).
Many studies have used changes in illumination, either alone or in combination with the inhibitors mentioned above, to induce redox signals. Dark–light shifts, low-light to high-light shifts or shifts between PSI- and PSII-light have commonly been used (reviewed in (Pfannschmidt, 2003). Each of these treatments induces different physiological responses that should be kept in mind when planning an experimental set-up. A dark–light transition of an etiolated seedling, for instance, will activate strong photomorphogenic effects controlled by photoreceptors, which include the build-up and assembly of the photosynthetic apparatus. Therefore, it is necessary to work with plants containing mature chloroplasts if studying photosynthetic redox signals. Typically, in dark–light illumination regimes plants grown in a defined light–dark cycle are used. With the onset of light, the activation of many light-dependent processes such as the Calvin–Benson cycle can be observed. Application of DCMU to control plants is helpful to distinguish between those processes activated by photoreceptors and those activated by photosynthetic electron flow, since only the latter are affected. Low-light to high-light shifts represent a different scenario, since under these conditions stress-dependent processes become activated. In general, the increase of incident light facilitates the generation of reactive oxygen species (ROS) which, in turn, activates antioxidant and repair processes. It is difficult to reconcile the responsible redox signals in these experiments directly as many different sites are possible for the generation of signals, e.g. reduced electron transport components, ROS and the antioxidant systems. However, considerable progress has been achieved in this field in recent years by using genetic strategies (see below). It should be mentioned that low-light to high-light shifts are strongly affected by the prevailing temperature (Huner et al., 1998; Ensminger et al., 2006). Low temperatures will decrease the activities of the Calvin–Benson cycle and the regeneration of the final electron acceptor NADP+, which reduces the photosynthetic electron transport efficiency even under low-light conditions. Stable and controlled temperature conditions are therefore required in all such experiments. Another important environmental factor in this context is water supply. Plants under water stress close their stomata in order to reduce the loss of water. However, this results in decreased CO2-uptake and hence slows down the Calvin–Benson cycle, which in turn affects the electron transport. Sufficient and stable watering is therefore a further requirement in order to study light-induced redox signals. Shifts between PSI- and PSII-light induce redox signals in the low-light range and are typically not connected to stress responses (Fey et al., 2005b; Piippo et al., 2006). The signals are generated within the photosynthetic electron transport and activate counterbalancing effects that are aimed to increase the efficiency of light absorption (see Introduction).
The use of photosynthesis mutants represents a third strategy to uncover possible origins of redox signals in the electron transport chain that control gene expression. In such mutants a genetic defect limits or blocks the electron transport at a distinct site, and thus generates reduced conditions before and oxidized conditions after the defect (Yang et al., 2001; Sherameti et al., 2002; Frigerio et al., 2007). The use of mutants avoids many of the problems mentioned above, such as side effects of inhibitors or the ambiguity of influences by photoreceptors, since the known location of the genetic defect allows for direct conclusions regarding photosynthesis as a signal generator. On the other hand, mutants with strong defects that directly affect the generation of the redox signal (e.g. absence of complexes of the photosynthetic electron transport) are of very limited use to study up- or down-regulation of gene expression. The reason for this is that the defect already exists in early development when the photosynthetic apparatus is established. Typically, mutants with a photosynthetic block display a pale-green or even white phenotype and have to be maintained on sugar-containing media (Leister and Schneider, 2003). Since photosynthesis in these mutants is disturbed in general, the electron transport is locked in a distinct state and the system loses its ability to sense environmental changes, and thus the ability for gene regulation (comparable to high concentrations of electron transport inhibitors; see above). Changes in gene expression observed in comparison with the wild-type actually show how the (remaining) signalling network of the mutant tries to compensate for the genetic defect. Furthermore, supplementing the mutants with sugar may cause cross-talk between carbohydrate and redox signalling (Oswald et al., 2001). Such mutants, therefore, can be helpful to identify photosynthetic electron transport as the origin for signals regulating genes; however, since they are static with respect to their defect conclusions on the precise site of signal generation and the regulation mode require additional experiments.
Solutions for many of problems described above are inducible experimental set-ups in which signals can be generated at a given time point; this is a general requirement for all studies focussed on regulation. Controlled induction of a redox signal requires either a regulated environmental system or a genetic system that can be controlled physiologically. Examples of environmental systems are light-shift experiments as described above, since they enable redox signals to be induced at a given time. Another experimental possibility is to change the ambient CO2 concentration, which enhances or represses the Calvin–Benson cycle and thus the electron flow through the photosynthetic transport chain (Wormuth et al., 2006). An example for an inducible genetic system is represented by the flu (fluorescent) mutant of Arabidopsis. Under continuous illumination the mutant is undistinguishable from wildtype; however, it accumulates free protochlorophyllide (Pchlide) when put into darkness. Upon re-illumination the mutant then produces high amounts of singlet oxygen and can be used to study specifically the responses to this particular ROS (Meskauskiene et al., 2001). Although the system is not very physiological, it provides a unique approach to test the impact of this particular ROS that is not possible with other systems such as high light since these also produce other ROS.
In summary, each of the strategies described above has its specific advantages or disadvantages. It is therefore highly recommended to combine several of these strategies in order to get as comprehensive a picture of the studied biological problem as possible.
POSSIBLE TRANSDUCTION OF REDOX SIGNALS
Transduction within plastids
While many studies have confirmed the existence of several redox signals controlling gene expression in plastids and the nucleus, little at present is known about the transduction of the signals to the level of gene expression. Within plastids redox signals have to be transduced only a short distance to the plastome (Bruick and Mayfield, 1999; Link, 2003; Pfannschmidt and Liere, 2005; Shiina et al., 2005). As outlined in the Introduction, the LTR controlling chloroplast genes for photosynthetic core proteins involves the action of the thylakoid membrane kinase STN7 (Bonardi et al., 2005). This kinase might therefore represent a crucial component for sensing PQ redox signals and transducing them via a (putative) phosphorylation cascade to the gene expression machinery (Fig. 2). The model of a phosphorylation cascade is consistent with models that describe the control of plastid transcription by phosphorylation of sigma factors of the RNA polymerase (Tiller and Link, 1993; Link, 1996). Whether sigma factors are involved in this regulation event or not is as yet unclear; therefore, we propose the existence of a putative redox-responsive factor (RRF, Fig. 2) that perceives the PQ redox signal. Another model for redox control of plastid gene expression involves thioredoxin regulation of thiol groups in the protein disulfide isomerase. It controls binding of a RNA-binding complex to the psbA mRNA and subsequent translation initiation (Danon and Mayfield, 1994; Kim and Mayfield, 1997). Because thioredoxins can move freely through the stroma they provide an easy way of signal transduction. The model has been proposed in Chlamydomonas reinhardtii, but some data suggest that it might also be valid for higher plants (Shen et al., 2001).
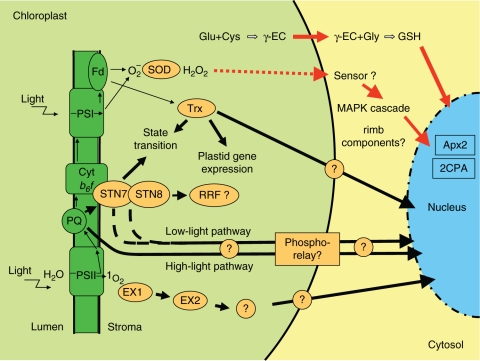
Fig. 2.
Possible signal transduction pathways of photosynthetic redox signals. The three plant cell compartments are depicted schematically: chloroplast (light green), cytosol (light yellow) and nucleus (blue). Redox signals generated within the electron transport chain (dark green) or by generation of ROS initiate signalling pathways that activate or repress specific target genes in the nucleus (for details see text). Thin black arrows represent electron transport. ROS are generated as by-products of photosynthesis, e.g. by transfer of electrons from PSI or reduced ferredoxin (Fd) to oxygen-generating superoxide (Mehler reaction). This is detoxified by superoxide dismutase (SOD) to hydrogen peroxide (H2O2). Hydrogen peroxide can be reduced to water by ascorbate peroxidases using ascorbate as the electron donor, and requiring glutathione (GSH) to restore the electron donor. Unscavenged H2O2 is able to diffuse across the chloroplast envelope and is thought to start MAP kinase cascades in the cytosol. Electrons from PSI are also transferred to thioredoxin (Trx), which can affect LHCII phosphorylation (state transitions) and plastid gene expression and possibly also nuclear gene expression. Glutathione synthesis might be another pathway by which stress signals can directly leave the plastid. Another ROS, singlet oxygen (1O2), is generated at PSII. Because of its short half-life it requires additional signalling components, Executer 1 and 2 (EX1, EX2). The plastoquinone pool is the origin for at least two redox signalling pathways that are active under low or high light and which are targeted to plastid and nuclear gene expression machineries. Perception of the redox signal requires a proposed redox responsive factor (RRF). An important sensor for the PQ redox state represents the thylakoid kinase STN7 (possibly in conjunction with STN8). Present data point to the involvement of a phosphorylation cascade in PQ redox signal transduction. Red arrows, redox signals by components that directly leave the plastid; black arrows, redox signals that are mediated by unknown components. Proteins are indicated by orange.
Transduction into the nucleus
Transduction of photosynthetic redox signals that control nuclear gene expression appears to be more complex than that within plastids since such signals have to leave the plastid, pass the cytosol and enter the nucleus. They therefore represent a novel type of retrograde signals (Rodermel, 2001; Beck, 2005; Nott et al., 2006; Bräutigam et al., 2007; Pesaresi et al., 2007). Two general types of signal-transduction mechanisms have been proposed: in the first the redox signal is sensed by a plastid-internal system and transduced over the envelope, whilst in the second a redox-regulated compound can leave the plastid directly.
Examples for the first scenario have been reported for thylakoid-located PQ molecules and short-lived singlet oxygen. The best candidate for sensing of PQ redox signals at present is the STN7 kinase (see above), but it is not clear whether it is involved in all mechanisms reported to be under PQ redox control. Some observations suggest a potential functional interaction of STN7 with its paralogue kinase STN8; however, further investigations are necessary in order to fully understand these relationships (reviewed in Dietzel et al., 2008). In addition, a recent array study proposed that under varying light qualities of low intensity the redox state of stromal components (potentially thioredoxin) may affect the expression of nuclear genes via a still-unknown pathway (Piippo et al., 2006). Beside the light-quality-dependent low-light pathway(s) mentioned above, another one has been described in Dunaliella tertiolecta, which represses Lhcb gene expression upon a high- to-low-light shift (Fig. 2). A current model assumes that a light-intensity-dependent pathway exists that involves the action of cytosolic kinase activities that eventually control the action of Lhcb gene repressors (Escoubas et al., 1995; Durnford and Falkowski, 1997; Chen et al., 2004). A completely different signalling pathway has been characterized by analysis of the flu mutant mentioned above. It was demonstrated that the response to singlet oxygen can be blocked genetically and that the activation of the activated-stress programme involves plastid-localized proteins called Executer1 and 2 (EX1, EX2; Fig. 2; Wagner et al., 2004; Lee et al., 2007). Their precise function is not known and neither are the further-downstream components of a signalling cascade. However, the singlet-oxygen-induced response was found to be promoted by ethylene-, salicylic acid- and jasmonic acid-dependent signalling pathways, whilst it was blocked by a jasmonic acid precursor (Danon et al., 2005).
Hydrogen peroxide is the principle ROS in plants that accumulates under various conditions such as excess excitation energy, drought, chilling and starvation. It activates a different response programme than singlet oxygen and is also more stable. It is therefore assumed that it can freely diffuse across the chloroplast envelope and that it activates a mitogen-activated protein kinase (MAPK) cascade in the cytosol that subsequently affects gene expression in the nucleus (Fig. 2; Kovtun et al., 2000; Vranova et al., 2002; Apel and Hirt, 2004; Mittler et al., 2004). By this means it represents an example for the second type of signal transduction.
Recent observations have suggested that glutathione may also act as a plastid signal that controls expression of stress-defence genes. Glutathione is an important component of the scavenging machinery that counteracts ROS. In Arabidopsis it has been proposed that the first step of its synthesis [cysteine and glutamate fused to γ-glutamylcysteine (γ-EC)] is confined to plastids while the second step (γ-glutamylcysteine and glycine fused to glutathione) is performed in the cytosol (Wachter et al., 2005; Rausch et al., 2007). Plastid γ-EC must therefore leave the plastid. Since its synthesis is affected by changes in photosynthesis, it can potentially report such changes to the nucleus (Mullineaux and Rausch, 2005).
VELOCITY OF REDOX SIGNALS
The time frames in which acclimation responses occur have been measured at various levels, e.g. by determination of changes in photosystem number, Chl and protein accumulation (Kim et al., 1993; Walters and Horton, 1994, 1995a, b; Pfannschmidt et al., 2001). However, the speed at which the respective genes respond to the redox signals is less well investigated. To do this requires that a distinct redox signal is induced at a given time and that the changes in gene expression are then followed during the course of the response. Light stress experiments indicated that the nuclear genes Apx1 and Apx2 (encoding cytosolic ascorbate peroxidases) are activated within 15–30 min upon a low-light to high-light shift in Arabidopsis (Karpinski et al., 1997); this was found to be part of a systemic response to excess excitation energy (Karpinski et al., 1999). In another study, kinetic experiments with isolated plastids showed that chloroplast gene expression is affected within 15–30 min after inducing a redox signal by light quality shifts (Pfannschmidt et al., 1999). First kinetic experiments suggest that the same redox signals induced by light quality are transduced to the nucleus within 30 min, which corresponds to the time frame within the plastids (K. Bräutigam and T. Pfannschmidt, unpubl. res.). These data suggest that the signal transduction cascades already exist when the signals are generated, and that the signals are processed as fast as other intracellular stimuli. It remains to be clarified as to how the components of the proposed signalling pathways (cf. Fig. 2) are integrated into the known parts of the signalling network.
PRIMARY TARGET GENES OF REDOX SIGNALS
The identification of (a) primary target gene(s) of a certain signal helps us to understand which cellular processes are under redox control and provide (a) molecular tool(s) that can be used to analyse the transduction pathway and its components. One possibility is to fuse the target gene or its respective promoter to a reporter gene and to transform plants (typically Arabidopsis) with such a construct. This transgenic line then can be used to analyse the expression of the target gene in space and time in planta: this has led to the discovery of systemic acclimation to excess excitation energy (Karpinski et al., 1999). In addition, after mutagenesis of such a stable transformed line, the progeny of it can be screened for a lack of a reaction of the reporter occurring to the signal of interest. The defective genes in mutants that are identified can be mapped and provide potential components of the signal transduction pathway, for instance in the case of rimb (redox imbalance) mutants (Heiber et al., 2007). Here, the promoter of the nuclear 2-Cys peroxiredoxin-A (2CPA) gene was used to search for these mutants. The 2CPA gene was shown earlier to be redox-regulated under light stress conditions. Kinase inhibitor experiments indicated that the signal transduction involves the action of MAP kinases and serine/threonine kinases, in accordance with the stress signal transduction mentioned above. Furthermore the involvement of abscisic acid could be shown (Konig et al., 2002; Baier et al., 2004).
The identification of real primary target genes is a difficult task. Many recent studies have used array techniques in order to investigate gene expression changes in responses to various redox signals such as hydrogen peroxide, singlet oxygen or redox signals from the photosynthetic electron transport chain. These studies indicate that several hundred genes are responsive to many such signals (Desikan et al., 2001; op den Camp et al., 2003; Davletova et al., 2005; Fey et al., 2005b; Vanderauwera et al., 2005; Piippo et al., 2006). The major problem in all these studies is to distinguish between genes that are controlled directly by a given signal (a true primary target gene) and genes that are controlled indirectly (the secondary or tertiary target genes), e.g. by the effects resulting from expression changes in primary target genes. The most effective way to identify potential primary targets is to perform a kinetic study in which gene expression changes are determined at different times after application of a defined environmental signal. Such a kinetic should range from short (minutes) to middle (hours) and late (days) stages to cover the complete extent of the physiological response. After a first survey, this time range can be refined according to the results. As a general assumption we propose that those genes that exhibit expression changes first and in a strong manner are the primary target genes of a given signal. Modern array techniques provide the technical possibility to observe the full genome (i.e. from Arabidopsis) in such a study. However, it should be noted that most probably many genes are affected by several parameters at the same time and that primary target genes responsive to only one environmental factor might be rare.
Several bioinformatics approaches can be performed using the data from such a kinetic array analysis. Most useful is analysis of gene expression patterns, which can unravel groups of co-regulated genes (Biehl et al., 2005). Such genes can have a common input signal for regulation, but do not necessarily need to have one. In any case, each potential target gene of interest should be checked for the reproducibility of its expression pattern by an independent experiment and approach (e.g. Northern analysis) before it is used as a target gene in a reporter-gene approach as mentioned above. However, array techniques provide the possibility to uncover potential expression signatures within a data set that are typical for a given redox signal. This signature can be compared with other array data and can lead to identification of genes that are only regulated under one certain condition. By this means redox-responsive promoters or promoter elements might be also identified.
DIFFERENTIATION AND INTEGRATION OF VARIOUS REDOX SIGNALS
A central question in plant cell biology is how the integration of various environmental signals at the same time is achieved in order to regulate expression of genes for appropriate responses (Bräutigam et al., 2007). Understanding of this complex task includes several plant-specific but also some general problems of modern molecular cell biology (summarized in Fig. 3).
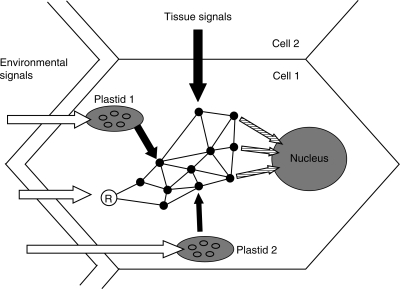
Fig. 3.
Model of integration of various signals in a plant cell. A plant cell and its environment including neighbouring cells are shown. Various environmental influences that are detected by cytosolic receptors (circle marked ‘R’) or photosynthesis in different chloroplasts are represented by white arrows. Due to their different positions in the cell, the impact on the plastids may vary (plastid 1 and 2). The perceiving systems have different impacts (black arrows of different thickness) on the intracellular signalling network (represented as black dots connected by lines), which integrates these signals as well as signals from other cells, resulting in integrated signals (hatched arrows) that affect gene expression events. The ‘gene-copy-number-problem’ is indicated by the numerous ovals within the plastids, representing multiple copies of the plastome (see text for details).
Under natural conditions many of the redox signals discussed in this article can occur at the same time or in a fast consecutive manner. The nucleus has to recognize all these different redox signals and, in addition, to distinguish one from another as well as from other environmental signals (mediated for instance by photoreceptors). Such a specific signal recognition and initiation of respective responses requires sophisticated mechanisms and interactions within the signalling networks, which we are just beginning to understand. Furthermore, higher plants possess up to 100 single plastids per cell and not all of them necessarily send the same signals. For example, strong light-quality gradients that affect photosynthesis occur even within a leaf or a single cell, and therefore the redox signals of the plastids may vary depending on their individual localization within the cell (Terashima and Saeki, 1985). The large number of plastids also introduces another consideration, the so-called gene-copy-number problem (for a review see Bräutigam et al., 2007). Higher plant plastids contain up to 100 copies of the plastid's own genome, the plastome. Since the photosynthetic apparatus and many other protein complexes in plastids are comprised of a patchwork of plastid and nuclear encoded protein subunits, up to a 10 000-fold excess of plastid over nuclear gene copies for one and the same protein complex has to be co-ordinated in expression. Interestingly, plastids of all known taxa universally encode the central proteins of the photosystems on the plastome, providing the possibility that the plastids themselves serve as the pacemakers in the co-ordination of photosynthesis gene expression within the two genetic compartments (Race et al., 1999). Redox regulation of chloroplast gene expression has been hypothesized as the evolutionary reason for the maintenance of a plastid genome (Allen, 1993).
It is a challenging task to understand how environmental signals of different intensity and quality – which in addition vary temporarily, spatially and in rate – are integrated into a cellular response that helps the plant to deal with environmental fluctuations and stresses. The cytosolic network of interacting signalling components most likely does most of this job, resulting in a unique combination of transcription factors that activate or repress a combination of genes within the nucleus and plastids (and mitochondria) that is appropriate for the given environmental situation (Fig. 3). In addition, the data presently available clearly indicate that many other levels of gene expression are also included in signal integration, e.g. post-transcriptional and post-translational events. Only approaches at the level of systems biology will provide enough data that can be used to generate an integrated view of all cellular responses at the same time. However, such studies will only be successful if the basic experimental problems outlined above are solved.
CONCLUSIONS
Photosynthetic redox signals generated in the electron transport chain are connected to many environmental influences either directly via illumination or indirectly via the Calvin–Benson cycle. Thus they are able to signal in a very sensible manner the prevailing environmental conditions to the level of gene expression. Interactions with the energy state of the cell, which also fluctuates in response to many environmental conditions, are very likely. Further studies of redox signalling networks that also include mitochondria will unravel these connections (Rhoads and Subbaiah, 2007).
ACKNOWLEDGEMENTS
Our work was supported by grants from the DFG, the ‘NWP’ and ‘Excellence in Science’ programmes of Thuringia to T.P., and to the DFG research groups FOR 387 and FOR804.
LITERATURE CITED
- Alfonso M, Perewoska I, Kirilovsky D. Redox control of psbA gene expression in the cyanobacterium Synechocystis PCC 6803. Involvement of the cytochrome b(6)/f complex. Plant Physiology. 2000;122:505–515. doi: 10.1104/pp.122.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JF. Control of gene expression by redox potential and the requirement for chloroplast and mitochondrial genomes. Journal of Theoretical Biology. 1993;165:609–631. doi: 10.1006/jtbi.1993.1210. [DOI] [PubMed] [Google Scholar]
- Allen JF, Forsberg J. Molecular recognition in thylakoid structure and function. Trends in Plant Science. 2001;6:317–326. doi: 10.1016/s1360-1385(01)02010-6. [DOI] [PubMed] [Google Scholar]
- Allen JF, Pfannschmidt T. Balancing the two photosystems: photosynthetic electron transfer governs transcription of reaction centre genes in chloroplasts. Philosophical Transactions of the Royal Society of London Series B. 2000;355:1351–1357. doi: 10.1098/rstb.2000.0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JM, Chow WS, Park Y-I. The grand design of photosynthesis: acclimation of the photosynthetic apparatus to environmental cues. Photosynthesis Research. 1995;46:129–139. doi: 10.1007/BF00020423. [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annual Review of Plant Biology. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Arnon DI. Sunlight, earth, life: the grand design of photosynthesis. The Sciences. 1982;22:22–27. [Google Scholar]
- Aro EM, Andersson B. Regulation of photosynthesis. Dordrecht: Kluwer Academic Publishers; 2001. [Google Scholar]
- Baier M, Dietz KJ. Chloroplasts as source and target of cellular redox regulation: a discussion on chloroplast redox signals in the context of plant physiology. Journal of Experimental Botany. 2005;56:1449–1462. doi: 10.1093/jxb/eri161. [DOI] [PubMed] [Google Scholar]
- Baier M, Stroher E, Dietz KJ. The acceptor availability at photosystem I and ABA control nuclear expression of 2-cys peroxiredoxin-alpha in Arabidopsis thaliana. Plant and Cell Physiology. 2004;45:997–1006. doi: 10.1093/pcp/pch114. [DOI] [PubMed] [Google Scholar]
- Beck CF. Signaling pathways from the chloroplast to the nucleus. Planta. 2005;222:743–756. doi: 10.1007/s00425-005-0021-2. [DOI] [PubMed] [Google Scholar]
- Bellafiore S, Bameche F, Peltier G, Rochaix JD. State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature. 2005;433:892–895. doi: 10.1038/nature03286. [DOI] [PubMed] [Google Scholar]
- Biehl A, Richly E, Noutsos C, Salamini F, Leister D. Analysis of 101 nuclear transcriptomes reveals 23 distinct regulons and their relationship to metabolism, chromosomal gene distribution and co-ordination of nuclear and plastid gene expression. Gene. 2005;344:33–41. doi: 10.1016/j.gene.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Blankenship RE. Molecular mechanisms of photosynthesis. Oxford: Blackwell Science Ltd; 2002. [Google Scholar]
- Bonardi V, Pesaresi P, Becker T, Schleiff E, Wagner R, Pfannschmidt T, Jahns P, Leister D. Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature. 2005;437:1179–1182. doi: 10.1038/nature04016. [DOI] [PubMed] [Google Scholar]
- Bräutigam K, Dietzel L, Pfannschmidt T. Plastid–nucleus communication: anterograde and retrograde signalling in development and function of plastids. In: Bock R, editor. Cell and molecular biology of plastids. Vol. 19. Berlin: Springer; 2007. pp. 409–455. [Google Scholar]
- Bruick RK, Mayfield SP. Light-activated translation of chloroplast mRNAs. Trends in Plant Science. 1999;4:190–195. doi: 10.1016/s1360-1385(99)01402-8. [DOI] [PubMed] [Google Scholar]
- op den Camp RGL, Przybyla D, Ochsenbein C, Laloi C, Kim CH, Danon A, et al. Rapid induction of distinct stress responses after the release of singlet oxygen in arabidopsis. Plant Cell. 2003;15:2320–2332. doi: 10.1105/tpc.014662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YB, Durnford DG, Koblizek M, Falkowski PG. Plastid regulation of Lhcb1 transcription in the chlorophyte alga Dunaliella tertiolecta. Plant Physiology. 2004;136:3737–3750. doi: 10.1104/pp.104.038919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A, Mayfield SP. Light-regulated translation of chloroplast messenger-RNAs through redox potential. Science. 1994;266:1717–1719. doi: 10.1126/science.7992056. [DOI] [PubMed] [Google Scholar]
- Danon A, Miersch O, Felix G, den Camp RGLO, Apel K. Concurrent activation of cell death-regulating signaling pathways by singlet oxygen in Arabidopsis thaliana. Plant Journal. 2005;41:68–80. doi: 10.1111/j.1365-313X.2004.02276.x. [DOI] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, et al. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell. 2005;17:268–281. doi: 10.1105/tpc.104.026971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depege N, Bellafiore S, Rochaix JD. Rote of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science. 2003;299:1572–1575. doi: 10.1126/science.1081397. [DOI] [PubMed] [Google Scholar]
- Desikan R, Mackerness SAH, Hancock JT, Neill SJ. Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiology. 2001;127:159–172. doi: 10.1104/pp.127.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzel L, Bräutigam K, Pfannschmidt T. Photosynthetic acclimation: state transitions and adjustment of photosystem stoichiometry – functional relationships between short-term and long-term light quality acclimation iin plants. Febs Journal. 2008;275:1080–1088. doi: 10.1111/j.1742-4658.2008.06264.x. [DOI] [PubMed] [Google Scholar]
- Durnford DG, Falkowski PG. Chloroplast redox regulation of nuclear gene transcription during photoacclimation. Photosynthesis Research. 1997;53:229–241. [Google Scholar]
- Ensminger I, Busch F, Huner NPA. Photostasis and cold acclimation: sensing low temperature through photosynthesis. Physiologia Plantarum. 2006;126:28–44. [Google Scholar]
- Escoubas JM, Lomas M, Laroche J, Falkowski PG. Light-intensity regulation of cab gene-transcription is signaled by the redox state of the plastoquinone pool. Proceedings of the National Academy of Sciences of the USA. 1995;92:10237–10241. doi: 10.1073/pnas.92.22.10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey V, Wagner R, Brautigam K, Pfannschmidt T. Photosynthetic redox control of nuclear gene expression. Journal of Experimental Botany. 2005;56:1491–1498. doi: 10.1093/jxb/eri180. [DOI] [PubMed] [Google Scholar]
- Fey V, Wagner R, Brautigam K, Wirtzt M, Hell R, Dietzmann A, et al. Retrograde plastid redox signals in the expression of nuclear genes for chloroplast proteins of Arabidopsis thaliana. Journal of Biological Chemistry. 2005;280:5318–5328. doi: 10.1074/jbc.M406358200. [DOI] [PubMed] [Google Scholar]
- Finazzi G, Zito F, Barbagallo RP, Wollman FA. Contrasted effects of inhibitors of cytochrome b(6)f complex on state transitions in Chlamydomonas reinhardtii – the role of Q(o) site occupancy in LHCII kinase activation. Journal of Biological Chemistry. 2001;276:9770–9774. doi: 10.1074/jbc.M010092200. [DOI] [PubMed] [Google Scholar]
- Frigerio S, Campoli C, Zorzan S, Fantoni LI, Crosatti C, Drepper F, et al. Photosynthetic antenna size in higher plants is controlled by the plastoquinone redox state at the post-transcriptional rather than transcriptional level. Journal of Biological Chemistry. 2007;282:29457–29469. doi: 10.1074/jbc.M705132200. [DOI] [PubMed] [Google Scholar]
- Haldrup A, Jensen PE, Lunde C, Scheller HV. Balance of power: a view of the mechanism of photosynthetic state transitions. Trends in Plant Science. 2001;6:301–305. doi: 10.1016/s1360-1385(01)01953-7. [DOI] [PubMed] [Google Scholar]
- Heiber I, Stroher E, Raatz B, Busse I, Kahmann U, Bevan MW, Dietz KJ, Baier M. The redox imbalanced mutants of arabidopsis differentiate signaling pathways for redox regulation of chloroplast antioxidant enzymes. Plant Physiology. 2007;143:1774–1788. doi: 10.1104/pp.106.093328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huner NPA, Oquist G, Sarhan F. Energy balance and acclimation to light and cold. Trends in Plant Science. 1998;3:224–230. [Google Scholar]
- Karpinski S, Escobar C, Karpinska B, Creissen G, Mullineaux PM. Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell. 1997;9:627–640. doi: 10.1105/tpc.9.4.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski S, Reynolds H, Karpinska B, Wingsle G, Creissen G, Mullineaux P. Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science. 1999;284:654–657. doi: 10.1126/science.284.5414.654. [DOI] [PubMed] [Google Scholar]
- Kim JM, Mayfield SP. Protein disulfide isomerase as a regulator of chloroplast translational activation. Science. 1997;278:1954–1957. doi: 10.1126/science.278.5345.1954. [DOI] [PubMed] [Google Scholar]
- Kim JH, Glick RE, Melis A. Dynamics of photosystem stoichiometry adjustment by light quality in chloroplasts. Plant Physiology. 1993;102:181–190. doi: 10.1104/pp.102.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Baier M, Horling F, Kahmann U, Harris G, Schurmann P, Dietz KJ. The plant-specific function of 2-Cys peroxiredoxin-mediated detoxification of peroxides in the redox-hierarchy of photosynthetic electron flux. Proceedings of the National Academy of Sciences of the USA. 2002;99:5738–5743. doi: 10.1073/pnas.072644999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs L, Wiessner W, Kis M, Nagy F, Mende D, Demeter S. Short- and long-term redox regulation of photosynthetic light energy distribution and photosystem stoichiometry by acetate metabolism in the green alga. Chlamydobotrys stellata. Photosynthesis Research. 2000;65:231–247. doi: 10.1023/A:1010650532693. [DOI] [PubMed] [Google Scholar]
- Kovtun Y, Chiu WL, Tena G, Sheen J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proceedings of the National Academy of Sciences of the USA. 2000;97:2940–2945. doi: 10.1073/pnas.97.6.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Kim C, Landgraf F, Apel K. EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the USA. 2007;104:10270–10275. doi: 10.1073/pnas.0702061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D, Schneider A. From genes to photosynthesis in Arabidopsis thaliana. In: Jeon K, editor. International review of cytology. A survey of cell biology. Vol. 228. London: Academic Press; 2003. pp. 31–83. [DOI] [PubMed] [Google Scholar]
- Link G. Green life: control of chloroplast gene transcription. Bioessays. 1996;18:465–471. [Google Scholar]
- Link G. Redox regulation of chloroplast transcription. Antioxidants and Redox Signaling. 2003;5:79–87. doi: 10.1089/152308603321223568. [DOI] [PubMed] [Google Scholar]
- Melis A, Murakami A, Nemson JA, Aizawa K, Ohki K, Fujita Y. Chromatic regulation in Chlamydomonas reinhardtii alters photosystem stoichiometry and improves the quantum efficiency of photosynthesis. Photosynthesis Research. 1996;47:253–265. doi: 10.1007/BF02184286. [DOI] [PubMed] [Google Scholar]
- Meskauskiene R, Nater M, Goslings D, Kessler F, den Camp RO, Apel K. FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the USA. 2001;98:12826–12831. doi: 10.1073/pnas.221252798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. Reactive oxygen gene network of plants. Trends in Plant Science. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Mullineaux PM, Rausch T. Glutathione, photosynthesis and the redox regulation of stress-responsive gene expression. Photosynthesis Research. 2005;86:459–474. doi: 10.1007/s11120-005-8811-8. [DOI] [PubMed] [Google Scholar]
- Murakami A, Fujita Y, Nemson JA, Melis A. Chromatic regulation in Chlamydomonas reinhardtii: time course of photosystem stoichiometry adjustment following a shift in growth light quality. Plant and Cell Physiology. 1997;38:188–193. [Google Scholar]
- Niyogi KK. Safety valves for photosynthesis. Current Opinion in Plant Biology. 2000;3:455–460. doi: 10.1016/s1369-5266(00)00113-8. [DOI] [PubMed] [Google Scholar]
- Noctor G, Foyer CH. Ascorbate and glutathione: keeping active oxygen under control. Annual Review of Plant Physiology and Plant Molecular Biology. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- Nott A, Jung HS, Koussevitzky S, Chory J. Plastid-to-nucleus retrograde signaling. Annual Review of Plant Biology. 2006;57:739–759. doi: 10.1146/annurev.arplant.57.032905.105310. [DOI] [PubMed] [Google Scholar]
- Oswald O, Martin T, Dominy PJ, Graham IA. Plastid redox state and sugars: interactive regulators of nuclear-encoded photosynthetic gene expression. Proceedings of the National Academy of Sciences of the USA. 2001;98:2047–2052. doi: 10.1073/pnas.021449998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P, Schneider A, Kleine T, Leister D. Interorganellar communication. Current Opinion in Plant Biology. 2007;10:600–606. doi: 10.1016/j.pbi.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T. Chloroplast redox signals: how photosynthesis controls its own genes. Trends in Plant Science. 2003;8:33–41. doi: 10.1016/s1360-1385(02)00005-5. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Liere K. Redox regulation and modification of proteins controlling chloroplast gene expression. Antioxidants and Redox Signaling. 2005;7:607–618. doi: 10.1089/ars.2005.7.607. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Nilsson A, Allen JF. Photosynthetic control of chloroplast gene expression. Nature. 1999;397:625–628. [Google Scholar]
- Pfannschmidt T, Schutze K, Brost M, Oelmuller R. A novel mechanism of nuclear photosynthesis gene regulation by redox signals from the chloroplast during photosystem stoichiometry adjustment. Journal of Biological Chemistry. 2001;276:36125–36130. doi: 10.1074/jbc.M105701200. [DOI] [PubMed] [Google Scholar]
- Piippo M, Allahverdiyeva Y, Paakkarinen V, Suoranta UM, Battchikova N, Aro EM. Chloroplast-mediated regulation of nuclear genes in Arabidopsis thaliana in the absence of light stress. Physiological Genomics. 2006;25:142–152. doi: 10.1152/physiolgenomics.00256.2005. [DOI] [PubMed] [Google Scholar]
- Race HL, Herrmann RG, Martin W. Why have organelles retained genomes? Trends in Genetics. 1999;15:364–370. doi: 10.1016/s0168-9525(99)01766-7. [DOI] [PubMed] [Google Scholar]
- Rausch T, Gromes R, Liedschulte V, Muller I, Bogs J, Galovic V, Wachter A. Novel insight into the regulation of GSH biosynthesis in higher plants. Plant Biology. 2007;9:565–572. doi: 10.1055/s-2007-965580. [DOI] [PubMed] [Google Scholar]
- Rhoads DM, Subbaiah CC. Mitochondrial retrograde regulation in plants. Mitochondrion. 2007;7:177–194. doi: 10.1016/j.mito.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Rintamaki E, Martinsuo P, Pursiheimo S, Aro EM. Cooperative regulation of light-harvesting complex II phosphorylation via the plastoquinol and ferredoxin-thioredoxin system in chloroplasts. Proceedings of the National Academy of Sciences of the USA. 2000;97:11644–11649. doi: 10.1073/pnas.180054297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodermel S. Pathways of plastid-to-nucleus signaling. Trends in Plant Science. 2001;6:471–478. doi: 10.1016/s1360-1385(01)02085-4. [DOI] [PubMed] [Google Scholar]
- Shen YX, Danon A, Christopher DA. RNA binding-proteins interact specifically with the Arabidopsis chloroplast psbA mRNA 5' untranslated region in a redox-dependent manner. Plant and Cell Physiology. 2001;42:1071–1078. doi: 10.1093/pcp/pce142. [DOI] [PubMed] [Google Scholar]
- Sherameti I, Sopory SK, Trebicka A, Pfannschmidt T, Oelmuller R. Photosynthetic electron transport determines nitrate reductase gene expression and activity in higher plants. Journal of Biological Chemistry. 2002;277:46594–46600. doi: 10.1074/jbc.M202924200. [DOI] [PubMed] [Google Scholar]
- Shiina T, Tsunoyama Y, Nakahira Y, Khan MS. Plastid RNA polymerases, promoters, and transcription regulators in higher plants. In: eon K, editor. International Review of Cytology. A Survey of Cell Biology. Vol. 244. London: Academic Press; 2005. pp. 1–68. [DOI] [PubMed] [Google Scholar]
- Terashima I, Saeki T. A new model for leaf photosynthesis incorporating the gradients of light environment and of photosynthetic properties of chloroplasts within a leaf. Annals of Botany. 1985;56:489–499. [Google Scholar]
- Tiller K, Link G. Phosphorylation and dephosphorylation affect functional characteristics of chloroplast and etioplast transcription systems from mustard (Sinapis alba L) EMBO Journal. 1993;12:1745–1753. doi: 10.1002/j.1460-2075.1993.tb05822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebst A. Inhibitors in electron flow: tools for the functional and structural localization of carriers and energy conservation sites. Methods in Enzymology. 1980;69:675–715. [Google Scholar]
- Vanderauwera S, Zimmermann P, Rombauts S, Vandenabeele S, Langebartels C, Gruissem W, Inze D, Van Breusegem F. Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiology. 2005;139:806–821. doi: 10.1104/pp.105.065896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vener AV, VanKan PJM, Rich PR, Ohad I, Andersson B. Plastoquinol at the quinol oxidation site of reduced cytochrome bf mediates signal transduction between light and protein phosphorylation: thylakoid protein kinase deactivation by a single-turnover flash. Proceedings of the National Academy of Sciences of the USA. 1997;94:1585–1590. doi: 10.1073/pnas.94.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranova E, Inze D, Van Breusegem F. Signal transduction during oxidative stress. Journal of Experimental Botany. 2002;53:1227–1236. [PubMed] [Google Scholar]
- Wachter A, Wolf S, Steininger H, Bogs J, Rausch T. Differential targeting of GSH1 and GSH2 is achieved by multiple transcription initiation: implications for the compartmentation of glutathione biosynthesis in the Brassicaceae. Plant Journal. 2005;41:15–30. doi: 10.1111/j.1365-313X.2004.02269.x. [DOI] [PubMed] [Google Scholar]
- Wagner D, Przybyla D, Camp ROD, Kim C, Landgraf F, Lee KP, et al. The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science. 2004;306:1183–1185. doi: 10.1126/science.1103178. [DOI] [PubMed] [Google Scholar]
- Walters RG. Towards an understanding of photosynthetic acclimation. Journal of Experimental Botany. 2005;56:435–447. doi: 10.1093/jxb/eri060. [DOI] [PubMed] [Google Scholar]
- Walters RG, Horton P. Acclimation of Arabidopsis thaliana to the light environment – changes in composition of the photosynthetic apparatus. Planta. 1994;195:248–256. [Google Scholar]
- Walters RG, Horton P. Acclimation of Arabidopsis thaliana to the light environment – changes in photosynthetic function. Planta. 1995;a 197 doi: 10.1007/BF00202652. [DOI] [PubMed] [Google Scholar]
- Walters RG, Horton P. Acclimation of Arabidopsis thaliana to the light environment – regulation of chloroplast composition. Planta. 1995;b 197:475–481. doi: 10.1007/BF00196669. [DOI] [PubMed] [Google Scholar]
- Wollman FA. State transitions reveal the dynamics and flexibility of the photosynthetic apparatus. EMBO Journal. 2001;20:3623–3630. doi: 10.1093/emboj/20.14.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormuth D, Baier M, K, lbinder A, Scheibe R, Hartung W, Dietz KJ. Regulation of gene expression by photosynthetic signals triggered through modified CO2 availability. BMC Plant Biology. 2006;6:15. doi: 10.1186/1471-2229-6-15. doi:10.1186/1471-2229-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DH, Andersson B, Aro EM, Ohad I. The redox state of the plastoquinone pool controls the level of the light-harvesting chlorophyll a/b binding protein complex II (LHC II) during photoacclimation – cytochrome b(6)f deficient Lemna perpusilla plants are locked in a state of high-light acclimation. Photosynthesis Research. 2001;68:163–174. doi: 10.1023/A:1011849919438. [DOI] [PubMed] [Google Scholar]
- Zito F, Finazzi G, Delosme R, Nitschke W, Picot D, Wollman FA. The Qo site of cytochrome b(6)f complexes controls the activation of the LHCII kinase. EMBO Journal. 1999;18:2961–2969. doi: 10.1093/emboj/18.11.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]