Abstract
The transcription of fatty acid synthase (FAS), a central enzyme in de novo lipogenesis, is dramatically induced by fasting/refeeding and insulin. We reported that upstream stimulatory factor binding to the −65 E-box is required for induction of the FAS transcription by insulin in 3T3-L1 adipocytes. On the other hand, we recently found that two upstream 5′ regions are required for induction in vivo by fasting/refeeding and insulin; one at −278 to −131 albeit at a low level, and the other at −444 to −278 with an E-box at −332 where upstream stimulatory factor functions for maximal induction. Here, we generated double transgenic mice carrying the chloramphenicol acetyltransferase reporter driven by the various 5′ deletions of the FAS promoter region and a truncated active form of the sterol regulatory element (SRE) binding protein (SREBP)-1a. We found that SREBP participates in the nutritional regulation of the FAS promoter and that the region between −278 and −131 bp is required for SREBP function. We demonstrate that SREBP binds the −150 canonical SRE present between −278 and −131, and SREBP can function through the −150 SRE in cultured cells. These in vivo and in vitro results indicate that SREBP is involved in the nutritional induction of the FAS promoter via the −278/−131 region and that the −150 SRE is the target sequence.
Fatty acid synthase (FAS) plays a central role in de novo lipogenesis in mammals (1). By the action of its seven active sites, FAS catalyzes all of the reaction steps in the conversion of acetyl-CoA and malonyl-CoA to palmitate. FAS activity is not known to be regulated by allosteric effectors or covalent modification. However, FAS concentration is exquisitely sensitive to nutritional and hormonal status in lipogenic tissues, liver, and adipose tissue (1–3). FAS mRNA is not detectable in livers of fasted mice but refeeding a high-carbohydrate, fat-free diet increases FAS mRNA levels dramatically, because of the changes in transcription (4, 5). Increased circulating insulin and decreased glucagon levels may participate in the regulation of FAS expression. FAS gene transcription was not detectable in fasted or refed streptozotocin-diabetic mice but increased by insulin administration (5). We mapped an insulin response sequence to −71 to −50 bp that contains a core E-box (CATGTG) by transfection of serial 5′ deletions of the FAS promoter fused to the luciferase (LUC) reporter gene into 3T3-L1 adipocytes (6, 7). Binding to the −65 E-box by upstream stimulatory factor (USF)1 and USF2, which belong to the basic helix–loop–helix leucine zipper family of transcription factors, was required for the insulin-mediated induction of the FAS gene (6–8). We also demonstrated that the 2.1-kb 5′ flanking sequence is sufficient for the tissue-specific and hormonal/nutritional regulation of the FAS gene in the in vivo context. Recently, by generating transgenic mice bearing various 5′ deletion FAS promoter-chloramphenicol acetyltransferase (CAT) genes, we found that, unlike in cultured cell system, two 5′ promoter regions were required for such regulation in vivo. The first region, located between −278 and −131 bp, confers FAS promoter activation upon refeeding/insulin treatment, but a second region, from −444 to −278 bp, is required for maximal activation similar to that observed in mice carrying the longer 2.1-kb FAS promoter region (9). We also demonstrated that USF binding to the −332 E-box may be responsible for high-level activation of the FAS promoter. Vaulont and coworkers (10) recently reported that in USF1 and USF2 knockout mice, induction of the FAS gene by a high-carbohydrate diet was severely impaired, demonstrating in vivo requirement for USFs for glucose/insulin regulation. These in vivo and in vitro studies clearly demonstrate that USF is a key transcription factor involved in the nutritional/hormonal regulation of the FAS gene.
In addition to USF, another basic helix–loop–helix transcription factor, sterol regulatory element (SRE) binding protein (SREBP), has been implicated in the regulation of the FAS gene. The Goldstein and Brown laboratory (11–13) showed higher mRNA levels for FAS, along with other lipogenic enzymes, in livers of transgenic mice expressing a truncated active form of SREBP, the induction being higher in SREBP-1a transgenic mice than in SREBP-2 transgenic mice. Furthermore, lipogenic enzyme gene induction during fasting/refeeding was abolished in SREBP-1 knockout mice (14). SREBP is an endoplasmic reticulum membrane-bound transcription factor that regulates various genes involved in cholesterol and fatty acid metabolism (15). SREBP must be proteolytically cleaved to release the amino-terminal segment to generate the mature form, which can enter the nucleus (16) and bind to SREs, 5′-ATCACCCCAC-3′, in various promoters (17–20). Osborne and coworkers (21, 25) reported that suppression of FAS transcription by sterol is mediated by SREBP binding to the two tandem copies of SREs overlapping with the −65 E-box, which we previously showed to be a USF binding site for insulin regulation. Spiegelman and coworkers (22), on the other hand, reported that SREBP can bind not only SRE but also E-box motif because of the presence of an atypical tyrosine residue in the conserved basic domain. By using a mutated SREBP that can bind only an E-box and not a consensus SRE, they reported that SREBP activated the FAS promoter by binding to the −65 E-box during nutritional and insulin regulation (23, 24). In our hands, however, mutations around the E-box, which dramatically reduced SREBP but not USF binding to this region, did not affect insulin regulation of the FAS promoter in 3T3-L1 adipocytes (6). Furthermore, as described above, the first 131 bp of the FAS promoter that contains this −65 sequence could not confer nutritional and insulin regulation of the CAT reporter gene in transgenic mice (9). Therefore, although studies on transgenic mice overexpressing SREBP suggest its role in the activation of the FAS promoter, SREBP binding site(s) in the FAS promoter or its precise role during the nutritional regulation of the FAS promoter are yet to be clarified.
To examine the potential role of SREBP in the nutritional regulation of the FAS promoter in vivo, we generated double transgenic mice containing a CAT reporter driven by the various 5′ deletion of the FAS promoter, −131, −278, −444, −644, and −2,100 bp, along with SREBP-1a driven by the phosphoenolpyruvate carboxykinase (PEPCK) promoter. We show that SREBP activates the FAS promoter in vivo. The −131 FAS promoter, which contains the −71/−54 region implicated for SREBP function in vitro (23, 25), is not activated by SREBP in transgenic mice. SREBP, when overexpressed in fasted mice, activates the FAS promoter to the level found in refeeding only in the presence of an upstream region between −278 and −131 containing an SRE at −150 where SREBP can bind and transactivate in vitro. Maximal activation observed in refeeding, as we have shown previously, can be achieved only via a further upstream region between −444 and −278 containing an E-box at −332 where USF functions.
Materials and Methods
Plasmid Construction and Production of Transgenic Mice.
Transgenic mice harboring the CAT gene driven by the various 5′ deletions of the FAS promoter were generated, and the transgenic mice were identified by PCR of the tail DNA as described (9, 26). We used a minimum of two independent founder lines for the different FAS promoter regions as previously described and essentially the same results were obtained. The B6SJL–SREBP-1a transgenic mice were obtained from The Jackson Laboratory (11) and bred with wild-type C57BL/6 mice. The progenies were bred to the different FAS transgenic mice to generate various double FAS-CAT/SREBP-1a transgenic mice. To identify the double-transgenic mice, the primers 5′-CATCCCTGTGACCCCTCC-3′ and 5′-CTCCAAACCACCCCCCTC-3′ were used to amplify a 151-bp fragment of the human growth hormone polyadenylation signal sequence, which is part of the SREBP-1a transgene. We used minimum of three F1 double-transgene-positive male mice of 8–12 weeks of age for each 5′ deletion FAS promoter-CAT for our studies.
The reporter gene constructs for −19 FAS-LUC, −136 FAS-LUC, −248 FAS-LUC, −318 FAS-LUC, and −2100 FAS-LUC, which contain, respectively, the −19/+67, −136/+67, −248/+67, −318/+67, and −2100/+67 sequences of the rat FAS promoter fused to the LUC reporter plasmid p0Luc, have been described (27–29). −2100(−65) FAS-LUC was generated by mutating −65/−60 sequence to 5′-GAATTC-3′ by site-directed mutagenesis (6). −444 FAS-LUC was constructed by ligating the DraIII–HindIII fragment from the −2100 FAS-LUC plasmid containing sequences −444 to +67 of the rat FAS gene into the promoterless vector p0LUC. −444(−65) FAS-LUC was constructed following the same procedure used for the −444 FAS-LUC construct but using the DraIII–HindIII fragment from −2100(−65) FAS-LUC plasmid. The constructs −136(−65)FAS-LUC and −184(−65)FAS-LUC were generated by ligating the AscI–BspDI fragment from −2100(65) FAS-LUC into the plasmids −136 FAS-LUC and −184 FAS-LUC previously cut with those same enzymes to eliminate the original E-box sequence. The −444/+67 FAS promoter fragment was subcloned into the pGL2 basic reporter vector by restriction digestion with XbaI. The plasmids −444(−150) FAS-LUC and −444(−65/−150) FAS-LUC were constructed by mutating the −150/−141 FAS sequence to 5′-ATCGATCCAC-3′ by site-directed mutagenesis. −444(−332) FAS-LUC, −444(−65/−332) FAS-LUC, −444(−150/−332) FAS-LUC and −444(−65/−150/−332) FAS-LUC plasmids were generated by mutating the −341/−327 FAS sequence to 5′-AAGCTTCCAACGCGT-3′ by using the same method used for the previous constructs. The heterologous promoter constructs were made by inserting one or six copies of the 5′-CGCGGGCATCACCCCACCGACGGC-3′ sequence into the pGL2 promoter reporter vector. The expression vector for the truncated active form of SREBP-1a has been described (6).
Animal Treatments.
The animals had access ad libitum to food pellets containing 58% carbohydrate. Before fasting/refeeding experiments, the mice were fed a synthetic low-carbohydrate protein diet (no. 5789C; Purina) containing 71% (wt/wt) casein and 4.25% (wt/wt) sucrose for 2 weeks (11). Mice were fasted for 24 h, or fasted mice were refed a high-carbohydrate (58%), fat-free diet for 16 h.
Cell Culture and Transient Transfection Assays.
3T3-L1 fibroblasts were cultured in DMEM containing 10% FBS. For transient transfection of 3T3-L1 fibroblasts, 5 μg of the experimental reporter plasmid with 25 ng of the SREBP-1a expression vector was cotransfected by using the calcium phosphate-DNA coprecipitation method along with 10 ng of the expression vector for cytomegalovirus-β-galactosidase, used as a control for transfection efficiency. Control cells were cotransfected with 25 ng of pcDNA3.1 (Invitrogen). Cells were washed twice with PBS 16–18 h after transfection. Forty-eight hours after transfection, cells were harvested, and LUC and β-galactosidase activities were measured by using a Tropix kit (Dual-Light Reporter Gene Assay). Transfections were carried out in quadruplicate, and at least two different preparations for each plasmid were used.
RNA Isolation, Northern Blot Analysis, and Ribonuclease Protection Assays.
Tissues excised by rapid dissection and frozen in liquid nitrogen were pulverized by using a ceramic mortar and pestle. Total RNA was isolated from the frozen tissues by using TRIzol (GIBCO/BRL) following the manufacturer's procedure. Northern blot and ribonuclease protection assays were carried out as described (9). The plasmid pSREBP-1a (kindly provided by Michael S. Brown and Joseph L. Goldstein, Univ. of Texas Southwestern Medical Center, Dallas) containing a 4,154-bp fragment of the human SREBP-1a cloned into the EcoRI/SalI restriction site in pBluescript II was used as a probe for Northern blot analysis.
In Vitro Transcription/Translation of SREBP-1a.
Plasmid DNA containing SREBP-1a sequence cloned in pcDNA3.1 (6) was digested with XbaI and in vitro-transcribed with T7 RNA polymerase. The RNA then was in vitro-translated by using rabbit reticulocyte lysates (Promega) by following the manufacturer's procedure. An in vitro translation reaction with no RNA added was used as the negative control. Parallel experiments were set up to allow one set of reactions to produce 35S-Met-labeled protein to monitor the translation product and the other set of reactions to produce unlabeled protein for gel mobility shift assays.
Gel Mobility Shift Assay.
The following single-stranded oligonucleotides were synthesized by GIBCO/BRL: −65 E-Box: 5′-AGCTGTCAGCCCATGTGGCGTGGCCGC-3′, 3′-AGTCGGGTACACCGCACCGGCGTCGAC-5′; −150 SRE: 5′-CGCGCGCGGGCATCACCCCACCGACGG-3′, 3′-GCCCGTAGTGGGGTGGCTGCCGCCG-5′; −150-M SRE: 5′-CGCGCGCGGGCATCGATCCACCGACGG-3′, 3′-GCCCGTAGCTAGGTGGCTGCCGCCG-5′; −332 E-Boxes: 5′-GCGCACAGTGCACACCTGG-3′, 3′-GTGTCACGTGTGGACCGGGG-5′; −332-M E-box: 5′-GCGAAGCTTCCAACGCGTG-3′, 3′-TTCGAAGGTTGCGCACGGGG-5′.
Double-stranded oligonucleotides were formed as described (6). All probes for gel shift assays were labeled by using [α-32P] dCTP in the presence of dATP, dGTP, and dTTP, and Klenow fragment of Escherichia coli DNA polymerase. Cold dCTP was added at the end of the labeling reaction to make full-length probes. Reactions (20 μl) containing the indicated amount of in vitro-translated SREBP-1a and 1 × 107 cpm oligonucleotide probes were carried out at room temperature for 1 h and subjected to 6% nondenaturing PAGE, and autoradiography was performed as described (7). For supershifting, specific SREBP-1 antibody (2 μl per reaction; Santa Cruz Biotechnology) was added to the binding reactions for the final 30 min.
Results and Discussion
Involvement of SREBP in Nutritional Regulation of the FAS Promoter in Vivo.
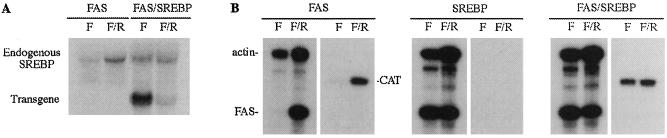
We previously reported that 2.1 kb of the 5′ flanking sequence is sufficient for tissue-specific and nutritional/hormonal regulation of the FAS gene in vivo. We also showed that two promoter regions are required for maximal nutritional/insulin regulation of the FAS gene: one from −278 to −131 mediates induction by feeding and by insulin albeit at a low level, and the other from −444 to −278 is required for maximal induction. Furthermore, we demonstrated that USF binds to the −332 E-box present in the distal region and mediates maximal activation of the FAS promoter (9). In addition to USF, SREBP may be involved in the regulation of the FAS promoter. The Goldstein and Brown laboratory (11, 12) showed that FAS mRNA levels, along with other lipogenic enzymes, are induced in livers of transgenic mice expressing a truncated active form of SREBP. SREBP-1 itself also is known to be induced in liver when animals were refed a high-carbohydrate diet, and SREBP-1c isoform recently has been shown to be selectively increased in liver by insulin treatment of diabetic animals (30, 31). To examine the involvement of SREBP in the nutritional regulation of the FAS promoter in vivo, we have generated double transgenic mice carrying the CAT reporter gene driven by the 2.1 kb of the FAS promoter and a truncated active form of the SREBP-1a driven by the phosphoenolpyruvate carboxykinase (PEPCK) promoter (11). Endogenous SREBP-1 mRNA levels were low in fasting and increased when animals were refed a high-carbohydrate, fat-free diet (the SREBP-1a sequence used as a probe detected both the SREBP-1a and −1c isoforms; ref. 32). The SREBP transgene was expressed at a high level only during fasting and was decreased to a very low level during feeding in FAS-CAT/SREBP double transgenic mice (Fig. 1A). This result was predicted because, contrary to the lipogenic enzymes, the PEPCK promoter driving the transgene is activated in fasting and repressed during feeding. In SREBP transgenic mice, the endogenous SREBP mRNA levels were slightly increased in fasting to the level observed in refeeding, possibly because of activation of the SREBP promoter by the SREBP transgene product. As we showed previously, RNase protection analysis demonstrates undetectable CAT mRNA levels in livers of mice carrying −2100-FAS-CAT in fasting, which are dramatically induced in refeeding, in a fashion similar to the endogenous FAS mRNA levels (Fig. 1B). In contrast, in SREBP or FAS-CAT/SREBP transgenic mice, FAS mRNA levels were high even in fasting, probably because of the activation of the FAS promoter by the SREBP transgene product, which is highly expressed only in fasting. The induction of the FAS mRNA levels in these SREBP and FAS-CAT/SREBP transgenic mice during feeding were similar to that of the FAS-CAT transgenic or wild-type mice. As expected, SREBP transgenic mice did not show any CAT expression. Similar to the FAS mRNA levels, FAS-CAT/SREBP double transgenic mice showed high CAT mRNA levels in fasting similar to those observed in refeeding. This finding indicates that, because SREBP transgene is highly expressed only in fasting but not during feeding, the activation of the FAS promoter and the resultant increase in CAT mRNA levels in fasting to those observed in refeeding was caused by the SREBP transgene product. This also indicates that the 2.1 kb of the 5′ flanking region of the FAS gene contains the regulatory elements required to achieve activation of the FAS promoter by SREBP.
Figure 1.
SREBP-1a activates the FAS promoter in vivo. (A) Northern blot analysis of both endogenous SREBP and SREBP transgene in −2100-FAS-CAT transgenic mice or −2100-FAS-CAT/SREBP-1a transgenic animals. Transgenic mice were either fasted for 24 h or fasted for 24 h then refed a high-carbohydrate/fat-free diet for 16 h. (B) mRNA levels for the endogenous FAS gene and CAT transgene analyzed by RNase protection assays using total liver RNA as described in Materials and Methods in transgenic mice carrying the −2100-FAS-CAT, the SREBP transgene, or both. Actin mRNA was used as a control. Essentially the same results were obtained from three independent experiments. F and R indicate fasted or refed states, respectively.
Region Responsible for Activation of the FAS Promoter by SREBP During Nutritional Regulation in Vivo.
To examine the 5′ flanking region(s) involved in the activation of the FAS gene by SREBP in vivo, transgenic mice carrying −644-, −444-, −278- and −131-FAS-CAT fusion genes (9) were bred to SREBP transgenic mice to obtain various FAS-CAT/SREBP-1a double transgenic animals. Multiple founder lines were used because transgene expression can be affected by the location of genomic integration and copy number. The estimation of copy numbers for each founder line for the various FAS promoter-CAT fusion genes is as follows: −2.1 FAS-CAT (2 lines: 3 copies each); −644 FAS-CAT (1 line: 3 copies); −444 FAS-CAT (2 lines: 4 or 5 copies); −278 FAS-CAT (3 lines: 2 or 3 copies); and −131 FAS-CAT (4 lines; 1, 3, 4, or 5 copies). As expected, none of the FAS-CAT transgenic mice showed detectable FAS mRNA levels during fasting but all showed dramatically increased levels when refed (Fig. 2 Left). As we reported previously and as in the 2.1-kb FAS-CAT transgenic mice shown in Fig. 1B, CAT mRNA levels in 5′ deletion FAS-CAT transgenic mice were not detectable in fasting. Upon refeeding, CAT mRNA levels were dramatically increased in −644- and −444-FAS-CAT transgenic mice. In −278-FAS-CAT transgenic mice, the induction of CAT expression by feeding was low, less than 10% of that observed in −644- and −444-FAS-CAT transgenic mice, whereas in −131-FAS-CAT transgenic mice CAT expression was not detectable even in feeding.
Figure 2.
Regulation by fasting/refeeding of CAT mRNA expression driven by the various FAS promoter regions. Total RNA isolated from liver was subjected to RNase protection assay to determine CAT, FAS, and actin mRNA levels as described in Materials and Methods and exposed to x-ray film (Fuji) for 1 day. * represents an autoradiogram on BioMax film (Kodak) exposed for 3 days, and F and R indicate fasted and refed states, respectively. Essentially the same results were obtained from three independent experiments.
As shown with the 2.1-kb FAS-CAT/SREBP transgenic mice, all FAS-CAT/SREBP double transgenic mice showed high levels for endogenous FAS mRNA whether fasted or refed (Fig. 2 Right). The high-level expression of the FAS mRNA during fasting, which is not observed in FAS-CAT transgenic or wild-type mice, is probably caused by the SREBP transgene, which is highly expressed only in fasting. Similarly, in −644- and −444-FAS-CAT/SREBP double transgenic mice, CAT mRNA was induced in fasting to the same high level observed in refed single −644 and −444-FAS-CAT transgenic mice. In −278-FAS-CAT/SREBP transgenic mice, however, CAT mRNA expression was still increased in fasting because of the SREBP transgene expression, but only to the low level observed in the refed −278-FAS-CAT transgenic mice. On the other hand, the −131-FAS-CAT/SREBP double transgenic mice did not show detectable CAT expression whether fasted or refed, indicating that deletion of the promoter sequence from −278 to −131 caused complete loss of CAT induction by SREBP transgene. As mentioned above, we used multiple founder lines and obtained similar results. This fact could be expected given the similar copies of incorporated transgene for −2.1, −644, −444, and −278 FAS-CAT. Even at higher copies of the transgene, the −131 FAS-CAT did not show any promoter activity, which indicates that the region up to −131 does not contain the site(s) sufficient for activation of the FAS promoter by SREBP and that sequences between −278 and −131 are required for FAS activation by SREBP. Interestingly, upon sequence examination, we found a consensus SRE at −150, although its function is unknown. As we have shown previously, however, an upstream regulatory region between −444 and −278, probably via USF binding to the −332 E-box, is responsible for the maximal increase in CAT expression similar to that observed in the endogenous FAS gene during feeding.
Transactivation of the FAS Promoter by SREBP in 3T3-L1 Fibroblasts.
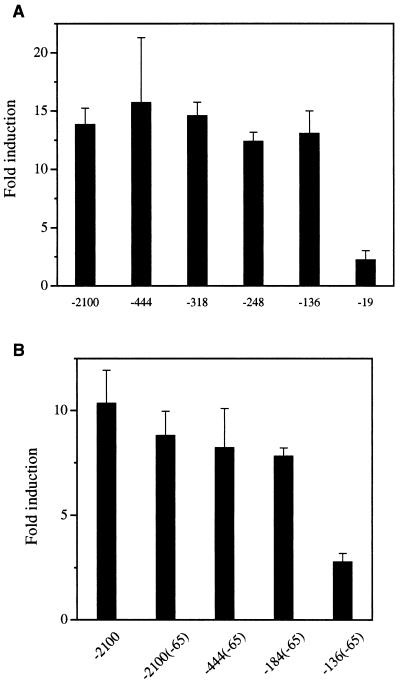
To demonstrate the role of the FAS promoter region between −278 and −131 in the regulation of the FAS gene by SREBP in vitro, we carried out transactivation studies of the FAS promoter in 3T3-L1 cells. The 2.1-kb FAS promoter was activated by a truncated active form of SREBP-1a (15) in a dose-dependent manner, and we used 25 ng of SREBP expression vector per 60-mm dish that gave on separate transfections 8- to 15-fold activation, which falls within the linear range of response (data not shown). To determine the region required for activation by SREBP, we cotransfected various constructs carrying 5′ deletion fragments ranging from −2100 to −136 fused to the LUC reporter gene along with the SREBP expression vector into 3T3-L1 cells. The −19 FAS-LUC was activated by SREBP slightly, probably because of cryptic SREBP binding site(s) in the vector itself because there is no significant element of the FAS promoter present in this construct. On the other hand, all of the FAS-LUC constructs, except −19 FAS-LUC, showed a similar degree of activation by SREBP, approximately 13-fold, suggesting the presence of a SRE between −136 and −19 of the FAS promoter (Fig. 3A). Because our transgenic mice experiments shown in Fig. 2 clearly demonstrate requirement of the region between −278 and −131 for SREBP function, it may be speculated that, in in vitro cotransfection studies, SREBP may activate and increase reporter activity if any single putative binding site is present, regardless of its physiological relevance. In this regard, previous in vitro studies reported that SREBP can bind to the proximal FAS promoter region from −71 to −50 for sterol or insulin regulation (21, 23, 25). On the other hand, we previously demonstrated that USF1/USF2 binds to the E-box located in this region and mediates insulin regulation in 3T3-L1 adipocytes in culture (6). We also found by site-directed mutagenesis of the sequences surrounding the −65 E-box that impaired SREBP binding had no effect on insulin regulation (6). Nevertheless, these studies suggested to us that the binding of SREBP to the −65 region could have masked the function of the putative SRE located between −278 and −131. We therefore mutated the −65 E-box sequence, thereby preventing SREBP binding (data not shown), within the context of the 2.1-kb FAS promoter. When we used the mutated reporter construct and the SREBP expression vector, FAS promoter was still activated to the same extent as the wild-type construct (Fig. 3B), suggesting the presence of other cis-acting regulatory sequences for SREBP. In addition, we tested the effect of −65 E-box mutation on the shorter −444, −184, and −136 FAS promoter context. Deletion of the FAS promoter up to −184 bp in the presence of the −65 mutation showed the same level of activation by SREBP as in −2100 FAS promoter. On the other hand, the SREBP activation was diminished to the level observed in the control −19 FAS-LUC when −136 FAS promoter containing the −65 mutation was used (Fig. 3B). This finding clearly demonstrates that deletion of sequences between −184 and −136 prevented FAS promoter activation by SREBP and, therefore, the presence of a response element for SREBP in this region. These in vitro studies demonstrating functional SREBP site between −136 and −184 support the in vivo results obtained in FAS-CAT/SREBP double transgenic mice in Fig. 2. We conclude that a functional SRE is present between −184 and −136.
Figure 3.
Localization of the FAS promoter region mediating FAS transactivation by SREBP-1a. (A) Five micrograms of the indicated 5′ deletion FAS promoter-LUC constructs was cotransfected with 25 ng of an expression vector for SREBP-1a into 3T3-L1 fibroblasts. (B) Five micrograms of either −2100 FAS-LUC, −2100(−65) FAS-LUC, −444(−65)FAS-LUC, −184(−65) FAS-LUC, or −136(−65) FAS-LUC plasmids was cotransfected with 25 ng of an expression vector for SREBP-1a into 3T3-L1 fibroblasts. The values represent the mean ± standard deviation. Essentially the same results were obtained from two independent experiments.
SREBP Activates the FAS Promoter via the SRE Located at −150.
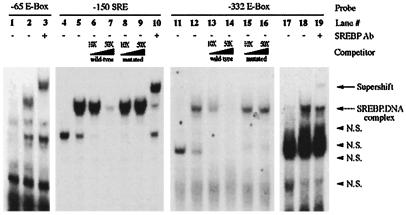
When we inspected the FAS promoter sequence from −184 to −136 for possible SREBP binding sites, we found a canonical SRE located between −150 and −141 bp. Interestingly, comparison among rat, human, and goose FAS promoter sequences indicates that this potential SRE is highly conserved among species, suggesting a potential role in the regulation of the FAS gene. This site has been previously reported to have a negative effect on the FAS promoter activity (33). To determine the potential function of the putative −150 SRE, we first examined the ability of SREBP to effectively bind this region by gel mobility shift assays. As shown previously, in vitro-transcribed and -translated SREBP-1a could bind oligonucleotides containing the −65 sequence effectively (Fig. 4). SREBP also was able to bind to oligonucleotides containing the −150 SRE. The protein-DNA complex formed was sequence-specific because its formation was blocked by competition with excess unlabeled oligonucleotides, whereas cold oligonucleotides bearing a mutated SRE were unable to compete. In addition, SREBP-1 antibody supershifted the band, indicating the presence of SREBP-1 in the −150 SRE-protein complex. Although we have previously described the function of USF at the −332 E-box, because SREBP can bind not only SRE but also E-box sequence (22), we further examined the possible function of SREBP at the −332 E-box. If this were the case, SREBP binding to the −332 E-box could be responsible for the maximal induction of FAS gene expression observed in the transgenic mice (9). In gel mobility shift assays, SREBP was able to bind to oligonucleotides containing the −332 E-box in a sequence-specific manner. Cold oligonucleotides were effective as a competitor (Fig. 4). To demonstrate the requirement of E-box for binding of SREBP to this sequence, we mutated the overlapping inverted repeat of an E-box present in this sequence. Oligonucleotides carrying this mutation were not effective as a competitor. SREBP-1 antibody supershifted the band, indicating the presence of SREBP-1 in the −332 SRE-protein complex. The SREBP binding to the −332 or −65 E-box, however, was weaker than the binding observed with the −150 SRE sequence (data not shown).
Figure 4.
Binding of SREBP-1a to the −150 SRE of the FAS promoter. Gel shift assay was performed with in vitro-translated SREBP-1a as described in Materials and Methods. Each reaction contained 0.1 μg of poly(dI-dC), 1 mM DTT, 0.1 ng of 32P-labeled oligonucleotide probe (the −65 E-box probe in lanes 1–3, −150 SRE probe in lanes 4–10, and −332 E-box probe in lanes 11–19), and cold wild-type and mutant oligonucleotides of −150 SRE and −332 E-box probes (lanes 6–9 and 13–16, respectively) as competitors. Lanes 1, 4, 11, and 17 are with unprogrammed control lysates, and lanes 2, 3, 5–10, 12–16, 18, and 19 are with in vitro-translated SREBP-1a. SREBP-1 antibody (lanes 3, 10, and 19) was added during the final 30 min of the DNA–protein complex formation for supershifting. N.S., nonspecific protein–DNA complex.
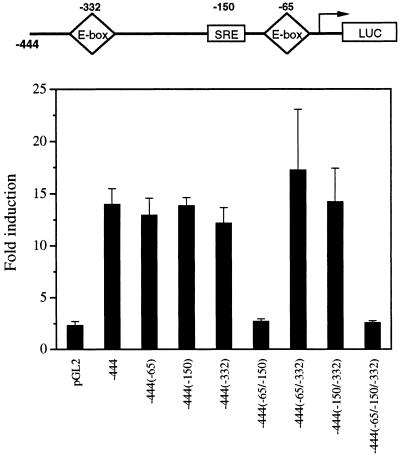
To examine specifically the contribution of the SREBP binding to the −150 SRE in the activation of the FAS promoter, we generated various FAS promoter-reporter constructs bearing single or double combinatorial mutations of the −65, −150, and −332 response elements. We tested the effects of these mutations in SREBP-1a function by transient cotransfection into 3T3-L1 cells. Our hypothesis is that a single potential SRE is sufficient for SREBP effect in transfection studies. As shown in Fig. 5, single mutations at either −65 or −332 E-boxes or a mutation at −150 SRE did not abolish transactivation of the FAS promoter by SREBP. Only when the FAS promoter carried mutations in both the −65 E-box and the −150 SRE was transactivation by SREBP abolished. We next showed that the −150 SRE is sufficient for activation by SREBP. A single copy of −150 SRE conferred 3-fold activation to a heterologous promoter by SREBP whereas six copies conferred 23-fold activation (Table 1). It appears that, in in vitro studies, either the −65 sequence or the −150 SRE can mediate full activation of the FAS promoter by SREBP, but our in vivo studies clearly demonstrate requirement of a functional SRE between −278 and −131. We therefore conclude that SREBP binding to the −150 SRE sequence contributes to the activation of the FAS promoter during feeding. It may be that, in in vitro transfection assays, SREBP could bind and transactivate the FAS promoter as long as a single potential SRE site exists, either the −65 or −150 sequence. Mutations at both sites, however, clearly demonstrate the significance of the −150 SRE in these in vitro studies. Furthermore, double mutation of −65 and −332 E-boxes could not abolish transactivation of the FAS promoter by SREBP. These studies show that SREBP binding to the −332 E-box has no functional role in FAS regulation by SREBP and further support our previous report of USF function at the −332 E-box (9).
Figure 5.
Role of the −150 SRE in activation of the FAS promoter by SREBP. Five micrograms of each of the indicated constructs containing −444 bp of the 5′ flanking sequence of the FAS gene bearing mutations at the indicated positions was cotransfected with 25 ng of an expression vector for SREBP-1a into 3T3-L1 fibroblasts. The values represent the mean ± standard deviation. Essentially the same results were obtained from two independent experiments.
Table 1.
The −150 SRE confers activation by SREBP
| Construct | Fold activation |
|---|---|
| pGL2 promoter | 0.8 ± 0.1 |
| p1x−150SRE | 3.5 ± 0.4 |
| p6x−150SRE | 23.0 ± 1.3 |
Five micrograms of each indicated construct was cotransfected with 25 ng of an expression vector for SREBP-1a into 3T3-L1 fibroblasts. The values represent the mean ± standard deviation. Essentially the same results were obtained from two different experiments.
In conclusion, our present transgenic mice studies in combination with transient transfection assays suggest that FAS promoter activation by SREBP occurs via the −150 SRE during nutritional regulation of the FAS gene. The fact that the SREBP level is increased by feeding supports these conclusions. Our present model is that during fasting SREBP is limiting, which prevents activation of the FAS promoter. During refeeding, the SREBP level is increased and by binding to the −150 SRE, in combination with USF binding to the −332 E-box, causes maximal activation of the FAS promoter. Transgenic mice bearing mutations of each of these sites at the FAS promoter will provide unequivocal evidence for this model.
Acknowledgments
We thank Drs. Joseph L. Goldstein and Michael S. Brown for SREBP-1a plasmid. This work was supported by National Institutes of Health Research Grant DK36264 to H.S.S.
Abbreviations
- SRE
sterol regulatory element
- SREBP
SRE binding protein
- LUC
luciferase
- CAT
chloramphenicol acetyltransferase
- FAS
fatty acid synthase
- USF
upstream stimulatory factor
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.180306597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.180306597
References
- 1.Wakil S J, Stoops J K, Joshi V C. Annu Rev Biochem. 1983;52:537–579. doi: 10.1146/annurev.bi.52.070183.002541. [DOI] [PubMed] [Google Scholar]
- 2.Hillgartner F B, Salati L M, Goodridge A G. Physiol Rev. 1995;75:47–76. doi: 10.1152/physrev.1995.75.1.47. [DOI] [PubMed] [Google Scholar]
- 3.Lakshmanan M R, Nepokroeff C M, Porter J W. Proc Natl Acad Sci USA. 1972;69:3516–3519. doi: 10.1073/pnas.69.12.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paulauskis J D, Sul H S. J Biol Chem. 1988;263:7049–7054. [PubMed] [Google Scholar]
- 5.Paulauskis J D, Sul H S. J Biol Chem. 1989;264:574–577. [PubMed] [Google Scholar]
- 6.Wang D, Sul H S. J Biol Chem. 1997;272:26367–26374. doi: 10.1074/jbc.272.42.26367. [DOI] [PubMed] [Google Scholar]
- 7.Wang D, Sul H S. J Biol Chem. 1995;270:28716–28722. doi: 10.1074/jbc.270.48.28716. [DOI] [PubMed] [Google Scholar]
- 8.Sul H S, Wang D. Annu Rev Nutr. 1998;18:331–351. doi: 10.1146/annurev.nutr.18.1.331. [DOI] [PubMed] [Google Scholar]
- 9.Moon Y S, Latasa M-J, Kim K-H, Wang D, Sul H S. J Biol Chem. 2000;275:10121–10127. doi: 10.1074/jbc.275.14.10121. [DOI] [PubMed] [Google Scholar]
- 10.Casado M, Vallet V S, Kahn A, Vaulont S. J Biol Chem. 1999;274:2009–2013. doi: 10.1074/jbc.274.4.2009. [DOI] [PubMed] [Google Scholar]
- 11.Shimano H, Horton J D, Hammer R E, Shimomura I, Brown M S, Goldstein J L. J Clin Invest. 1996;98:1575–1584. doi: 10.1172/JCI118951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimano H, Horton J D, Shimomura I, Hammer R E, Brown M S, Goldstein J L. J Clin Invest. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horton J D, Shimomura I, Brown M S, Hammer R E, Goldstein J L, Shimano H. J Clin Invest. 1998;101:2331–2339. doi: 10.1172/JCI2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimano H, Yahagi N, Amemiya-Kudo M, Hasty A H, Osuga J, Tamura Y, Shionoiri F, Iizuka Y, Ohashi K, Harada K, et al. J Biol Chem. 1999;274:35832–35839. doi: 10.1074/jbc.274.50.35832. [DOI] [PubMed] [Google Scholar]
- 15.Brown M S, Goldstein J L. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Sato R, Brown M S, Hua X, Goldstein J L. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 17.Ericsson J, Jackson S M, Lee B C, Edwards P A. Proc Natl Acad Sci USA. 1996;93:945–950. doi: 10.1073/pnas.93.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yokoyama C, Wang X, Briggs M R, Admon A, Wu J, Hua X, Goldstein J L, Brown M S. Cell. 1993;75:187–197. [PubMed] [Google Scholar]
- 19.Lopez J M, Bennett M K, Sanchez H B, Rosenfeld J M, Osborne T F. Proc Natl Acad Sci USA. 1996;93:1049–1053. doi: 10.1073/pnas.93.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan G, Dai P H, Osborne T F, Kim J B, Shechter I. J Biol Chem. 1997;272:10295–10302. doi: 10.1074/jbc.272.15.10295. [DOI] [PubMed] [Google Scholar]
- 21.Magana M M, Osborne T F. J Biol Chem. 1996;271:32689–32694. doi: 10.1074/jbc.271.51.32689. [DOI] [PubMed] [Google Scholar]
- 22.Kim J B, Spotts G D, Halvorsen Y D, Shih H M, Ellenberger T, Towle H C, Spiegelman B M. Mol Cell Biol. 1995;15:2582–2588. doi: 10.1128/mcb.15.5.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J B, Sarraf P, Wright M, Yao K M, Mueller E, Solanes G, Lowell B B, Spiegelman B M. J Clin Invest. 1998;101:1–9. doi: 10.1172/JCI1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foretz M, Pacot C, Dugail I, Lemarchand P, Guichard C, Le Liepvre X, Berthelier-Lubrano C, Spiegelman B, Kim J B, Ferre P, Foufelle F. Mol Cell Biol. 1999;19:3760–3768. doi: 10.1128/mcb.19.5.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett M K, Lopez J M, Sanchez H B, Osborne T F. J Biol Chem. 1995;270:25578–25583. doi: 10.1074/jbc.270.43.25578. [DOI] [PubMed] [Google Scholar]
- 26.Soncini M, Yet S F, Moon Y, Chun J Y, Sul H S. J Biol Chem. 1995;270:30339–30343. doi: 10.1074/jbc.270.51.30339. [DOI] [PubMed] [Google Scholar]
- 27.Moustaid N, Beyer R S, Sul H S. J Biol Chem. 1994;269:5629–5634. [PubMed] [Google Scholar]
- 28.Misra S, Sakamoto K, Moustaid N, Sul H S. Biochem J. 1994;298:575–578. doi: 10.1042/bj2980575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moustaid N, Sakamoto K, Clarke S, Beyer R S, Sul H S. Biochem J. 1993;292:767–772. doi: 10.1042/bj2920767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horton J D, Bashmakov Y, Shimomura I, Shimano H. Proc Natl Acad Sci USA. 1998;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimomura I, Bashmakov Y, Ikemoto S, Horton J D, Brown M S, Goldstein J L. Proc Natl Acad Sci USA. 1999;96:13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hua X, Wu J, Goldstein J L, Brown M S, Hobbs H H. Genomics. 1995;25:667–673. doi: 10.1016/0888-7543(95)80009-b. [DOI] [PubMed] [Google Scholar]
- 33.Boizard M, Le Liepvre X, Lemarchand P, Foufelle F, Ferre P, Dugail I. J Biol Chem. 1998;273:29164–29171. doi: 10.1074/jbc.273.44.29164. [DOI] [PubMed] [Google Scholar]