Abstract
Jasmonic acid (JA) and its precursor 12-oxophytodienoic acid (OPDA) act as plant growth regulators and mediate responses to environmental cues. To investigate the role of these oxylipins in anther and pollen development, we characterized a T-DNA-tagged, male-sterile mutant of Arabidopsis, opr3. The opr3 mutant plants are sterile but can be rendered fertile by exogenous JA but not by OPDA. Cloning of the mutant locus indicates that it encodes an isozyme of 12-oxophytodienoate reductase, designated OPR3. All of the defects in opr3 are alleviated by transformation of the mutant with an OPR3 cDNA. Our results indicate that JA and not OPDA is the signaling molecule that induces and coordinates the elongation of the anther filament, the opening of the stomium at anthesis, and the production of viable pollen. Just as importantly, our data demonstrate that OPR3 is the only isoform of OPR capable of reducing the correct stereoisomer of OPDA to produce JA required for male gametophyte development.
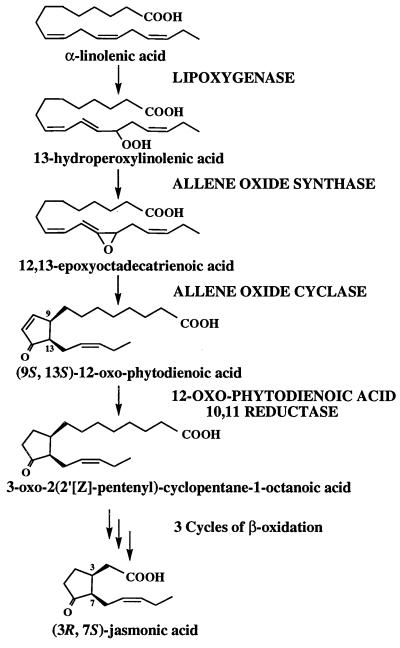
Jasmonic acid (JA) is a lipid-derived signaling compound that is involved in the regulation of many different processes in plants. The role of JA in protecting plants against insects is long established (1–3), and it is now known that JA also acts, together with ethylene, to activate plant defenses against fungal pathogens (4, 5). In healthy, unwounded plant tissues, jasmonate is involved in carbon partitioning (6), mechanotransduction (7), and the maturation and release of pollen (8). The critical requirement for jasmonate in plant reproduction was revealed through characterization of a triple mutant of Arabidopsis, fad3-2 fad7-2 fad8, that lacks the hexadecatrienoic (16:3) and linolenic (18:3) fatty acid precursors of the jasmonate pathway (8) (Fig. 1). The triple-mutant plants are male sterile but can be rendered fertile by treatment of flower buds with jasmonate (or 18:3). The Arabidopsis coi1 mutant, which is unresponsive to jasmonate and related compounds, is also male sterile (9).
Figure 1.
The biosynthesis of jasmonic acid from linolenic acid. The projections shown represent the absolute stereoconfiguration of the side chains.
Understanding the physiological roles of JA in these many different processes is complicated by the observation that intermediates in the synthesis of JA are also active in at least some of these processes. For example, 12-oxophytodienoic acid (OPDA) has been shown to be a far more effective inducer of tendril coiling in Bryonia dioica than JA itself (7, 10) and to act as an elicitor of alkaloid biosynthesis in Eschscholtzia californica cell cultures (11). The levels of JA, OPDA, and other intermediates of oxylipin synthesis vary considerably among species (1, 7, 12, 13), giving rise to the suggestion that the relative and absolute concentrations of different compounds—the oxylipin signature—may provide flexibility to this multifunctional chemical signaling system (14). In unwounded Arabidopsis leaves, the levels of OPDA are 10-fold higher than the levels of JA: 340–1,000 ng/g fresh weight versus 30–50 ng/g (2, 12, 13). Although measurements on flower tissues have not been made, these data beg the question of whether OPDA as well as JA can induce pollen maturation and anther dehiscence and indeed whether OPDA might be the true physiological effector in anther tissue.
The relatively sparse information about the role of OPDA is matched by limited knowledge about the enzymes that convert OPDA to JA. The first step in this conversion is catalyzed by 12-oxophytodienoic acid reductase (OPR) (15) (Fig. 1), and a cDNA encoding an OPR was first cloned from Arabidopsis, using sequence of the enzyme purified from Corydalis sempervirens (16). However, subsequent studies (17) showed that the Arabidopsis gene, OPR1, encodes a protein with almost no activity against 9S,13S-OPDA, which is believed to be the principal isomer in plants (18). At the same time, a second, separable OPR activity from C. sempervirens was shown to have the characteristics expected of an isozyme involved in JA synthesis in that it efficiently converted 9S,13S-OPDA to 3-oxo-2(2′[Z]-pentenyl)-cyclopentane-1-octanoic acid (OPC:8) (17), which is hypothesized to be converted to 3R,7S-JA [(+)-7-iso-JA] by three cycles of β-oxidation (19). A second OPR gene, OPR2, was identified in Arabidopsis (20). Originally, the observation by Biesgen and Weiler (20) that the OPR2 promoter directed β-glucuronidase expression in a late stage of pollen development led these authors to suggest that OPR2 might be the gene involved in producing JA during pollen development. More recently, biochemical studies performed on purified OPR1 and OPR2 showed that these two isoforms were able to reduce 9S,13S-OPDA only very poorly (21), whereas OPR3, a third isoform identified as a cDNA representing a brassinosteroid-up-regulated gene (22), was highly effective in reducing 9S,13S-OPDA (21), the naturally occurring isomer in plants. These biochemical results suggest that OPR3 rather than OPR1 and OPR2 is the enzyme involved in JA biosynthesis in A. thaliana, although the contributions of both OPR1 and OPR2 cannot be ruled out.
In this paper, we report the cloning by T-DNA tagging of a gene encoding the third isoform of OPR, OPR3. Characterization of the mutant phenotype indicates that JA but not OPDA acts to ensure pollen fertility and that the OPR3 gene product is the only isozyme of OPR that can provide the JA in anther tissue required for pollen maturation and release.
Materials and Methods
Screening of the Arabidopsis Male-Sterile T-DNA Population.
Twenty-eight male-sterile T-DNA mutagenized lines of Arabidopsis thaliana ecotype Wassilewskija (WS) (23) were obtained from the Arabidopsis Biological Resource Center at Ohio State University and screened for their response to JA. ABRC numbers CS2321–2329, CS2331, CS2332, CS2336–2338, CS2340–2343, CS2346, CS2347, CS2349, CS2350, CS2352, CS2811, CS2812, CS2817, CS2819, and CS2821 were germinated on soil and grown under a 12-h photoperiod (130 μmol quanta⋅m−2⋅sec−2) at 22°C. After bolting, flower buds of sterile plants from each line were painted with a 100 μM solution of 7-epi JA (Cayman Chemicals, Ann Arbor, MI) in 0.1% Tween 20 (Bio-Rad) every 2 days over a period of 2 weeks. Mutant plants were also tested for their response to a 0.1% solution of linolenic acid (18:3) soap (Nu Chek Prep, Elysian, MN), a 100 μM solution of OPDA (Cayman Chemicals) (both painted on flower buds), and a 450 μM solution of methyl jasmonate (MeJA) (Bedoukian Research, Danbury, CT) in 0.1% Tween 20 sprayed on plants during flowering.
Seeds were collected from a single kanamycin-resistant fertile individual of line CS2338. To test for segregation of kanamycin resistance, seeds were sown in Petri dishes on Murashige and Skoog basal medium (GIBCO/BRL) solidified with 0.8% agar (Difco) containing 100 μg/ml kanamycin (ICN). After 10 days, green seedlings were scored as kanamycin-resistant, and chlorotic seedlings not expanding beyond the cotyledon stage were scored as kanamycin-sensitive.
Plasmid Rescue.
Genomic DNA from homozygous mutant plants was digested to completion with either EcoRI (right border rescue) or BamHI (left border rescue) restriction enzyme and allowed to religate at a dilute concentration (0.2 μg⋅ml−1) to promote self-ligation. The ligation products were used to transform Escherichia coli cells that were then subjected to ampicillin selection. No ampicillin-resistant colony was obtained from the experiment with EcoRI-digested DNA, but 22 colonies were identified from the plasmid rescue of BamHI-digested DNA. Based on the results of restriction analysis using BamHI/EcoRI double digestion, these 22 colonies included two different DNA constructs that contained the expected 14.2-kb portion of the T-DNA and one or more additional restriction fragments. Additional restriction fragments thus obtained were gel-purified (Prep-A-Gene; Bio-Rad), radiolabeled (Decaprime II; Ambion, Austin, TX), and used as probes in a Southern blot. The Southern blot was performed by digesting 5 μg of wild-type or mutant genomic DNA with BamHI and EcoRI, separation in 0.8% agarose gel, and transfer to Hybond N+ nylon membrane (Amersham Pharmacia). Hybridization and washes were carried out at 65°C. When the additional DNA fragments from one class of plasmids were used as probes in Southern analysis, hybridization only occurred with DNA from mutant plants and not with wild-type DNA, suggesting that these plasmids might be derived from internal rearrangements within multiple copies of the T-DNA at the insertion site. The second class of plasmids (represented by only a single clone) provided DNA fragments of 1.3 and 0.65 kb, which hybridized to DNA from both mutant and wild-type (ecotype WS) plants but labeled different-sized bands in the mutant relative to wild type (data not shown). This observation identified the fragments as plant DNA flanking the left border of the T-DNA insert.
These two 650-bp EcoRI–EcoRI and 1.3-kb EcoRI–BamHI fragments of plant sequence were subcloned into pBluescript SK (Stratagene) (designated pSK650 and pSK1300, respectively) and sequenced on both strands with an Applied Biosystem automated sequencer. Sequence analysis was performed using Genetic Computer Group (Madison, WI) software. Database searches were carried out (12/28/99) using the blast servers (24) at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) and the Arabidopsis Information Resource (http://www.arabidopsis.org).
Isolation of cDNA and Genomic Clones Spanning the Site of T-DNA Insertion.
Nucleotides 1–1087 from pSK1300 showed 98% similarity to nucleotides 28,326–29,414 from A. thaliana chromosome II (accession number AC006413), which lie within intron 3 of a putative OPDA reductase gene. Gene-specific primers were designed to allow amplification by reverse transcription–PCR (RT-PCR) of the entire coding sequence for the putative reductase. Primer 1 (5′-CCCGGGTCTAGAATTCATGACGGCGGCACAAGGGAACTC-3′) encompassed the translational start ATG and nucleotides 29,727–29,749 of AC006413, flanked by an EcoRI restriction site (underlined), and primer 2 (5′-CCCGGGATCGATTCAGAGGCGGGAAGAAGGAGCCAAG-3′) encompassed the sequence complementary to the stop codon and nucleotides 27,018–27,042 of AC006413, flanked by a ClaI restriction site (underlined). RT-PCR on 500 ng of wild-type WS anther total RNA was performed using the Superscript 1 step RT-PCR system (GIBCO/BRL). After an initial incubation at 50°C for 30 min, 30 cycles of PCR (95°C, 20 sec; 63°C, 40 sec; 72°C, 80 sec) were performed.
Using primers 1 and 2, a cDNA amplicon of 1.18 kb was obtained, digested with EcoRI and ClaI, cloned into pART7 (25) as pART7-OPR3, and sequenced. The WS sequence differed by four nucleotides from the Columbia allele, which has recently been designated OPR3 (22). Primers 1 and 2 were also used to generate a genomic fragment by PCR from wild-type WS DNA in a 500-μl PCR reaction containing 50 ng of DNA as a template. After 38 cycles (95°C, 20 sec; 63°C, 40 sec; 72°C, 225 sec), the PCR reaction was separated on a 0.8% agarose gel, and a predominant 4-kb fragment was gel purified and ethanol precipitated. Direct sequencing of this fragment was performed using 60 ng of DNA for each sequencing reaction. Finally, primer 3 (5′-TTGTAACTTATTGAGCATGCCAAC-3′), encompassing nucleotides 27,390–27,413 of AC006413, and primer 4 (5′-GCTTTGAATGCGATTCGAGCTGG-3′), encompassing nucleotides 27,657–27,680 of AC006413, were used in combination with primers T3 or T7 to screen by PCR a flower cDNA library (CD4–6, ABRC). A 650-bp and a 800-bp amplicon, corresponding, respectively, to the 5′ and the 3′ ends of the OPR3 cDNA, were cloned into TOPO TA (Invitrogen) and sequenced.
RNA Extraction and Gel-Blot Analysis.
Total RNA was obtained (26) from whole flowers or anthers harvested at stage 12 (27) or rosette stage leaves from both wild-type WS and mutant CS2338. Poly(A)+ RNA was purified from flower and leaf total RNA (PolyATtract; Promega), following the manufacturer's protocol. RNA samples were separated on a 1.2% agarose/formaldehyde denaturing gel, transferred to HyBond N+, and probed with 32P-labeled OPR3 cDNA. Washes were performed at 65°C and 0.2× SSC.
Gene Constructs and Plant Transformation.
The 1.18-kb OPR3 cDNA cloned in pART7-OPR3 is under the control of the cauliflower mosaic virus 35S promoter and the transcriptional termination region of the octopine synthase gene. The entire expression cassette was released with NotI and inserted into the binary vector pBART, a derivative of pART27 (25) in which the neomycin phosphotransferase gene has been replaced by the bialaphos resistance gene (bar) coding for phosphinothricin acetyltransferase to create pBART-35SOPR3. Agrobacterium tumefaciens GV3101 transfected with pBART-35SOPR3 by electroporation were used to transform 2-month-old homozygous mutant plants by the floral dip method (28). After transformation, T0 plants were sprayed every second day with MeJA to allow seed set, and the resulting T1 seeds were germinated on potting mix wetted with 25 mg/liter of glufosinate ammonium (Finale) to select for transformants.
Pollen Germination.
Pollen germination tests were carried out according to the method of Preuss et al. (29). In the case of the mutant, for which dehiscence of the anther did not occur at anthesis, the locule was opened manually to release pollen. Pollen was incubated for 24 h at room temperature and then analyzed for pollen tube formation. In the representative experiment reported herein, a total of 2,900 pollen grains from untreated mutant flowers in 60 microscope fields were scored along with 1195 wild-type pollen (32 microscope fields) and 1193 pollen (28 microscope fields) from mutant flowers that had been treated with MeJA.
Flower Development and JA Responsiveness.
To determine the stage of flower development at which JA is responsible for the complementation of the male-sterile phenotype in the mutant, opened flowers (beyond anthesis and JA responsiveness) were removed from 63 inflorescences belonging to 24 plants. The remaining young flower buds were sprayed and left in contact with MeJA in a confined growth cabinet for 2 h, then transferred to a growth chamber without MeJA. Simultaneously, buds on 12 inflorescences were staged according to the method of Smyth et al. (27). After 5 days, nonelongated followed by elongated siliques from JA-treated mutant plants were counted and averaged and compared with the average of the cumulative number of flowers at each stage of development along the flower bud.
Light Microscopy.
Bright-field photographs of individual flowers were taken using a dissecting microscope (Model Orthoplan; Leitz). Photographs of germinating pollen grains were taken with a compound microscope (model B202; Olympus, New Hyde Park, NY).
Results
Identification of a Jasmonate-Responsive Male-Sterile Mutant.
The original population of Arabidopsis lines in which mutations have been created by T-DNA insertion (23) contain 28 lines that have been characterized as male sterile (see Materials and Methods). A total of 30–90 plants from each line were grown in the course of five experiments, and male-sterile segregants (identified by their inability to set seed) were treated by painting flower buds with either 100 μM JA in 0.1% aqueous Tween-20 or with a control detergent solution without JA. One line, CS2338, segregated male-sterile plants that consistently produced progeny seed after treatment with JA but remained completely sterile after treatment with the control solution.
The absence of progeny from male-sterile plants would have resulted in a progressive loss of the mutation during propagation of the CS2338 line before we obtained it. For this reason, seeds were germinated on agar containing 100 μg/ml kanamycin, and surviving seedlings were transferred to soil. At maturity, a single fertile plant was identified, and seeds from this plant were harvested. When 153 of these seeds were germinated on kanamycin-containing media, 118 were kanamycin resistant and 35 were sensitive. These data are a good approximation to a 3:1 ratio expected for a single T-DNA insertion. A further 365 progeny were germinated on soil and grown to maturity, and 94 of these were sterile. This proportion of sterile plants is a good fit to the 3:1 hypothesis (χ2 = 0.094; P > 0.5), indicating that sterility is caused by a single, recessive nuclear mutation. For reasons described below, the mutant locus was designated opr3. After confirmation of the sterile phenotype, plants were sprayed with a 450 μM methyl jasmonate (MeJA) solution to render them fertile, and seeds were harvested. Approximately 60–100 seeds from each plant were germinated on agar containing 100 μg/ml kanamycin. All progeny from all 94 sterile plants rescued with MeJA were kanamycin resistant, indicating that these plants were also homozygous for the T-DNA insert. This experiment indicated cosegregation of the opr3 mutation with the T-DNA insert.
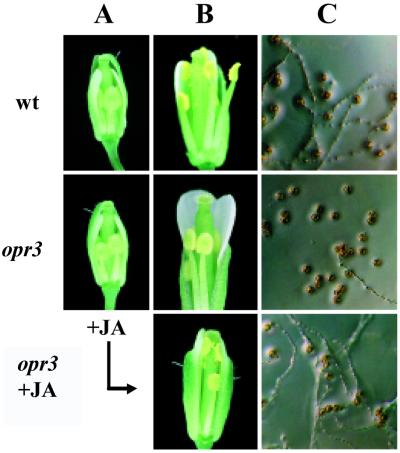
Our previous investigation of the JA-responsive, male-sterile mutant fad3-2 fad7-2 fad8 (8) identified three characteristics of the male-sterile phenotype, and each of these characteristics is also observed in the opr3 mutant. (i) Floral organs develop normally within the closed bud, but the anther filaments do not elongate sufficiently to position the locules above the stigma at anthesis (Fig. 2 A and B). (ii) The anther locules do not dehisce at the time of flower opening (Fig. 2B) (although limited dehiscence occurs later). (iii) Even though pollen on mutant plants develops to the trinucleate stage, as determined by staining with 4′,6-diamino-2-phenylindole (8) (data not shown), the pollen grains are predominantly inviable (Fig. 2C). Irrespective of the stage at which pollen was taken from opr3 plants, germination of the pollen was <4%, compared with 97.6% for mature pollen from wild-type plants. Application of JA to flower buds corrected all three of these defects in opr3 plants (Fig. 2B), resulting in rates of pollen germination equivalent to wild type (97.2%) in in vitro tests (Fig. 2C) and abundant seed set on treated plants. By staging flowers (27) and monitoring seed production after a single application of JA, we established that only flower buds corresponding to stage 12 in floral development (27) responded to JA; flowers at earlier and later stages of development could not be rendered fertile by JA treatment. Stage 12 corresponds to the final stage before the bud opens; it starts when the petals reach the top of the long stamens (Fig. 2A) and ends when the sepals open. At this stage, the second mitotic division occurs in the anthers.
Figure 2.
Phenotypes of wild-type and opr3 mutant flowers. (A) Flowers at stage 12. (B) Flowers at anthesis. Pollination did not occur in opr3 plants unless buds had previously been treated with jasmonate (Bottom). (C) Germination of pollen harvested at anthesis.
In all of these respects, the phenotype of the opr3 mutants was similar to those of fad3-2 fad7-2 fad8 plants that are deficient in 18:3 and 16:3 fatty acids, which are the precursors for JA synthesis (8). However, opr3 plants contained normal levels of 18:3 and 16:3 fatty acids, and application of 18:3 did not restore fertility to this mutant. These results suggest that the opr3 mutation blocks conversion of 18:3 and 16:3 fatty acids to oxylipin(s) (Fig. 1) that initiate the processes of anther and pollen maturation. To more closely define the mutant lesion within the pathway of JA synthesis, we treated opr3 plants and fad3-2 fad7-2 fad8 plants with OPDA. As expected, fad3-2 fad7-2 fad8 plants responded to exogenous OPDA by becoming fertile and producing selfed seeds, thus indicating that OPDA could enter the floral tissue and restore fertility, either by acting directly or by being converted to JA. In contrast, application of OPDA did not restore fertility to opr3 plants. We concluded that the mutant lesion most probably affected OPDA reduction or the β-oxidation of 3-oxo-2(2′[Z]-pentenyl)cyclopentane-1-octanoic acid (OPC:8). Just as importantly, these results indicated that OPDA itself cannot act as a signal in anther development and pollen maturation.
Cloning of a cDNA Encoding Isoform 3 of 12-Oxophytodienoic Acid Reductase.
The modified T-DNA used to generate the mutant population contains the origin of replication and the ampicillin resistance gene of plasmid pBR322 (23). This feature allows recovery of T-DNA/plant DNA junction fragments by the method of plasmid rescue (30). Left border plasmid rescue (see Materials and Methods) identified a 1.9-kb plant DNA fragment subcloned as pSK 650 and pSK 1300.
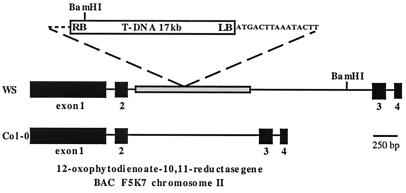
Comparison of the plant-derived DNA in pSK650 and pSK1300 with the database available from the Arabidopsis Genome Initiative (AGI) (http://www.arabidopsis.org) showed that the sequence from 16 to 771 bp (relative to the left border/plant DNA junction) is a rigorous match (blast score 1499) to a sequence on overlapping cosmids T5E21 and F10B6 at 23.6 cM on chromosome I. The sequence on chromosome I does not appear to be part of an active gene but is homologous to sequences scattered throughout the genome that are related to the tandem repeat of the putative nonautonomous transposable element Tnat1 (31). In contrast, the sequence from 766 to the BamHI site at 1852 bp corresponds closely to the sequence from 28,326 to 29,414 of cosmid F5K7 on chromosome II. This is part of an intron within a predicted ORF (GenBank accession AAD19764) that encodes a homolog of OPR (Fig. 3) now identified as OPR3 (21, 22). The AGI sequence that is derived from the Columbia ecotype (Col-O) does not contain the Tnat1-homologous sequence in the predicted OPR intron on cosmid F5K7. To resolve this discrepancy, we designed oligonucleotide primers to sequences within exon 1 and exon 4 of the predicted AGI coding sequence and carried out PCR on both Col-O and WS wild-type genomic DNA. The size of the Col-O PCR product (estimated after agarose-gel electrophoresis) was 2.8 kb, which is consistent with the AGI sequence for cosmid F5K7 (2,702 bp). In contrast, the WS product was 4.0 kb, and sequencing of this DNA confirmed that the WS predicted intronic sequence contains a 1,284-bp insert (relative to the AGI Col-O), which corresponds to Tnat1 (Fig. 3). Over its entire length, this insert is 99% identical to AGI cosmids T5E21 and F10B6 on chromosome I and shows lower sequence identity to other, shorter Tnat1 sequences (31). The difference between Col-O and WS at the OPR3 locus can most easily be explained by a transposition event occurring after divergence of the Col-O and WS ecotypes.
Figure 3.
Genome structure of the opr3 locus. The T-DNA (not to scale) with an additional 15 bp adjacent to the left border is inserted into a Tnat1 sequence (shaded), which is part of an intron in the OPR3 gene of ecotype WS. The four exons of OPR3 are shown, as are the BamHI sites used for plasmid recovery. The structure of the remainder of the T-DNA insert was not determined. The Col-O allele of OPR3 is shown for comparison.
Interestingly, the 15 bp immediately adjacent to the left border of the T-DNA (Fig. 3) is not present in either the Col-O or WS allele of OPR3. A possible explanation for these observations is that the T-DNA first integrated elsewhere in the genome and was then excised together with 15 bp or more of plant DNA adjacent to the left border. This plant DNA then acted as a de facto left border (32) during integration into chromosome II. There is not a perfect match to this 15-bp sequence in the current AGI database.
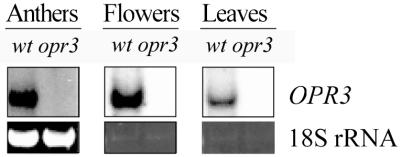
Using information from the AGI sequence, we designed oligonucleotide primers and amplified two independent RT-PCR products, using anther RNA from WS wild type as a template. We also isolated a cDNA clone from a library derived from the Landsberg erecta wild type. RT-PCR products and the cDNA provided identical sequences, with the exception of a single base pair change; the peptide sequence predicted for the WS wild type is shown in Fig. 4. The OPR3 RT-PCR product was used as a probe to estimate transcript levels by gel blot analysis (Fig. 5). In lanes containing total RNA from wild-type anthers or polyA RNA from wild-type flower buds harvested at stages 11–12, strong hybridization occurred to a band of the predicted size (∼1.3 kb). The corresponding lanes of mutant RNA did not contain any detectable band of this or any other size, indicating that any (truncated) mRNA transcribed from OPR3 was rapidly degraded. OPR3 transcript was readily detected in leaf tissue (Fig. 5) and other parts of wild-type plants (not shown). There was no indication that the OPR3 probe cross-hybridized to any related transcript at the high stringency used in these experiments.
Figure 4.
Amino acid alignment of the A. thaliana OPR1 (GenBank accession number Y10617), OPR2 (U92460) and OPR3 (AF293653). Identical residues in at least two of three sequences are indicated in reverse print. The peroxisomal targeting signal in OPR3 is underlined.
Figure 5.
Expression of OPR3 in anthers, flowers, and leaves of wild-type and opr3 plants. Gel-blot analysis was performed, using 10 μg of total RNA from anthers and 5 μg of poly(A)+ RNA from flowers and leaves, with an OPR3 cDNA as a probe. Ethidium bromide staining of the 18S rRNA band was used to confirm equal loading.
Expression of OPR3 Restores Fertility to opr3 Plants.
The Identification of OPR3 as one of the loci disrupted by T-DNA insertion in line CS2338 provided a persuasive explanation of the male-sterile phenotype and of the results obtained in our attempts to restore fertility to opr3 plants, with OPDA in place of JA. However, in light of the complex structure of the opr3 locus, it was particularly important to demonstrate that expression of the OPR3 protein could complement the mutant phenotype. We transformed opr3 mutant plants with a cDNA expressed under control of the cauliflower mosaic virus 35S promoter. Treatment of T0 plants with MeJA during flowering allowed for abundant seed set. When 50,000 T1 seeds were germinated in the presence of glufosinate ammonium, a total of 123 resistant plants were identified and allowed to grow to maturity. All but one of these plants were partially or fully fertile. Twelve of the lines were chosen for further analysis, and a single leaf from each of these was used to prepare genomic DNA. When tested by PCR using appropriate primers, all 12 plants were positive for both the T-DNA allele of opr3 and the 35SOPR3 transgene construct. The number of T2 seeds harvested from each of these 12 plants ranged from 1,000 to 20,000. Because T1 plants are typically hemizygous for the transgene insert, it was possible to test for cosegregation between complementation of the opr3 mutation and expression of the OPR3 cDNA. To do this, we germinated 128 T2 progeny from a single T1 transgenic parent. Of these, 92 were fertile at maturity and 36 were sterile. These data are a good fit (χ2 = 0.594, P > 0.2) to the 3:1 hypothesis. When RNA from 15 fertile and 13 sterile plants was analyzed by gel blot analysis using an OPR3 probe, all of the fertile plants showed high expression of the corresponding transcript, whereas the sterile plants showed no detectable transcript (data not shown). These results confirm that OPR3 expression is required for fertility in Arabidopsis.
Discussion
The production of the male gametophyte in angiosperm plants is a complex series of developmental processes that has been studied by genetic techniques for several decades (33, 34). The discovery that a jasmonate-deficient mutant of Arabidopsis was male sterile and was rendered fertile by applications of JA (8) provided new insight into the chemical regulation of some of the later processes in both pollen and anther development. The stringent requirement for JA signaling potentially has great practical significance, since male sterility and the ability to restore fertility are prerequisites for the development of hybrid breeding strategies in many crop plants (34). opr3 was isolated as a JA-responsive male-sterile mutant in which a distinctive member of the OPR gene family, designated OPR3, is disrupted by a T-DNA insertion. The phenotype of the opr3 mutant is strikingly similar to that described for the Arabidopsis triple mutant fad3-2 fad7-2 fad8 (8) in several different respects. The anther filaments on opr3 plants do not elongate before anthesis, and anther locules do not dehisce at anthesis, so that mutant stigmas typically remain unpollinated. Just as importantly, pollen taken from opr3 anthers at the time of anthesis showed less than 4% germination compared with more than 97% germination for pollen from either wild-type or JA-treated opr3 plants. Mutant analysis suggests that pollen maturation and anther dehiscence are uncoupled processes (8, 35), so our characterization of the opr3 mutant confirms that JA independently activates at least two separate maturation processes in anthers. The fad3-2 fad7-2 fad8 mutant lacks 18:3 and 16:3 fatty acids that are precursors for JA synthesis but is assumed to be wild type for all of the enzymes of the JA pathway. Accordingly, JA, OPDA, or 18:3 restored fertility to fad3-2 fad7-2 fad8 plants when applied. Application of 18:3 to unopened flower buds of triple-mutant plants measurably increased 18:3 levels in sepals and petals, but this fatty acid could not be detected in the anthers after application. This result was interpreted as indicating that JA (or other oxylipins) produced in the petals and sepals could meet the requirements for JA signaling in the anthers (8). In contrast, only JA, not OPDA or 18:3, could render opr3 plants fertile. This result demonstrates that OPDA, which is more abundant than JA in vegetative tissues of Arabidopsis (2, 12), cannot replace JA as the chemical signal triggering anther and pollen maturation.
These experiments on chemical complementation of the opr3 male-sterile phenotype, together with measurements of OPR3 transcript levels in mutant and wild-type plants and the successful molecular complementation of opr3 by transgenic expression of an OPR3 cDNA, indicate that OPR3 encodes the only OPR isozyme capable of producing the JA required for pollen maturation and release. This conclusion is supported by the recent finding by Schaller et al. (21) that the recombinant Arabidopsis OPR3 protein reduces 9S,13S-OPDA as efficiently as it does the 9R,13R isomer. The previously described OPR1 and OPR2 genes are strongly expressed in at least some flower tissues, as determined by transcript analysis and promoter–β-glucuronidase fusions (20). The fact that 18:3 applied to fad3-2 fad7-2 fad8 flower buds restores fertility apparently without penetrating to the anthers suggests that the failure of OPR1 and OPR2 to maintain fertility in opr3 plants is not likely to be due to insufficient expression in a particular organ or tissue of the flower. The critical determinant may instead be the substrate specificity of the OPR1 and OPR2 gene products. Characterization of the recombinant OPR1 and OPR2 proteins has determined that they show little activity against 9S,13S-OPDA (21), the principal isomer in plants and the isomer believed to be involved in JA synthesis. Instead, the recombinant enzymes reduced mostly other OPDA isomers and thus correspond to the biochemical activity designated OPRI (17). A distinct OPRII activity from C. sempervirens has been separated from OPRI on a chromatofocusing column, and this enzyme reduces 9S,13S-OPDA as well as other isomers (17). Our results are consistent with OPR3 encoding an OPRII activity in anthers and other tissues of Arabidopsis. They also suggest that OPR1 and OPR2 may not be involved in the synthesis of biologically active JA. The physiological function of the additional OPR genes and enzymes remains to be elucidated. While this suggestion is substantially consistent with the biochemical data discussed above, it will require extensive additional studies of the OPR genes and proteins to determine whether this is the case.
OPR3 is a quite distant relative of OPR1 and OPR2, showing 53% identity with both of these proteins (Fig. 4). Two other putative OPR genes are identified in the Arabidopsis database (GenBank accession numbers AAC33200 and ABO10695). The putative coding sequence of ABO10695 (located on P1 clone MDK4 of chromosome 5) predicts a protein somewhat shorter than other OPR isozymes and may represent a pseudogene. Regardless of this possibility, both protein sequences are more closely related to OPR1 and OPR2 (69–73% identity in pairwise comparisons) than to OPR3 (40–50% identity). Because mutation of OPR3 induces male sterility, it can be concluded that these additional genes do not contribute to JA production required for fertility, either because they are not expressed in appropriate floral organs or because they do not encode an enzyme active on 9S,13S-OPDA.
The presence of the SRL peptide sequence at the carboxyl terminus of OPR3 indicates that the protein is probably resident in the peroxisome (36, 37), although this conclusion will require experimental confirmation. There is no such peroxisomal targeting sequence in OPR1, OPR2, or the two additional putative OPR homologs in Arabidopsis. The initial steps in JA synthesis up to the production of OPDA (Fig. 1) are catalyzed by plastid enzymes (38, 39). The cellular location of the subsequent reactions is not known, but the peroxisome is the only known site of β-oxidation in plant cells, and it is assumed that OPDA or 3-oxo-2(2′[Z]-pentenyl)cyclopentane-1-octanoic acid (OPC:8) is transferred from the plastid to the peroxisome. Given the demonstrated importance of OPR3 to JA synthesis in floral tissue and the established sufficiency of SRL as a peroxisomal targeting sequence (36, 37), our data support the suggestion that the peroxisome is the site of final JA synthesis and imply that OPDA is the intermediate transported from the plastid. However, several steps in the biosynthesis of JA require further investigation, and additional evidence for this model of pathway organization is also necessary. Further studies of the opr3 mutant would provide useful information in this respect and help to elucidate the respective roles of OPDA and JA in other oxylipin responses.
Acknowledgments
We are grateful to Steve Fischer and Chris Skidmore for their help with some of the experiments and to John Crock for reviewing part of the manuscript. This work was supported by the United States Department of Energy (Grant DE-FG06-92ER20077) and by the Agricultural Research Center, Washington State University.
Abbreviations
- AGI
Arabidopsis Genome Initiative
- JA
jasmonic acid
- 16:3
hexadecatrienoic acid
- 18:3
linolenic acid
- MeJA
methyl jasmonate
- OPDA
12-oxophytodienoic acid
- OPR
12-oxophytodienoate-10,11-reductase
- RT-PCR
reverse transcription–PCR
- WS
Wassilewskija
Note Added in Proof.
A paper describing some aspects of the opr3 mutant was published recently by Sanders et al. (40).
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AC006413, AAD19764, AAC33200, and AB010695).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.190264497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.190264497
References
- 1.Howe G A, Lightner J, Browse J, Ryan C A. Plant Cell. 1996;8:2067–2077. doi: 10.1105/tpc.8.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McConn M, Creelman R A, Bell E, Mullet J E, Browse J. Proc Natl Acad Sci USA. 1997;94:5473–5477. doi: 10.1073/pnas.94.10.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryan C A. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:651–674. [Google Scholar]
- 4.Xu Y, Chang P-F L, Liu D, Narasimhan M L, Raghothama K G, Hasegawa P M, Bressan R A. Plant Cell. 1994;6:1077–1085. doi: 10.1105/tpc.6.8.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vijayan P, Shockey J, Lévesque C A, Cook R J, Browse J. Proc Natl Acad Sci USA. 1998;95:7209–7214. doi: 10.1073/pnas.95.12.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mason H S, Mullet J E. Plant Cell. 1990;2:569–579. doi: 10.1105/tpc.2.6.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiler E W, Albrecht T, Groth B, Xia Z Q, Luxem M, Liss H, Andert L, Spengler P. Phytochemistry. 1993;32:591–600. [Google Scholar]
- 8.McConn M, Browse J. Plant Cell. 1996;8:403–416. doi: 10.1105/tpc.8.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feys B J F, Benedetti C E, Penfold C N, Turner J G. Plant Cell. 1994;6:751–759. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiler E W, Kutchan T M, Gorba T, Brodschelm W, Niesel U, Bublitz F. FEBS Lett. 1994;345:9–13. doi: 10.1016/0014-5793(94)00411-0. [DOI] [PubMed] [Google Scholar]
- 11.Blechert S, Brodschelm W, Hölder S, Kammerer L, Kutchan T M, Mueller M J, Xia Z-Q, Zenk M H. Proc Natl Acad Sci USA. 1995;92:4099–4105. doi: 10.1073/pnas.92.10.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stelmach B A, Müller A, Hennig P, Laudert D, Andert L, Weiler E W. Phytochemistry. 1998;47:539–546. doi: 10.1016/s0031-9422(97)00547-5. [DOI] [PubMed] [Google Scholar]
- 13.Weber H, Vick B A, Farmer E E. Proc Natl Acad Sci USA. 1997;94:10473–10478. doi: 10.1073/pnas.94.19.10473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reymond P, Weber H, Damond M, Farmer E E. Plant Cell. 2000;12:707–719. doi: 10.1105/tpc.12.5.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vick B A, Zimmerman D C. Plant Physiol. 1986;80:202–205. doi: 10.1104/pp.80.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaller F, Weiler E W. J Biol Chem. 1997;272:28066–28072. doi: 10.1074/jbc.272.44.28066. [DOI] [PubMed] [Google Scholar]
- 17.Schaller F, Hennig P, Weiler E W. Plant Physiol. 1998;118:1345–1351. doi: 10.1104/pp.118.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laudert D, Hennig P, Stelmach B A, Müller A, Andert L, Weiler E W. Anal Biochem. 1997;246:211–217. doi: 10.1006/abio.1997.2012. [DOI] [PubMed] [Google Scholar]
- 19.Vick B A, Zimmerman D C. Plant Physiol. 1984;75:458–461. doi: 10.1104/pp.75.2.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biesgen C, Weiler E W. Planta. 1999;208:155–165. doi: 10.1007/s004250050545. [DOI] [PubMed] [Google Scholar]
- 21.Schaller F, Biesgen C, Müssig C, Altmann T, Weiler E W. Planta. 2000;210:979–984. doi: 10.1007/s004250050706. [DOI] [PubMed] [Google Scholar]
- 22.Müssig, C., Biesgen, C., Lisso, J., Uwer, U., Weiler, E. W. & Altmann, T. (2000) J. Plant Physiol., in press. [DOI] [PubMed]
- 23.Feldmann K A, Marks M D, Christianson M L, Quatrano R S. Science. 1989;243:1351–1354. doi: 10.1126/science.243.4896.1351. [DOI] [PubMed] [Google Scholar]
- 24.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Gleave A P. Plant Mol Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- 26.Heitz T, Geoffroy P, Stintzi A, Fritig B, Legrand M. J Biol Chem. 1993;268:16987–16992. [PubMed] [Google Scholar]
- 27.Smyth D R, Bowman J L, Meyerowitz E M. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clough S J, Bent A F. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 29.Preuss D, Lemieux B, Yen G, Davis R W. Gene Dev. 1993;7:974–985. doi: 10.1101/gad.7.6.974. [DOI] [PubMed] [Google Scholar]
- 30.Behringer F J, Medford J I. Plant Mol Biol Rep. 1992;10:190–194. [Google Scholar]
- 31.Noma K, Ohtsubo E. DNA Res. 2000;7:1–7. doi: 10.1093/dnares/7.1.1. [DOI] [PubMed] [Google Scholar]
- 32.Tinland B. Trends Plant Sci. 1996;1:178–190. [Google Scholar]
- 33.Goldberg R B, Beals T P, Sanders P M. Plant Cell. 1993;5:1217–1229. doi: 10.1105/tpc.5.10.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaul M. Male Sterility in Higher Plants. Berlin: Springer; 1988. [Google Scholar]
- 35.Sanders P M, Bui A Q, Weterings K, McIntire K N, Hsu Y-C, Lee P Y, Truong M T, Beals T P, Goldberg R B. Sex Plant Reprod. 1999;11:297–322. [Google Scholar]
- 36.Hayashi M, Aoki M, Kato A, Kondo M, Nishimura M. Plant J. 1996;10:225–234. doi: 10.1046/j.1365-313x.1996.10020225.x. [DOI] [PubMed] [Google Scholar]
- 37.Mullen R T, Lee M S, Flynn C R, Trelease R N. Plant Physiol. 1997;115:881–889. doi: 10.1104/pp.115.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blée E. Prog Lipid Res. 1998;37:33–72. doi: 10.1016/s0163-7827(98)00004-6. [DOI] [PubMed] [Google Scholar]
- 39.Ziegler J, Stenzel I, Hause B, Maucher H, Hamberg M, Grimm R, Ganal M, Wasternack C. J Biol Chem. 2000;275:19132–19138. doi: 10.1074/jbc.M002133200. [DOI] [PubMed] [Google Scholar]
- 40.Sanders P M, Lee P Y, Biesgen C, Boone J D, Beals T P, Weiler E W, Goldberg R B. Plant Cell. 2000;12:1041–1061. doi: 10.1105/tpc.12.7.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]