Abstract
The activity-dependent transcription factor zif268 is re-activated in sleep following hippocampal long-term potentiation (LTP). However, the activation of secondary genes, possibly involved in modifying local synaptic strengths and ultimately stabilizing memory traces during sleep, has not yet been studied. Here, we investigated changes in hippocampal and cortical gene expression at a time point subsequent to the previously reported initial zif268 re-activation during sleep. Rats underwent unilateral hippocampal LTP and were assigned to SLEEP or AWAKE groups. Eighty minutes after a long rapid-eye-movement sleep (REMS) episode (or an equivalent amount of time for awake group) animals had their hippocampi dissected and processed for gene microarray hybridization. Prefrontal and parietal cortices were also collected for qRT-PCR analysis. The microarray analysis identified 28 up-regulated genes in the hippocampus: 11 genes were enhanced in the LTPed hemisphere of sleep animals; 13 genes were enhanced after sleep, regardless of hemisphere; and 4 genes were enhanced in LTPed hemisphere, regardless of behavioral state. qRT-PCR analysis confirmed the upregulation of aif-1 and sc-65 during sleep. Moreover, we observed a down-regulation of the purinergic receptor, P2Y4R in the LTP hemisphere of awake animals and a trend for the protein kinase, CaMKI to be up-regulated in the LTP hemisphere of sleep animals. In the prefrontal cortex, we showed a significant LTP-dependent down-regulation of gluR1 and spinophilin specifically during sleep. Zif268 was downregulated in sleep regardless of the hemisphere. No changes in gene expression were observed in the parietal cortex. Our findings indicate that a set of synaptic plasticity-related genes have their expression modulated during sleep following LTP, which can reflect biochemical events associated with reshaping of synaptic connections in sleep following learning.
Key Words: Rapid-Eye Movement (REM) sleep, Long-Term Potentiation (LTP), Hippocampus, Prefrontal Cortex (PFC), Gene Expression, Microarray Analysis
1. Introduction
Sleep provides a time in which the nervous system can process acquired information without the interference of sensory inputs. Both slow-wave sleep (SWS) and rapid-eye movement sleep (REMS) have been shown to be important for the consolidation of memories involving different modalities including visual, verbal, emotional and motor [29,47,50,52]. In addition, new experiences and learning modify sleep architecture and promote changes in the neuronal firing patterns, gene expression and oxygen/blood supply in specific brain areas [13,31,36,38,55]. These changes may reflect the re-activation of specific neuronal groups within distinct brain regions required for stabilization or strengthening of synapses previously used during wakefulness.
In general, gene expression is reduced during sleep, when compared to wakefulness [10,15,37]. This observation supported the view that gene expression in sleep was of little relevance for the enhancement of neuronal plasticity. However, studies from our laboratory reported that the activity-dependent transcription factor zif268 (also known as NGFI-A, egr-1, ZENK, Krox-24) is up-regulated during sleep when animals are exposed to an enriched environment or submitted to hippocampal long-term potentiation (LTP) prior to sleep [38,39]. Compared to animals kept in their home cages, enriched environment animals show an increase in zif268 mRNA levels in the hippocampus, amygdala, and cortex, specifically during REMS. Following hippocampal LTP, zif268 up-regulation during REMS involves extra-hippocampal areas. More specifically, in early REMS, zif268 up-regulation is observed in the auditory and entorhinal cortices, whereas in late REMS (at least 2h after sleep onset) it is observed in the temporal, motor and somatosensory cortices, besides the amygdala. Both of these studies analyzed zif268 gene expression 30 minutes after the criteria for wakefulness, SWS or REMS were met.
As a transcription factor, zif268 is involved in the activation or repression of target genes, such as synapsins I and II, activity-regulated cytoskeletal protein (arc), serum-glucocorticoid regulated kinase (SGK) and components of the neuronal proteasome [20,26,51] some of which are involved in synaptic plasticity. It is possible, therefore that zif268 up-regulation during REMS following LTP promotes changes in the expression of specific effector genes important for sleep-dependent synaptic plasticity. In the present study, we used a gene microarray approach to screen for secondary gene expression changes in the hippocampus during REMS that follows hippocampal LTP induction during waking. We also investigated the expression patterns of a selected group of well-known plasticity-related genes in the prefrontal (PFC) and parietal cortex (PC), regions where we had previously observed the extra-hippocampal post-LTP up-regulation of zif268 [39]. Zif-268 mRNA levels peak at about 30 min after a given stimulus or significant behavioral experience [12,32,38]. Zif268 protein has been shown to peak between 1–2h after dentate gyrus LTP [1,2,40]. We, therefore, chose the 80min time proint after REM sleep which should correspond to the peak of a secondary gene mRNA expression.
2. Materials and Methods
2.1 Animals and surgery
Male Sprague-Dawley rats (300–350 g) were housed individually in standard rodent cages in a vivarium maintained at 24°C, and with a 12h/12h light-dark cycle – lights on at 0700h. Food and water were available ad libitum during all phases of the experiment. Eight animals were implanted with chronic stimulating electrodes in the medial perforant path (mPP) and recording electrodes in the dentate gyrus (DG) for the recording of evoked local field potentials (LFP). Briefly, tungsten micro-electrodes (100 μm cross-section diameter) were implanted bilaterally in the DG and mPP of all animals under deep sodium pentobarbital (Nembutal®; 40 mg/kg, i.p., Abbot Labs, IL, USA) anesthesia. The animals were placed in a stereotaxic frame and the skull was exposed and cleaned under aseptic conditions. The electrodes were lowered into the brain through holes made in the skull at the following coordinates: mPP (7.9 mm posterior to the bregma, 4.1 mm lateral to the midline and 3.0 mm ventral to the dura-mater) and DG (3.8 mm posterior to the bregma, 2.1 mm lateral to the midline and 3.3 mm ventral to the dura-mater). The final positions of the electrodes in the DG were determined by audio monitoring of unit firing and recording of evoked responses elicited after test stimulations of the mPP (80 μA, 250 μs, 0.05 Hz). A screw positioned in the frontal bone served as ground and a screw above the PC served as the stimulus/recording indifferent reference. Two additional screws, over the parietal and occipital cortices were used as anchors for the assembly. The electrodes were assembled in a connector, which was cemented to the skull.
All animals were allowed at least 5 days to recover from surgery before the experiments started. On each of the recovery days, they were allowed to have full sleep cycles inside the sleep chamber during the lights-on period. The sleep chamber consisted of a wooden box (in cm; 45W×45L×80H) and was illuminated by a light bulb (2 lux floor light intensity). A small fan fixed to one side of the chamber provided both ventilation and constant low-intensity white noise to muffle external sounds. The chamber was completely enclosed and viewing of the animals was accomplished by means of two one-way mirrors, set up on two of the chamber walls. A recording cable was attached to a commutator on the ceiling in order to allow for free movement of the rats during the electrophysiological recordings. All animals showed habituation to the experimental conditions as revealed by the reduction of exploratory behavior and establishment of regular sleep cycles. All procedures were performed according to NIH guidelines for animal research (Guide for the Care and Use of Laboratory Animals, NRC, 1996) and approved by the IACUC committee at The Rockefeller University. All efforts were made to minimize animal suffering.
2.2 Experimental paradigm
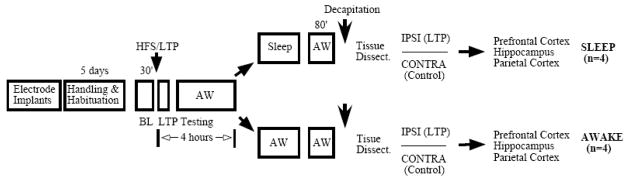
The experimental paradigm is depicted in Figure 1. Briefly, field evoked potentials (fEPSP) elicited in the DG following mPP stimulation were initially recorded in both hemispheres of all animals, during the 2 days preceding the LTP protocol. Monophasic test pulses of 250 μs (mPP-DG) were delivered every 20 s at increasing intensities, 20–300 μA (S88 stimulator, Grass Technologies, RI, USA) while the animals were in a quiet awake behavioral state, characterized by low amplitude/high frequency desynchronized EEG. fEPSP slope and population spike were calculated by averaging four responses per stimulus intensity and then, used to plot input-output (I/O) curves for each brain hemisphere for all animals (PowerLab 8SP and Scope 5.0 software, ADInstruments, CO, USA).
Figure 1. Experimental design.
Stimulating and recording electrodes were implanted bilaterally in the mPP and DG for stimulation and recording of fEPSPs, respectively. After recovery from surgery, the animals underwent 5 days of habituation and handling, during which baseline recordings were performed for both hemispheres. Following this, HFS (2 series of 10 trains at 400 Hz, 5 min apart) was applied to the mPP unilaterally in order to induce LTP in the DG. The contralateral hemisphere was not tetanized. LTP was assessed for 15 min in each hemisphere after HFS. The animals were kept awake for 3 h and were either allowed to sleep (SLEEP group) or kept awake (AWAKE or AW group). After a long REMS episode (>120 s), they were kept awake for an additional 80 min. The AWAKE animals were kept awake for a comparative amount of time. All animals were then decapitated and the prefrontal cortex, hippocampus and parietal cortex were dissected from the LTP and control (ipsi- and contralateral) hemispheres for microarray and qRT-PCR analysis. Trunk blood samples from each animal were also collected for CORT measurements.
On experimental days, the animals were placed in the sleep chamber at approximately 1000h for 10 min and baseline (BL) recordings were taken bilaterally for 30 min. Ipsi- and contra-lateral evoked responses were simultaneously recorded following unilateral stimulation. Test stimulation was set at half-maximum intensity calculated from the fEPSP slope I/O curves and applied every 20 s. LTP was unilaterally induced by applying high-frequency stimulation (HFS) to the mPP. HFS consisted of 2 series of 10 trains (50 ms duration; 20 pulses) at 400 Hz, 5 min apart. This protocol has previously been shown to induce late-phase LTP and zif268 gene re-expression during sleep [39]. The hemisphere to be tetanized was randomized between animals. As a control, the contralateral hemisphere was not stimulated and no LTP was induced. Immediately after LTP, ipsi- and contralateral potentials were monitored in two 30 min sessions (one test pulse every 20 s).
The animals were kept awake for 4 h and then, assigned to two groups – SLEEP and AWAKE (AW). The SLEEP group was allowed to have SWS and REMS while the AWAKE group was kept awake for the same time period, by gentle tapping of the sleeping box. Vigilance states were determined by combining electrophysiological (hippocampal EEG) and behavioral parameters: AW was characterized by exploratory or resting behavior with opened eyes and variable low-amplitude EEG oscillations; SWS was characterized by stillness, eyes closed, and high-amplitude slow wave activity and spindle EEG oscillations; and REM sleep was characterized by stillness and eyes closed with a gradual loss of muscle tonus accompanied by whisker twitches, associated with strong theta-wave EEG oscillations. As the hippocampal EEG undergoes a clear-cut change in the transition from SWS to REM sleep along with muscle atonia and whisker twitches, the combination of the electrophysiology and behavioral assessment was a very reliable method to discriminate vigilance states. Group assignment was further confirmed by offline spectral analysis of EEG oscillations and behavioral data. To monitor LTP decay, evoked responses were recorded immediately after sleep (approximately 1800h). All recordings were performed while the animals were in a quiet awake state, based on hippocampal EEG and the animal’s behavior. Special care was taken to avoid recording evoked responses during drowsiness [as described in 42], due to behavioral-state dependent modulation of hippocampal responses [6,56]. SLEEP animals (n=4) were decapitated 80 min after their first long REM sleep bout (> 120 s duration). AWAKE animals (n=4) were decapitated at times matching those of the SLEEP animals. The reason for keeping the animals awake following REM-sleep was based on our previous findings that LTP induction [39], or exposure to an enriched environment [38], enhanced zif268 expression 30 min later in animals allowed to go to REM sleep, but not SWS or AWAKE. It was hypothesized that genes regulated by zif268 would then appear at the time of sacrifice - since all groups of animals were kept awake during the 80 min interval following sleep, differential gene expression would reflect the behavioral state of each group. The within group design ensured that extraneous variables (e.g., stress) would have minimum effects on gene expression.
The hippocampus of each hemisphere (ipsi- and contralateral to LTP; LTP and control (CTL) hemispheres, respectively) was dissected on ice-cold Petri dish under RNAse-free conditions and individually stored in RNAlater® solution (Ambion, Austin, TX, USA) at −20°C. The PFC and PC were also dissected and stored in RNAlater® solution (Ambion, Austin, TX, USA) at −20°C for posterior qRT-PCR analysis. In addition, blood samples were collected for plasma corticosterone quantification. LTP, sleep time and corticosterone concentration were compared using Welch’s t-test with significance level set to p<0.05.
2.3 RNA isolation and microarray hybridization
Sixteen samples were processed for microarray screening: control and LTP hippocampi from all animals in the sleep and awake groups (4 animals per group). Each sample was hybridized on individual gene chips. Total RNA was extracted by the guanidine thiocyanate method using the Trizol® protocol (Invitrogen, Carlsbad, CA, USA), which involved extraction by phase separation and alcohol precipitation. The RNA pellet was re-suspended in DEPC-treated water and its concentration and quality were determined by optical densitometry, gel electrophoresis and subsequent array parameters. Microarray samples were prepared according to the Affymetrix protocol. In brief, 15 μg of total RNA was reverse-transcribed using SuperScript II (Life Technologies, Rockville, MD) and T7-oligo(dT)24 primer (Geneset, La Jolla, CA, USA). After second-strand DNA synthesis, biotinylated complementary RNA (cRNA) was in vitro transcribed using the T7 RNA polymerase promoter site (Enzo BioArray High Yield RNA Transcript labeling Kit; Affymetrix and Enzo Diagnostics, Farmingdale, NY, USA). Biotin-labeled cRNA was purified, fragmented (in 100 mM MES pH 7.4/1 M NaCl/20 mM EDTA/0.01% Tween 20), and hybridized on RU34A rat oligonucleotide microarrays using standard protocols with the Affymetrix oven and fluidics station. Hippocampal cRNA samples from LTP and control hemispheres were individually hybridized on separate gene chips (16 gene chips: sleep, n=8 samples; awake, n=8 samples). Approximately 8799 annotated genes including expressed sequence tag (EST) clusters were represented in the RU34A microarray. Microarray quality control parameters were as follows: noise (RawQ) less than 5, background signal less than 100, actin and GAPDH 3′/5′ signal ratio less than 3, and consistent detection of bioB and bioC hybridization spiked controls. Probeset signal intensities were calculated with the robust multi-array average (RMA) algorithm [19]. Group (SLEEP vs. AWAKE), Treatment (LTP vs. control) and Interaction (Group vs. Treatment) effects were calculated by multiple-test one-way ANOVA (MANOVA). In order to select the most promising candidate genes whose expression patterns changed during sleep, we used a combination of objective criteria: 1) probeset p-calls per group ≥ 2: hybridization signal for each probe set had to be detected in at least 2 out of 4 chips hybridized for each experimental condition; 2) two-way ANOVA p-values < 0.01; 3) consistency among biological replicates: probe sets with the lowest variance among samples from the same experimental condition; 4) significantly high expression level changes (fold change) compared to control conditions (SLEEP vs. AWAKE and, LTP vs. control hemisphere).
2.4 Quantitative Real-Time PCR (qRT-PCR)
To confirm the microarray data, we used quantitative real-time PCR (qRT-PCR) to quantify mRNA levels in the same hippocampal samples used in the microarray analysis. We also analyzed gene expression changes in PFC and PC samples obtained from the same animals. PFC and PC cDNAs were prepared from total RNA extracted by the guanidine thiocyanate method followed by DNAse treatment using the standard Affymetrix reverse transcription protocol with SuperScript II (Life Technologies, Rockville, MD, USA) and T7-oligo(dT)24 primer (Genset, San Diego, CA, USA). Small PCR products (100–200 base-pairs) were amplified in quadruplets using an Opticon real-time PCR machine (MJ Research, Waltham, MA, USA) and SYBR green dye. Primers for 20 genes were designed using Primer 3 software [43]. We used universal PCR conditions: 65°C to 95°C touchdown cycle followed by 35 cycles [15 s at 95°C, 10 s at 59°C and 10 s at 72°C] to amplify 150 pg of cDNA in 20 μl reactions (0.3X Sybr-green, 3 mM MgCl2, 200 μM dNTPs, 200 μM primers, 0.5 unit Platinum Taq DNA polymerase from Invitrogen, Carlsbad, CA). Experimental and control samples were run on the same 96-well plates. Dissociation curves were calculated for each target gene at the end of each qRT-PCR run and all primers showed a single peak in the dissociation curve. ΔΔCt method was used to calculate experimental vs. control gene expression differences.
To analyze selected plasticity-related genes in cortical and hippocampal samples, small PCR products (100–200 base-pairs) were amplified in triplicates on an ABI Prism 7000 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) using the manufacturer’s SYBR green master mix solution. Primer-dimers were assessed by amplification in the absence of cDNA. Primers were retained if no primer-dimers or non-specific signals occurred before the 35th cycle, and primers amplified with efficiency higher than 85%. The amplification rate and efficiency of each gene were compared to those of β-actin through standard calibration curves (6 serial dilutions; 20–1000 pg) using rat brain cDNA and blank controls. Only genes whose primers amplified at the same rate as the reference gene (β-actin) were analyzed. The HPRT gene was also tested as reference and showed the same rate of amplification as β-actin. The standard qRT-PCR conditions used were as follows: initial activation step, 10 min at 95°C followed by 40 cycles [denaturation: 15 s at 94°C; annealing: 20–30 s at 60°C; extension: 10–30 s at 72°C] to amplify 200 pg of cDNA in 21 μl reactions (10 μl of 2x SYBR Green PCR Master Mix, 5 μl of nuclease-free water, 2.5 μl of forward primer (100 μM), 2.5 μl of reverse primer (100 μM), and 1 μl of cDNA). Experimental and control samples were run on the same 96-well plates. All primers showed a single peak in the dissociation curve. The ΔΔCt method was used to calculate experimental vs. control gene expression differences.
Table 1 shows the primers designed for gluR1, gluR2, spinophilin, BDNF, L-VGCC, zif268, NR2B, HPRT and β-actin using Primer3 software. The results were analyzed using Prism 7000 SDS software (Applied Biosystems). Gene expression changes were calculated as the difference in amplification cycles between experimental and biological control samples after normalizing to rat β-actin. We used the formula %change = 2−Δ ΔCt, where ΔΔCt=(Ctgene-Ctactin) experimental group/(Ctgene-Ctactin) control group. Ct is the measured qRT-PCR cycle for a specific gene. The final results were analyzed by two-way ANOVA for repeated measures followed by Bonferroni post-hoc test and Welch’s t-test when appropriate. Two-way ANOVA was performed on LTP hemisphere measurements (LTP vs. control) as repeated measures and behavioral state measurements (SLEEP vs. AWAKE) as independent measures. The significance level was set to p<0.05.
Table 1. Primers used in the qRT-PCR of hippocampus, prefrontal and parietal cortex samples.
Genes: zif268; BDNF; gluR1; L-VGCC; spinophilin; gluR2; NR2B.
| Gene Name | ID | Forward Primer | Size | Reverse Primer | Size | Product Size |
|---|---|---|---|---|---|---|
| zif268 | M18416 | gagcatgtgtcagagtgttgt | 21 | cacacaaaggcaccaaga | 18 | 174 |
| Spinophilin | AF016252 | agaccaaactgccagacttt | 20 | aatgcaacagacagcacaat | 20 | 186 |
| BDNF | D10938 | cagtattagcgagtgggtca | 20 | gggattacacttggtctcgt | 20 | 139 |
| Glu R1 | X17184 | gtttgttcggaccacagag | 19 | ggctttcgttgctcaatatac | 21 | 108 |
| Glu R2 | M85035 | gactccagaatttcccaaag | 20 | aggcactcagaaggttccta | 20 | 112 |
| NMDA-NR2B | NM012574 | atctgtccaccattcctgtt | 20 | gaacaaagagagcagcatca | 20 | 161 |
| L-typeVGCC | NM012517 | tcagtttagataaggcggtgt | 21 | acacacaagacgacccacta | 20 | 134 |
| HPRT | M86443 | ggccagtaaagaactagcaga | 21 | cctacaggctcatagtgcaa | 20 | 146 |
| β-act in | XM577559 | agccttccttcctgggta | 18 | cactgtgttggcatagaggt | 20 | 107 |
2.5 Corticosterone assay
To determine the level of stress induced during the experimental paradigm, plasma corticosterone (CORT) concentration was determined in both sleep and awake animals from blood samples collected at the time of the decapitation. The samples were initially stored at −20°C and corticosterone concentrations were determined by radioimmunoassay using a commercial kit (Coat-a-Count; Diagnostic Products Co., Los Angeles, CA, USA) and its standard protocol.
3. Results
3.1 LTP, sleep patterns and CORT levels
LTP induction levels were similar between SLEEP and AWAKE groups when evaluated by population spike (427.8 ± 143.3% and 307.8 ± 106.0%, for SLEEP and AWAKE groups, respectively; t(3)=0.67, p=0.27) and fEPSP slope (18.4 ± 4.6% and 12.7 ± 4.9% for SLEEP and AWAKE groups, respectively; t(3)=0.86, p=0.23). The hippocampus in the control hemisphere did not show significant changes in the evoked responses following tetanization of the experimental hemisphere, as measured either by the fEPSP slope or the population spike (data not shown). Following LTP induction, animals in the SLEEP group had on-average 3–4 min of SWS followed by at least 2 min of REMS, whereas AWAKE animals were kept fully awake to match the total time (Fig. 1). Plasma CORT levels from blood samples collected immediately after decapitation (which was performed early in the evening), did not differ between SLEEP (143.5 ± 36.3 ng/ml) and AWAKE groups (139.1 ± 34.1 ng/ml; t(5)=0.09, p=0.93). These levels were in the range observed in non-stressed animals at their daily peak time early in the evening (100–200 ng/ml) [44].
3.2 Microarray analysis of hippocampal gene expression
A total of 28 genes were observed to be up-regulated in the hippocampus and were selected from the gene microarray results for subsequent analysis. Table 2 shows a list of 11 genes whose expression level ratios in the LTP over control hemispheres were significantly higher in the SLEEP, as compared to the AWAKE group (“sleep-LTP genes”). These were genes identified by MANOVA analysis, where the interactions between group and treatment were significant. They could be grouped as: transcription factors (1); signal transduction proteins (4); neurotransmitter receptors (2); lipoproteins (1); cell adhesion molecules (1); and metabolism proteins (2). Table 3 shows a list of 13 genes selected for being specifically over expressed during sleep, regardless of hemisphere (“sleep genes”; Table 3A) and, 4 genes over expressed in the LTP hemisphere, regardless of the behavioral state (“LTP genes”; Table 3B). Sleep genes were grouped as: transcription factors (3); signal transduction proteins (1); membrane receptors (1); cell adhesion molecules (1); metabolism proteins (2); transport proteins (1); injury response molecules (2); chromosomal proteins (1); and synaptic proteins (1). Two of them were detected with two different probe sets. LTP genes were grouped as: growth factors (1); metabolism proteins (1); translation factors (1); and cell cycle proteins (1). Based on our selection criteria (see Material and Methods), we did not detect any down-regulated genes in the hippocampus.
Table 2. Microarray genes for LTP-dependent up-regulation during sleep in the hippocampus.
P-calls, number of microarrays in which the gene reached detection level, from a total of 4 arrays per group: awake-control (I), awake-LTP (II), sleep-control (III) and sleep-LTP (IV). P-value, ANOVA p-values for group vs. treatment interaction.
| Accession Number | Gene Name | P-calls | Fold Change | P-value | ||||
|---|---|---|---|---|---|---|---|---|
| Transcription Factor | I | II | III | IV | AW (LIP/−) | Sleep (LTP/−) | ||
| L26292 | Kruppel-like factor 4(KIf-4) | 4 | 4 | 4 | 4 | 0.98±0.004 | 1.09±0.008 | 0.00002 |
| Signal Transduction | ||||||||
| D86557 | Calcium-Calmodulin dependent protein Kinase I (CaMK-I) | 4 | 4 | 4 | 4 | 0.96±0.018 | 1.16±0.033 | 0.00124 |
| U13396 | Jannus Tyrosine Kinase 2 (JAK-2) | 4 | 4 | 4 | 4 | 1.00±0.012 | 1.14±0.018 | 0.00055 |
| U97146 | Calcium-Independent phospholipase A2 (PLA-2) | 4 | 4 | 4 | 4 | 0.88±0.022 | 1.14±0.076 | 0.00853 |
| D85435 | Protein kinase C Delta-Bindig Protein | 3 | 3 | 2 | 4 | 0.98±0.014 | 1.10±0.019 | 0.00161 |
| Neurotransmitter Receptor | ||||||||
| Y14706 | Metabotropic Purinergic Receptor (P2Y4) | 2 | 4 | 4 | 3 | 0.97±0.012 | 1.12± 0.027 | 0.00218 |
| U01227 | Serotonin Receptor (5HT-3) | 4 | 4 | 4 | 4 | 0.97±0.025 | 1.09±0.012 | 0.00490 |
| Lipoprotein | ||||||||
| J02596 | Apolipoprotein C-III | 2 | 3 | 2 | 2 | 0.96±0.009 | 1.08±0.016 | 0.00091 |
| Cell Adhesion | ||||||||
| L00686 | Carcinoembryonic Antigen-Related protein | 4 | 3 | 4 | 3 | 1.01±0.025 | 1.16±0.026 | 0.00571 |
| Metabolism | ||||||||
| S68245 | Carbonic Anhydrase IV | 4 | 3 | 4 | 3 | 0.98±0.025 | 1.10±0.017 | 0.00803 |
| E07296 | N-Acetylglucosamine Transferase-I | 4 | 4 | 4 | 4 | 0.95±0.026 | 1.09±0.022 | 0.00653 |
Table 3. (A) Microarray genes for sleep-dependent up-regulation in the hippocampus. (B) Microarray genes for LTP-dependent up-regulation in the hippocampus.
P-calls, number of microarrays in which the gene reached detection level from a total of 4 arrays per group: awake-control (I), awake-LTP (II), sleep-control (III) and sleep-LTP (IV). P-value, ANOVA p-values for group vs. treatment interaction.
| A | |||||||
|---|---|---|---|---|---|---|---|
| Accession Number | Gene Name | P-calls | Fold Change | P- value | |||
| Transcription Factor | I | II | III | IV | Sleep/AW | ||
| AI176662 | Early Growth Response 1 (egr-1) or zif 268 | 2 | 2 | 4 | 4 | 1.36 | 0.0012 |
| M18416 | Early Growth Response 1 (egr-1) or zif 268 | 4 | 4 | 4 | 4 | 1.19 | 0.0075 |
| AF054826 | Vesicle-Associated Protein (VAMP-5) | 3 | 2 | 4 | 4 | 1.11 | 0.0072 |
| J03179 | D-site Albumin Promoter Binding Protein (DBP) | 4 | 4 | 4 | 4 | 1.10 | 0.0003 |
| Signal Transduction | |||||||
| U17901 | Phospholipase A2-Activating Protein | 4 | 4 | 4 | 4 | 1.10 | 0.0048 |
| Membrane Receptor | |||||||
| M55291 | Neurotrophin Receptor (TrkB) | 4 | 4 | 4 | 4 | 1.08 | 0.0052 |
| Cell Adhesion | |||||||
| M84488 | Vascular Cell Adhesion Molecule 1 (vCAM-1) | 4 | 4 | 4 | 4 | 1.13 | 0.0022 |
| Metabolism | |||||||
| D00569 | 2,4-dienoyl-CoA reductase (EC 1.3.1.34) | 4 | 4 | 4 | 4 | 1.10 | 0.0064 |
| AI012589 | Glutathione S-Transffrase, pi 2 | 4 | 4 | 4 | 4 | 1.10 | 0.0050 |
| Transport | |||||||
| AA957777 | Nuclear Transport Factor 2 (Nutf-2) | 4 | 4 | 4 | 4 | 1.12 | 0.0031 |
| Injury Response | |||||||
| U17919 | Allograft Inflammatory Factor 1 (AIF-1) | 4 | 4 | 4 | 4 | 1.27 | 0.0014 |
| U10894 | Allograft Inflammatory factor 1 (AIF-1) | 4 | 4 | 4 | 4 | 1.24 | 0.0037 |
| U07619 | Thromboplastin | 4 | 4 | 4 | 4 | 1.15 | 0.0068 |
| Cromosomal Protein | |||||||
| X65454 | Synaptonemal Complex Protein (SC-65) | 4 | 4 | 4 | 4 | 1.15 | 0.0064 |
| Synaptic Proteins | |||||||
| AFO52596 | Synaptosomal-Associated Protein (SNAP-23) | 4 | 4 | 4 | 4 | 1.10 | 0.0099 |
|
B | |||||||
| Accession Number | Gene Name | P-calls | Fold Change | P-value | |||
|
| |||||||
| Growth Factor | I | II | III | IV | LTP/− | ||
| M32167 | Vascular Endothelial Growth Factor A (vEGF) | 4 | 4 | 4 | 4 | 1.14 | 0.0084 |
| Metabolism | |||||||
| AA799889 | Cyctophytin homologue | 4 | 4 | 4 | 4 | 1.12 | 0.0011 |
| Translation Factor | |||||||
| AF096835 | eIF-2 Kinase (PEK) alpha-subunit | 3 | 4 | 4 | 3 | 1.12 | 0.0030 |
| Cell Cycle | |||||||
| D14013 | CycHn C | 2 | 2 | 2 | 3 | 1.17 | 0.0095 |
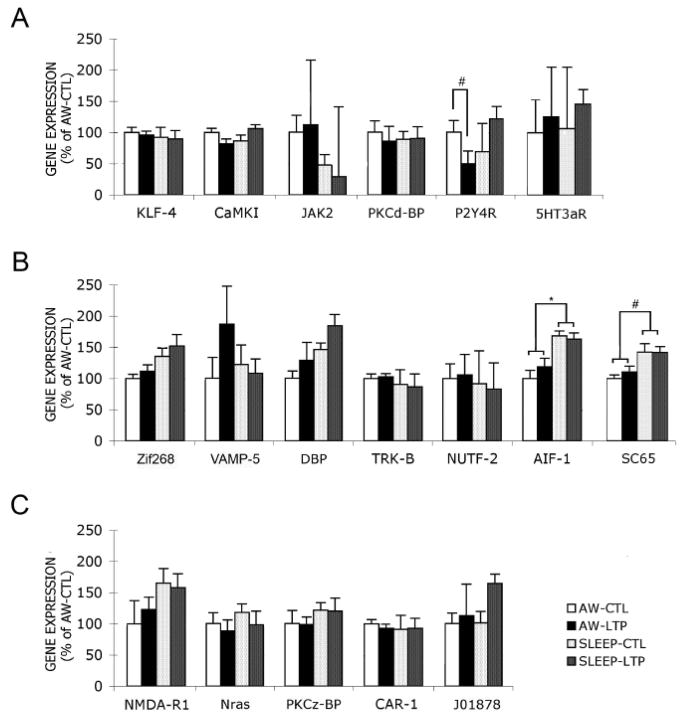
We used qRT-PCR to confirm the observed differences in the microarray analysis. The reactions were run on the same 16 hippocampal DNA samples used in the microarray hybridizations (LTP and control hippocampi from each of 8 animals: 4 in the awake and 4 in the sleep group). The candidate genes tested comprised 6 sleep-LTP genes, 7 sleep genes and 5 ‘other’ genes, selected based on their involvement in synaptic plasticity. The results are shown in figure 2. Among sleep-LTP genes, CaMKI and P2Y4R showed significant interaction effects between LTP and behavioral state (F1,6=6.79, p<0.05 and F1,6=5.92, p<0.05, respectively). Paired t-tests were then applied to detect specific differences. CaMKI showed a trend towards lower expression in the LTP hemisphere of AWAKE animals compared to the control hemisphere (t(3)=1.92, p=0.08) and to be upregulated in the LTP hemisphere of SLEEP animals, compared to its control hemisphere (t(3)=1.77, p=0.09). On the other hand, P2Y4R did not show inter-hemispheric differences in SLEEP animals but was significantly less expressed in the LTP hemisphere of AWAKE animals (t(3)=4.79, p<0.01; −50%). Two out of 7 sleep genes were confirmed to be upregulated during sleep regardless of hemisphere: aif-1 (F1,6=10.96, p<0.02; 50%) and sc-65 (F1,6=18.19, p<0.01; 35%). In addition, the transcription factors zif268 and dbp showed a trend to be upregulated during sleep (F1,6=3.93, p=0.09; 36% and F1,6=4.04, p=0.09; 44%, respectively), as also observed in the microarray analysis. From the other 5 selected genes, the brain-specific marker J01878 showed similar results when comparing microarray and qRT-PCR analysis. Although not significant, J01878 showed a trend towards up-regulation in the LTP hemisphere during sleep (qRT-PCR, 62%; microarray, 14%).
Figure 2. QRT-PCR analysis of hippocampal gene expression.
A, Sleep-LTP interaction. CaMKI and P2Y4R showed a significant interaction between LTP and behavioral state (F1,6=6.79 p<0.05 and F1,6=5.92 p<0.05, respectively). CaMKI showed a trend towards lower expression in the LTP hemisphere of AWAKE animals and to be upregulated in the LTP hemisphere of SLEEP animals, compared to the control hemisphere. P2Y4R was significantly less expressed in the LTP hemisphere of AWAKE animals (50%), although it did not change in SLEEP animals. B, Sleep effect. Aif-1 and sc-65 were confirmed to be upregulated during sleep: aif-1 (F1,6=10.96 p<0.02; 50%) and sc-65 (F1,6=18.19 p<0.01; 35%). In addition, the transcription factors, zif268 and dbp showed a trend towards upregulation during sleep (F1,6=3.93 p=0.09; 36% and F1,6=4.04 p=0.09; 44%, respectively) (supporting the microarray results). C, Other - genes selected based on functional relevance or expression patterns, which did not meet microarray screening criteria (see Material and Methods). J01878 showed a trend towards up-regulation in the LTP hemisphere in SLEEP group (62%). Data represent mean (± SEM) percent change, compared to AWAKE-Control samples, following β-actin normalization. Two-way ANOVA and Welch t-test: #, p<0.01; *, p<0.05.
3.3 Gene expression changes in the prefrontal and parietal cortex during sleep
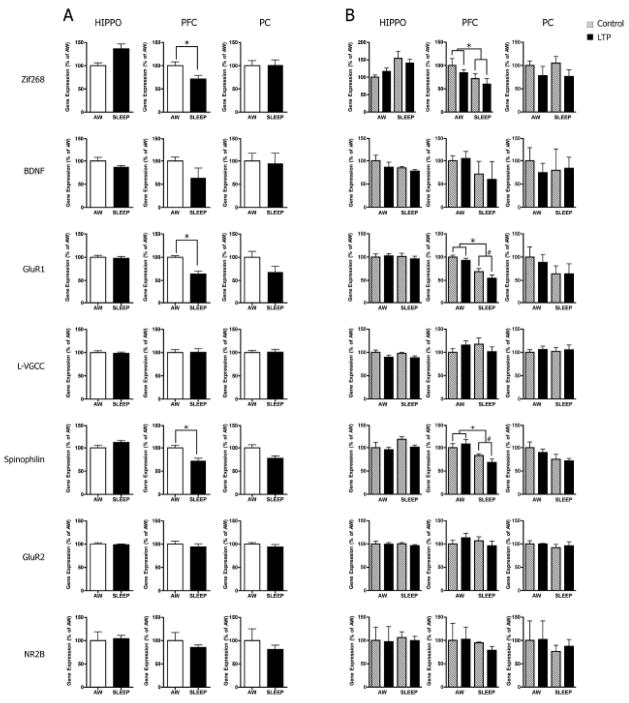
To extend our qRT-PCR analysis of gene expression in sleep to extra-hippocampal areas, we investigated two regions of the neocortex - the PFC and PC. We used cDNA samples from the same eight animals (sixteen samples) from the microarray experiment and analyzed 7 selected genes known to be involved in synaptic plasticity both in the hippocampus and neocortex: zif268; BDNF; gluR1; gluR2; NR2B; L-VGCC; and spinophilin [3,11,21,23,28,49]. We detected a number of significant changes in gene expression in the PFC that were not present either in the hippocampus or in the PC (Fig. 3). Specifically, we observed significant group effects for zif268, gluR1 and spinophilin (AWAKE vs. SLEEP; zif268: F1,5= 7.16, p<0.05; gluR1: F1,5= 43.49, p=0.001 and spinophilin: F1,5= 22.34, p<0.01), reflecting their down-regulation in SLEEP animals, regardless of hemisphere (t(11)=3.12, p<0.01, −40%; t(8)=4.72, p<0.0001, −61% and t(11)=4.02, p=0.001, −21%, respectively - Fig. 3A). Moreover, we observed that gluR1 and spinophilin were down-regulated during sleep specifically in the LTP hemisphere (t(3)=3.84, p<0.05, −25% and t(3)=3.72, p<0.05, −21%, respectively), suggesting LTP-dependent modulation of gene expression in sleep in the PFC (Fig. 3B). Although not significant, there was a trend for zif268 to be down-regulated in the PFC during sleep in the LTP hemisphere (t(3)=1.88, p=0.08). AWAKE animals did not show any inter-hemispheric differences either in gluR1 or in spinophilin expression (t(2)=0.72, p>0.05 and t(2)=0.03, p>0.05, respectively.
Figure 3. QRT-PCR analysis of gene expression in the hippocampus, prefrontal cortex (PFC) and parietal cortex (PC) of AWAKE (n=4) and REMS (n=4) animals following unilateral hippocampal LTP.
A, Sleep effect. In the hippocampus, the qRT-PCR results were consistent with the microarray findings. We did not observe significant group effects for any of the 6 genes analyzed. Zif268 showed a non-significant up-regulation trend during sleep in the hippocampus. In the PFC, sleep induced down-regulation of zif268 (−40%), gluR1 (−61%) and spinophilin (−21%). The parietal cortex showed a non-significant trend for gluR1 (−47%) to be down-regulated during sleep. B, Sleep-LTP effect. In the hippocampus, we did not observe any significant gene expression changes during sleep in the LTP hemisphere. In the PFC, however, gluR1 (−25%) and spinophilin (−21%) were specifically down-regulated in the LTP hemisphere during sleep. No significant gene expression changes were observed in the PC. Data are presented as the mean (± SEM) ΔCt after β-actin normalization. Two-way ANOVA and Welch t-test: #, p<0.01; *, p<0.05.
In the hippocampus, our findings were consistent with the microarray results for all genes analyzed. We did not observe any significant effects for zif268, BDNF, gluR1, L-VGCC, spinophilin, gluR2 and NR2B. In the PC, we also did not observe any significant changes in gene expression either due to sleep, LTP or to the interaction between sleep and LTP. However, we observed a trend in spinophilin expression towards down-regulation in SLEEP animals regardless of the LTP hemisphere (F1,6= 4.98, p=0.07; AWAKE vs. SLEEP, t(13)=3.02, p<0.01).
4. Discussion
Gene expression during sleep may be an important molecular mechanism to reshape local synaptic connections activated during learning. For example, a recent study by Datta, et al. [13] reported that two-way active avoidance training activated the pontine-wave generator during REM sleep and enhanced expression of a number of genes, including pCREB, arc, BDNF and Egr-1 in the dorsal hippocampus and amygdala. We have also reported sleep-dependent re-expression of zif268 in the entorhinal and temporal cortex of rats subjected to unilateral hippocampal LTP [39]. In that study, zif268 mRNA levels were analyzed in brain tissues obtained 30 min after the offset of SWS or REM sleep that followed LTP induction. Considering that zif268 is a transcription factor and its expression in sleep could affect the expression of a variety of downstream target genes, our present study was aimed at determining LTP-dependent changes in gene expression in brain areas 80 min after sleep offset. It is relevant to also state that we employed a within subject design in which we compared LTPed vs. control hemispheres in the same animal, thus minimizing effects of behavioral state, stress levels, etc. Furthermore, we used separate gene chips for each sample (i.e., LTPed/control hemispheres from each animal – tissue were not pooled) and adhered to strict analysis criteria for the selection of active genes. Our main findings are: (1) the microarray analysis identified 28 genes upregulated in the hippocampus - 11 sleep-LTP genes, 13 sleep genes and 4 LTP genes. Among sleep-LTP genes, qRT-PCR analysis showed up-regulation of CaMKI in the LTP hemisphere of SLEEP animals and, a down-regulation of P2Y4R in the LTP hemisphere of AWAKE animals. We also observed the up-regulation of J01878, which was not in the original selection from the microarray analysis. Among the thirteen sleep genes, qRT-PCR analysis confirmed the up-regulation of aif-1 and sc-65. It also showed an up-regulation trend for zif268 and dbp, similar to that observed in the microarray analysis. LTP genes were not tested in the qRT-PCR; (2) in the PFC, qRT-PCR analysis revealed that zif268, gluR1 and spinophilin are down-regulated during sleep, while gluR1 and spinophilin are specifically down-regulated in the LTP hemisphere during sleep and, (3) in the PC, we only observed a trend for spinophilin to be down-regulated during sleep.
The fact that enriched environment experience and hippocampal LTP induce zif268 re-expression during subsequent REMS [38,39], raised the question of what other genes could be affected during sleep following learning and how they could contribute to memory consolidation in sleep. The data on J01878 and CaMKI indicate up-regulation of these genes in the potentiated hippocampus during sleep. Although of unknown function, J01878 may be targeted to dendrites as it contains a dendritic targeting element (DTE) typically seen in the 3′ UTR of mRNAs of dendritic localization. The DTE in the 3′ UTR of J01878 shares 94% identity with a DTE present in the translational regulator BC1 [53], which is translocated to dendrites upon synaptic activity [33]. CaMKI is an important mediator of activity-dependent dendritic remodeling and Ca2+ signaling in ERK-dependent LTP induction [45,48,54]. Its up-regulation indicates that it is also part of the re-activation response during sleep that follows relevant brain activation. The sleep-dependent up-regulation of aif-1 and sc-65 suggests a role for sleep in brain immunity and cell proliferation in the hippocampus, since these proteins are expressed during microglial activation and cell division, respectively [8,46]. Moreover, the up-regulation trend of the transcription factor dbp is consistent with previous studies reporting higher levels during sleep compared to wakefulness [9,27] and supports findings suggesting its role in sleep dependent memory consolidation and synaptic plasticity [22,41]. Similar to previous studies, we also observed an up-regulation of zif268 during sleep. However, in our study, zif268 was up-regulated in the hippocampus both in the LTP and control hemispheres of sleep animals, in contrast to the LTP-dependent re-expression of zif268 previously observed in the cortex and amygdala [39]. One possible explanation for this might be that zif268 expression in the hippocampus occurs at a later time frame, since we analyzed brain tissues collected 80 min after sleep criteria instead of 30 min used in the earlier study. Given that zif268 is an activity-dependent transcription factor required for long-term memory and late-LTP maintenance [5,16,17,21], it could synergistically function with dbp controlling activity-dependent gene expression throughout the sleep-wake cycle. Their expression in both potentiated and non potentiated hemispheres could play permissive roles on synapses that were previously activated.
The fact that we could detect hippocampal up-regulation of several genes, including J01878, CaMKI, aif-1, sc-65, dbp and zif268 80 min after waking up the animals, strongly suggests that sleep has a pronounced and lasting modulatory effect on hippocampal gene expression. However, the number of sleep-LTP genes we described is probably greatly underestimated, due to limitations of the microarray methodology such as compression effects (i.e., the fold-induction detected on the arrays being less than the actual induction values in the brain). In addition, the tissue dissection procedure used may have diluted higher expression differences present in specific hippocampal subfields. Finally, other genes might have been expressed at different temporal windows.
Recent findings indicate that the PFC is also sensitive to sleep deprivation and that its function can be modulated by sleep [34,57]. In contrast to the hippocampus, where neuronal reactivation and gene re-expression seems to occur in sleep, our data suggest that the PFC decreases its activity possibly in preparation for proper functioning upon wakening. We observed a general down-regulation of zif268, gluR1 and spinophilin during sleep as compared to the awake state. Such down-regulation could be analogous to the human frontal cortex deactivation observed at the onset of sleep, in which the cortex shifts to a hypometabolic state. Several studies have shown that the human dorsolateral PFC has its metabolism deeply reduced at the initiation of SWS and maintains a relative deactivation during REMS [18,30,35]. The PFC gene down-regulation observed in our study supports the idea of a strong need for synaptic or biochemical quiescence after its intense activity during wakefulness. Another important result was that gluR1 and spinophilin expression in the PFC were modulated in an LTP-dependent manner during sleep. GluR1 and spinophilin are important components of dendritic spines, which are major sites of excitatory synapses undergoing continuous activity-dependent remodeling [4,24,25,49]. Their modulation during sleep could contribute to synaptic mechanisms of plasticity in the PFC.
Human fMRI studies have shown that sleep deprivation reverses the PFC deactivation observed during sleep, and is correlated with memory impairment [7,14]. In addition, short-term REMS deprivation delays the normal decay of PFC LTP in freely moving rats, possibly precluding further PFC plasticity [42]. In contrast to the PFC, our study showed that the PC had a more limited pattern of gene expression changes following sleep. That included only a trend to down-regulation of spinophilin during sleep, suggesting a functional dissociation between cortical areas.
In conclusion, our study has identified a set of genes whose expression levels are altered in the hippocampus and PFC at 80 min after sleep offset. Their functions include activity-dependent synaptic plasticity and memory, as well as transcriptional control, homeostatic control of sleep, immunity and cell proliferation. Distinct gene expression patterns were seen in the hippocampus, PFC and PC, suggesting their functional dissociation during sleep. Future studies should focus on the implications of such molecular events during sleep that follows learning, including brain region specificity and temporal activation patterns.
Acknowledgments
The authors wish to thank Benjamin Lee and Christie McPherson for their technical assistance. We also like to thank Tomoko Soga and Aaron Jasnow for helpful discussions on the experiments. Supported by NHLBI grant HL69699 to CP.
Abbreviations
- 5HT3aR
Serotonin receptor subtype 3a
- aif-1
Allograft inflammatory factor 1
- BDNF
Brain-derived neurotrophic factor
- CaMKI
Ca2+-calmodulin dependent kinase 1
- Car-1
Cell adhesion regulator 1
- CORT
Corticosterone
- dbp
Albumin D-binding protein
- EST
Expressed sequence tag
- fEPSP
Field excitatory postsynaptic potential
- gluR12
AMPA-type glutamate receptor subunit 1, 2
- HFS
High frequency stimulation
- HPRT
Hypoxanthine-guanine phosphoribosyl transferase
- IEG
Immediate early genes
- J01878
Brain-specific sequence identifier
- JAK2
Jannus kinase 2
- klf-4
Kruppel-like factor 4
- L-VGCC
L-type voltage-gated calcium channel
- LTP
Long-term potentiation
- NR2B
NMDA glutamate receptor subunit 2B
- NMDA-R1
NMDA glutamate receptor subunit 1
- Nras
Neuronal ras protein
- nutf-2
Nuclear transport factor 2
- P2Y4R
Metabotropic purine receptor
- PC
Parietal cortex
- PFC
Prefrontal cortex
- PKCd-BP
Protein kinase c delta binding protein
- PKCz-BP
Protein kinase c zeta binding protein
- PS
Population spike
- qRT-PCR
Quantitative real-time polymerase chain reaction
- REMS
Rapid eye movement sleep
- sc-65
Synaptonemal complex 65
- SWS
Slow wave sleep
- Trk-B
Tyrosine kinase (BDNF) receptor
- VAMP-5
Vesicular-associated protein 5
- zif268
Nerve growth factor induced gene A (also NGFI-A, egr-1, krox-24, ZENK)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abraham WC, Dragunow M, Tate WP. The role of immediate early genes in the stabilization of long-term potentiation. Mol Neurobiol. 1991;5:297–314. doi: 10.1007/BF02935553. [DOI] [PubMed] [Google Scholar]
- 2.Abraham WC, Mason SE, Demmer J, Williams JM, Richardson CL, Tate WP, Lawlor PA, Dragunow M. Correlations between immediate early gene induction and the persistence of long-term potentiation. Neuroscience. 1993;56:717–727. doi: 10.1016/0306-4522(93)90369-q. [DOI] [PubMed] [Google Scholar]
- 3.Bauer EP, Schafe GE, LeDoux JE. NMDA receptors and L-type voltage-gated calcium channels contribute to long-term potentiation and different components of fear memory formation in the lateral amygdala. J Neurosci. 2002;22:5239–49. doi: 10.1523/JNEUROSCI.22-12-05239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boehm J, Malinow R. AMPA receptor phosphorylation during synaptic plasticity. Biochem Soc Trans. 2005;33:1354–6. doi: 10.1042/BST0331354. [DOI] [PubMed] [Google Scholar]
- 5.Bozon B, Davis S, Laroche S. A requirement for the immediate early gene zif268 in reconsolidation of recognition memory after retrieval. Neuron. 2003;40:695–701. doi: 10.1016/s0896-6273(03)00674-3. [DOI] [PubMed] [Google Scholar]
- 6.Bramham CR, Srebro B. Synaptic plasticity in the hippocampus is modulated by behavioral state. Brain Res. 1989;493:74–86. doi: 10.1016/0006-8993(89)91001-9. [DOI] [PubMed] [Google Scholar]
- 7.Chee MW, Choo WC. Functional imaging of working memory after 24 hr of total sleep deprivation. J Neurosci. 2004;24:4560–7. doi: 10.1523/JNEUROSCI.0007-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Q, Pearlman RE, Moens PB. Isolation and characterization of a cDNA encoding a synaptonemal complex protein. Biochem Cell Biol. 1992;70:1030–8. doi: 10.1139/o92-147. [DOI] [PubMed] [Google Scholar]
- 9.Cirelli C, Gutierrez CM, Tononi G. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron. 2004;41:35–43. doi: 10.1016/s0896-6273(03)00814-6. [DOI] [PubMed] [Google Scholar]
- 10.Cirelli C, Tononi G. Differences in gene expression between sleep and waking as revealed by mRNA differential display. Mol Brain Res. 1998;56:293–305. doi: 10.1016/s0169-328x(98)00057-6. [DOI] [PubMed] [Google Scholar]
- 11.Clayton DA, Mesches MH, Alvarez E, Bickford PC, Browning MD. A hippocampal NR2B deficit can mimic age-related changes in long-term potentiation and spatial learning in the Fischer 344 rat. J Neurosci. 2002;22:3628–37. doi: 10.1523/JNEUROSCI.22-09-03628.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cullinan WE, Herman JP, Battaglia DF, Akil H, Watson SJ. Pattern and time course of immediate early gene expression in rat brain following acute stress. Neuroscience. 1995;64:477–505. doi: 10.1016/0306-4522(94)00355-9. [DOI] [PubMed] [Google Scholar]
- 13.Datta S, Li G, Auerbach S. Activation of phasic pontine-wave generator in the rat: a mechanism for expression of plasticity-related genes and proteins in the dorsal hippocampus and amygdala. Eur J Neurosci. 2008;27:1876–92. doi: 10.1111/j.1460-9568.2008.06166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drummond SP, Brown GG, Gillin JC, Stricker JL, Wong EC, Buxton RB. Altered brain response to verbal learning following sleep deprivation. Nature. 2000;403:655–7. doi: 10.1038/35001068. [DOI] [PubMed] [Google Scholar]
- 15.Grassi-Zucconi G, Menegazzi M, De Prati AC, Bassetti A, Montagnese P, Mandile P, Cosi C, Bentivoglio M. c-fos mRNA is spontaneously induced in the rat brain during the activity period of the circadian cycle. Eur J Neurosci. 1993;5:1071–8. doi: 10.1111/j.1460-9568.1993.tb00960.x. [DOI] [PubMed] [Google Scholar]
- 16.Guzowski JF, Setlow B, Wagner EK, McGaugh JL. Experience-dependent gene expression in the rat hippocampus after spatial learning: a comparison of the immediate-early genes Arc, c-fos, and zif268. J Neurosci. 2001;21:5089–98. doi: 10.1523/JNEUROSCI.21-14-05089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herry C, Mons N. Resistance to extinction is associated with impaired immediate early gene induction in medial prefrontal cortex and amygdala. Eur J Neurosci. 2004;20:781–90. doi: 10.1111/j.1460-9568.2004.03542.x. [DOI] [PubMed] [Google Scholar]
- 18.Hofle N, Paus T, Reutens D, Fiset P, Gotman J, Evans AC, Jones BE. Regional cerebral blood flow changes as a function of delta and spindle activity during slow wave sleep in humans. J Neurosci. 1997;17:4800–8. doi: 10.1523/JNEUROSCI.17-12-04800.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 20.James AB, Conway AM, Morris BJ. Regulation of the neuronal proteasome by Zif268 (Egr1) J Neurosci. 2006;26:1624–34. doi: 10.1523/JNEUROSCI.4199-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones MW, Errington ML, French PJ, Fine A, Bliss TV, Garel S, Charnay P, Bozon B, Laroche S, Davis S. A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat Neurosci. 2001;4:289–96. doi: 10.1038/85138. [DOI] [PubMed] [Google Scholar]
- 22.Klugmann M, Leichtlein CB, Symes CW, Klaussner BC, Brooks AI, Young D, During MJ. A novel role of circadian transcription factor DBP in hippocampal plasticity. Mol Cell Neurosci. 2006;31:303–14. doi: 10.1016/j.mcn.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 23.Korte M, Kang H, Bonhoeffer T, Schuman E. A role for BDNF in the late-phase of hippocampal long-term potentiation. Neuropharmacology. 1998;37:553–9. doi: 10.1016/s0028-3908(98)00035-5. [DOI] [PubMed] [Google Scholar]
- 24.Lee HK, Takamiya K, Han JS, Man H, Kim CH, Rumbaugh G, Yu S, Ding L, He C, Petralia RS, Wenthold RJ, Gallagher M, Huganir RL. Phosphorylation of the AMPA receptor GluR1 subunit is required for synaptic plasticity and retention of spatial memory. Cell. 2003;112:631–43. doi: 10.1016/s0092-8674(03)00122-3. [DOI] [PubMed] [Google Scholar]
- 25.Li C, Brake WG, Romeo RD, Dunlop JC, Gordon M, Buzescu R, Magarinos AM, Allen PB, Greengard P, Luine V, McEwen BS. Estrogen alters hippocampal dendritic spine shape and enhances synaptic protein immunoreactivity and spatial memory in female mice. Proc Natl Acad Sci U S A. 2004;101:2185–90. doi: 10.1073/pnas.0307313101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Carter J, Gao X, Whitehead J, Tourtellotte WG. The neuroplasticity-associated arc gene is a direct transcriptional target of early growth response (Egr) transcription factors. Mol Cell Biol. 2005;25:10286–300. doi: 10.1128/MCB.25.23.10286-10300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez-Molina L, Conquet F, Dubois-Dauphin M, Schibler U. The DBP gene is expressed according to a circadian rhythm in the suprachiasmatic nucleus and influences circadian behavior. Embo J. 1997;16:6762–71. doi: 10.1093/emboj/16.22.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci. 2002;25:103–26. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- 29.Maquet P. The role of sleep in learning and memory. Science. 2001;294:1048–52. doi: 10.1126/science.1062856. [DOI] [PubMed] [Google Scholar]
- 30.Maquet P, Degueldre C, Delfiore G, Aerts J, Peters JM, Luxen A, Franck G. Functional neuroanatomy of human slow wave sleep. J Neurosci. 1997;17:2807–12. doi: 10.1523/JNEUROSCI.17-08-02807.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maquet P, Laureys S, Peigneux P, Fuchs S, Petiau C, Phillips C, Aerts J, Del Fiore G, Degueldre C, Meulemans T, Luxen A, Franck G, Van Der Linden M, Smith C, Cleeremans A. Experience-dependent changes in cerebral activation during human REM sleep. Nat Neurosci. 2000;3:831–6. doi: 10.1038/77744. [DOI] [PubMed] [Google Scholar]
- 32.Mello CV, Clayton DF. Differential induction of the ZENK gene in the avian forebrain and song control circuit after metrazole-induced depolarization. J Neurobiol. 1995;26:145–61. doi: 10.1002/neu.480260112. [DOI] [PubMed] [Google Scholar]
- 33.Muslimov IA, Santi E, Homel P, Perini S, Higgins D, Tiedge H. RNA transport in dendrites: a cis-acting targeting element is contained within neuronal BC1 RNA. J Neurosci. 1997;17:4722–33. doi: 10.1523/JNEUROSCI.17-12-04722.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muzur A, Pace-Schott EF, Hobson JA. The prefrontal cortex in sleep. Trends Cogn Sci. 2002;6:475–481. doi: 10.1016/s1364-6613(02)01992-7. [DOI] [PubMed] [Google Scholar]
- 35.Nofzinger EA, Price JC, Meltzer CC, Buysse DJ, Villemagne VL, Miewald JM, Sembrat RC, Steppe DA, Kupfer DJ. Towards a neurobiology of dysfunctional arousal in depression: the relationship between beta EEG power and regional cerebral glucose metabolism during NREM sleep. Psychiatry Res. 2000;98:71–91. doi: 10.1016/s0925-4927(00)00045-7. [DOI] [PubMed] [Google Scholar]
- 36.Pavlides C, Winson J. Influences of hippocampal place cell firing in the awake state on the activity of these cells during subsequent sleep episodes. J Neurosci. 1989;9:2907–18. doi: 10.1523/JNEUROSCI.09-08-02907.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pompeiano M, Cirelli C, Ronca-Testoni S, Tononi G. NGFI-A expression in the rat brain after sleep deprivation. Mol Brain Res. 1997;46:143–53. doi: 10.1016/s0169-328x(96)00295-1. [DOI] [PubMed] [Google Scholar]
- 38.Ribeiro S, Goyal V, Mello CV, Pavlides C. Brain gene expression during REM sleep depends on prior waking experience. Learn Mem. 1999;6:500–8. doi: 10.1101/lm.6.5.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ribeiro S, Mello CV, Velho T, Gardner TJ, Jarvis ED, Pavlides C. Induction of hippocampal long-term potentiation during waking leads to increased extrahippocampal zif-268 expression during ensuing rapid-eye-movement sleep. J Neurosci. 2002;22:10914–23. doi: 10.1523/JNEUROSCI.22-24-10914.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richardson CL, Tate WP, Mason SE, Lawlor PA, Dragunow M, Abraham WC. Correlation between the induction of an immediate early gene, zif/268, and long-term potentiation in the dentate gyrus. Brain Res. 1992;580:147–154. doi: 10.1016/0006-8993(92)90938-6. [DOI] [PubMed] [Google Scholar]
- 41.Ripperger JA, Shearman LP, Reppert SM, Schibler U. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 2000;14:679–89. [PMC free article] [PubMed] [Google Scholar]
- 42.Romcy-Pereira R, Pavlides C. Distinct modulatory effects of sleep on the maintenance of hippocampal and medial prefrontal cortex LTP. Eur J Neurosci. 2004;20:3453–62. doi: 10.1111/j.1460-9568.2004.03808.x. [DOI] [PubMed] [Google Scholar]
- 43.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 44.Sage D, Maurel D, Bosler O. Involvement of the suprachiasmatic nucleus in diurnal ACTH and corticosterone responsiveness to stress. Am J Physiol Endocrinol Metab. 2001;280:E260–9. doi: 10.1152/ajpendo.2001.280.2.E260. [DOI] [PubMed] [Google Scholar]
- 45.Schmitt JM, Guire ES, Saneyoshi T, Soderling TR. Calmodulin-dependent kinase kinase/calmodulin kinase I activity gates extracellular-regulated kinase-dependent long-term potentiation. J Neurosci. 2005;25:1281–90. doi: 10.1523/JNEUROSCI.4086-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwab JM, Frei E, Klusman I, Schnell L, Schwab ME, Schluesener HJ. AIF-1 expression defines a proliferating and alert microglial/macrophage phenotype following spinal cord injury in rats. J Neuroimmunol. 2001;119:214–22. doi: 10.1016/s0165-5728(01)00375-7. [DOI] [PubMed] [Google Scholar]
- 47.Smith C. Sleep states and memory processes. Behav Brain Res. 1995;69:137–145. doi: 10.1016/0166-4328(95)00024-n. [DOI] [PubMed] [Google Scholar]
- 48.Soderling TR. CaM-kinases: modulators of synaptic plasticity. Curr Opin Neurobiol. 2000;10:375–80. doi: 10.1016/s0959-4388(00)00090-8. [DOI] [PubMed] [Google Scholar]
- 49.Stafstrom-Davis CA, Ouimet CC, Feng J, Allen PB, Greengard P, Houpt TA. Impaired conditioned taste aversion learning in spinophilin knockout mice. Learn Mem. 2001;8:272–8. doi: 10.1101/lm.42101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stickgold R, Hobson JA, Fosse R, Fosse M. Sleep, learning, and dreams: offline memory reprocessing. Science. 2001;294:1052–7. doi: 10.1126/science.1063530. [DOI] [PubMed] [Google Scholar]
- 51.Thiel G, Schoch S, Petersohn D. Regulation of synapsin I gene expression by the zinc finger transcription factor zif268/egr-1. J Biol Chem. 1994;269:15294–301. [PubMed] [Google Scholar]
- 52.Walker MP, Stickgold R. Sleep-dependent learning and memory consolidation. Neuron. 2004;44:121–33. doi: 10.1016/j.neuron.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 53.Wang H, Tiedge H. Translational control at the synapse. Neuroscientist. 2004;10:456–66. doi: 10.1177/1073858404265866. [DOI] [PubMed] [Google Scholar]
- 54.Wayman GA, Impey S, Marks D, Saneyoshi T, Grant WF, Derkach V, Soderling TR. Activity-dependent dendritic arborization mediated by CaM-kinase I activation and enhanced CREB-dependent transcription of Wnt-2. Neuron. 2006;50:897–909. doi: 10.1016/j.neuron.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 55.Wilson MA, McNaughton BL. Reactivation of hippocampal ensemble memories during sleep. Science. 1994;265:676–9. doi: 10.1126/science.8036517. [DOI] [PubMed] [Google Scholar]
- 56.Winson J, Abzug C. Gating of neuronal transmission in the hippocampus: efficacy of transmission varies with behavioral state. Science. 1977;196:1223–1225. doi: 10.1126/science.193192. [DOI] [PubMed] [Google Scholar]
- 57.Yoo SS, Hu PT, Gujar N, Jolesz FA, Walker MP. A deficit in the ability to form new human memories without sleep. Nat Neurosci. 2007;10:385–92. doi: 10.1038/nn1851. [DOI] [PubMed] [Google Scholar]