Abstract
Lipid A structural modifications can substantially impact the host’s inflammatory response to bacterial LPS. Bacteroides fragilis, an opportunistic pathogen associated with life-threatening sepsis and intra-abdominal abscess formation, and Bacteroides thetaiotaomicron, a symbiont pivotal for proper host intestinal tissue development, both produce an immunostimulatory LPS comprised of penta-acylated lipid A. Under defined conditions, Porphyromonas gingivalis, an oral pathogen associated with periodontitis, also produces an LPS bearing a penta-acylated lipid A. However, this LPS preparation is 100–1000 times less potent than Bacteroides LPS in stimulating endothelial cells. We analyzed Bacteroides and P. gingivalis lipid A structures using MALDI-TOF MS and gas chromatography to determine the structural basis for this phenomenon. Even though both Bacteroides and P. gingivalis lipid A molecules are penta-acylated and mono-phosphorylated, subtle differences in mass and fatty acid content could account for the observed difference in LPS potency. This fatty acid heterogeneity is also responsible for the peak “clusters” observed in the mass spectra and obfuscates the correlation between LPS structure and immunostimulatory ability. Further, we show the difference in potency between Bacteroides and P. gingivalis LPS is TLR4-dependent. Altogether, the data suggest subtle changes in lipid A structure may profoundly impact the host’s innate immune response.
Keywords: LPS, lipid A, TLR4, Bacteroides fragilis, Bacteroides thetaiotaomicron, Porphyromonas gingivalis
1. Introduction
The Bacteroides consists of anaerobic Gram-negative rods and is the most numerous genus found in the human ileum and large intestine, outnumbering E. coli 100 to 1 [1, 2]. B. thetaiotaomicron, an important member of the host’s normal flora, lives in a symbiotic relationship with its human host. B. fragilis, on the other hand, poses a potential threat as an opportunistic pathogen. Due to the presence of several different virulence factors, this organism can cause systemic infection when it leaves its intestinal niche and enters the bloodstream [3, 4]. Indeed, despite constituting only 0.5% of the entire intestinal microbiota, B. fragilis is the most common anaerobic isolate from patients with sepsis or intra-abdominal abscesses [5]. Porphyromonas gingivalis, an anaerobic Gram-negative rod formerly classified as Bacteroides, is an oral pathogen associated with periodontitis [6].
Gram-negative bacteria contain lipopolysaccharide (LPS) in the outer leaflet of their outer membrane. While LPS contains large, variable polysaccharide and oligosaccharide regions, the relatively conserved lipid region (lipid A) is the endotoxic and biologically active moiety responsible for septic shock. The “canonical” structure is represented by that from Escherichia coli and is the most potent lipid A species known (Fig. 1A). It consists of a 1,4′-biphosphorylated glucosamine disaccharide bearing six fatty acids which are unbranched chains 12 or 14 carbon units long [7]. Other lipid A species, however, show variability in the number, length, and composition of attached fatty acids, as well as variability in the level of phosphorylation and number and types of substituted groups found attached to the phosphate residues [1, 7]. For instance, lipid A isolated from B. fragilis is penta-acylated and mono-phosphorylated [8]. Additionally, the fatty acids present in B. fragilis LPS are branched and 15, 16, or 17 carbon units long (Fig. 1B) [8]. Although not identical, the penta-acylated and mono-phosphorylated lipid A structure of P. gingivalis, generated under the condition of low hemin concentration, closely resembles that of Bacteroides (Fig. 1C) [9, 10]. Significantly, deviations from the canonical lipid A structure are known to have a profound impact on the host innate immune response [7].
Fig. 1.
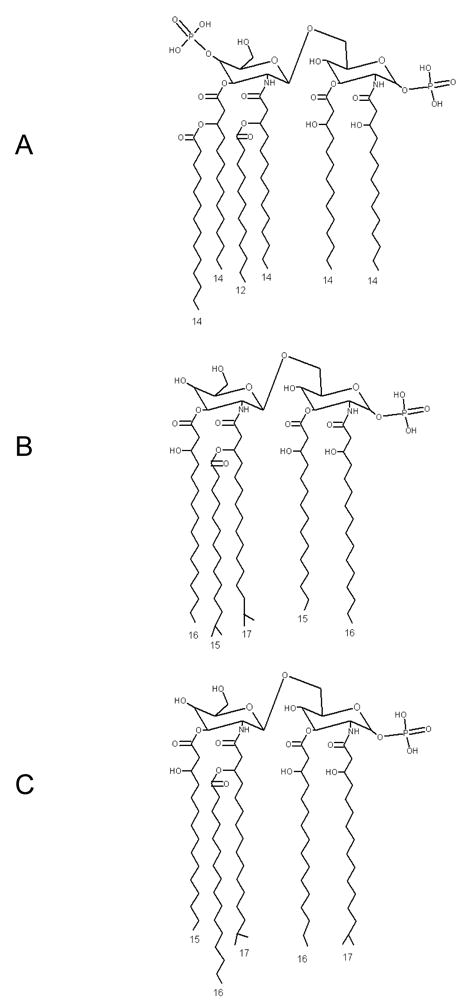
The published structures of (A) E. coli, (B) B. fragilis, and (C) P. gingivalis lipid A. As calculated from their published structures, the masses of each lipid A moiety are 1798, 1662, and 1690 amu, respectively.
One source of variation between the Bacteroides and P. gingivalis lipid A structures is the distribution of attached fatty acids. Previously, we demonstrated P. gingivalis lipid A heterogeneity by revealing the presence of multiple lipid A structures in LPS preparations isolated from a single strain [11]. The lipid A structures differed by 14 amu, giving rise to the peak “clusters” observed on the MALDI-TOF mass spectra, and we speculated that this variation arose from the differential distribution of attached fatty acids [11]. We showed the fatty acid content of P. gingivalis lipid A primarily depended on the acyl chain length specificity of LpxA and LpxD—which attach the primary ester- and amide-linked fatty acids, respectively—and the fatty acid substrate pool available to the biosynthetic enzymes [11].
Much is still unclear about the potency of penta-acylated, mono-phosphorylated LPS as determined by its ability to activate an innate immune response via TLR4. For instance, in the current literature, the potency of Bacteroides LPS remains a controversial issue. Some research indicates that Bacteroides LPS exhibits substantial potency, while other research shows it exhibits little to no potency [12–15]. On the other hand, penta-acylated and mono-phosphorylated P. gingivalis LPS has been consistently shown to exhibit weak endotoxicity [16–19].
Therefore, in the present work, we sought to clarify the confusion in the literature regarding the recognition of Bacteroides LPS by TLR4. To achieve this, we conducted side-by-side studies of the LPS isolated from Bacteroides and compared it to the more highly characterized TLR4 agonist LPS isolated from P. gingivalis. Two different LPS isolation methods were employed, and the resulting preparations were characterized structurally. Additionally, the preparations were analyzed functionally by assessing their ability to stimulate an innate immune response via TLR4 using two different methods. Our results demonstrate that Bacteroides LPS is a potent stimulator of innate immunity through TLR4.
2. Results
2.1. TRI Reagent-extracted and phenol-extracted Bacteroides lipid A structures are very similar, but both are slightly smaller than P. gingivalis lipid A
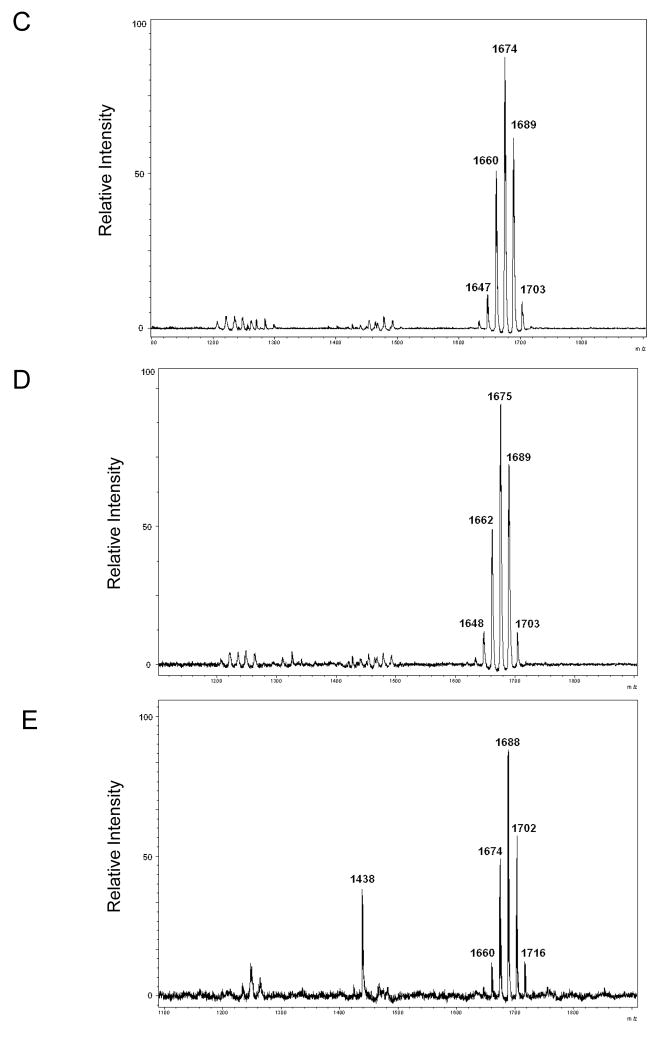
LPS isolation methods often vary between laboratories, and we have shown different LPS isolation methods can selectively purify different types of LPS. For instance, the EtOH/MgCl2 (Darveau/Hancock) method is known to selectively isolate tetra-acylated lipid A [17, 20, 21]. Because of this, we used MALDI-TOF MS to compare the Bacteroides lipid A structures obtained after LPS purification using one of two different LPS isolation techniques: the TRI Reagent (a commercial preparation of phenol and guanidine thiocyanate) and hot phenol-water methods [22, 23]. The TRI Reagent method yielded a penta-acylated lipid A molecule with a cluster of mass peaks centered at m/z 1676 for both B. fragilis and B. thetaiotaomicron (Figs. 2A and 2C). (The minor peaks centered at m/z 1235 and 1454 may be trace amounts of tri- and tetra-acylated lipid A.) Similarly, the hot phenol-water method yielded the exact same pattern (Figs. 2B and 2D).
Fig. 2.
The MALDI-TOF MS profiles of lipid A isolated from Bacteroides and P. gingivalis. B. fragilis lipid A isolated using (A) the TRI Reagent method or (B) the hot phenol-water method yielded nearly identical MALDI-TOF MS profiles, indicating both methods isolate similar molecules. Comparable results were observed with B. thetaiotaomicron lipid A isolated using (C) the TRI Reagent method or (D) the hot phenol-water method. (E) The MALDI-TOF MS profile of the lipid A isolated from a P. gingivalis mutant capable of only synthesizing a penta-acylated moiety is shown.
We then compared the lipid A mass peak profile of Bacteroides with that of P. gingivalis, since P. gingivalis was once classified as, and is thus closely related to, Bacteroides [24, 25]. Confounding this comparison was the discovery that P. gingivalis produces penta-acylated lipid A molecules when the hemin concentration is relatively low and tetra-acylated lipid A molecules when the hemin concentration is relatively high [10]. To eliminate this source of variation, all of the procedures reported in this study utilized a strain designated “P. gingivalis 1626,” a mutant we constructed that exclusively synthesizes penta-acylated lipid A species regardless of hemin concentration (unpublished data). Previously, we have shown that the penta-acylated, mono-phosphorylated structure of P. gingivalis lipid A is not affected by the LPS isolation method; therefore, we used only the TRI Reagent method to isolate LPS from the P. gingivalis mutant [17]. When the lipid A of this mutant was analyzed by MALDI-TOF MS, it produced a mass peak cluster centered at m/z 1690 (Fig. 2E). (The single peak at m/z 1438 is known to be a minor, non-lipid A contaminant.) Notably, the lipid A produced by P. gingivalis 1626 (termed “Pg 1690” lipid A) is, on average, 14 amu (one methylene subunit) larger than that produced by Bacteroides. Taken together, the data indicate the TRI Reagent and hot phenol-water methods both isolate similar LPS species in terms of lipid A structure as measured by MALDI-TOF MS. Further, the data indicate P. gingivalis lipid A is slightly larger than that of Bacteroides.
2.2. Bacteroides LPS contains a distribution of fatty acids that is different from P. gingivalis LPS
To further examine potential structural differences between Bacteroides and P. gingivalis lipid A, we analyzed the fatty acid content of their respective LPS molecules by gas chromatography. We found the distribution of fatty acids to be quite different for each of the three LPS species examined (see Table 1). For instance, B. fragilis had a preference for attaching an iso-C15 fatty acid, while B. thetaiotaomicron preferred an anteiso-C15 fatty acid. Another major difference was that P. gingivalis attached a straight-chain C16 fatty acid, while both Bacteroides species attached almost no C16 fatty acids, but instead preferred a 3-OH-C16 fatty acid. Finally, P. gingivalis and B. fragilis attached a high percentage of a 3-OH-C17 fatty acid, but B. thetaiotaomicron attached the fatty acid at a much lower percentage. Altogether, our results demonstrate that the peak clusters observed in the mass spectra of Bacteroides and P. gingivalis lipid A are due to subtle differences in mass and fatty acid content between the lipid A moieties isolated from each organism and between the lipid A moieties isolated from the same organism. In other words, each bacterium produces a heterogeneous population of lipid A molecules that differ in mass and fatty acid content, and the populations of lipid A molecules also differ between the bacterial species. Further, our results are relatively consistent with the published structures for B. fragilis and P. gingivalis lipid A [8, 9].
Table 1.
The fatty acid proportions present in the LPS isolated from B. fragilis, B. thetaiotaomicron, and P. gingivalis 1626 given as a percent of the total identified fatty acidsa.
| B. fragilis | B. thetaiotaomicron | P. gingivalis 1626 | |
|---|---|---|---|
| Iso-C15 | 19.7 % | 8.5 % | 14.1 % |
| Anteiso-C15 | 6.6 % | 20.4 % | 12.5 % |
| C16 | 1.9 % | 3.2 % | 24.1 % |
| 3-OH-C15 | 0.0 % | 0.0 % | 0.0 % |
| 3-OH-C16 | 31.6 % | 40.8 % | 8.0 % |
| 3-OH-C17 | 40.3 % | 27.1 % | 41.4 % |
The percents were calculated as follows: The amount of each fatty acid was determined by dividing the area (in mm2) under the GC trace by the amount of dry weight bacteria (in mg) used to isolate the LPS. These quotients were added, and the resulting sum (total area/mg) was set equivalent to 100%. Therefore, each fatty acid (measured in area/mg) could be calculated as a percent of the total area/mg. The results are representative of two independent determinations.
2.3. Bacteroides LPS exhibits immune potency independent of the LPS extraction method
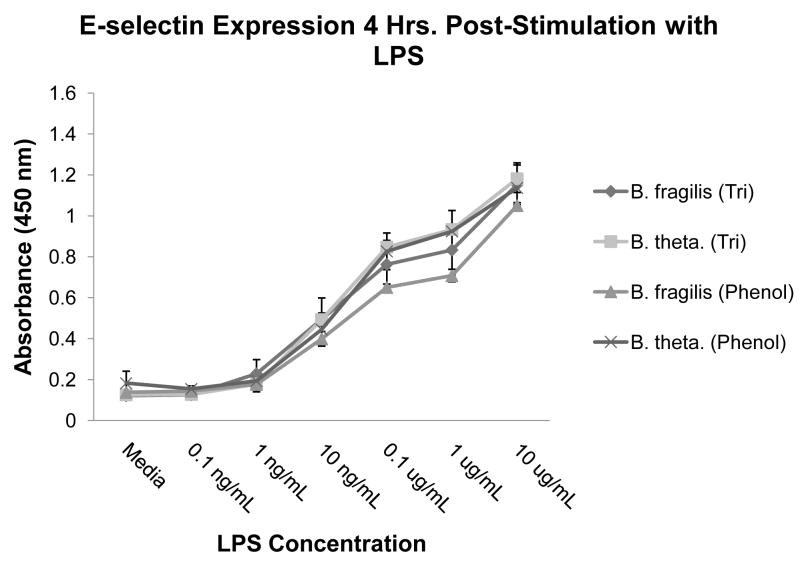
Since we demonstrated slight structural variations existed between B. fragilis and B. thetaiotaomicron lipid A, we wanted to determine if these variations resulted in differing abilities to stimulate E-selectin expression from human umbilical vein endothelial cells (HUVECs), which predominantly express TLR4 [26]. Although the LPS extraction method did not appear to affect the lipid A structures, we also wanted to ensure that the extraction method did not affect the potency of the LPS preparations. Therefore, we stimulated HUVECs with both TRI-extracted and phenol-extracted LPS from B. fragilis and B. thetaiotaomicron, and no appreciable difference in E-selectin expression (as measured by ELISA) was detected (Fig. 3). (Henceforth, all cell stimulation assays were performed using LPS extracted using the TRI Reagent method.) Importantly, our results indicate the potency of Bacteroides LPS independent of the extraction method used.
Fig. 3.
E-selectin expression profile for LPS-stimulated HUVECS. HUVECs were stimulated with B. fragilis or B. thetaiotaomicron LPS that had been isolated using either the TRI Reagent method or the hot phenol-water method, and E-selectin protein expression was measured by ELISA. Results are means and standard deviations of triplicate wells and are representative of at least two independent determinations.
2.4. Bacteroides LPS is less potent than E. coli LPS
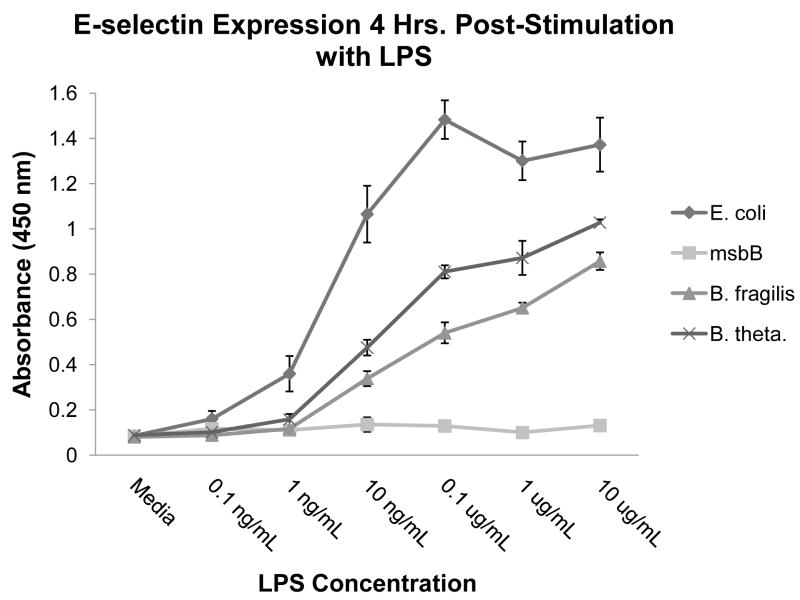
Because Bacteroides LPS exhibited potent endotoxicity, we compared its ability to stimulate E-selectin expression on HUVECs with that of E. coli LPS, the most potent endotoxin known, and with that of E. coli msbB LPS, a non-stimulatory LPS produced by a mutant which only synthesizes penta-acylated lipid A [27]. E. coli msbB lacks the ability to attach its secondary C14 fatty acid and, as a result, produces a non-toxic LPS molecule. This LPS was used as a negative control in cell stimulation assays [21]. As shown in Figure 4, both Bacteroides LPS preparations were more potent than the msbB LPS negative control, but were less potent than E. coli LPS. Interestingly, B. thetaiotaomicron LPS was consistently and significantly more potent than B. fragilis LPS at 0.1 μg/mL (p < 0.05) and at 1 μg/mL (p < 0.05), despite both species producing extremely similar lipid A moieties. Altogether, our results demonstrate the potency of Bacteroides LPS, with B. thetaiotaomicron LPS being slightly more immunostimulatory than B. fragilis LPS.
Fig. 4.
E-selectin expression profile for LPS-stimulated HUVECs. HUVECs were stimulated with E. coli, E. coli msbB, B. fragilis, or B. thetaiotaomicron LPS, and E-selectin protein expression was measured by ELISA. Results are means and standard deviations of triplicate wells and are representative of at least two independent determinations.
2.5. Bacteroides LPS and lipid A more potently activate E-selectin expression from HUVECs than P. gingivalis LPS and lipid A
It has been reported that a few select proteins may stimulate an immune response via TLR4 [28]. Therefore, a possibility existed that protein contamination may have contributed to the immune potency observed for the Bacteroides LPS preparations. To examine this, we ran 10 μg of the TRI-extracted B. thetaiotaomicron LPS preparation on an SDS-PAGE gel, transferred the contents to a PVDF membrane, and then subjected the membrane to colloidal gold staining (Fig. 5A, lane 1). As shown on the gel, the only contaminant present was a low molecular weight protein. Most likely, this minor contaminant is an endotoxin-associated lipoprotein that has been shown to activate TLR2, but not TLR4 [29, 30]. Additionally, by comparing the intensity of the lipoprotein band (lane 1) to the much darker band corresponding to 100 ng of BSA (lane 5), we calculated that the B. thetaiotaomicron LPS sample contained less than 1% lipoprotein.
Fig. 5.

(A) PVDF membrane stained using colloidal gold. LPS or lipid A samples (10 μg) were run on an SDS-PAGE gel, transferred to a PVDF membrane, and stained with colloidal gold to detect protein contamination. The contents of the lanes were as follows: Protein ladder (lane L); B. thetaiotaomicron LPS (lane 1) or lipid A (lane 2); Pg 1690 LPS (lane 3) or lipid A (lane 4); and 100 ng (lane 5), 10 ng (lane 6), or 1 ng (lane 7) of BSA. (B) E-selectin expression profile for LPS- or lipid A-stimulated HUVECs. HUVECs were stimulated with B. thetaiotaomicron or Pg 1690 LPS or lipid A, and E-selectin protein expression was measured by ELISA. Results are means and standard deviations of triplicate wells and are representative of at least two independent determinations.
Even though this lipoprotein does not stimulate through TLR4, we attempted to eliminate it from the LPS preparation using the “Vogel re-purification” [31] method. However, LPS preparations we subjected to this re-purification procedure gave inconclusive and inconsistent structural and functional data (not shown). Alternatively, we attempted to reduce the lipoprotein content of the LPS preparations by isolating purified lipid A. As shown in Fig. 5A, the B. thetaiotaomicron lipid A preparation (lane 2) contained slightly less lipoprotein than the B. thetaiotaomicron LPS preparation (lane 1).
After carefully characterizing the B. thetaiotaomicron LPS and lipid A preparations, we compared their ability to stimulate E-selectin expression from HUVECs with that of the P. gingivalis LPS and lipid A preparations. (Additionally, the lipoprotein content of the P. gingivalis LPS and lipid A preparations was also determined to be ~1%; see lanes 3 and 4 in Fig. 5A.) Strikingly, we observed that—despite possessing only subtle differences in lipid A structure—P. gingivalis 1626 LPS (“Pg 1690” LPS) was 100–1000 times less potent at activating E-selectin expression than Bacteroides LPS (Fig. 5B). However, it is thought that the presence of the O-Antigen in LPS can lead to aggregation, potentially influencing the ability of TLR4/MD-2 to properly recognize and respond to the LPS [32]. Additionally, the presence of the O-Antigen makes it difficult to determine if equal amounts of the bioactive moiety lipid A are being applied to the cells. To address these concerns, we isolated LPS from B. thetaiotaomicron and P. gingivalis and cleaved off the O-Antigen using a mild acid hydrolysis [33]. The resulting lipid A preparations were used to stimulate HUVECs. (It should be noted that both lipid A preparations contained less lipoprotein than the LPS preparations; compare lane 1 with lane 2 and lane 3 with lane 4.) Consistent with our previous results using LPS, B. thetaiotaomicron lipid A was approximately 1000 times more potent than P. gingivalis lipid A (Fig. 5B). Taken together, these results indicate that the lipid A moiety of LPS is primarily responsible for the differences in immune potency observed between Bacteroides and P. gingivalis LPS.
2.6. The difference in immunostimulatory ability between Bacteroides and Pg 1690 LPS is TLR4-dependent
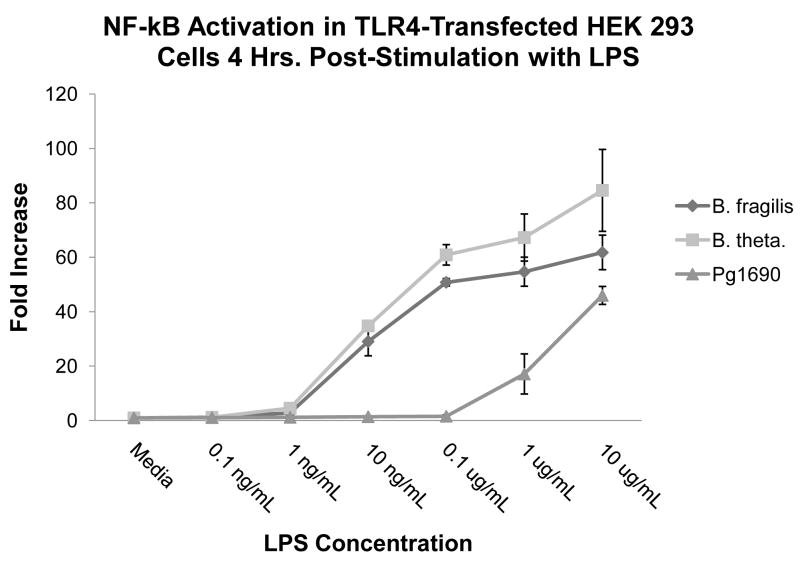
Since HUVECs might express a small amount of TLR1, -2, and -3, we wanted to ensure that the potency differences observed between Bacteroides and Pg 1690 LPS were indeed due to TLR4 [26]. To determine this, we stimulated human TLR4/MD-2-transfected HEK 293 cells with B. fragilis, B. thetaiotaomicron, and Pg 1690 LPS preparations. In addition to human TLR4 and MD-2 plasmid constructs, HEK 293 cells were transfected with plasmid constructs containing ELAM-1 firefly luciferase (which is responsive to NF-κB production) and β-actin Renilla luciferase (as a transfection control). Following LPS stimulation, the fold increase of relative luciferase units was determined by calculating the ratio of firefly/Renilla luciferase production for the stimulated samples to the firefly/Renilla luciferase production for the non-stimulated samples. Consistent with our previous results, we observed B. fragilis and B. thetaiotaomicron LPS species to be approximately 100- to 1000-fold more potent than Pg 1690 LPS (Fig. 6). Thus, it appears that differential interaction at the TLR4-MD-2 complex is responsible for the observed differences in the immunostimulatory ability of the LPS species. Taken together, our results suggested P. gingivalis LPS, which contains subtle differences in its lipid A structure as compared to that of Bacteroides, is greatly reduced in its ability to stimulate an immune response in a TLR4-dependent manner.
Fig. 6.
NF-κB activation profile of TLR4-transfected HEK 293 cells following LPS stimulation. TLR4-transfected HEK 293 cells were stimulated with B. fragilis, B. thetaiotaomicron, or Pg 1690 LPS, and the relative amount of firefly luciferase activity was determined. Values are reported as the fold increase of relative luciferase units (firefly luciferase/Renilla luciferase) for the LPS-stimulated samples compared to the non-stimulated control response, whose ratio was set at 1. Results are means and standard deviations of triplicate wells and are representative of at least two independent determinations.
3. Discussion and conclusions
The structure-function relationship that has defined the interaction of LPS—and more specifically, lipid A—with the TLR4-MD-2 complex is becoming increasingly complicated. Although lipid A is considered to be the most conserved moiety of the LPS molecule, much evidence indicates that the ability of Gram-negative bacteria to modify this structure for the purpose of manipulating the host innate immune response appears to be a ubiquitous phenomenon. At first, relatively major changes to the lipid A structure were observed to confer special properties on the bacteria. For instance, 4-aminoarabinose modification of Salmonella typhimurium lipid A confers the organism with increased resistance to cationic antimicrobial peptides [34]. In addition, it was found that Yersinia pestis, the etiologic agent of bubonic plague, modified its hexa-acylated lipid A to a tetra-acylated form when the organism underwent a temperature shift from 27° C. (ambient temperature) to 37° C. (mammalian body temperature) [35]. This deacylation caused the organism’s LPS to be greatly reduced in its ability to elicit an innate immune response as measured by TNF-α secretion [35]. Perhaps most strikingly, when Y. pestis was genetically modified to always produce a hexa-acylated LPS, regardless of temperature, the organism was completely avirulent in a murine model of infection [36].
Recently, relatively small changes in lipid A structure have been observed to greatly influence the innate immune response of the host. For example, we showed that when the C12 fatty acid normally present in the lipid A structure of E. coli (Fig. 1A) was replaced by a C16 fatty acid, the LPS was greatly reduced in its ability to trigger an innate immune response from Mono-Mac-6 cells as measured by IL-8 secretion [37]. The difference in immunostimulatory ability between the two molecules rested solely in their fatty acid chain lengths. Thus, the extremely subtle change of replacing a single fatty acid with a slightly longer fatty acid directly impacted LPS recognition by Mono-Mac-6 cells.
In the present study, we examined the ability of Bacteroides and P. gingivalis LPS—both containing structurally similar penta-acylated and mono-phosphorylated lipid A moieties—to stimulate an innate immune response. Overall, our results demonstrated that subtle, often unappreciated differences in lipid A structure may substantially impact the host’s innate immune response. Confounding the issue, however, is the heterogeneous nature of the lipid A molecules present within certain bacterial species. This heterogeneity can be observed in the mass spectra, where lipid A molecules—despite having been isolated from the same organism—do not give rise to single peaks, but to peak “clusters,” which represent lipid A molecules that differ in mass by 14 amu (Fig. 2). The cause of these “lipid A clusters” has been attributed to variations in fatty acid content [11]. These variations arise due to relaxed acyl chain length specificity of the lipid A biosynthetic enzymes LpxA and LpxD and due to varying substrate availability [11]. The advantage gained by a bacterium producing a heterogeneous pool of lipid A molecules, as opposed to a single lipid A structure, is currently unclear.
Further confounding the issue is the possibility that differences in phosphate placement between Bacteroides and P. gingivalis lipid A could also contribute to the difference in their immunostimulatory abilities. Currently, the published structure of P. gingivalis lipid A places a phosphate in the 1 position [9]. However, recent preliminary analyses in our laboratory indicate that the lipid A from Pg 1690 LPS likely bears a 4′-phosphate rather than a 1-phosphate as is currently accepted (S. Coats and R. Darveau, unpublished data). Therefore, we conclude that the 100- to 1000-fold difference in immune potency observed between the highly similar penta-acylated and mono-phosphorylated Bacteroides and P. gingivalis LPS preparations may be due to one or more subtle lipid A structural differences including fatty acid content and phosphate placement.
Because of lipid A structural heterogeneity, traditional structure-function relationships are becoming more difficult to define in LPS preparations. Currently, there is no literature on the “lipid A cluster-function” relationship. Since the lipid A molecules present in Bacteroides and P. gingivalis exist as a heterogeneous mixture, the best way to define such a relationship is by observing correlations. For instance, in this study, the presence of a 3-OH-C16 fatty acid and a smaller lipid A moiety correlated with greater immune potency. We propose that future research use such correlations when assessing heterogeneous lipid A structure-function relationships.
Another potential solution would be to construct synthetic lipid A molecules. The usefulness of this approach is two-fold: (1) This approach allows the more traditional single structure-function relationship to be analyzed; and (2) It eliminates potential contaminants. Indeed, despite demonstrating that our LPS preparations contain only a trace amount of lipoprotein contamination, it cannot be ruled out that as of yet unidentified molecules are contaminating the LPS preparations. In the case of Bacteroides LPS, it is possible that a contaminating molecule is enhancing its immunostimulatory ability; in the case of P. gingivalis LPS, it is possible that a contaminating molecule is antagonizing its immunostimulatory ability. However, the technical difficulty of constructing synthetic lipid A molecules makes this approach impractical for most laboratories.
The impact of lipid A heterogeneity on the host innate immune response is also unclear. The conventional wisdom is that hexa-acylated lipid A is transferred from LBP to CD14 and then to the TLR4/MD-2 complex, triggering oligomerization and the subsequent translocation of NF-κB into the nucleus followed by expression of pro-inflammatory mediators [38]. However, there is much controversy surrounding the immunostimulatory ability of LPS with lipid A containing fewer than six fatty acids—so-called “under-acylated” LPS. In the case of Bacteroides, some evidence indicates its LPS stimulates cells exclusively via TLR4, stimulates cells exclusively via TLR2, or exhibits almost no stimulatory ability [12–15]. P. gingivalis LPS has also been observed to stimulate via TLR2 [20]. Critics of these findings claim TLR2 activation is due to lipoprotein contamination that may be a result of improper LPS purification [30, 31, 39, 40]. While this may be true for TLR2 activation by hexa-acylated LPS preparations (such as that from E. coli), ample evidence indicates under-acylated LPS still retains the ability to activate TLR2 following extensive re-purification [13, 14, 41, 42]. Due to the contentious nature of the issue and the presence of trace lipoprotein in our LPS preparations, we chose to limit this study to examining the interaction between heterogeneous lipid A clusters and TLR4.
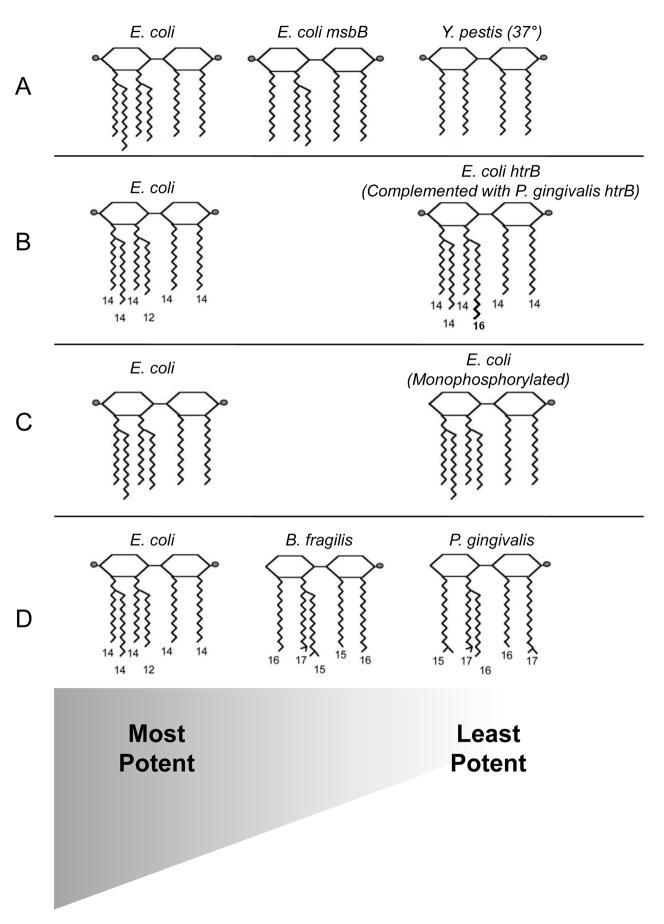
Based on our findings and the current state of the literature, we diagrammatically summarize the present understanding of the relationship between lipid A structure and immune potency in Fig. 7. While exceptions to the current paradigm are certain to exist, the diagram is simply intended to represent the overall trend in the literature. The first pattern to emerge from the literature seems to indicate that lipid A molecules bearing six fatty acids correlate with optimum LPS potency, while lipid A molecules containing fewer fatty acids leads to a decrease in potency (Fig. 7A). Indeed, lipid A isolated from the E. coli msbB mutant, which is identical to wild-type E. coli lipid A with the exception of lacking a single secondary C14 fatty acid, has dramatically decreased immunostimulatory ability [27]. Similarly, Y. pestis grown at 37° produces a non-stimulatory, tetra-acylated lipid A that is identical in structure (with the exception of lacking both secondary fatty acids) to the strongly immunostimulatory, hexa-acylated lipid A produced when grown at 27° [35, 43]. Evidence from the literature also indicates lipid A moieties containing fatty acids that are 12–14 carbon units long are most potent, while deviations from these carbon-chain lengths is associated with decreased potency (Fig. 7B). Indeed, as mentioned previously, we demonstrated when the secondary C12 fatty acid in E. coli lipid A was replaced by a C16 fatty acid, the LPS was greatly reduced in potency [37]. Finally, a pattern which is widely acknowledged is lipid A molecules that have two phosphate groups are usually much more potent than lipid A molecules containing only one (or no) phosphate groups [44] (Fig. 7C). Indeed, E. coli lipid A lacking either its 1- or 4′-phosphate is substantially reduced in its immunostimulatory ability [45].
Fig. 7.
Proposed model for the relationship between lipid A structure and potency. While exceptions to our proposed model certainly exist, the model is meant to reflect general trends in the literature. See Sections 1 and 3 for specific details about each of the lipid A structures. (The lipid A diagrams were graciously provided by Erridge et al.[1]). In each row, the lipid A molecules are arranged from most potent (left) to least potent (right). (A) E. coli, with six fatty acids, is the most potent lipid A structure known. Existing evidence indicates that as the number of attached fatty acids decreases, so does the potency. (B) Lipid A appears to be most potent when the attached fatty acids are 12–14 carbon units long. Deviations from these carbon-chain lengths decrease the potency. (C) Current research indicates the most potent lipid A molecules contain two phosphate groups, and lipid A molecules with one (or no) phosphate groups are much less potent. (D) Data from this work shows that Bacteroides LPS is less potent than E. coli LPS, and P. gingivalis LPS is the least potent of all three. A possible explanation for this is that although both Bacteroides and P. gingivalis possess penta-acylated and mono-phosphorylated lipid A molecules, the fatty acid chain lengths of Bacteroides lipid A are closer in length to those of E. coli lipid A than the fatty acid chain lengths of P. gingivalis lipid A are to those of E. coli lipid A.
Therefore, according to the paradigm, a possible explanation emerges for why P. gingivalis LPS is less potent than Bacteroides LPS. Both LPS species are penta-acylated and mono-phosphorylated, however, P. gingivalis LPS may have a greater proportion of longer-chain fatty acids (as indicated by the mass spectra, gas chromatography data, and previously published structure) (Fig. 7D). Phosphate position may also differ between Bacteroides and P. gingivalis LPS, but further research must be done to confirm this. Ultimately, these subtle differences may explain why P. gingivalis LPS—despite sharing much lipid A structural similarity—is 100 to 1000 times less potent than Bacteroides LPS.
This study has provided an insight on the complexity underlying the structure-function relationship as manifested by the interaction of LPS and TLR4. Indeed, the heterogeneity of lipid A molecular structures in certain Gram-negative bacteria confounds traditional structure-function studies. Further research into the molecular mechanisms behind the production of multiple lipid A species and their impact on the innate immune response hopefully will clarify the increasingly complex interactions observed between host and microbe.
4. Materials and methods
4.1. Bacterial strains and growth conditions
P. gingivalis ATCC 33277 and B. fragilis NCTC 9343 (ATCC 25285) were obtained from the American Type Culture Collection, Rockville, MD. B. thetaiotaomicron VPI-5482 was obtained from the Virginia Polytechnic Institute, Blacksburg, VA. P. gingivalis 1626, a mutant strain that only synthesizes penta-acylated lipid A, was constructed by S. Coats in our lab (unpublished data), and this strain was used for all procedures involving P. gingivalis LPS and lipid A. Cultures were made from frozen bacterial stocks to avoid repetitive subculture. For LPS isolation, bacterial strains were grown anaerobically at 37° C. in TYHK media, which consists of trypticase soy broth (BBL # 211768) at 30 g/L, yeast extract (BD #212750) at 5 g/L, and vitamin K3 (menadione) (Sigma #M-5625) at 1 μg/mL. After autoclaving, filter-sterilized hemin (Fluka #51280) at 5 μg/mL was added. Stationary phase cells were employed for LPS isolation.
4.2. LPS purification
LPS was purified using one of two different methods. Briefly, in the TRI Reagent method, as described previously by Yi and Hackett [22], 50 mL of bacterial culture were centrifuged at 6500 rpm, and the bacterial pellet was rinsed once with sterile water. After this, the pellet was re-suspended in 1 mL of TRI Reagent (MRC #TR 118), a commercial preparation of phenol and guanidine thiocyanate. After thorough mixing, 1/5 volume of chloroform was added and mixed. The mixture was centrifuged for 10 min. at 12 000 rpm in a standard tabletop microcentrifuge, and the top, aqueous layer was retained. LPS-free water was used to re-extract the mixture twice more, and the pooled aqueous phases were lyophilized to produce “crude LPS.” Crude LPS was then washed once with cold 0.375 M MgCl2 in 95% EtOH, centrifuged at 4° C. at 5000 rpm (in a microcentrifuge) for 5 min., and washed thrice more with cold 95% EtOH followed by a final wash with cold 100% EtOH. After drying, the LPS was subjected to a modified Folch extraction to remove contaminating phospholipids [46]. Briefly, LPS was re-suspended to 1% (w/v) in Folch reagent (2:1 ratio of chloroform: MeOH), centrifuged at 4° C. at 10 000 rpm (in a microcentrifuge) for 5 min., and dried. The final product was re-suspended in water and ~1% TEA (to aid in solubility) and considered to be “purified LPS.”
The other procedure used to purify LPS was a modification of the hot phenol-water extraction [23]. Briefly, lyophilized bacterial cells re-suspended in water at 20 mg/mL were mixed with an equal volume of 90% phenol (w/v) and incubated at 70° C for 1 hr. Then, after the mixture was cooled to room temperature, it was centrifuged at approximately 2000 × g for 30 min. The top aqueous layer was retained, and the remaining mixture was extracted with an equal volume of LPS-free water twice more. The pooled aqueous phases were dialyzed against distilled water, frozen, and lyophilized, and the dried product was referred to as “crude LPS.” Crude LPS was then re-suspended in 10 mL of 10 mM Tris (pH 5) and incubated at 37° C. for 2 hrs. in the presence of RNase A at 25 μg/mL and DNase I at 100 μg/mL. Proteinase K was then added at 100 μg/mL, and the mixture was again incubated at 37° C. for 1–2 hrs. After 5 mL of water-saturated phenol were added, the mixture was vortexed and centrifuged at room temperature at 2000 × g for 30 min. Once again, the aqueous phase was retained and dialyzed against distilled water, frozen, and lyophilized. The final product was considered to be “purified LPS.”
4.3. Gas chromatography (GC) of LPS fatty acids
LPS was isolated using a crude hot phenol-water extraction. Briefly, after approximately 20 mg of lyophilized bacteria were re-suspended in 500 μL of water, 500 μL of 90% phenol was added, and the mixture was incubated for 1 hr. in a 70° C. water bath. The mixture was cooled and then centrifuged at room temperature at 10 000 rpm for 10 min., after which the aqueous (top) layer was removed and set aside. The extraction was repeated twice more each with 500 μL of water, and the aqueous phases were pooled. Following the extractions, 2 mL of diethyl ether was added to the pooled aqueous phase, then the mixture was vortexed and centrifuged at 3000 rpm for 5 min. The ether (top) layer was removed and discarded, and the diethyl ether extraction was performed once more. Finally, the aqueous (lower) phase was removed and lyophilized. The fatty acids from this crude LPS preparation were then analyzed by GC as previously described [27]. Briefly, the crude LPS preparation was subjected to methanolysis in 2 M methanolic HCl at 90° C. for 18 hrs. in order to derivatize the fatty acids to methyl esters. After the addition of 200 μL of a saturated NaCl solution, the methyl esters were extracted twice with 400 μL of hexane and analyzed by GC.
4.4. Lipid A purification
Lipid A was purified by a mild acid hydrolysis, as described previously [33]. Briefly, 500 μL of 1% SDS in 10 mM sodium acetate (pH 4.5) was added to 1–3 mg of purified LPS and incubated at 100° C. for 1 hr. After this, the solution was frozen and lyophilized. Next, the dried material was re-suspended in 100 μL LPS-free water and 1 mL of cold, acidified 95% EtOH (made by adding 100 μL of 4 N HCl to 20 mL of 95% EtOH) and centrifuged at 4° C. at 5000 rpm (in a microcentrifuge) for 5 min. The supernatant was discarded, and the lipid A was washed thrice more with cold 95% EtOH. All four washes were repeated, followed by a final wash with cold 100% EtOH. The purified lipid A was then dried and used for MALDI analysis or was re-suspended in water and ~1% TEA for HUVEC stimulation.
4.5. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) of lipid A
Negative ion MALDI-TOF MS of purified lipid A was performed using a Bruker Bioflex III mass spectrometer (Bruker Daltonics, Billerica, MA). Approximately 0.5–1 mg of lipid A was re-suspended in 5–10 μL of CMBT (5-chloro-2-mercaptobenzothiazole) matrix solution (which consisted of 10 mg of CMBT combined with 250 μL of chloroform and 250 μL methanol). The re-suspended lipid A was spotted onto a polished steel target plate (Bruker Daltonics) and analyzed. Detected ion peaks were rounded to the nearest whole number.
4.6. Colloidal gold staining
Colloidal gold staining was used to detect potential protein contamination in the LPS preparations. LPS samples (10 μg) were run on a 4–12% gradient polyacrylamide gel (Invitrogen) at a constant 35 mA for approximately 1 hr. The samples were then transferred to a PVDF membrane at a constant 100 V for 1 hr. The membrane was then stained with the Colloidal Gold Total Protein Stain (Bio-Rad) according to the manufacturer’s instructions. An image was obtained by scanning the membrane with an Epson Perfection 3200 Photo scanner, and the image was converted to grayscale using Adobe Photoshop.
4.7. HUVEC culture, stimulation, and E-selectin detection
Human umbilical vein endothelial cells (HUVECs) were grown and maintained in HUVEC growth media and stimulated with LPS or lipid A as described previously [17, 47]. Following stimulation, E-selectin protein expression was detected by ELISA, as described previously, with a few modifications [17, 47]. Briefly, cells were fixed with 0.5% glutaraldehyde in PBS, after which the cells were washed thrice with PBS. The fixed cells were then incubated at 4° C. overnight with blocking reagent (0.02 M EDTA and 3% goat serum in PBS) mixed with anti-human E-selectin antibody (R & D Systems #BBA1) at 0.25 μg/mL. Cells were rinsed thrice with PBS, and then incubated at room temperature for at least 2 hrs. with horseradish peroxidase-conjugated AffiniPure F(ab′)2 fragment goat anti-mouse IgG antibody (Jackson ImmunoResearch #115-036-071) at 0.4 μg/mL in blocking reagent. Next, cells were again washed thrice with PBS. Finally, cells were briefly incubated at room temperature with 100 μL of TMB (3, 3′, 5, 5′-tetramethylbenzidene), after which 50 μL of 1 N H2SO4 was added to stop the reaction. Color was detected by light absorbance at 450 nm in an ELISA plate reader.
4.8. HEK 293 cell culture, stimulation, and luciferase assays
Human embryonic kidney (HEK) 293 cells were maintained and stimulated with LPS as previously described [20, 48, 49]. Following stimulation, the cells were rinsed with PBS and lysed with 50 μL of 1x passive lysis buffer (PLB) (Promega). Reporter gene expression in 12 μL of each lysate was measured using the Dual-Luciferase Reporter 1000 Assay System (Promega) in a Centro LB 960 Microplate Luminometer (Berthold Technologies) as described previously [20, 48].
4.9. Statistical tests for significance
Significance tests were performed using Microsoft Excel 2007. P-values were determined using the two-sample, two-tailed Student’s T test assuming unequal variance.
Acknowledgments
We would like to thank Brian W. Bainbridge for his many invaluable contributions to this study. We would also like to thank Ilana Cohen for operating the gas chromatograph. This research was supported in part by the National Institute of Health Grant # 5 R01 DE12768.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Erridge C, Bennett-Guerrero E, Poxton IR. Structure and function of lipopolysaccharides. Microbes Infect. 2002;4:837–51. doi: 10.1016/s1286-4579(02)01604-0. [DOI] [PubMed] [Google Scholar]
- 2.Wilson M. Microbial Inhabitants of Humans. Cambridge, UK: Cambridge University Press; 2005. [Google Scholar]
- 3.Kuwahara T, Yamashita A, Hirakawa H, Nakayama H, Toh H, Okada N, et al. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc Natl Acad Sci U S A. 2004;101:14919–24. doi: 10.1073/pnas.0404172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fille M, Mango M, Lechner M, Schaumann R. Bacteroides fragilis group: trends in resistance. Curr Microbiol. 2006;52:153–7. doi: 10.1007/s00284-005-0249-x. [DOI] [PubMed] [Google Scholar]
- 5.Falagas ME, Siakavellas E. Bacteroides, Prevotella, and Porphyromonas species: a review of antibiotic resistance and therapeutic options. Int J Antimicrob Agents. 2000;15:1–9. doi: 10.1016/s0924-8579(99)00164-8. [DOI] [PubMed] [Google Scholar]
- 6.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 7.Dixon DR, Darveau RP. Lipopolysaccharide heterogeneity: innate host responses to bacterial modification of lipid a structure. J Dent Res. 2005;84:584–95. doi: 10.1177/154405910508400702. [DOI] [PubMed] [Google Scholar]
- 8.Weintraub A, Zahringer U, Wollenweber HW, Seydel U, Rietschel ET. Structural characterization of the lipid A component of Bacteroides fragilis strain NCTC 9343 lipopolysaccharide. Eur J Biochem. 1989;183:425–31. doi: 10.1111/j.1432-1033.1989.tb14945.x. [DOI] [PubMed] [Google Scholar]
- 9.Kumada H, Haishima Y, Umemoto T, Tanamoto K. Structural study on the free lipid A isolated from lipopolysaccharide of Porphyromonas gingivalis. J Bacteriol. 1995;177:2098–106. doi: 10.1128/jb.177.8.2098-2106.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Qutub MN, Braham PH, Karimi-Naser LM, Liu X, Genco CA, Darveau RP. Hemin-dependent modulation of the lipid A structure of Porphyromonas gingivalis lipopolysaccharide. Infect Immun. 2006;74:4474–85. doi: 10.1128/IAI.01924-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bainbridge BW, Karimi-Naser L, Reife R, Blethen F, Ernst RK, Darveau RP. Acyl chain specificity of the acyltransferases LpxA and LpxD and substrate availability contribute to lipid A fatty acid heterogeneity in Porphyromonas gingivalis. J Bacteriol. 2008;190:4549–58. doi: 10.1128/JB.00234-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mancuso G, Midiri A, Biondo C, Beninati C, Gambuzza M, Macri D, et al. Bacteroides fragilis-derived lipopolysaccharide produces cell activation and lethal toxicity via toll-like receptor 4. Infect Immun. 2005;73:5620–7. doi: 10.1128/IAI.73.9.5620-5627.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erridge C, Pridmore A, Eley A, Stewart J, Poxton IR. Lipopolysaccharides of Bacteroides fragilis, Chlamydia trachomatis and Pseudomonas aeruginosa signal via toll-like receptor 2. J Med Microbiol. 2004;53:735–40. doi: 10.1099/jmm.0.45598-0. [DOI] [PubMed] [Google Scholar]
- 14.Erridge C, Spickett CM, Webb DJ. Non-enterobacterial endotoxins stimulate human coronary artery but not venous endothelial cell activation via Toll-like receptor 2. Cardiovasc Res. 2007;73:181–9. doi: 10.1016/j.cardiores.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Magnuson DK, Weintraub A, Pohlman TH, Maier RV. Human endothelial cell adhesiveness for neutrophils, induced by Escherichia coli lipopolysaccharide in vitro, is inhibited by Bacteroides fragilis lipopolysaccharide. J Immunol. 1989;143:3025–30. [PubMed] [Google Scholar]
- 16.Chen C, Coats SR, Bumgarner RE, Darveau RP. Hierarchical gene expression profiles of HUVEC stimulated by different lipid A structures obtained from Porphyromonas gingivalis and Escherichia coli. Cell Microbiol. 2007;9:1028–38. doi: 10.1111/j.1462-5822.2006.00849.x. [DOI] [PubMed] [Google Scholar]
- 17.Reife RA, Coats SR, Al-Qutub M, Dixon DM, Braham PA, Billharz RJ, et al. Porphyromonas gingivalis lipopolysaccharide lipid A heterogeneity: differential activities of tetra- and penta-acylated lipid A structures on E-selectin expression and TLR4 recognition. Cell Microbiol. 2006;8:857–68. doi: 10.1111/j.1462-5822.2005.00672.x. [DOI] [PubMed] [Google Scholar]
- 18.Coats SR, Reife RA, Bainbridge BW, Pham TT, Darveau RP. Porphyromonas gingivalis lipopolysaccharide antagonizes Escherichia coli lipopolysaccharide at toll-like receptor 4 in human endothelial cells. Infect Immun. 2003;71:6799–807. doi: 10.1128/IAI.71.12.6799-6807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawada N, Ogawa T, Asai Y, Makimura Y, Sugiyama A. Toll-like receptor 4-dependent recognition of structurally different forms of chemically synthesized lipid As of Porphyromonas gingivalis. Clin Exp Immunol. 2007;148:529–36. doi: 10.1111/j.1365-2249.2007.03346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darveau RP, Pham TT, Lemley K, Reife RA, Bainbridge BW, Coats SR, et al. Porphyromonas gingivalis lipopolysaccharide contains multiple lipid A species that functionally interact with both toll-like receptors 2 and 4. Infect Immun. 2004;72:5041–51. doi: 10.1128/IAI.72.9.5041-5051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darveau RP, Hancock RE. Procedure for isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella typhimurium strains. J Bacteriol. 1983;155:831–8. doi: 10.1128/jb.155.2.831-838.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi EC, Hackett M. Rapid isolation method for lipopolysaccharide and lipid A from gram-negative bacteria. Analyst. 2000;125:651–6. doi: 10.1039/b000368i. [DOI] [PubMed] [Google Scholar]
- 23.Fischer W, Koch HU, Haas R. Improved preparation of lipoteichoic acids. Eur J Biochem. 1983;133:523–30. doi: 10.1111/j.1432-1033.1983.tb07495.x. [DOI] [PubMed] [Google Scholar]
- 24.Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, et al. Complete genome sequence of the oral pathogenic Bacterium porphyromonas gingivalis strain W83. J Bacteriol. 2003;185:5591–601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jousimies-Somer H, Summanen P. Recent taxonomic changes and terminology update of clinically significant anaerobic gram-negative bacteria (excluding spirochetes) Clin Infect Dis. 2002;35:S17–21. doi: 10.1086/341915. [DOI] [PubMed] [Google Scholar]
- 26.Faure E, Equils O, Sieling PA, Thomas L, Zhang FX, Kirschning CJ, et al. Bacterial lipopolysaccharide activates NF-kappaB through toll-like receptor 4 (TLR-4) in cultured human dermal endothelial cells. Differential expression of TLR-4 and TLR-2 in endothelial cells. J Biol Chem. 2000;275:11058–63. doi: 10.1074/jbc.275.15.11058. [DOI] [PubMed] [Google Scholar]
- 27.Somerville JE, Jr, Cassiano L, Bainbridge B, Cunningham MD, Darveau RP. A novel Escherichia coli lipid A mutant that produces an antiinflammatory lipopolysaccharide. J Clin Invest. 1996;97:359–65. doi: 10.1172/JCI118423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sacre SM, Drexler SK, Foxwell BM. Toll-like receptors and rheumatoid arthritis: is there a connection? In: O’Neill LAJ, Brint E, editors. Toll-like Receptors in Inflammation. Birkhauser; Basel, Switzerland: 2005. pp. 19–40. [Google Scholar]
- 29.Asai Y, Hashimoto M, Fletcher HM, Miyake K, Akira S, Ogawa T. Lipopolysaccharide preparation extracted from Porphyromonas gingivalis lipoprotein-deficient mutant shows a marked decrease in toll-like receptor 2-mediated signaling. Infect Immun. 2005;73:2157–63. doi: 10.1128/IAI.73.4.2157-2163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogawa T, Asai Y, Makimura Y, Tamai R. Chemical structure and immunobiological activity of Porphyromonas gingivalis lipid A. Front Biosci. 2007;12:3795–812. doi: 10.2741/2353. [DOI] [PubMed] [Google Scholar]
- 31.Manthey CL, Vogel SN. Elimination of trace endotoxin protein from rough chemotype LPS. J Endotoxin Res. 1994;1:84–91. [Google Scholar]
- 32.Alexander C, Rietschel ET. Bacterial lipopolysaccharides and innate immunity. J Endotoxin Res. 2001;7:167–202. [PubMed] [Google Scholar]
- 33.Caroff M, Tacken A, Szabo L. Detergent-accelerated hydrolysis of bacterial endotoxins and determination of the anomeric configuration of the glycosyl phosphate present in the “isolated lipid A” fragment of the Bordetella pertussis endotoxin. Carbohydr Res. 1988;175:273–82. doi: 10.1016/0008-6215(88)84149-1. [DOI] [PubMed] [Google Scholar]
- 34.Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, et al. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol. 1998;27:1171–82. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 35.Kawahara K, Tsukano H, Watanabe H, Lindner B, Matsuura M. Modification of the structure and activity of lipid A in Yersinia pestis lipopolysaccharide by growth temperature. Infect Immun. 2002;70:4092–8. doi: 10.1128/IAI.70.8.4092-4098.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montminy SW, Khan N, McGrath S, Walkowicz MJ, Sharp F, Conlon JE, et al. Virulence factors of Yersinia pestis are overcome by a strong lipopolysaccharide response. Nat Immunol. 2006;7:1066–73. doi: 10.1038/ni1386. [DOI] [PubMed] [Google Scholar]
- 37.Bainbridge BW, Coats SR, Pham TT, Reife RA, Darveau RP. Expression of a Porphyromonas gingivalis lipid A palmitylacyltransferase in Escherichia coli yields a chimeric lipid A with altered ability to stimulate interleukin-8 secretion. Cell Microbiol. 2006;8:120–9. doi: 10.1111/j.1462-5822.2005.00605.x. [DOI] [PubMed] [Google Scholar]
- 38.Miyake K. Innate recognition of lipopolysaccharide by Toll-like receptor 4-MD-2. Trends Microbiol. 2004;12:186–92. doi: 10.1016/j.tim.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 39.Kumada H, Haishima Y, Watanabe K, Hasegawa C, Tsuchiya T, Tanamoto K, et al. Biological properties of the native and synthetic lipid A of Porphyromonas gingivalis lipopolysaccharide. Oral Microbiol Immunol. 2008;23:60–9. doi: 10.1111/j.1399-302X.2007.00392.x. [DOI] [PubMed] [Google Scholar]
- 40.Zahringer U, Lindner B, Inamura S, Heine H, Alexander C. TLR2 - promiscuous or specific? A critical re-evaluation of a receptor expressing apparent broad specificity. Immunobiology. 2008;213:205–24. doi: 10.1016/j.imbio.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 41.Hirschfeld M, Ma Y, Weis JH, Vogel SN, Weis JJ. Cutting edge: repurification of lipopolysaccharide eliminates signaling through both human and murine toll-like receptor 2. J Immunol. 2000;165:618–22. doi: 10.4049/jimmunol.165.2.618. [DOI] [PubMed] [Google Scholar]
- 42.Hirschfeld M, Weis JJ, Toshchakov V, Salkowski CA, Cody MJ, Ward DC, et al. Signaling by toll-like receptor 2 and 4 agonists results in differential gene expression in murine macrophages. Infect Immun. 2001;69:1477–82. doi: 10.1128/IAI.69.3.1477-1482.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rebeil R, Ernst RK, Gowen BB, Miller SI, Hinnebusch BJ. Variation in lipid A structure in the pathogenic yersiniae. Mol Microbiol. 2004;52:1363–73. doi: 10.1111/j.1365-2958.2004.04059.x. [DOI] [PubMed] [Google Scholar]
- 44.Takada H, Kotani S. Structure-function relationships of lipid A. In: Morrison DC, Ryan JL, editors. Bacterial Endotoxic Lipopolysaccharides. CRC Press; Boca Raton, FL: 1992. pp. 107–34. [Google Scholar]
- 45.Loppnow H, Brade H, Durrbaum I, Dinarello CA, Kusumoto S, Rietschel ET, et al. IL-1 induction-capacity of defined lipopolysaccharide partial structures. J Immunol. 1989;142:3229–38. [PubMed] [Google Scholar]
- 46.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 47.Darveau RP, Cunningham MD, Bailey T, Seachord C, Ratcliffe K, Bainbridge B, et al. Ability of bacteria associated with chronic inflammatory disease to stimulate E-selectin expression and promote neutrophil adhesion. Infect Immun. 1995;63:1311–7. doi: 10.1128/iai.63.4.1311-1317.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hajjar AM, Ernst RK, Tsai JH, Wilson CB, Miller SI. Human Toll-like receptor 4 recognizes host-specific LPS modifications. Nat Immunol. 2002;3:354–9. doi: 10.1038/ni777. [DOI] [PubMed] [Google Scholar]
- 49.Hajjar AM, O’Mahony DS, Ozinsky A, Underhill DM, Aderem A, Klebanoff SJ, et al. Cutting edge: functional interactions between toll-like receptor (TLR) 2 and TLR1 or TLR6 in response to phenol-soluble modulin. J Immunol. 2001;166:15–9. doi: 10.4049/jimmunol.166.1.15. [DOI] [PubMed] [Google Scholar]