Abstract
Immunogenicity caused by the use of non-human enzymes in Antibody Directed Enzyme Prodrug Therapy (ADEPT) has limited its clinical application. To overcome this problem, we have developed a mutant human purine nucleoside phosphorylase (PNP), which unlike the wild-type enzyme, accepts (deoxy)adenosine-based prodrugs as substrates. Amongst the different mutants of human PNP tested, a double mutant with amino acid substitutions E201Q:N243D (hDM) is most efficient in cleaving (deoxy)adenosine-based prodrugs. While hDM is capable of utilizing multiple prodrugs as substrates, it is most effective at cleaving 2-fluoro-2′-deoxyadenosine to a cytotoxic drug. To target hDM to the tumor site, the enzyme was fused to an Anti-HER2/neu Peptide mimetic (AHNP). Treatment of HER2/neu expressing tumor cells with hDM-AHNP results in cellular localization of enzyme activity. As a consequence, harmless prodrug is converted to a cytotoxic drug in the vicinity of the tumor cells, resulting in tumor cell apoptosis. Unlike the non-human enzymes, the hDM should have minimal immunogenicity when used in ADEPT thus providing a novel promising therapeutic agent for the treatment of tumors.
Keywords: ADEPT, Immunogenicity, Enzyme, Prodrug, Cytotoxicity
Introduction
Antibody Directed Enzyme Prodrug Therapy (ADEPT) is a two step cancer therapy. First an antibody-enzyme fusion is injected intravenously. This fusion protein binds selectively to tumor antigens via its antibody component, and localizes the enzyme to the tumor site (1–4). Secondly, after the unbound fraction is cleared from circulation, a non-toxic prodrug is systemically administered. The prodrug is converted to a potent cytotoxic drug by the antibody-enzyme fusion protein only at the tumor site. To date, only enzymes of non-human origin have been utilized in ADEPT, and despite promising results, their immunogenicity has restricted ADEPT to phase I clinical trials (5–7). However, if a human enzyme is used, the prodrug will be converted to a cytotoxic drug not only in the vicinity of the tumor, but also at sites where the endogenous enzyme is present, causing systemic toxicity. One approach to overcoming this problem is to engineer a human enzyme with altered specificity, so that it can cleave a prodrug that is not cleaved by the naturally occurring enzyme. If the engineered human enzyme contains minimal changes in its structure and amino acid composition compared to the wild-type, it should be far less immunogenic than the currently used bacterial enzymes. Therefore, we have developed mutants of human purine nucleoside phosphorylase (hPNP) with altered substrate specificity for use in ADEPT.
hPNP, a ubiquitously expressed homotrimer, catalyzes the reversible phosphorolysis of 6-oxo purine nucleosides to the corresponding free purine base and ribose 1-phosphate, but does not accept adenosine or adenosine-based prodrugs as substrates (8–10). In contrast, homohexameric E. coli PNP (ePNP) efficiently converts adenosine-based prodrugs to adenine containing drugs (11) that can freely diffuse across cell membranes and are toxic to both dividing and non-dividing cells (11–15), including the stromal cells that support tumor growth. Although these characteristics make ePNP an attractive candidate for ADEPT, the immunogenicity resulting from its bacterial origin limits the number of treatments that can be administered to cancer patients.
In the present study we have used available crystallographic and enzyme-substrate studies (8–10, 16–22) to rationally design hPNP mutants that can cleave adenosine-based prodrugs not recognized by wild-type hPNP. In particular, a double mutant of hPNP, E201Q:N243D (hDM) that is fused to an Anti-HER2/neu Peptide mimic (23) (AHNP) is able to utilize (deoxy)adenosine-based prodrugs Cladribine, 2-fluoroadenosine, and 2-fluoro-2'-deoxyadenosine as substrates, with the highest enzymatic efficiency seen with 2-fluoro-2'-deoxyadenosine. hDM-AHNP localizes to HER2/neu-expressing tumors, and produces the toxic metabolite 2-fluoro-adenine by the phosphorolysis of 2-fluoro-2'-deoxyadenosine. Generation of cytotoxic drug results in tumor cell apoptosis, making hDM-AHNP and 2-fluoro-2'-deoxyadenosine a novel enzyme-prodrug combination for treating HER2/neu expressing tumors.
Materials and Methods
Materials
Adenosine, guanosine, xanthine oxidase from buttermilk, Cl-dAdo, F-Ado, and F-Ade were purchased from Sigma-Aldrich (St. Louis, MO). F-dAdo was purchased from Berry & Associates (Dexter, MI) and Fludarabine was from Berlex (Alameda, California). CT26 cell line was purchased from ATCC (Manassas, VA). Construction and characterization of CT26HER2/neu is described previously (24). MCF7-HER2 was a gift from Dr. Dennis Slamon (University of California, Los-Angeles). Cells were cultured in ISCOVE’s Modified Dulbecco’s Medium (IMDM; GIBCO, Carlsbad, CA) containing 5% calf-serum (GIBCO) for CT26 and CT26HER2/neu and IMDM containing 10% fetal bovine serum (GIBCO), 1% non-essential amino acids (GIBCO) and 1% sodium pyruvate (GIBCO) for MCF-7HER2 cells. hPNP was purchased from Calbiochem (Los Angeles, CA). ECDHER2-Fc was purchased from R & D SYSTEMS (Minneapolis, MN). Expression vectors for expression of TEV enzyme and ECDHER2 were gifts from Dr. James Bowie (University of California, Los-Angeles) and Dr. James Marks (University of California, San-Francisco), respectively.
Cloning, expression, and purification of all proteins, as well as binding assays using ELISA and flow cytometry are described in Supplementary Materials and Methods
Determination of kinetic parameters
For all enzyme reactions, the concentration of the enzyme was adjusted such that product formation was linear with respect to time. Unless stated, all enzyme reactions were performed in triplicate in 96-well UV plates at 37°C in a final volume of 100 µl containing 125 mM KH2PO4 (pH 7.4) and 50 mM HEPES. Following addition of substrates, a SpectraMax M5 spectrophotometer (Molecular Devices; Sunnyvale, CA) was used to monitor the enzymatic reactions. The Michaelis-Menten kinetic parameters were determined using Lineweaver-Berk plots of mili-units of absorbance/min versus 1/concentration of substrate. Units of absorbance/min were converted to µM/min using the extinction coefficient of either substrate consumed or product formed. Vmax was then converted to kcat.
Enzymatic activity of PNPs with adenosine, and guanosine
Phosphorolysis of adenosine to adenine was followed in a continuous reaction containing xanthine oxidase (XOD) by following the decrease in absorbance at 260 nm and the increase in absorbance at 301 nm. Adenosine was converted to adenine by PNP, and adenine was hydrolyzed to 2,8-dihydroxyadenine by XOD. Both adenosine and adenine absorb at 260 nm, but 2,8-dihydroxyadenine absorbs at 301 nm. Molar extinction coefficients for adenosine and 2,8-dihydroxyadenine are 15,400 and 15,200 M−1cm−1, respectively. Phosphorolysis of guanosine to guanine was followed by the decrease in absorbance at 257 nm using a molar extinction coefficient of 13,700 M−1cm−1 for guanosine.
Enzymatic activity of PNPs with F-dAdo, Cl-dAdo, F-Ado, and Fluradabine
Enzymatic cleavage of F-dAdo to F-Ade by PNPs was followed by a decrease in absorbance at 260 nm and a concurrent increase in absorbance at 280 nm. The conversion of Cl-dAdo and F-Ado to the corresponding products was followed in a coupled enzyme reaction containing XOD with product formation monitored by a decrease in absorbance at 250 nm and a simultaneous increase in absorbance at 280 nm. The molar extinction coefficients of F-dAdo were determined to be 16,300 M−1cm−1 at 260 nm and 1,300 M−1cm−1 at 280 nm. The molar extinction coefficients of Cl-dAdo and F-Ado were 10,000 M−1cm−1 and 12,000 M−1cm−1 at 250 nm, respectively. The ability of PNPs to cleave Fluradabine was determined in a cell-based assay. First, the toxicity of Fluradabine with CT26-HER2/neu cells was determined. Following overnight growth of cells seeded at 5×103 cells/well in a 96-well tissue culture plate, different concentrations of Fluradabine alone or Fluradabine and 2 µM of ePNP were added and the level of cell proliferation following incubation at 37°C for 72 hours was determined by MTS assay (CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay, Promega; Madison, WI). At Fluradabine concentrations exceeding 17 µM, a dose dependent decrease in cellular proliferation was observed, but at lower concentrations there was no inhibition of growth. When both Fluradabine and ePNP were added, inhibition of cellular proliferation was observed at Fluradabine concentrations of 8 µM. Therefore, it was concluded that at concentrations below 17 µM, Fluradabine does not impair cell proliferation, but when present at 8 µM, it is converted to a cytotoxic product by ePNP that inhibits cell proliferation. Therefore, 8 µM Fluradabine was used with varying concentrations of ePNP and hPNPs to determine their enzymatic activity.
SPR analysis of the interaction of hDM-αH-AHNP with ECDHER2
Binding of hDM-αH-AHNP to ECDHER2-Fc was studied using surface plasmon resonance on a BIAcore T100. ECDHER2-Fc was immobilized on the surface of a CM5 sensor chip following the standard amine coupling procedure based on the manufacturer’s recommendation, achieving a surface density of 1,136 resonance units. The remaining active groups were blocked by injecting ethanolamine. The reference surface was generated following the same procedure, but without protein. Following each binding cycle, the chip was regenerated using 0.2% DMSO (v/v). hDM-αH-AHNP was injected at various concentration at 20 µl/min. hPNP was injected under the same condition, but only at a concentration equal to the highest concentration of hDM-αH-AHNP. Binding of hDM-αH-AHNP to the immobilized ligand was monitored in real-time by following association and dissociation phases on the experimental surface subtracted from the blank surface using BIAevaluation 3.0 software.
Association of hDM-αH-AHNP and 6XHis-ePNP-GS-AHNP enzymatic activity with CT26-HER2/neu expressing cells
CT26 and CT26-HER2/neu cells were seeded at 5×103 cells per well in a 96-well microtiter plate in 50 µl of growth medium. The next day 50 µl of different dilutions of hDM-αH-AHNP were added in triplicate to cells and incubated for one hour at room temperature. The unbound proteins were carefully pipetted out, and each well was washed twice with 200 µl of cold medium. Different dilutions of 6XHis-ePNP-GS-AHNP were incubated with cells at 4°C for one hour and washed five times with ice cold medium containing 20% calf-serum. The cells were then resuspended in 100 µl of growth medium containing F-dAdo at a final concentration of 1.5 and 6 µM for CT26 or CT26HER2/neu cells, and MCF-7HER2 cells respectively, and incubated at 37°C. Inhibition of cell growth (after 72 hours) or induction of apoptosis (after 60 hours) was determined by using a MTS assay, or an Apoptosis Assay Kit (cat # V13241, Molecular Probes, Invitrogen; Carlsbad, CA), respectively. Induction of DNA fragmentation due to apoptosis was examined 72 hours post addition of F-dAdo as described previously (25). To block the interaction of AHNP in hDM-αH-AHNP with HER2/neu expressed on the surface of CT26-HER2/neu cells, different concentrations of soluble ECDHER2 were incubated overnight at 4°C with hDM-αH-AHNP with mild shaking. The mix was then added to cells that had been seeded the night before and the assay was repeated as described above. For controls, F-dAdo, hDM-αH-AHNP, hPNP-GS-AHNP, hPNP-GS-AHNP plus F-dAdo, 6XHis-ePNP-GS-AHNP or ECDHER2 at the highest concentrations used was added to cells and the degree of cell proliferation determined.
Molecular modeling
The structure of hPNP with guanine (1ULB) was superimposed on the structure of ePNP in complex with F-dAdo (1PKE), and viewed by PyMol (26). The two proteins share a conserved subunit fold; hence their superimposition was possible.
Results
Substrate specificity of mutant hPNPs with adenosine, F-dAdo, and guanosine
In order to change the substrate specificity of hPNP so that it could cleave adenosine and adenosine-containing prodrugs, the single mutant N243D (hSM) or the double mutant E201Q:N243D (hDM) were introduced. Both mutants were fused to an S-tag at their N-terminus to facilitate purification.
Asn243 in hPNP was previously recognized as the residue responsible for its substrate specificity (9, 10, 17, 18). In contrast to hPNP, ePNP, which is highly active with adenosine and adenosine-based prodrugs, has Asp204 that corresponds to Asn243 of hPNP (11, 17, 27). This Asp residue is conserved in all prokaryotic PNPs that phosphorolyse adenosine to adenine (17, 27–29). The S-tagged hSM (S-hSM) converted adenosine to adenine with a Km value of 501 µM, and an overall efficiency of 296 M−1s−1 (Table 1). At the concentrations used, S-tagged wild-type human PNP (S-hPNP) did not show any activity with adenosine (Table 1). Importantly, it was previously shown that hSM was able to convert the cytotoxic drug 2-amino adenine to 2-amino-2’-deoxyadenosine (2-amino-2’-dA) (10). Given that the reaction is reversible, we hypothesized that hSM would also be able to catalyze the reaction in which an adenosine-based prodrug is converted to a toxic metabolite. Indeed, S-hSM utilized the relatively non-toxic deoxyadenosine-based prodrug, 2-fluoro-2’-deoxyadenosine (F-dAdo) as a substrate with a Km value of 3,735 µM and an overall efficiency of 26 M−1s−1 (Table 2). Therefore, hSM is capable of cleaving both adenosine and F-dAdo, with 100-fold lower efficiency for F-dAdo than for adenosine.
Table 1.
Kinetic constants of engineered PNP fusion proteins for adenosine and guanosine
| Substrate: Adenosine | Substrate: Guanosine | |||||
|---|---|---|---|---|---|---|
| Km (µM) | kcat (s−1) | kcat/Km (M−1 s−1) | Km (µM) | kcat (s−1) | kcat/Km (M−1 s−1) | |
| S-hPNP | NDA | NDA | NDA | 32 ± 4 | 37 ± 4 | 1.16×106 |
| S-hSM (N243D) | 501 ± 55 | 0.148 ± 0.018 | 296 | 133 ± 12 | 0.092 ± 0.007 | 689 |
| S-hDM (E201Q+N243D) | 679 ± 50 | 0.050 ± 0.003 | 74 | 476 ± 54 | 0.0077 ± 0.0006 | 16 |
| hPNP-GS-AHNP | NDA | NDA | NDA | - | - | - |
| hDM-GS-AHNP ( E201Q+N243D) | 83 ± 6 | 0.050 ± 0.001 | 651 | - | - | - |
| 6XHis-ePNP-GS-AHNP | 22 ± 4 | 2 ± 1 | 90,554 | - | - | - |
-, not determined. NDA, no detectable activity.
Values determined from Lineweaver-Berk plots. Assays were conducted with variable concentrations of nucleoside substrates and fixed concentrations of phosphate and enzyme. Kinetic parameters are determined from at least three independent studies with each study performed in triplicate.
Table 2.
Kinetic constants for engineered PNP fusion proteins for 2-fluoro-2'-deoxyadenosine (F-dAdo)
| Substrate: F-dAdo | |||
|---|---|---|---|
| Km (µM) | kcat (s−1) | kcat/Km (M−1 s−1) | |
| S-hPNP | NDA | NDA | NDA |
| S-hSM (N243D) | 3,735 ± 220 | 0.099 ± 0.005 | 26 |
| S-hDM ( E201Q:N243D) | 1,178 ± 98 | 0.044 ± 0.001 | 37 |
| hPNP-GS-AHNP | NDA | NDA | NDA |
| hDM-GS-AHNP ( E201Q+N243D) | 174 ± 15 | 0.05 ± 0.004 | 285 |
| hDM-αH -AHNP ( E201Q+N243D) | 121 ± 14 | 0.065 ± 0.009 | 533 |
| 6XHis-ePNP-GS-AHNP | 33 ± 8 | 0.64 ± 0.1 | 19,461 |
NDA, no detectable activity.
Values determined from Lineweaver-Berk plots. Assays were conducted with variable concentrations of nucleoside substrates and fixed concentrations of phosphate and enzyme. Kinetic parameters are determined from at least three independent studies with each study performed in triplicate.
F-dAdo has a highly electronegative fluorine atom at the C2-position of the purine base. Based on the crystallographic studies, the double mutant of hPNP, E201Q:N243D (hDM), is predicted to tolerate the fluorine substitution at the C2 position of F-dAdo better than hSM. Although S-hDM was 4-fold less efficient than S-hSM in phosphorolysis of adenosine, (Table 1), it was somewhat more effective than S-hSM in cleaving F-dAdo to a cytotoxic drug, largely as a consequence of decreased Km (Table 2).
Conversion of F-dAdo to the cytotoxic drug, F-Ade by S-hDM was verified using a cell based assay in which CT26-HER2/neu tumor cells were incubated with the enzyme in the presence or absence of F-dAdo. Cells proliferated normally when incubated with S-hPNP and F-dAdo; however, in the presence of S-hDM or S-tagged ePNP, and F-dAdo, the toxic end product generated by either enzyme resulted in an inhibition of cell proliferation comparable to that observed in the presence of equal molar concentrations of F-Ade (Supplementary Fig. S1).
Guanosine is a natural substrate of hPNP (21). The wild-type enzyme catalyzes the reversible phosphorolysis of guanosine to guanine and ribose-1-phosphate with high efficiency. While the mutant PNPs were capable of utilizing guanosine as a substrate, they exhibited lower enzymatic efficiency than the hPNP. The S-hSM with a Km value of 133 µM shows a 4-fold reduced affinity for phosphorolysis of guanosine and an overall reduction of 1,700-fold in the catalytic efficiency when compared to wild-type enzyme (Table 1). Compared to S-hPNP, S-hDM showed an almost 15-fold reduced affinity for guanosine, indicating that the residue E201 plays an important role in phosphorolysis of guanosine (Table 1).
In an attempt to further increase the activity of hDM with (deoxy)adenosine-based prodrugs, additional mutations were introduced within either the base- or ribose-binding pocket of the enzyme (28). However, none of the additional mutations resulted in enhanced enzyme activity with F-dAdo (Supplementary Table S1), and overall, hDM was the most efficient of the enzymes tested in cleaving the prodrug, F-dAdo.
hDM is active when fused to AHNP
To target the enzyme to the tumor site, hDM was fused at its C-terminus to a 12-amino acid exocyclic peptide called Anti-HER2/neu Peptide mimic (AHNP) (23). AHNP was designed based on the structure of the CDR-H3 loop of the anti-HER2/neu recombinant monoclonal antibody 4D5 and binds specifically to the extracellular domain of HER2/neu (23, 30–32) with an affinity of 150 nM (32). We hypothesized that by fusing AHNP to the C-terminus of hDM, active enzyme would be specifically targeted to HER2/neu expressing cells via the AHNP.
When AHNP was fused at the C-terminus of hDM via a flexible Gly3SerGly linker (GS) (yielding hDM-GS-AHNP), its kcat using adenosine as substrate was comparable to S-hDM. However, an 8-fold increase in affinity of hDM-GS-AHNP for adenosine resulted in an 8-fold improvement in overall kinetic efficiency (Table 1), suggesting that the N-terminal S-tag might interfere with enzyme-substrate binding. The N-terminus of hPNP is in close proximity to the enzyme’s binding pocket and the N-terminal fusion may negatively influence enzyme activity through steric interference. ePNP and hPNP were also fused at their C-terminus to AHNP via a GS linker, producing 6XHis-ePNP-GS-AHNP, and hPNP-GS-AHNP, as positive and negative controls, respectively. When the enzymatic activities of the AHNP fusion proteins were compared using adenosine as substrate, hDM-GS-AHNP remained 140 fold less efficient than 6XHis-ePNP-GS-AHNP (Table 1). At the concentration used, hPNP-GS-AHNP was not active (Table 1). Importantly, hDM-GS-AHNP utilized F-dAdo with kcat values comparable to S-hDM, but with ~7-fold lower Km, making hDM-GS-AHNP a more efficient enzyme (Table 2). When compared to 6XHis-ePNP-GS-AHNP, hDM-GS-AHNP was approximately 70-fold less active with F-dAdo as substrate (Table 2).
When the flexible GS linker was replaced with a rigid α-helical linker (αH) (33, 34), hDM-αH-AHNP was found to be 2-fold more active using F-dAdo as substrate (Table 2). The increased activity was possibly due to a better spatial separation that eliminated unfavorable interactions between AHNP and hDM.
AHNP targets hDM to the extracellular domain of HER2/neu
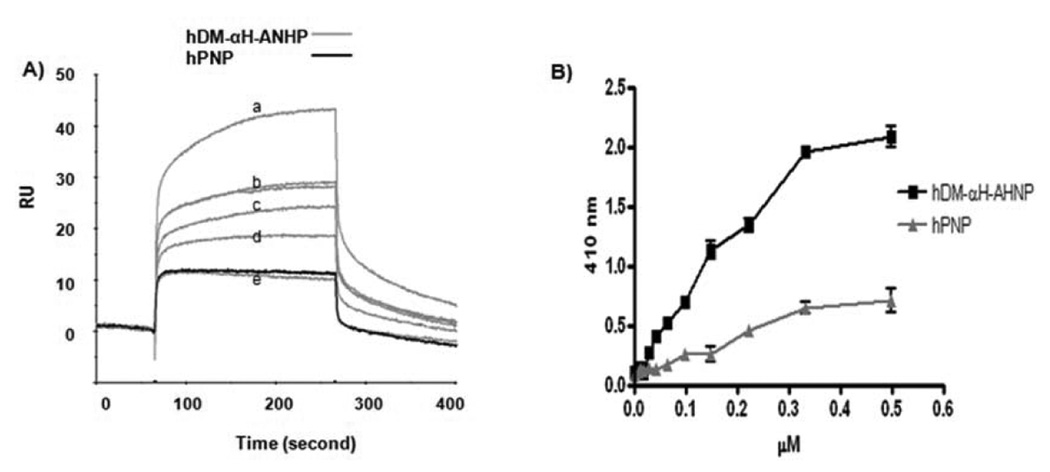
In order to demonstrate that AHNP in the fusion protein hDM-αH-AHNP specifically targets the enzyme to HER2/neu, binding of hDM-αH-AHNP to ECDHER2-Fc was examined by surface plasmon resonance as described above. Analysis by global fitting using a 1:1 Langmuir model, with the binding of each AHNP to ECDHER2-Fc assumed to be independent of the others, yielded a KD of 430 nM (Fig. 1A), 3-fold higher than the published value for AHNP alone (150 nM) (32). Binding of hPNP (Calbiochem) at a concentration equal to the highest concentration of hDM-αH-AHNP to ECDHER2-Fc immobilized on the sensor chip was negligible (Fig. 1A). In addition, biotinylated hDM-αH-AHNP showed a dose dependent binding to ECDHER2 immobilized on microtiter plates, with half maximum binding at ~ 0.17 µM (Fig. 1B). Specific binding by hDM-αH-AHNP greatly exceeded the non-specific binding seen by biotinylated hPNP (Fig. 1B).
Figure 1.
Specific binding of hDM-αH-AHNP to ECDHER2-Fc immobilized on the surface of a SPR chip or a microtiter plate. A, hDM- αH-ANP at concentrations of a) 0.5, b) 0.25, c) 0.2, d) 0.15, and e) 0.1 µM was flowed across the sensor chip at 20 µl/min for 220 second. Binding of hPNP at a concentration of 0.5 µM to immobilized ECDHER2-Fc is shown in black. B, Binding of biotinylated hDM-αH-AHNP and hPNP to ECDHER2 immobilized on microtiter plates was detected using streptavidin-AP. Error bars represent standard deviation within each set of values. These experiments were repeated two independent times with each study performed in triplicate.
Hexameric 6XHis-ePNP-GS-AHNP bound ECDHER2-Fc immobilized on the CM5 chip with an affinity of ~ 2.3 nM, a strong interaction owing to the slow off-rate (~ 8.5×10−5 1/s, data not shown). Specific binding of 6XHis-ePNP-GS-AHNP to ECDHER2 and to cells expressing HER2/neu was also seen using ELISA and flow-cytometry (Supplementary Fig. S2).
hDM-αH-AHNP is capable of using multiple prodrugs as substrate
In addition to F-dAdo, two other (deoxy)adenosine-based prodrugs, Cladribine (Cl-dAdo) and 2-fluoroadenosine (F-Ado) were evaluated as substrates for hDM-αH-AHNP. F-Ado and Cl-dAdo are adenosine and deoxyadenosine analog prodrugs, respectively, with Cl-dAdo currently being used to treat leukemia (35). hDM-αH-AHNP was active with both Cl-dAdo and F-Ado as substrates, with Km values of 838 and 447 µM, respectively (Table 3). Although hDM-αH-AHNP had a 3-fold higher kcat value for F-Ado compared to F-dAdo, the overall kinetic efficiency of the enzyme was slightly higher for F-dAdo. 6XHis-ePNP-GS-AHNP had the highest enzyme efficiency with F-Ado, followed by F-dAdo and Cl-dAdo (Table 2 and Table 3). Enzymatic activity of hPNPs with Fludarabine as substrate was tested by their ability to convert Fludarabine to a cytotoxic drug that would inhibit cell proliferation. CT26-HER2/neu cells grow normally in the presence of 8 µM of Fludarabine, but when 6XHis-ePNP-GS-AHNP was also present, generation of toxic product due to enzymatic activity of ePNP resulted in 90% inhibition of cell-growth. In contrast to ePNP, addition of Hpnp mutants did not result in growth inhibition of either CT26-HER2/neu or CT26 cells, indicating that these mutant enzymes could not use Fludarabine as substrate.
Table 3.
Comparison of kinetic constants for adenosine-based prodrugs of hPNP-GS-AHNP, hDM-αH-AHNP, and 6XHis-ePNP-GS-AHNP
| Substrate: Cl-dAdo | Substrate: F-Ado | |||||
|---|---|---|---|---|---|---|
| Km (µM) | kcat (s−1) | kcat/Km (M−1 s−1) | Km (µM) | kcat (s−1) | kcat/Km (M−1 s−1) | |
| hPNP-GS-AHNP | NDA | NDA | NDA | NDA | NDA | NDA |
| hDM-αH –AHNP ( E201Q:N243D) | 838 ± 77 | 0.12 ± 0.01 | 147.3 | 447 ± 33 | 0.19 ± 0.009 | 429 |
| 6X His-ePNP-GS-AHNP | 54 ± 9 | 0.89 ± 0.03 | 16,435 | 66 ± 3 | 1.95 ± 0.4 | 29,525 |
NDA, no detectable activity.
Values determined from Lineweaver-Berk plots. Assays were conducted with variable concentrations of nucleoside substrates and fixed concentrations of phosphate and enzyme. Kinetic parameters are determined from at least three independent studies with each study performed in triplicate.
In vitro cytotoxicity of F-dAdo in the presence of localized hDM-αH-AHNP or 6XHis-ePNP-GS-AHNP
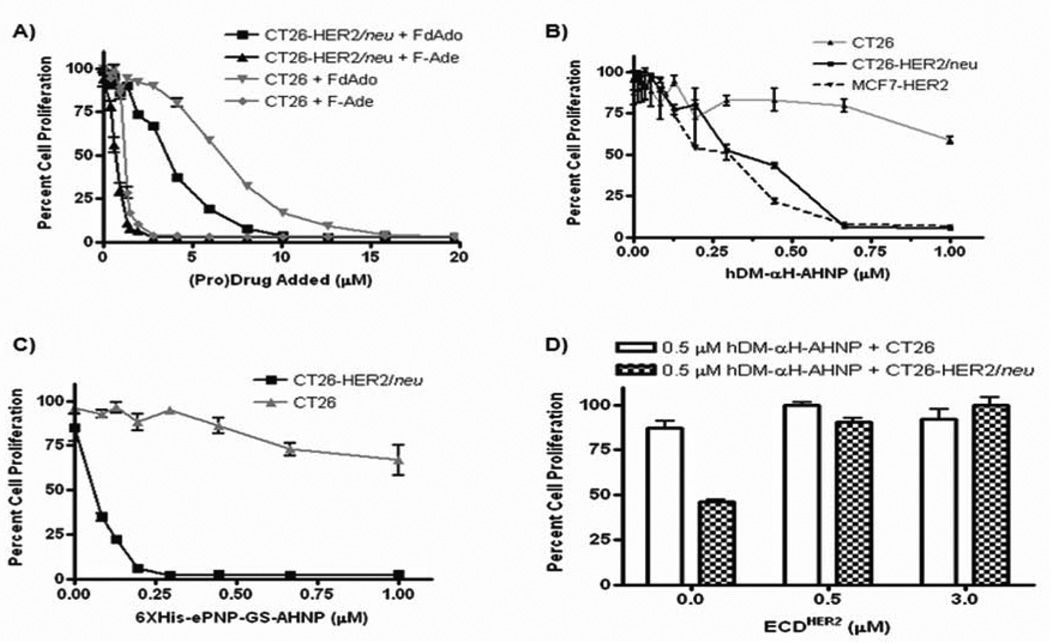
To determine if AHNP association with HER2/neu can specifically target the enzyme to HER2/neu expressing tumor cells, the fusion protein was added to CT26-HER2/neu or CT26 cells, unbound protein washed away, and F-dAdo added. If the fusion protein remained bound to the cells, the enzyme would catalyze phosphorolysis of F-dAdo to F-Ade, resulting in inhibition of cell proliferation. To determine the optimal prodrug concentration, different dilutions of F-dAdo were added to cells and after 72 hours the degree of cell proliferation determined by MTS assay. CT26-HER2/neu or CT26 cells grow normally if F-dAdo was added at concentrations of 2 µM or lower (Fig. 2A). In contrast, addition of 1.5 µM F-Ade inhibited CT26-HER2/neu or CT26 proliferation by at least 85% (Fig. 2A). Therefore addition of 1.5 µM F-dAdo will result in significant cytotoxicity only if it is converted to F-Ade by the enzymatic activity of PNP. Different concentrations of hDM-αH-AHNP or 6XHis-ePNP-GS-AHNP were added to cells and the unbound fraction washed away, followed by addition of F-dAdo at 1.5 µM. This resulted in a dose-dependent inhibition of proliferation with CT26-HER2/neu but not with CT26 (Figs. 2B and 2C). Cytotoxicity was seen only when fusion protein and F-dAdo were added, since addition of enzyme or prodrug alone (at the highest concentration) did not cause any cytotoxicity resulting in inhibition of cell proliferation (data not shown). Consistent with its greater enzyme activity, cytotoxicity was seen at lower concentrations of 6XHis-ePNP-GS-AHNP than hDM-αH-AHNP (Figs. 2B and 2C). Incubation of the hDM-αH-AHNP with soluble ECDHER2 prior to addition to CT26-HER2/neu cells inhibited the interaction of the fusion protein with HER2/neu expressed on the cell surface and restored cell proliferation to normal levels (Fig. 2D).
Figure 2.
AHNP specifically targets hDM or ePNP to the extracellular domain of HER2/neu. Error bars represent standard deviation within each set of values. A, Inhibition of proliferation of CT26 and CT26-HER2/neu cells was examined in the presence of different concentrations of F-dAdo and F-Ade. Different concentrations of (pro)drug were added to cells and inhibition of cell proliferation was determined by MTS. B, hDM-αH-AHNP and C, 6XHis-ePNP-GS-AHNP associate with HER2/neu expressing cells resulting in cytotoxicity upon addition of F-dAdo to CT26, CT26HER2/neu or MCF-7HER2 cells. Different concentrations of either hDM-αH-AHNP or 6XHis-ePNP-GS-AHNP were incubated with cells and unbound enzyme washed away. Then F-dAdo (1.5 µM for CT26 or CT26HER2/neu cells and 6 µM for MCF7-HER2 cells) was added and 72 hours later cellular proliferation was determined by MTS assay. D, Soluble ECDHER2 inhibits the cell-association of hDM-αH-AHNP and generation of cytotoxicity upon F-dAdo addition. hDM-αH-AHNP was incubated overnight with different concentrations of ECDHER2 prior to addition to cells. The assay was completed as described above. All studies, except for MCF-7HER2 cells in B were repeated at least three independent times, each time in triplicate.
hDM-αH-AHNP was also capable of associating with MCF-7HER2, a human breast cancer cell line that was transfected with full-length HER2/neu cDNA to increase its expression of HER2/neu. MCF-7HER2 cells proliferated normally in the presence F-dAdo or hDM-αH-AHNP alone (data not shown). However, the enzymatic activity of the hDM-αH-AHNP that remained bound to HER2/neu expressed on MCF7-HER2 cells resulted in conversion of F-dAdo to F-Ade and inhibition of cell proliferation (Fig. 2B).
Inhibition of cellular proliferation as a result of conversion of F-dAdo to F-Ade was also seen with non-HER2/neu expressing tumor cells if the cells were not washed after addition of hDM-αH-AHNP. Proliferation of 38C13 cells, a mouse B lymphoma cell line, was inhibited in a dose-dependent manner with different concentrations of hDM-αH-AHNP in the presence of 1 µM F-dAdo (Supplementary Fig. S3). hDM-αH-AHNP at its highest concentration or 1 µM of F-dAdo alone had no affect on proliferation of 38C13 cells (data not shown).
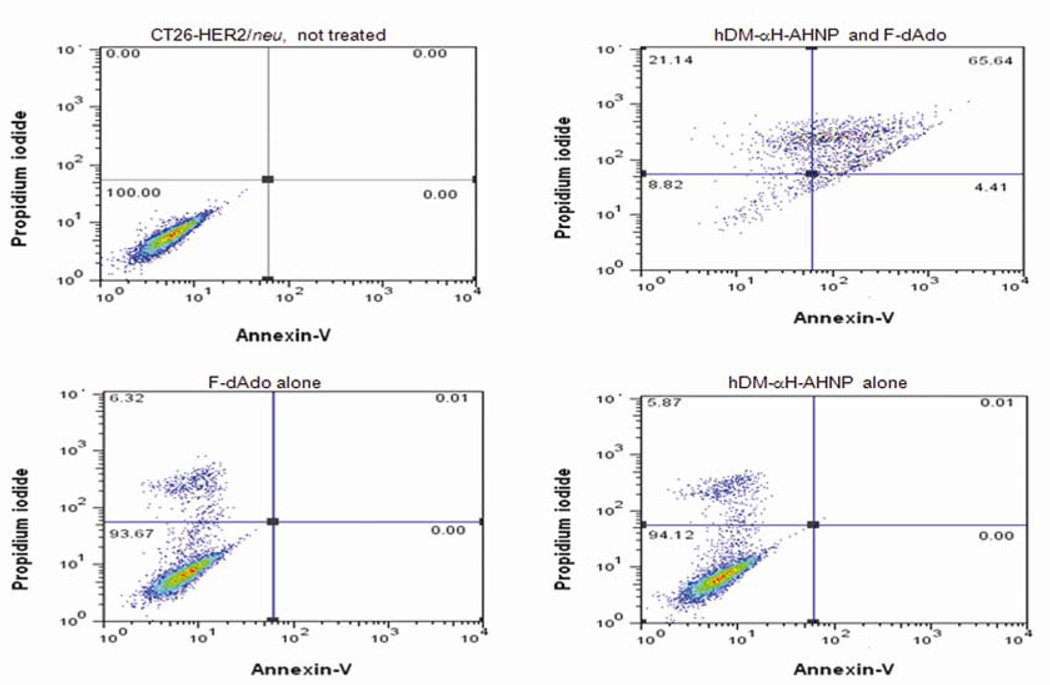
Importantly, to determine if generation of cytotoxic drug due to hDM enzymatic activity causes only inhibition of cell proliferation or if it results in tumor cell death, CT26-HER2/neu cells were incubated with hDM-αH-AHNP in the presence or absence of F-dAdo. Cells were then examined for DNA fragmentation or stained with propidium iodide and annexin-V and examined by flow cytometry, 72 or 60 hours post addition of F-dAdo, respectively. Staining at later times resulted in a strong signal only with PI. Association of CT26-HER2/neu tumor cells with 1 µM of hDM-αH-AHNP resulted in apoptosis only in the presence of F-dAdo (Fig 3). Cells grow normally when only hDM-αH-AHNP or F-dAdo was present (Fig. 3). When cells were examined for DNA fragmentation, DNA was completely degraded to small fragments (Supplementary Fig. S4). DNA fragmentation was seen only when both hDM-αH-AHNP and F-dAdo or F-Ade were present. When only hDM-αH-AHNP or F-dAdo was added, only high molecular weight DNA was recovered (Supplementary Fig. S4). In summary, the enzymatic activity of hDM or ePNP fused to AHNP is localized in the vicinity of HER2/neu expressing tumors, resulting in induction of apoptosis as a consequence of the conversion of F-dAdo to the cytotoxic drug, F-Ade.
Figure 3.
Generation of the cytotoxic drug, F-Ade due to enzymatic activity of hDM causes apoptosis. Association of hDM-αH-AHNP at a concentration of 1 µM with CT26HER2/neu cells results in induction of apoptosis in the presence of 1.5 µM F-dAdo. No toxicity was observed in the presence of hDM-αH-AHNP or F-dAdo alone. Cells were stained with propidium iodide and annexin-V and examined by flow cytometry 60 hours post addition of hDM-αH-AHNP and F-dAdo. This study was repeated two independent times in duplicate.
Discussion
Cancer treatment using ADEPT has remained limited due to immunogenicity caused by the use of non-human components. Even with the administration of immune-suppressants such as cyclosporine, generation of antibodies against the bacterial enzyme and the murine targeting component has prevented continued therapy (6). To address issues of immunogenicity, we have now shown that mutants of hPNP can utilize (deoxy)adenosine-based prodrugs as substrate. We focused on hPNP for two reasons: first, the endogenous wild-type hPNP cannot use (deoxy)adenosine-based prodrugs as substrate; second, multiple preclinical models have shown that the bacterial homologue of the enzyme is highly effective in cleaving (deoxy)adenosine-based prodrugs and generating high levels of cytotoxic drugs (11–13, 15). Among the mutations made within the purine binding pocket of the enzyme, hDM (E201Q:N243D) was most efficient in the phosphorolysis of F-dAdo. However, changing Asn243 to Asp (hSM) was significant for the enzyme to use adenosine-based prodrugs as substrates, probably due to hydrogen bond formation between Asp and 6-amino group and N-7 of the purine (10) (Supplementary Fig. S5). Enzymatic efficiency of the hSM with adenosine-based prodrugs was further increased when the second mutation of Glu201 to Gln was introduced. Crystal structures of hPNP in complex with either guanosine or inosine indicate that the N1-C2 side of these nucleotides faces Glu201 of hPNP (8, 16, 22). In fact, the carboxyl side chain of Glu201 forms hydrogen bonds with the amino group at C2 in guanosine and with the N1-hydrogen of both guanosine and inosine (16, 18, 21). Since the prodrugs contain highly electronegative fluorine or chlorine at the C2 position, replacing Glu201 with glutamine should allow the amide group in Gln to form hydrogen bonds with the C2-fluorine (or C2-Chlorine) and N1 of the prodrugs, which may enable the enzyme to accept the prodrugs with higher efficiency (Supplementary Fig. S5).
hDM is capable of using several (deoxy)adenosine-based prodrugs as substrate. Among the prodrugs tested, hDM had the highest kinetic efficiency with F-dAdo, followed by F-Ado, and Cl-dAdo, indicating that the hDM accepts (deoxy)adenosine prodrugs that contain fluorine more readily than those containing chorine. Fluorine is smaller than chlorine, and may fit better in the nucleotide binding pocket of the enzyme. hDM had no activity with Fludarabine, which contains arabinose as opposed to (deoxy)ribose present in the other prodrugs tested. In general, PNPs favor ribonucleosides as substrates, due to the stability of the oxocarbenium intermediate that forms upon glycosidic bond breakage during nucleoside phosphorolysis (11). Even ePNP which utilizes Fludarabine as substrate, has 3,000-fold less activity with Fludarabine than with F-dAdo (36).
To target the enzymes to tumors, they were fused to AHNP. AHNP binds specifically to HER2/neu on the cell surface (23, 30, 32, 37, 38). Treatment of cells expressing HER2/neu, but not the parental CT26 cells, with hDM-αH-AHNP or 6XHis-ePNP-GS-AHNP followed by washing and the addition of prodrug resulted in inhibition of cell proliferation (Fig. 2). Although it has been shown that AHNP alone inhibits growth of HER2/neu expressing tumor cells such as SKBR3 or T6-17 in vitro and in vivo (23, 30, 31), no growth inhibition of CT26-HER2/neu was seen following treatment with hPNP-GS-AHNP (data not shown). Therefore, the observed cytotoxicity is due to conversion of prodrug to cytotoxic drug as a consequence of PNP enzymatic activity, and is unrelated to AHNP function (39, 40). Although proliferation of MCF7-HER2 cells is inhibited by anti-HER2/neu antibodies, at the concentrations used hDM-αH-AHNP had no affect by itself. However, at higher concentrations of hDM-αH-AHNP, the cytotoxicity generated as a consequence of prodrug conversion by the PNPs may combine with the anti-tumor activity of AHNP and result in greater inhibition of tumor growth.
hDM was fused to AHNP at its C-terminus via two different linkers. The activity of hDM was increased when the rigid αH linker, instead of a flexible GS linker, was used to fuse the enzyme to AHNP. The rigid αH linker may result in a greater spatial separation of hDM from AHNP (33, 34), thereby reducing unfavorable interactions between the two domains (enzyme and peptide) in the bifunctional hDM-αH-AHNP fusion protein. In addition, the rigidity of the αH linker would render the whole molecule less flexible, and may make it less immunogenic than hDM-GS-AHNP (41, 42).
The binding affinity of hDM-αH-AHNP for ECDHER2-Fc determined by surface plasmon resonance was found to be similar to the published binding affinity of the free peptide, AHNP, to ECDHER2-Fc (30). Although the enzyme is a trimer, it remains unclear if more than one peptide on each trimer is accessible for binding simultaneously. Examination of the crystal structure of hPNP (Supplementary Fig. S6A) suggests that the C-termini, where each AHNP is fused, point in different directions; hence it may not be possible for the AHNPs in hDM-αH-AHNP to interact simultaneously with the immobilized ECDHER2. In contrast, 6XHis-ePNP-GS-AHNP interacted with ECDHER2-Fc with a ~ 200-fold stronger affinity than hDM-αH-AHNP. The difference may reflect the different availability of multiple AHNP for simultaneous interaction within the two enzymes. 6XHis-ePNP-GS-AHNP is a hexamer consisting of a trimer of dimers. Within each dimer, the two C-termini face in the same direction and are only 26 Å apart (Supplementary Fig. S6B). Additionally, the ePNP hexamer contains six AHNP peptides compared to the three present in hPNP. Moreover, different linkers connect the AHNP in the two proteins. With the flexible GS linker it may be easier for multiple AHNP molecules to simultaneously bind the immobilized HER2/neu.
When compared to the exceedingly efficient ePNP (6XHis-ePNP-GS-AHNP), hDM-αH-AHNP showed only a 36-fold reduced activity with F-dAdo as substrate (Table 2). Since it has been shown that expression of ePNP in only 0.1–1% of tumor cells results in complete eradication of tumors when prodrug is administered (13, 15), the enzymatic activity of hDM-αH-AHNP should be sufficient to generate enough cytotoxic drug to result in complete tumor eradication. Importantly, we showed that conversion of F-dAdo to F-Ade due to enzymatic activity of hDM causes tumor cell apoptosis, most probably due to inhibition of DNA, RNA and protein synthesis (43).
When compared to the currently leading therapeutics such as HER2/neu specific antibodies or small molecule inhibitors, hDM-αH-AHNP-F-dAdo has multiple advantages. First, specific antibodies only target HER2/neu expressing cells. In contrast, the cytotoxic drug generated by hDM enzymatic activity freely diffuses through the cell membrane and causes apoptosis in the heterogeneous tumor mass, irrespective of the cell surface expression of HER2/neu (15). Second, antibodies show variable efficacy in different patient groups, possibly because of patient specific differences such as Fc receptor polymorphism (44). No such differences in patient responses would be expected with hDM-αH-AHNP-F-dAdo treatment. Finally, it has been shown that small molecule inhibitor treatment results only in decreased cellular proliferation, and, unlike hDM-αH-AHNP therapy, fails to induce tumor cell apoptosis (45).
We have now shown that alteration of only two amino acids within hPNP is sufficient to produce an enzyme that utilizes (deoxy)adenosine-based prodrugs as substrate. Use of this mutant enzyme, which should be structurally similar to the naturally occurring human enzyme, is expected to result in greatly reduced immunogenicity in comparison to the currently used bacterial enzymes. We have demonstrated that hDM fused to a peptide mimetic that binds the HER2/neu is targeted to HER2/neu expressing tumors. Moreover, fusion with other tumor specific peptides, such as the peptide against prostate cancer cells (46) or with antibodies of different specificities, should make it possible to target hDM to many different cancers. We anticipate that targeting hDM to tumors followed by administration of a prodrug will induce tumor cell apoptosis, with minimal systemic toxicity and immunogenicity.
Supplementary Material
Acknowledgments
The authors thank Dr. Steven Clarke, and Dr. Jonathan Lowenson for the valuable discussions on enzyme kinetics. The authors also wish to thank Dr. Michael Sawaya for his excellent assistance with protein structural modeling.
Grant support: The work was supported by National Institute of Health RO1 GM074051 Grant and by the National Institute of Health Clinical & Fundamental Immunology Training Grant, NIH T32AI07126.
Abbreviations List
- ADEPT
Antibody Dependent Enzyme Prodrug Therapy
- PNP
purine nucleoside phosphorylase
- hPNP
human PNP
- hSM
hPNP with single mutation of N243D
- hDM
hPNP with double mutations of E201Q:N243D
- ePNP
E. coli PNP
- S-hDM
hDM fused to an S-tag at N-terminus
- S-hPNP
hPNP fused to an S-tag at N-terminus
- GS linker
Gly3SerGly linker
- αH linker
α-helical linker
- hDM-αH-AHNP
hDM fused to AHNP via a αH linker
- 6XHis-ePNP-GS-AHNP
His tagged ePNP fused to AHNP via a GS linker
- hPNP-GS-AHNP
hPNP fused to AHNP via a GS linker
- F-dAdo
2-fluoro-2’-deoxyadenosine
- Cl-dAdo
Cladribine
- F-Ado
2-fluoroadenosine
- F-Ade
2-fluoroadenine
Footnotes
The authors have no conflict of interest.
References
- 1.Bagshawe KD, Sharma SK, Springer CJ, Rogers GT. Antibody directed enzyme prodrug therapy (ADEPT). A review of some theoretical, experimental and clinical aspects. Ann Oncol. 1994;5:879–891. doi: 10.1093/oxfordjournals.annonc.a058725. [DOI] [PubMed] [Google Scholar]
- 2.Schrama D, Reisfeld RA, Becker JC. Antibody targeted drugs as cancer therapeutics. Nat Rev Drug Discov. 2006;5:147–159. doi: 10.1038/nrd1957. [DOI] [PubMed] [Google Scholar]
- 3.Xu G, McLeod HL. Strategies for Enzyme/Prodrug Cancer Therapy. Clin Cancer Res. 2001;7:3314–3224. [PubMed] [Google Scholar]
- 4.Springer CJ, Niculescu-Duvaz I. Prodrug-activating systems in suicide gene therapy. J Clin Invest. 2000;105:1161–1167. doi: 10.1172/JCI10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Francis RJ, Sharma SK, Springer C, et al. A phase I trial of antibody directed enzyme prodrug therapy (ADEPT) in patients with advanced colorectal carcinoma or other CEA producing tumours. Br J Cancer. 2002;87:600–607. doi: 10.1038/sj.bjc.6600517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Napier MP, Sharma SK, Springer CJ, et al. Antibody-directed Enzyme Prodrug Therapy: Efficacy and Mechanism of Action in Colorectal Carcinoma. Clin Cancer Res. 2000;6:765–772. [PubMed] [Google Scholar]
- 7.Bagshawe KD, Sharma SK, Springer CJ, et al. Antibody directed enzyme prodrug therapy (ADEPT): clinical report. Dis Markers. 1991;9:233–238. [PubMed] [Google Scholar]
- 8.Canduri F, Fadel V, Dias M, et al. Crystal structure of human PNP complexed with hypoxanthine and sulfate ion. Biochem Biophys Res Commun. 2005;326:335–338. doi: 10.1016/j.bbrc.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 9.Erion MD, Takabayashi K, Smith HB, et al. Purine Nucleoside Phosphorylase. 1. Structure-Function Studies. Biochemistry. 1997;36:11725–11734. doi: 10.1021/bi961969w. [DOI] [PubMed] [Google Scholar]
- 10.Stoeckler JD, Poirot AF, Smith RM, et al. Purine Nucleoside Phosphorylase. 3. Reversal of Purine Base Specificity by Site-Directed Mutagenesis. Biochemistry. 1997;36:11749–11756. doi: 10.1021/bi961971n. [DOI] [PubMed] [Google Scholar]
- 11.Bennett EM, Li C, Allan PW, Parker WB, Ealick SE. Structural Basis for Substrate Specificity of Escherichia coli Purine Nucleoside Phosphorylase. J Biol Chem. 2003;278:47110–47118. doi: 10.1074/jbc.M304622200. [DOI] [PubMed] [Google Scholar]
- 12.Kikuchi E, Menendez C, Ohori C, et al. Delivery of replication-competent retrovirus expressing Escherichia coli purine nucleoside phosphorylase increases the metabolism of the prodrug, fludarabine phosphate and suppresses the growth of bladder tumor xenografts. Cancer Gene Ther. 2007;14:279–286. doi: 10.1038/sj.cgt.7701013. [DOI] [PubMed] [Google Scholar]
- 13.Voeks D, Martiniello-Wilks R, Madden V, et al. Gene therapy for prostate cancer delivered by ovine adenovirus and mediated by purine nucleoside phosphorylase and fludarabine in mouse models. Gene Ther. 2002;2002:759–768. doi: 10.1038/sj.gt.3301698. [DOI] [PubMed] [Google Scholar]
- 14.Martiniello-Wilks R, Dane A, Voeks DJ, et al. Gene-directed enzyme prodrug therapy for prostate cancer in a mouse model that imitates the development of human disease. J Gene Med. 2004;6:43–54. doi: 10.1002/jgm.474. [DOI] [PubMed] [Google Scholar]
- 15.Hong JS, Waud WR, Levasseur DN, et al. Excellent In vivo Bystander Activity of Fludarabine Phosphate against Human Glioma Xenografts that Express the Escherichia coli Purine Nucleoside Phosphorylase Gene. Cancer Res. 2004;64:6610–6615. doi: 10.1158/0008-5472.CAN-04-0012. [DOI] [PubMed] [Google Scholar]
- 16.Ealick SE, Rule SA, Carter DC, et al. Three-dimensional Structure of Human Erythrocytic Purine Nucleoside Phosphorylase at 3.2 Å Resolution. J Biol Chem. 1990;265:1812–1820. doi: 10.2210/pdb2pnp/pdb. [DOI] [PubMed] [Google Scholar]
- 17.Bzowska A, Kulikowska E, Shugar D. Purine nucleoside phosphorylases: properties, functions, and clinical aspects. Pharmacol Ther. 2000;88:349–425. doi: 10.1016/s0163-7258(00)00097-8. [DOI] [PubMed] [Google Scholar]
- 18.Canduri F, Silva RG, Dos Santos DM, et al. Structure of human PNP complexed with ligands. Acta Crystallogr D Biol Crystallogr. 2005;61:856–862. doi: 10.1107/S0907444905005421. [DOI] [PubMed] [Google Scholar]
- 19.Mao C, Cook WJ, Zhou M, Federov AA, Almo SC, Ealick SE. Calf Spleen Purine Nucleoside Phosphorylase Complexed with Substrates and Substrate Analogues. Biochemistry. 1998;37:7135–7146. doi: 10.1021/bi9723919. [DOI] [PubMed] [Google Scholar]
- 20.Maynes J, Yam WS, Jenuth JP, et al. Design of an adenosine phosphorylase by active-site modification of murine purine nucleoside phosphorylase. Biochemical J. 1999;344:585–592. [PMC free article] [PubMed] [Google Scholar]
- 21.Cacciapuoti G, Gorassini G, Mazzeo MF, et al. Biochemical and structural characterization of mammalian like purine nucleoside phosphorylase from the Archaeon. Pyrococcus furiosus. 2007;274:2482–2495. doi: 10.1111/j.1742-4658.2007.05784.x. FEBS J. [DOI] [PubMed] [Google Scholar]
- 22.Silva RG, Pereira JH, Canduri F, de Azevedo WF, Jr, Basso LA, Santos DS. Kinetics and crystal structure of human purine nucleoside phosphoylase in complex with 7-methyl-6-thio-guanosine. Arch Biochem Biophys. 2005;442:49–58. doi: 10.1016/j.abb.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 23.Park BW, Zhang HT, Wu C, et al. Rationally designed anti-HER2/neu peptide mimetic disables P185HER2/neu tyrosine kinases in vitro and in vivo. Nat Biotechnol. 2000;18:194–198. doi: 10.1038/72651. [DOI] [PubMed] [Google Scholar]
- 24.Dela Cruz JS, Trinh KR, Morrison SL, Penichet ML. Recombinant anti-human HER2/neu IgG3-(GM-CSF) fusion protein retains antigen specificity and cytokine function and demonstrates antitumor activity. J. Immunol. 2000;165:5112–5121. doi: 10.4049/jimmunol.165.9.5112. [DOI] [PubMed] [Google Scholar]
- 25.Zhivotovsky B, Orrenius S. Assessment of Apoptosis and Necrosis by DNA Fragmentation and Morphological Criteria. Current Protocols in Cell Biology. 2001;UNIT 18.3:1–23. doi: 10.1002/0471143030.cb1803s12. [DOI] [PubMed] [Google Scholar]
- 26.WL D. The PyMOL molecular graphics system on worldwide web. 2005 http://www.pymol.org. [Google Scholar]
- 27.Zang Y, Wang W, Wu S, Ealick SE, Wang CC. Identification of a Subversive Substrate of Trichomonas vaginalis Purine Nucleoside Phosphorylase and the Crystal Structure of the Enzyme-Substrate Complex. J. Biol. Chem. 2005;280:22318–22325. doi: 10.1074/jbc.M501843200. [DOI] [PubMed] [Google Scholar]
- 28.Munagala N, Wang CC. The Purine Nucleoside Phosphorylase from Trichomonas Vaginalis Is a Homologue of the Bacterial Enzyme. Biochemistry. 2002;41:10382–10389. doi: 10.1021/bi026025n. [DOI] [PubMed] [Google Scholar]
- 29.Chaudhary K, Ting LM, Kim K, Roos DS. Toxoplasma gondii Purine Nucleoside Phosphorylase Biochemical Characterization, Inhibitor Profiles, and Comparison with the Plasmodium falciparum Ortholog. J Biol Chem. 2006;281:25652–25658. doi: 10.1074/jbc.M602624200. [DOI] [PubMed] [Google Scholar]
- 30.Berezov A, Chen J, Liu Q, Zhang HT, Greene MI, Murali R. Disabling Receptor Ensembles with Rationally Designed Interface Peptidomimetics. J Biol Chem. 2002;277:28330–28339. doi: 10.1074/jbc.M202880200. [DOI] [PubMed] [Google Scholar]
- 31.Masuda K, Richter M, Song X, et al. AHNP-streptavidin: a tetrameric bacterially produced antibody surrogate fusion protein against p185her2/neu. Oncogene. 2006;25:7740–7746. doi: 10.1038/sj.onc.1209745. [DOI] [PubMed] [Google Scholar]
- 32.Berezov A, Zhang HT, Greene MI, Murali R. Disabling ErbB Receptors with Rationally Designed Exocyclic Mimetics of Antibodies: Structure-Function Analysis. J Med Chem. 2001;44:2565–2574. doi: 10.1021/jm000527m. [DOI] [PubMed] [Google Scholar]
- 33.Marqusee S, Robbins VH, Baldwin RL. Unusually stable helix formation in short alanine-based peptides. Proc Natl Acad Sci USA. 1989;86:5286–5290. doi: 10.1073/pnas.86.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arai R, Ueda H, Kitayama A, Kamiya N, Nagamune T. Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng. 2001;14:529–532. doi: 10.1093/protein/14.8.529. [DOI] [PubMed] [Google Scholar]
- 35.Kearns CM, Blakley RL, Santana VM, Crom WR. Pharmacokinetics of Cladribine (2-Chlorodeoxyadenosine) in Children with Acute Leukemia. Cancer Res. 1994;54:1235–1239. [PubMed] [Google Scholar]
- 36.Parker WB, Allan PW, Hassan AE, Secrist JA, Sorscher EJ, Waud WR. Antitumor activity of 2-fluoro-2'-deoxyadenosine against tumors that express Escherichia coli purine nucleoside phosphorylase. Cancer Gene Ther. 2003;10:23–29. doi: 10.1038/sj.cgt.7700520. [DOI] [PubMed] [Google Scholar]
- 37.Fantin VR, Berardi MJ, Babbe H, Michelman MV, Manning CM, Leder PH. A Bifunctional Targeted Peptide that Blocks HER-2 Tyrosine Kinase and Disables Mitochondrial Function in HER-2-Positive Carcinoma Cells. Cancer Res. 2005;65:6891–6900. doi: 10.1158/0008-5472.CAN-05-0395. [DOI] [PubMed] [Google Scholar]
- 38.Guillemard V, Nedev HN, Berezov A, Murali R, Saragovi HU. HER2-Mediated Internalization of a Targeted Prodrug Cytotoxic Conjugate Is Dependent on the Valency of the Targeting Ligand. DNA Cell Biol. 2005;24:350–358. doi: 10.1089/dna.2005.24.351. [DOI] [PubMed] [Google Scholar]
- 39.Dela Cruz JS, Lau SY, Ramirez EM, et al. Protein vaccination with the HER2/neu extracellular domain plus anti-HER2/neu antibody-cytokine fusion proteins induces a protective anti-HER2/neu immune response in mice. Vaccine. 2003;21:1317–1326. doi: 10.1016/s0264-410x(02)00741-7. [DOI] [PubMed] [Google Scholar]
- 40.Cho HM, Rosenblatt JD, Kang YS, et al. Enhanced inhibition of murine tumor and human breast tumor xenografts using targeted delivery of an antibody-endostatin fusion protein. Mol Cancer Ther. 2005;2005:956–967. doi: 10.1158/1535-7163.MCT-04-0321. [DOI] [PubMed] [Google Scholar]
- 41.Carmicle S, Dai G, Steede NK, Landry SJ. Proteolytic Sensitivity and Helper T-cell Epitope Immunodominance Associated with the Mobile Loop in Hsp10s. J Biol Chem. 2002;277:155–160. doi: 10.1074/jbc.M107624200. [DOI] [PubMed] [Google Scholar]
- 42.Landry SJ. Local protein instability predictive of helper T-cell epitopes. Immunol Today. 1997;18:527–532. doi: 10.1016/s0167-5699(97)01152-3. [DOI] [PubMed] [Google Scholar]
- 43.Parker WB, Allan PW, Shaddix SC, et al. Metabolism and Metabolic Actions of 6-Methylpurine and 2-Fluoroadenine in Human Cells. Biochemical Pharmacology. 1998;55:1673–1681. doi: 10.1016/s0006-2952(98)00034-3. [DOI] [PubMed] [Google Scholar]
- 44.Cartron G, Dacheux L, Salles G, et al. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgRIIIa gene. BLOOD. 2002;99:754–758. doi: 10.1182/blood.v99.3.754. [DOI] [PubMed] [Google Scholar]
- 45.Harari PM, Huang SM. Combining EGFR inhibitors with radiation or chemotherapy: Will preclinical studies predict clinical results? Int. J. Radiat. Oncol. Biol. Phys. 2004;58:976–983. doi: 10.1016/j.ijrobp.2003.09.097. [DOI] [PubMed] [Google Scholar]
- 46.Zitzmann S, Kramer S, Mier W, et al. Identification of a New Prostate-Specific Cyclic Peptide with the Bacterial FliTrx System. J Nucl Med. 2005;46:782–785. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.