Abstract
Background
Using human brain microvascular endothelial cells (HBMECs) as an in vitro model for how African trypanosomes cross the human blood-brain barrier (BBB) we recently reported that the parasites cross the BBB by generating calcium activation signals in HBMECs through the activity of parasite cysteine proteases, particularly cathepsin L (brucipain). In the current study, we examined the possible role of a class of protease stimulated HBMEC G protein coupled receptors (GPCRs) known as protease activated receptors (PARs) that might be implicated in calcium signaling by African trypanosomes.
Methodology/Principal Findings
Using RNA interference (RNAi) we found that in vitro PAR-2 gene (F2RL1) expression in HBMEC monolayers could be reduced by over 95%. We also found that the ability of Trypanosoma brucei rhodesiense to cross F2RL1-silenced HBMEC monolayers was reduced (39%–49%) and that HBMECs silenced for F2RL1 maintained control levels of barrier function in the presence of the parasite. Consistent with the role of PAR-2, we found that HBMEC barrier function was also maintained after blockade of Gαq with Pasteurella multocida toxin (PMT). PAR-2 signaling has been shown in other systems to have neuroinflammatory and neuroprotective roles and our data implicate a role for proteases (i.e. brucipain) and PAR-2 in African trypanosome/HBMEC interactions. Using gene-profiling methods to interrogate candidate HBMEC pathways specifically triggered by brucipain, several pathways that potentially link some pathophysiologic processes associated with CNS HAT were identified.
Conclusions/Significance
Together, the data support a role, in part, for GPCRs as molecular targets for parasite proteases that lead to the activation of Gαq-mediated calcium signaling. The consequence of these events is predicted to be increased permeability of the BBB to parasite transmigration and the initiation of neuroinflammation, events precursory to CNS disease.
Author Summary
Human African trypanosomiasis, or sleeping sickness, occurs when single-cell trypanosome protozoan parasites spread from the blood to brain over the blood-brain barrier (BBB). This barrier is composed of brain microvascular endothelial cells (BMECs) especially designed to keep pathogens out. Safe drugs for treating sleeping sickness are lacking and alternative treatments are urgently required. Using our human BMEC BBB model, we previously found that a parasite protease, brucipain, induced calcium activation signals that allowed this barrier to open up to parasite crossing. Because human BMECs express protease-activated receptors (PARs) that trigger calcium signals in BMECs, we hypothesized a functional link between parasite brucipain and BMEC PARs. Utilizing RNA interference to block the production of one type of PAR called PAR-2, we hindered the ability of trypanosomes to both open up and cross human BMECs. Using gene-profiling methods to interrogate candidate BMEC pathways specifically triggered by brucipain, several pathways that potentially link brain inflammatory processes were identified, a finding congruent with the known role of PAR-2 as a mediator of inflammation. Overall, our data support a role for brucipain and BMEC PARs in trypanosome BBB transmigration, and as potential triggers for brain inflammation associated with the disease.
Introduction
Human African trypanosomiasis (HAT), commonly called sleeping sickness, is a vector-borne disease for which death is inevitable if the patient is untreated [1],[2],[3]. HAT is caused by two subspecies of African trypanosomes, Trypanosoma brucei rhodesiense and T. b. gambiense causing East African and West African sleeping sickness, respectively. In classical late stage HAT (stage 2), the parasites invade the central nervous system (CNS) and the infected individuals suffer from progressive neurologic deterioration with concomitant psychiatric disorders, sleep disturbances, stupor, and coma. The role of the parasites in the pathogenesis of CNS lesions is not completely understood [4].
Using an in vitro model of the blood-brain barrier (BBB) consisting of human brain microvascular endothelial cells (HBMEC), we showed that human infective T. b. rhodesiense have a high potential for transendothelial migration, while animal infective T. b. brucei cross inefficiently [5]. We initially proposed that African trypanosome associated protease(s) could mediate the process of parasite traversal across the BBB [6]. In vitro studies utilizing both cysteine protease inhibitors and RNA interference (RNAi) have identified a key role for the T. brucei Clan CA (papain) family of cysteine proteases in the lifecycle of T. b. brucei [7],[8]. The T. brucei genome encodes two distinct Clan CA cysteine proteases. Brucipain (aka trypanopain-Tb, rhodesain) is a cathepsin L-like protease responsible for the bulk of protease activity in the organism [7]. T. brucei cathepsin B (TbCatB) has activity that is upregulated in the bloodstream form (BSF) of the parasites [7]. Remarkably, it was found that the ability of the parasites to cross HBMECs through the generation of PLC/PKC mediated Ca2+ activation signals correlated with levels of brucipain activity [9],[10]. RNAi studies investigating the roles played by TbCatB and brucipain in the pathogenesis of T. b. brucei in vivo also suggested brucipain possibly facilitates parasite entry into the CNS [11].
Cysteine proteases can activate a class of G protein coupled receptors (GPCR) known as protease activated receptors (PARs). In neutrophils and oral epithelial cells PAR activation by gingipain, a cysteine protease of Porphyromonas gingivalis, leads to calcium signaling and IL-6 production [12],[13],[14]. HBMEC express all 4 protease activated receptors (PARs), and activation of PAR-1 and PAR-2 has been shown to trigger calcium mediated BMEC transmembrane signaling [15]. Since we observed activating signals (increases in [Ca2+]i and phospholipase C activation) during the interaction of African trypanosomes with HBMECs [5],[9],[10], we hypothesized that the activation of PARs by the parasite could also play a role in increasing HBMEC permeability, enabling subsequent crossing [4]. We now show that African trypanosome traversal across the human BBB requires, at least in part, the participation of a PAR-2-mediated calcium signaling pathway(s).
Materials and Methods
Chemicals
N-methyl-Pip-F-homoF-vinyl sulfonyl phenyl (K11777) [16],[17], an irreversible inhibitor of Trypanosoma cruzi Clan A cysteine protease cruzipain [17] was a gift from Dr. James McKerrow (University of California at San Francisco).
Purification of recombinant Pasteurella multocida toxin
Recombinant Pasteurella multocida toxin (PMT) was cloned, expressed, purified, quantified, and tested for biological activity as previously described [18]. The toxin samples were stored at −80°C until use.
HBMEC and trypanosomes
Primary HBMECs (≤ passage 13) were maintained as previously described [5],[9],[15]. The bloodstream form (BSF) T. b. rhodesiense used was originally obtained from the CSF from a Kenyan patient with sleeping sickness [5],[9],[10]. This parasite was formerly classified as T. b. gambiense IL1852, but has been reclassified as T. b. rhodesiense IL1852 based on the presence of the SRA gene [10]. The bloodstream form (BSF) trypanosomes were maintained in culture in HMI-9 [19]. For the microarray study, to inactivate brucipain activity, the parasites were pretreated for 30 min with 5 µM of the cathepsin-L inhibitor K11777 The parasites were then washed with medium to remove excess inhibitor prior to incubation with HBMEC monolayers [9],[10].
Electrical cell-substrate impedance sensing for real-time transendothelial electrical resistance measurement and cell signaling
The elevated transendothelial electrical resistance (TEER) and the lower paracellular permeability of the brain microvasculature is a characteristic feature that distinguishes it from non-brain endothelium. Measurement of TEER is a one of the most straightforward methods to access the barrier tightness using in vitro models [20]. Electrical Cell-Substrate Impedance Sensing (ECIS) gathers TEER data as an electrical method for assessing barrier function that detect changes in endothelial cell shape in real-time [21]. The Model 1600R ECIS system (Applied Biophysics) [22],[23],[24] was used to measure HBMEC TEER changes in real-time during exposure to African trypanosomes and their secreted products. HBMECs were grown on collagen-coated 8-well single (8W1E) or multiple (8W10E+) gold electrode ECIS arrays until confluent. While changes in relative TEER recorded by the single and multiple arrays were similar, absolute TEER values differ. Steady state TEER >10,000 ohms and >1,000 ohms were used for the 8W1E and 8W10E+ arrays respectively (Applied Biophysics). The multiple electrode arrays in which HBMEC resistances are >1,000 ohms [4],[5],[25],[26] record the activities of more cells over a larger region of the substrate. However, the 10-fold lower capacitance of the 8W1E array leads to increased resistances (more than 10 times that of the multiple electrode arrays) and a higher signal to noise ratio (ECIS 1600R instruction manual). Changes in resistance of HBMEC monolayers were monitored every 80 sec in response to experimental variables. For the PMT experiments, HBMECs were simultaneously incubated with PMT (30 ng/ml) or pretreated with the toxin for 90 min then washed with fresh medium prior to incubation with parasites.
HBMEC gene silencing by RNAi and analysis by laser capture microdissection (LCM)
In single cell studies, when stimulated with PAR-2 agonists strong Ca2+ signals are induced ([15] in >60% of HBMEC (YV Kim, unpublished) suggesting a role for PAR-2 in parasite HBMEC traversal. We silenced the F2RL1 expression by co-transfecting a pre-designed siRNA for F2RL1 and a GFP-expressing plasmid into subconfluent HBMECs using Lipofectamine 2000 and standard protocols. A matched negative control siRNA (Ambion) was used as control. To determine the efficiency of gene silencing, HBMECs were grown on 35 mm LCM dishes (PALM) prior to RNAi silencing. Using laser capture microdissection (LCM), 15 individual GFP-HBMECs that expressed GFP in the F2RL1-silenced and control siRNA cultures were collected. The GFP-HBMECs were marked and catapulted using the following settings on the P.A.L.M. Microlaser (Bernied, Germany): energy-cut of 60; energy-lpc of 86; focus-cut of 75 and focus-lpc of 92 [27]. The RNA extracted using Ambion's RNAqueous®-Micro kit was then amplified using Ambion's MessageAmp™II kit. qRT-PCR was done using pre-designed F2RL1 primers (Invitrogen) and the data were normalized to ACTB transcripts.
Co-incubation of HBMECs with trypanosomes
HBMECs grown to confluency in Transwell inserts, ECIS arrays or 6-well microtiter plates were incubated at 37°C in 5% CO2 in Experimental Medium (HMI-9 and Medium 199 mixed 1∶1) containing 10% FBS [5],[9],[10]. HBMEC were incubated in triplicate with BSF T. b. rhodesiense IL1852 (5×105 to 1×106/ml) under the indicated study conditions.
Transcriptome microarray analysis
Samples from 2 independent experiments containing duplicate sets of infected (wild type or K11777 pretreated trypanosomes) and uninfected HBMECs were rapidly dissociated with trypsin/EDTA, than washed. Total cellular RNA was isolated using the RNAeasy kit (QIAGEN) following the manufacturer's instructions. Purified RNA was treated with RNase-free DNase to remove contaminating genomic DNA. The integrity of RNA transcripts was verified by electrophoresis through denaturing agarose-formaldehyde gels followed by ethidium bromide staining [28].
cDNAs were radiolabeled with 32P α-dCTP Isoblue (ICN) using SuperScript II Reverse Transcriptase (Invitrogen). Unbound label was separated using a Biospin P-30 spin column (Bio-Rad). Each cDNA probe was adjusted to 106 cpm/mL and hybridized to separate nylon MGC-1 microarrays [29] at 68°C overnight in Microhyb hybridization solution (Research Genetics). The MGC-1 microarray represents 9,600 different human gene features including those encoding cytokines and other immunological regulatory proteins such as chemokines, growth factors, and cellular receptors [29]. Membranes were washed three times in 2×SSC (1×SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-1% SDS for 30 min at 68°C and twice in 0.1× SSC-0.5% SDS for 30 min at 68°C. Membranes were exposed overnight and scanned on a Molecular Dynamics STORM phosphoimager set to 50-micron resolution. mRNA expression levels were analyzed by scanning densitometry using ArrayPro imaging software. Differential patterns of gene expression were assessed by preparing RNA from both uninfected control HBMEC or HBMEC that were co-incubated with wild-type (WT) T. b. rhodesiense IL1852 or T. b. rhodesiense K11777 inhibited for brucipain activity for 3 and 6 hours. These cDNAs were hybridized in parallel to pairs of identical microarrays.
Raw microarray data were subjected to Z normalization and tested for significant changes as previously described [30]. Genes were determined to be differentially expressed after calculating the Z ratio, which indicates the fold-difference between experimental groups, and false discovery rate (fdr), which controls for the expected proportion of falsely rejected hypotheses. Individual genes with p value ≤0.05, absolute value of Z ratio>1.5 and fdr<0.3 were considered significantly changed. Hierarchical cluster method with complete algorithm and K-mean cluster were employed to identify clustering within groups. Array data for each experimental condition were hierarchically clustered with the DIANE 6.0 software in JMP 6.0 environment and R programs. Pilot studies showed no signal detected when trypanosome cDNA was hybridized to the microarrays.
Probability scores for each network or functional Parametric analysis of gene set enrichment (PAGE ) [31] gene set analysis was performed by using the PAGE algorithm and PERL/R code with MYSQL database. The archive of gene sets used with this algorithm for this analysis is from the Molecular Signatures Database (MSigDB) (http://www.broad.mit.edu/gsea/msigdb/genesets.jsp?collection=CP) [32]. Significance of functions and pathways was calculated using the z-test between each function group genes and the genes in the whole sample. The p-value was calculated by comparing the number of user-specified genes of interest participating in a given function or pathway relative to the total number of occurrences of these genes in all functional/pathway annotations stored in the knowledge base. The Enrichment score, which is called pathway z-scores, were calculated by the difference of the mean z-ratio of the selected function groups with the mean of z-ratio of the whole sample genes to represent the pathway change altitude.
Array data from infected samples are presented as relative changes to uninfected controls in mRNA expression following normalization of gene signals to total signal and levels of housekeeping gene mRNA to ensure analysis of equivalent amounts of RNA. This approach facilitated the direct comparison of mRNA levels between control and the parasite-treated HBMEC.
Results/Discussion
The role of PAR-2 in trypanosome transmigration across HBMECs
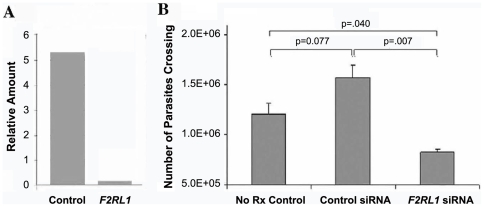
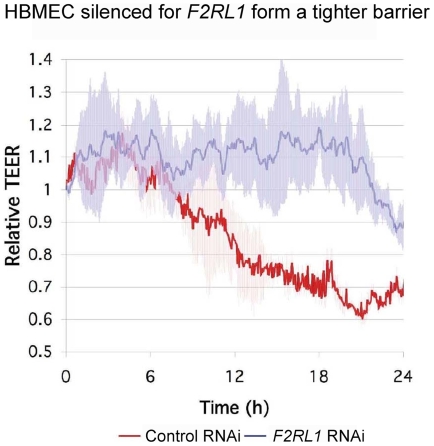
After silencing of F2RL1 by RNAi, based on qRT-PCR targeting F2RL1 transcripts normalized to ACTB (β-actin transcripts), we found that PAR-2 expression was reduced by over 95% (Fig 1A) in LCM-isolated HBMECs. To verify whether the ability of T. b. rhodesiense to cross HBMECs requires host cell PAR-2 signaling, we incubated T. b. rhodesiense IL1852 for 16h with HBMEC monolayers silenced for F2RL1 expression (F2RL1 siRNA), transfected with a matched scrambled siRNA control (control siRNA), or with untreated HBMECs (no Rx control) (Fig 1B) and examined parasite transmigration. There was no statistical difference in parasite transmigration between the 2 control samples: in medium alone and or medium with control siRNA (p = 0.077; 2-tail Student t-test). Considering a 95% reduction in F2RL1 expression in the ∼60% of the HBMEC that express PAR-2, significant differences were observed between F2RL1-silenced HBMECs compared to both control conditions: 39% inhibition (p = 0.040) versus untreated HBMECs (no Rx control) and 49% inhibition (p = 0.007) versus control siRNA-treated HBMECs. In the absence of PAR-2-induced signaling, we predicted that F2RL1-silenced primary HBMECs would maintain a tighter barrier upon stimulation by T. b. rhodesiense. ECIS was used to monitor real-time TEER changes in HBMEC monolayer integrity. F2RL1 RNAi-transfected HBMECs grown in ECIS chambers were incubated overnight with T. b. rhodesiense IL1852 (1.2×106/mL). Fig 2 shows that unlike the matched HBMEC scrambled siRNA control (red line), HBMECs silenced for PAR-2 maintained control TEER levels (about 10,000 ohms in the 8W1E arrays used) (blue line) for at least 20 hours even in the presence of a high parasite load.
Figure 1. T. b. rhodesiense transmigration across HBMEC silenced for F2RL1 expression by RNAi.
Using laser capture microdissection 15 individual HBMEC expressing GFP in the PAR2-silenced and control siRNA cultures were collected and the RNA extracted. (A) Based on qRT-PCR using pre-designed F2RL1 primers and normalized to ACTB (β-actin transcripts), PAR-2 gene expression was reduced by over 95%. (B) T. b. rhodesiense IL1852 was incubated for 16h in triplicate with HBMEC monolayers silenced for PAR-2 expression (F2RL1 siRNA), with a matched scrambled siRNA control (control siRNA) construct, or with untreated HBMEC (no Rx control) and examined parasite transmigration.
Figure 2. Real-time TEER changes in HBMEC silenced for PAR-2 gene expression.
F2RL1 RNAi transfected HBMEC grown in 8W1E ECIS chambers were incubated overnight with T. b. rhodesiense IL1852. The changes in TEER relative to matched HBMEC control containing scrambled siRNA construct (red line) and HBMEC silenced for F2RL1 (blue line) in the presence of parasites are shown.
The Gα-specific toxin from Pasteurella multocida blocks trypanosome-induced changes in TEER in HBMEC
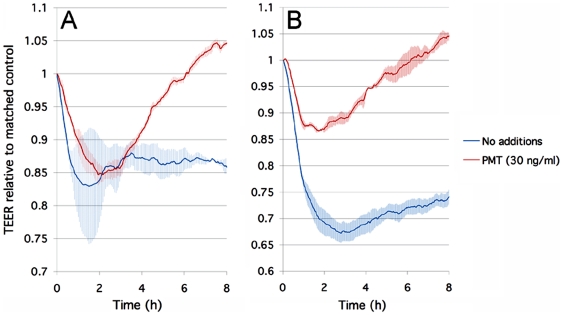
PARs, including PAR-2, are GPCRs known to mediate their cellular effects through the activation of Gαq/11, Gα12/13 and Gαiβγ signaling pathways [33],[34],[35],[36]. The protein toxin from Pasteurella multocida (PMT) has been shown to target Gαq [37],[38],[39], Gα12/13 [40] and Gαi [41] heterotrimeric G proteins in eukaryotic cells. PMT potentiates Gαq protein-mediated GPCR responses to ligands by primarily activating phospholipase C (i.e. PLC-β1, 3, 4) [37],[38],[39],[42]. Accordingly, this leads to calcium mobilization and activation of PKCs, as well as activation of mitogenic pathways, including MAPK (ERK1/2, p38) activation [39],[42],[43]. PMT enters cells via receptor-mediated endocytosis and acts intracellularly to activate Gαq [37],[38],[39],[44]. This is subsequently followed by uncoupling of Gαq signaling when cellular Gαq-mediated responses then become refractory to further stimulation [45],[46]. This also occurs when HBMECs are incubated with PMT. As shown in Fig. 3A, when incubated together with HBMECs and T. b. rhodesiense, PMT (30 ng/ml) initially does not inhibit the parasite-induced drop in TEER by ECIS. However, TEER increases above control levels with parasites after 2 hours, consistent with uncoupling of Gαq signaling by PMT. When HBMECs are pretreated for 3h with PMT to allow for Gαq uncoupling prior to trypanosome addition, the toxin clearly inhibited the ability of the parasites to compromise the HBMEC monolayers (Fig 3B). Since PAR-2 is a GPCR that can act via Gαq signaling, taken together, the data strongly suggest a role for PAR-2 and host Gαq-mediated calcium signaling in parasite interactions with the human BBB. While not yet tested would it be interesting to see the effect of PMT treatment on PAR-2 RNAi treated cells; i.e. are there additive effects of PMT treatment in these cells or is the PMT effect occluded by PAR-2 knockdown.
Figure 3. PMT from Pasteurella multocida blocks T. b. rhodesiense induced changes in HBMEC TEER.
HBMEC grown in 8W10E+ ECIS chambers were incubated with T. b. rhodesiense IL1852. Shown are the changes in real-time TEER measured by ECIS. A) HBMECs incubated with T. b. rhodesiense IL1852 in the absence (blue line) or continuous presence of PMT (30 ng/ml) (red line). B) HBMECs incubated with T. b. rhodesiense IL1852 was incubated with HBMECs untreated (blue line) or pretreated with PMT (30 ng/ml) (red line). The data are represented as the average change in TEER±EM (n = 3). The changes in TEER are represented as the average change in TEER±SEM (n = 2).
Gene expression pathways in HBMEC in response to African trypanosomes lacking brucipain activity
PAR-2 signaling has been shown in other systems to have neuroinflammatory and neuroprotective roles [47],[48],[49],[50] and our data implicate a role for brucipain and PAR-2 in African trypanosome/HBMEC interactions. Therefore, we used transcription-profiling methods to interrogate candidate HBMEC pathways with particular attention paid to pathways that are specifically triggered by brucipain and that potentially link cellular processes to physiologic (i.e. CNS passage across the BBB) and pathophysiologic (neuroinflammation) processes associated with CNS HAT [4]. T. b. rhodesiense inhibited for brucipain activity via pretreatment with K11777 (a cell-permeable class-specific irreversible inhibitor of brucipain) are defective in crossing HBMECs [5],[9],[10],[16],[17]. We interrogated candidate HBMEC pathways whose expression was modulated by exposure to T. b. rhodesiense pretreated with the brucipain inhibitor K11777. Because the half-life of brucipain is not known, the experimental time frame was kept to 6 hours, the approximate doubling time of the parasite. This was done to minimize the potential problems in data interpretation because of the contribution of decreasing drug within the parasites that were doubling.
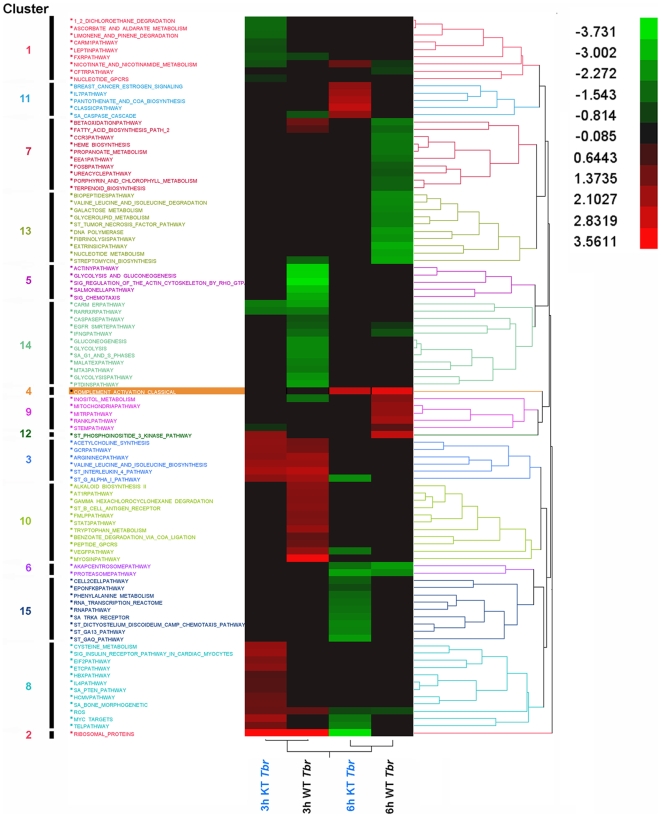
Gene set enrichment analysis of HBMEC based on the known canonical pathways showed that WT and modified African trypanosomes differentially altered the expression of genes represented in 99 pathways relative to the uninfected controls (Fig 4; Table 1). While the 99 pathways clustered into 15 groups according to their gene expression profiles, 28 pathways clustered into 4 unique cluster groups that were specifically expressed by HBMECs only after exposure to the K11777-pretreated trypanosomes (Table 1). A functional overview of the MSigDB gene sets was then done to categorize a small number of selected “gene families” whose members shared a common feature such as homology or biochemical activity, although not necessarily having common origins. Analysis of the gene functions within the HBMEC pathways showed 47 genes differentially expressed by HBMECs in response to trypanosome infection (Table 2). From this gene subset, 30 genes were expressed exclusively by HBMEC in response to brucipain-inhibited parasites (Tables 2 and 3). Of these, 30 genes functionally i) 3 encoded cytokines, ii) 16 encoded transcription factors, iii) 8 encoded kinases, iv) 3 encoded for translocated genes, and v) 4 encoded for oncogenes. Interestingly, HBMEC genes encoding for cell surface markers and tumor suppressors in response to wild-type trypanosomes, were not expressed by brucipain-inhibited parasites (Table 2).
Figure 4. Pathway changes altered by T. b. rhodesiense inhibited for brucipain activity.
Significantly regulated, functional pathway clusters were generated from wild-type (WT Tbr) T. b. rhodesiense or parasites K11777 inhibited for brucipain activity (KT Tbr) gene sets using PAGE gene set analysis. Pathways that were significantly up-regulated (red) or down-regulated (green) are shown. Cluster group numbers are shown on the left, while the grading scale is in the upper right.
Table 1. Gene expression in pathways altered only by T. b. rhodesiense inhibited for brucipain activity.
| Time | Pathway Name | Cluster | Annotation | z-Score | |
| 3 h | 6 h | ||||
| After 3 h | 1_2_DICHLOROETHANE_ DEGRADATION | 1 | [GenMAPP] | −1.55 | n.s. |
| ASCORBATE_AND_ALDARATE_ METABOLISM | 1 | [GenMAPP] | −1.55 | n.s. | |
| CARM1_PATHWAY | 1 | The methyltransferase CARM1 interacts with transcription factors such as CBP/p300 and methylates histones H3 and H4. [BioCarta] | −1.10 | n.s. | |
| LEPTIN PATHWAY | 1 | Leptin is a peptide secreted by adipose tissue that, in skeletal muscle, promotes fatty acid oxidation, decreases cells' lipid content, and promotes insulin sensitivity. [BioCarta] | −1.04 | n.s. | |
| LIMONENE_AND_PINENE_ DEGRADATION | 1 | [GenMAPP] | −1.47 | n.s. | |
| NUCLEOTIDE_GPCRS | 1 | [GenMAPP] | −0.49 | n.s. | |
| CYSTEINE_METABOLISM | 8 | [GenMAPP] | 1.89 | n.s. | |
| EIF2_PATHWAY | 8 | Eukaryotic initiation factor 2 (EIF2) initiates translation by transferring Met-tRNA to the 40S ribosome in a GTP-dependent process. [BioCarta] | 1.52 | n.s. | |
| ETC_PATHWAY | 8 | Energy is extracted from carbohydrates via oxidation and transferred to the mitochondrial electron transport chain, which couples ATP synthesis to the reduction of oxygen to water. [BioCarta] | 1.74 | n.s. | |
| HBX_PATHWAY | 8 | Hbx is a hepatitis B protein that activates a number of transcription factors, possibly by inducing calcium release from the mitochondrion to the cytoplasm. [BioCarta] | 0.83 | n.s. | |
| HCMV_PATHWAY | 8 | Cytomegalovirus activates MAP kinase pathways in the host cell, inducing transcription of viral genes. [BioCarta] | 1.18 | n.s. | |
| IL4_PATHWAY | 8 | IL4 promotes Th2 cell differentiation via a heterodimeric receptor that activates Stat6/JAK and MAP kinase pathways. [BioCarta] | 0.89 | n.s. | |
| SA_BONE_MORPHOGENETIC | 8 | Bone morphogenetic protein binds to its receptor to induce ectopic bone formation and promote development of the viscera. [SigmaAldrich] | 1.24 | n.s. | |
| SA_PTEN_PATHWAY | 8 | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. [SigmaAldrich] | 0.90 | n.s. | |
| SIG_INSULIN_RECEPTOR_ PATHWAY_IN_CARDIAC_ MYOCYTES | 8 | Genes related to the insulin receptor pathway. [SIGNALINGAlliance] | 1.98 | n.s. | |
| After 6 h | BREAST_CANCER_ESTROGEN_SIGNALING | 11 | Genes preferentially expressed in breast cancers, especially those involved in estrogen-receptor-dependent signal transduction. [GEArray] | n.s. | 1.82 |
| CLASSIC_PATHWAY | 11 | The classic complement pathway is initiated by antibodies and promotes phagocytosis and lysis of foreign cells as well as activating the inflammatory response. [BioCarta] | n.s. | 2.62 | |
| IL7_PATHWAY | 11 | IL7 is required for B and T cell development and proliferation and may contribute to activation of VDJ recombination. [BioCarta] | n.s. | 2.05 | |
| PANTOTHENATE_AND_COA_ BIOSYNTHESIS | 11 | [GenMAPP] | n.s. | 2.27 | |
| CELL_2_CELL PATHWAY | 15 | Epithelial cell adhesion proteins such as cadherins transduce signals into the cell via catenins, which alter cell shape and motility. [BioCarta] | n.s. | −1.31 | |
| EPO_NFKB_PATHWAY | 15 | The cytokine erythropoietin (Epo) prevents stress-induced neuronal apoptosis by stimulating anti-apoptotic pathways through JAK2 kinase and NFkB. [BioCarta] | n.s. | −1.02 | |
| PHENYLALANINE_ METABOLISM | 15 | [GenMAPP] | n.s. | −1.52 | |
| RNA_TRANSCRIPTION_ REACTOME | 15 | [GenMAPP] | n.s. | −1.61 | |
| RNA_PATHWAY | 15 | dsRNA-activated protein kinase phosphorylates elF2a, which generally inhibits translation, and activates NFkB to provoke inflammation. [BioCarta] | n.s. | −1.69 | |
| SA_TRKA_RECEPTOR | 15 | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. [SigmaAldrich] | n.s. | −1.84 | |
| ST_DICTYOSTELIUM_ DISCOIDEUM_CAMP_ CHEMOTAXIS_PATHWAY | 15 | The fungus Dictyostelium discoideum is a model system for cytoskeletal organization during chemotaxis. [SIGNALING Transduction KE] | n.s. | −2.04 | |
| ST_GA13_PATHWAY | 15 | G-alpha-13 influences the actin cytoskeleton and activates protein kinase D, PI3K, and Pyk2. [SIGNALING Transduction KE] | n.s. | −1.99 | |
| ST_GAQ_PATHWAY | 15 | G-alpha-q activates phospholipase C, resulting in calcium influx and increasing protein kinase C activity. [SIGNALING Transduction KE] | n.s. | −2.45 | |
These gene sets are canonical representations of a biological process compiled by domain experts. PAGE gene set analysis was performed using the DIANE 6.0 software using the PAGE algorithm. The archive of gene sets used with this algorithm for this analysis is from the MSigDB. Significance of functions and pathways was calculated using the right-tailed Fisher's Exact Test. The p-value was calculated by comparing the number of user-specified genes of interest participating in a given function or pathway relative to the total number of occurrences of these genes in all functional/pathway annotations stored in the knowledge base.
n.s.: not significant
Table 2. Functional overview of significant categorized MSigDB gene sets based on the pathways altered by both wild-type and T. b. rhodesiense inhibited for brucipain activity.
| Cytokines | Transcription Factors | Cell Surface Markers | Kinases | Translocated Genes | Oncogenes | Tumor Suppressors | |
| Tumor suppressors | TP53 | CDKN2A, CFL1, PTEN, TP53 | |||||
| Oncogenes | CDK4, RAF1 | CCND1 | CCND1, CDK4, HRAS, MDM2, PTPN1, RAF1 | ||||
| Translocated genes | NFKB2 | TRFC | LCK | CCND1, LCK, NFKB2, TRFC | |||
| Kinases | ACTR2, CAMK2B, CDK2, CDK4, FYN, LCK, MAP2K2, MAP3K3P, PAK4, RAF1, RPS6KA1, RPS6KA2, RPS6KB2 | ||||||
| Cell surface Markers | CD38, TRFC | ||||||
| Transcription factors | ERCC3, GATA3, GTF2F 1, HDAC1, HMGB1, KLF5, MEF2A, MEF2B, MYOD1, NFATC3, NFKB2, NFKBIB, NFKBIE, NFYB, NROB, NR1H3, RELA, STAT1, TAF6, TAF9, TP53 | ||||||
| Cytokines | IFNGR1, IFNGR2, IL2RG, IL6 |
A functional overview of the MSigDB gene sets categorized into a small number of selected gene families whose members a common feature such as homology or biochemical activity. They do not necessarily have common origins. Annotation of pathway genes significantly expressed in HBMEC only in response to T. b. rhodesiense inhibited for brucipain activity are shown in bold font.
Table 3. Annotation of pathway genes significantly expressed in HBMEC only in response to T. b. rhodesiense inhibited for brucipain activity.
| Gene Family [MSigDB] | Symbol | MsigDB Accession No. | Gene Names | GO Definitions | z-Ratio | |
| 3 h | 6 h | |||||
| cytokine | IFNGR2 | BC003624 | interferon gamma receptor 2 (interferon gamma transducer 1) | cell surface receptor linked signal transduction | 0.81 | n.s. |
| integral to plasma membrane | ||||||
| antiviral response protein activity | ||||||
| cytokine | IL2RG | BC014972 | interleukin 2 receptor, gamma (severe combined immunodeficiency) | cell proliferation | n.s. | 1.12 |
| integral to plasma membrane | ||||||
| interleukin-2 receptor activity | ||||||
| cytokine | IL6 | BC015511 | interleukin 6 (interferon, beta 2) | acute-phase response | n.s. | 0.84 |
| extracellular space | ||||||
| IL-6 receptor ligand activity | ||||||
| kinase | CAMK2B | BC019070 | calcium/calmodulin-dependent protein kinase (CaM kinase) II beta | protein amino acid phosphorylation | 0.80 | n.s. |
| ATP binding activity | ||||||
| kinase | CDK2 | BC003065 | cyclin-dependent kinase 2 | G2/M transition of mitotic cell cycle | 0.75 | n.s. |
| cytoplasm | ||||||
| ATP binding activity | ||||||
| kinase | MAP2K2 | BC000471 | mitogen-activated protein kinase kinase 2 | protein amino acid phosphorylation | 1.98 | n.s. |
| extracellular | ||||||
| ATP binding activity | ||||||
| kinase | PAK4 | BC002921 | p21(CDKN1A)-activated kinase 4 | cell motility | −1.25 | n.s. |
| Golgi apparatus | ||||||
| ATP binding activity | ||||||
| kinase | RPS6KA2 | BC002363 | ribosomal protein S6 kinase, 90kDa, polypeptide 2 | protein amino acid phosphorylation | 1.36 | n.s. |
| nucleus | ||||||
| ATP binding activity | ||||||
| kinase | RPS6KB2 | BC000094 | ribosomal protein S6 kinase, 70kDa, polypeptide 2 | protein amino acid phosphorylation | 1.46 | n.s. |
| ATP binding activity | ||||||
| kinase, translocated gene | LCK | BC013200 | lymphocyte-specific protein tyrosine kinase | RAS protein signal transduction | n.s. | 1.65 |
| membrane fraction | ||||||
| ATP binding activity | ||||||
| oncogene | MDM2 | BC009893 | Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) | negative regulation of cell proliferation | −1.69 | n.s. |
| nucleus | ||||||
| ligase activity | ||||||
| oncogene | PTPN1 | BC015660 | protein tyrosine phosphatase, non-receptor type 1 | protein amino acid dephosphorylation | 0.66 | n.s. |
| cytoplasm | ||||||
| prenylated protein tyrosine phosphatase activity | ||||||
| oncogene, kinase | CDK4 | BC010153 | cyclin-dependent kinase 4 | G1/S transition of mitotic cell cycle | 1.11 | n.s. |
| ATP binding activity | ||||||
| oncogene, translocated gene | CCND1 | BC014078 | cyclin D1 (PRAD1: parathyroid adenomatosis 1) | G1/S transition of mitotic cell cycle | −1.13 | n.s. |
| cellular_component unknown | ||||||
| transcription factor | ACTR2 | BC014546 | ARP2 actin-related protein 2 homolog (yeast) | cell motility | −1.58 | n.s. |
| Arp2/3 protein complex | ||||||
| structural constituent of cytoskeleton | ||||||
| transcription factor | ERCC3 | BC008820 | excision repair cross-complementing rodent repair deficiency, complementation group 3 (xeroderma pigmentosum group B complementing) | regulation of transcription\, DNA-dependent nucleus ATP dependent DNA helicase activity | −1.62 | n.s. |
| transcription factor | FYN | NM_002037 | FYN oncogene related to SRC, FGR, YES | cell growth and/or maintenance ATP binding activity | −0.36 | 0.61 |
| transcription factor | GATA3 | BC006793 | GATA binding protein 3 | defense response (activates Th2 cytokine gene expression) nucleus transcription factor activity | 1.44 | n.s. |
| transcription factor | GTF2F1 | BC000120 | general transcription factor IIF, polypeptide 1, 74kDa | regulation of transcription\, DNA-dependent | n.s. | 0.77 |
| transcription factor TFIIF complex | ||||||
| DNA binding activity | ||||||
| transcription factor | HMGB1 | BC003378 | high-mobility group box 1 | DNA unwinding | 0.97 | n.s. |
| chromatin | ||||||
| single-stranded DNA binding activity | ||||||
| transcription factor | MEF2A | BC013437 | MADS box transcription enhancer factor 2, polypeptide A (myocyte enhancer factor 2A) | muscle development | −1.34 | n.s. |
| nucleus | ||||||
| transcription co-activator activity | ||||||
| transcription factor | MYOD1 | BC000353 | myogenic factor 3 | cell differentiation | 1.86 | n.s. |
| nucleus | ||||||
| RNA polymerase II transcription factor activity\, enhancer binding | ||||||
| transcription factor | NFATC3 | BC001050 | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 | inflammatory response | −0.90 | n.s. |
| nucleus | ||||||
| transcription co-activator activity | ||||||
| transcription factor | NFKBIE | BC011676 | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon | cytoplasm | 1.79 | n.s. |
| transcription factor binding activity\, cytoplasmic sequestering | ||||||
| transcription factor | NFYB | BC005316 | nuclear transcription factor Y, beta | regulation of transcription\, DNA-dependent | 0.82 | n.s. |
| nucleus | ||||||
| transcription factor activity | ||||||
| transcription factor | NR1H3 | BC008819 | nuclear receptor subfamily 1, group H, member 3 | regulation of transcription\, DNA-dependent | −0.94 | n.s. |
| nucleus | ||||||
| steroid hormone receptor activity | ||||||
| transcription factor | RELA | BC014095 | v-rel reticuloendotheliosis viral oncogene homolog A, nuclear factor of kappa light polypeptide gene enhancer in B-cells 3, p65 (avian) | anti-apoptosis | 1.40 | n.s. |
| transcription factor complex | ||||||
| protein binding activity | ||||||
| transcription factor | TAF6 | BC018115 | TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa | regulation of transcription\, DNA-dependent | 1.18 | n.s. |
| transcription factor TFIID complex | ||||||
| DNA binding activity | ||||||
| transcription factor | TAF9 | BC007349 | TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa | regulation of transcription\, DNA-dependent | 0.51 | n.s. |
| transcription factor TFIID complex | ||||||
| DNA binding activity | ||||||
| translocated gene, transcription factor | NFKB2 | BC002844 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) | cell growth and/or maintenance | −2.37 | n.s. |
| nucleus | ||||||
| transcription co-activator activity | ||||||
Raw microarray data were subjected to Z normalization and tested for significant changes. Genes were determined to be differentially expressed after calculating the Z ratio and fdr.
n.s.: not significant
Analysis of the HBMEC pathways specifically altered by the brucipain-inhibited parasites included those found only within down-regulated Clusters-1 and Cluster-15, or up-regulated Cluster-8 and Cluster-11. Cluster-1 consisted of 6 different pathways that had negative Z-score values (and were therefore positively effected by brucipain) by 3-hours, and that returned to control levels by 6h (Figure 4, Table 1). While roles for the metabolic/degradation pathways in Cluster-1 in HAT are not clear, the data suggest brucipain alters the CARM1 and Leptin pathways. Leptin is a protein hormone typically produced by adipose tissue that is known to regulate appetite via binding to the leptin receptor (LEPR) in the hypothalamus after crossing the BBB [51],[52]. It has been suggested that compromised leptin transport into the CNS resulting in low leptin levels in the hippocampus could lead to cognitive deficits [53]. There is evidence that the secretion of photoperiodic hormones such as melatonin is inversely regulated by leptin [54]. It is also tempting to speculate a role for leptin in HAT considering that disturbance in the circadian rhythm of the melatonin-generating systems in the pathogenesis of African sleeping sickness has been demonstrated [55].
Brucipain may also alter processes linked to arginine methylation. CARM1 is a protein arginine N-methlytransferase that plays a role in protein arginine methylation, a process that is implicated in signal transduction, nascent pre-RNA metabolism and transcriptional activation [56]. These data suggest that CARM1, as a promoter-specific regulator of NF-κB-dependent gene expression [57], could play a role in the inflammatory responses associated with CNS HAT.
In contrast to the Cluster-1 pathways, 3 hours exposure to the brucipain-inhibited trypanosomes upregulated 9 pathways represented in Cluster-8 (Figure 1, Table 1). Inhibiting brucipain activity appeared to activate the eukaryotic initiation factor-2 (EIF2)-pathway. EIF2 binding to GTP and Met-tRNA would in turn initiate translation by transferring the Met-tRNA to the 40S ribosomal subunit. If brucipain plays a role in downregulating this pathway, the event could shut-down cellular protein synthesis and could have consequences to overall cell viability [58].
The response to activation of PAR-2 is the elevation of intracellular Ca2+ via the PLC/IP3 pathway [59],[60], which leads to downstream increases in intracellular Ca2+, activation of PKC, mitogen-activated protein kinases (MAPK) and/or stress-activated protein kinases [61],[62],[63]. It has been also shown in other systems that direct PAR-2 activation of extracellular signal-regulated kinase (ERK) subfamily of MAPK [64] may be neuroprotective [47],[48],[49]. Because of the above characteristics associated with PAR-2 activation, it is remarkable that the most dramatic changes that happened between 3 and 6 hours in the pathways were associated with Cluster-11 and Cluster-15 (Table 1). Cluster-11 contained 4 pathways with strong positive Z-scores in response to the brucipain-inhibited trypanosomes. Of these 4 pathways, 3 were involved either with cell signaling (BREAST_CANCER_ESTROGEN_SIGNALING) or inflammatory responses (CLASSIC_PATHWAY, IL7_PATHWAY). In contrast to Cluster 11, all pathways in Cluster 15 displayed negative Z-scores, indicating downregulation. In some respects, Cluster 15 is the most interesting as it contained pathways that conceivably play a role in parasite transmigration of the BBB as an early event, and in the subsequent later inflammatory responses associated with HAT. The involvement of the ST_GAQ_PATHWAY (Fig. 4, Table 1) is interesting given that calcium signaling may play an important role in trypanosome / BBB associated events. Furthermore, the changes in the ST_GA13- SA_TRKA_RECEPTOR, ST_DICTOYOSTELIUM_DISCOIDEUM_CAMP_CHEMOTAXIS, and CELL_2_CELL-pathways also parallel roles for PI3K, MAPK and cell cytoskeleton. A role for brucipain as an inducer of inflammatory responses [4] is also predicted (i.e., EPO_NFKB_PATHWAY, RNA_PATHWAY).
Conclusions
We studied parasite proteases in the interaction of T. b. rhodesiense with HBMECs. Overall, our RNAi, PMT and DNA-microarray data support an important role for brucipain, HBMEC PAR-2 (and possibly other PARs) and Gq signaling for trypanosome transmigration across the BBB. Owing to PAR-2's role in neuroinflammation, these data also suggest a role for this GPCR in CNS HAT. In murine models of HAT the neuroinflammatory response [1],[65] is likely a balance between pro-inflammatory cytokines such as interferon-γ (IFN-γ), interleukin-1(IL-1) and tumour necrosis factor-α (TNF-α), and counter-inflammatory cytokines such as IL-10 [66]. A role for cytokines in determining entry of trypanosomes into the CNS was provided by a seminal study in knockout mice in which the gene for IFN-γ had been disrupted [67]. Following systemic infection, it was found that trypanosomes accumulated in the perivascular regions, ‘trapped’ between the endothelial and the parenchymal basement membranes. While these findings suggested that lymphocyte-derived IFN-γ is required for trypanosome traversal across cerebral blood vessels [67], precisely how IFN-γ facilitates BBB traversal by the parasites has yet to be determined. Interestingly, minocycline, a tetracycline antibiotic, was also found to impede the penetration of leukocytes and trypanosomes into the brain parenchyma [68]. It is tempting to speculate a role for PAR-2 in these processes. It has been shown that the inflammatory response in mouse colonic tissue mediated by PAR-2 activation (using PAR-2 agonists) that leads to i) increased tissue permeability, ii) increased IFN-γ and TNF-α expression, and iv) decreased IL-10 expression, are abolished in IFN-γ deficient B6 mice [69]. More recently, minocycline has been shown to block the PAR-2-mediated TNF-α-induced production of IL-8 proinflammatory response in epidermal keratinocytes (Ishikawa) [70].
Our previous work strongly suggested that T. b. rhodesiense crosses the human BBB by generating Ca2+ activation signals in HBMECs through the activity of parasite cysteine proteases [5],[9]. Using T. b. brucei silenced for (RNAi) it was later found that parasite cathepsin L (brucipain) could be the parasite factor initiating transmigration and increased vascular permeability [11]. While a singular role has not been established for this process of parasite transmigration in vivo, GPCR PAR2 is one of the molecular targets for brucipain as an activator of Gq-mediated calcium-signaling involving the downstream effectors PLC and PKC [5],[9]. The in vivo consequence of these events, similar to our in vitro findings, is predicted to be increased permeability of the BBB to parasite transmigration, an event precursory to CNS disease [4],[11]. The critical role of brucipain raises the possibility for this protein as an attractive drug or vaccine target.
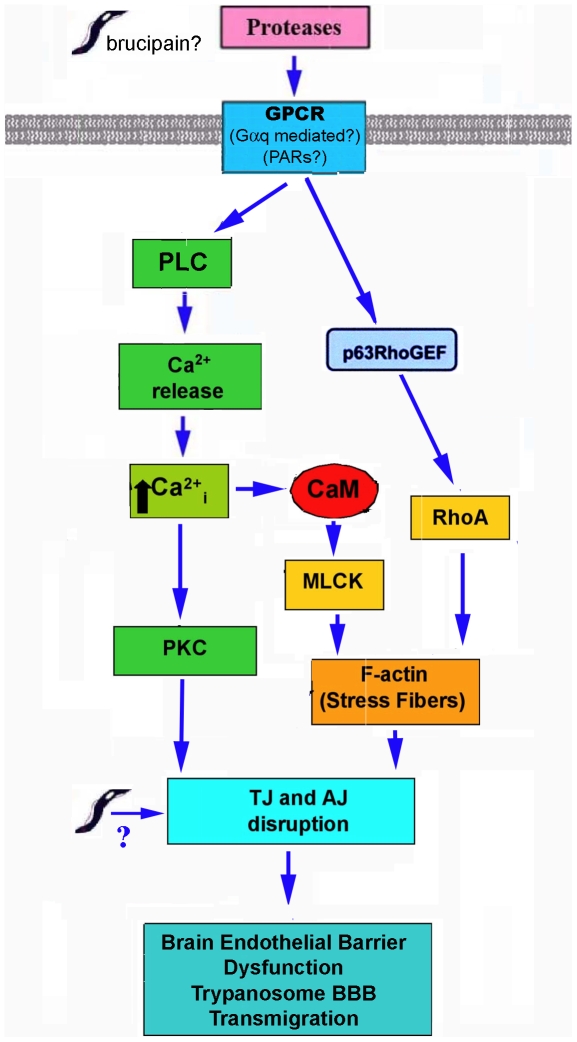
Based on our published and findings reported here [4],[5],[9],[10], we hypothesize that African trypanosome-mediated BBB dysfunction is linked to the interactions of parasite and/or parasite protease activity with BMEC GPCRs (i.e. PARs) (Fig. 5). Activation of downstream effectors then enable African trypanosome transmigration through the BBB after cytoskeletal rearrangements that induce cell retraction and loosening of junctional complexes. We predict that trypanosomes/parasite/proteases trigger Gαq activation of PLC-β, in turn generating inositol-1,4,5-triphosphate (IP3) and diacylglycerol (DAG) from phosphatidylinositol-4,5-bisphosphate (PIP2). Binding of IP3 to its receptor on the endoplasmic reticulum releases Ca2+ from intracellular stores. The increase in intracellular calcium leads to calmodulin (CaM) activation of myosin light chain (MLC) kinase (MLCK) and/or other effectors ultimately leading to cytoskeletal changes and barrier dysfunction. Ca2+-independent activation of the cytoskeleton by Ras-superfamily GTPases (i.e RhoA) traditionally by Gα12/13 GPCRs, is also possible via Gαq activation of p63RhoGEF [42],[71]. Although not yet determined, parasite and/or host-derived proteases may also contribute by degrading or altering adherens junction (AJ) and tight junction (TJ) proteins.
Figure 5. Proposed model for African trypanosome-induced BBB dysfunction.
We hypothesize that parasite proteases trigger GPCRs (i.e. PARs?) via Gαq activation, which leads to PLC-mediated Ca2+ release from intracellular stores. The increase in intracellular calcium leads to calmodulin (CaM) activation of myosin light chain kinase (MLCK), ultimately leading to cytoskeletal changes and barrier dysfunction. Ca2+-independent activation of the cytoskeleton mediated by Ras-superfamily GTPases (i.e. RhoA) is also possible via p63RhoGEF. Parasite and/or host-derived proteases may also contribute by degrading or altering adherens junction (AJ) and tight junction (TJ) proteins.
Clearly, a study on the contribution of the products of gene expression identified with the biochemical pathways needs to be investigated as well as testing using established animal models of HAT [4]. An understanding of these responses at a molecular level will help identify candidates for the early diagnosis, treatment, and prevention of CNS invasion with HAT.
Acknowledgments
The authors wish to thank Dr. J. McKerrow and Dr. Maha Abdulla, University of California San Francisco, for providing K11777 and for critical reading of the manuscript. We also thank Joseph Crompton, Emily Clemens, Donna Pearce and Nicole Barat for expert technical assistance, and Applied Biophysics (Troy, NY) for loan of a 1600R ECIS instrument. DJG dedicates this work in remembrance of Justina Brincko.
Footnotes
The authors have declared that no competing interests exist.
This research was supported in part by grants from the National Institutes of Health (RO1AI051464) to DJG and (R01AI038936) to BAW, the Fogarty International Foundation (FIRCA) to DJG/JS, the American Heart Association (SDG 0435177N) to YVK, and by the Intramural Research Program of the NIH, National Institute on Aging (KGB). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kennedy PG. Human African trypanosomiasis of the CNS: current issues and challenges. J Clin Invest. 2004;113:496–504. doi: 10.1172/JCI21052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molyneux DH, Pentreath V, Doua F. African trypanosomiasis in man. London: Saunders WB; 1996. pp. 1171–1196. [Google Scholar]
- 3.Rogers DJ, Williams BG. Monitoring trypanosomiasis in space and time. Parasitology. 1993;106(Suppl):S77–92. doi: 10.1017/s0031182000086133. [DOI] [PubMed] [Google Scholar]
- 4.Grab DJ, Kennedy PE. Traversal of human and animal trypanosomes across the blood-brain barrier. J Neurovirology. 2008;14:344–351. doi: 10.1080/13550280802282934. [DOI] [PubMed] [Google Scholar]
- 5.Grab DJ, Nikolskaia O, Kim YV, Lonsdale-Eccles JD, Ito S, et al. African trypanosome interactions with an in vitro model of the human blood-brain barrier. J Parasitol. 2004;90:970–979. doi: 10.1645/GE-287R. [DOI] [PubMed] [Google Scholar]
- 6.Lonsdale-Eccles JD, Grab DJ. Trypanosome hydrolases and the blood-brain barrier. Trends Parasitol. 2002;18:17–19. doi: 10.1016/s1471-4922(01)02120-1. [DOI] [PubMed] [Google Scholar]
- 7.Mackey ZB, O'Brien TC, Greenbaum DC, Blank RB, McKerrow JH. A cathepsin B-like protease is required for host protein degradation in Trypanosoma brucei. J Biol Chem. 2004;279:48426–48433. doi: 10.1074/jbc.M402470200. [DOI] [PubMed] [Google Scholar]
- 8.Scory S, Caffrey CR, Stierhof YD, Ruppel A, Steverding D. Trypanosoma brucei: killing of bloodstream forms in vitro and in vivo by the cysteine proteinase inhibitor Z-phe-ala-CHN2. Exp Parasitol. 1999;91:327–333. doi: 10.1006/expr.1998.4381. [DOI] [PubMed] [Google Scholar]
- 9.Nikolskaia OV, Lima APCA, Kim YV, Lonsdale-Eccles JD, Fukuma T, et al. Blood-brain barrier traversal by African trypanosomes requires calcium signaling induced by parasite cysteine protease. J Clin Invest. 2006;116:2739–2747. doi: 10.1172/JCI27798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nikolskaia OV, de ALAP, Kim YV, Lonsdale-Eccles JD, Fukuma T, et al. Erratum. J Clin Invest. 2008;118:1974. doi: 10.1172/JCI27798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdulla MH, O'Brien T, Mackey ZB, Sajid M, Grab DJ, et al. RNA Interference of Trypanosoma brucei Cathepsin B and L Affects Disease Progression in a Mouse Model. PLoS Negl Trop Dis. 2008;2:e298. doi: 10.1371/journal.pntd.0000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lourbakos A, Yuan YP, Jenkins AL, Travis J, Andrade-Gordon P, et al. Activation of protease-activated receptors by gingipains from Porphyromonas gingivalis leads to platelet aggregation: a new trait in microbial pathogenicity. Blood. 2001;97:3790–3797. doi: 10.1182/blood.v97.12.3790. [DOI] [PubMed] [Google Scholar]
- 13.Lourbakos A, Potempa J, Travis J, D'Andrea MR, Andrade-Gordon P, et al. Arginine-specific protease from Porphyromonas gingivalis activates protease-activated receptors on human oral epithelial cells and induces interleukin-6 secretion. Infect Immun. 2001;69:5121–5130. doi: 10.1128/IAI.69.8.5121-5130.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lourbakos A, Chinni C, Thompson P, Potempa J, Travis J, et al. Cleavage and activation of proteinase-activated receptor-2 on human neutrophils by gingipain-R from Porphyromonas gingivalis. FEBS Lett. 1998;435:45–48. doi: 10.1016/s0014-5793(98)01036-9. [DOI] [PubMed] [Google Scholar]
- 15.Kim YV, Di Cello F, Hillaire CS, Kim KS. Differential Ca2+ signaling by thrombin and protease-activated receptor-1-activating peptide in human brain microvascular endothelial cells. Am J Physiol Cell Physiol. 2004;286:C31–42. doi: 10.1152/ajpcell.00157.2003. [DOI] [PubMed] [Google Scholar]
- 16.Palmer JT, Rasnick D, Klaus JL, Bromme D. Vinyl sulfones as mechanism-based cysteine protease inhibitors. J Med Chem. 1995;38:3193–3196. doi: 10.1021/jm00017a002. [DOI] [PubMed] [Google Scholar]
- 17.Doyle PS, Zhou YM, Engel JC, McKerrow JH. A cysteine protease inhibitor cures Chagas' disease in an immunodeficient-mouse model of infection. Antimicrob Agents Chemother. 2007;51:3932–3939. doi: 10.1128/AAC.00436-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aminova LR, Wilson BA. Calcineurin-independent inhibition of 3T3-L1 adipogenesis by Pasteurella multocida toxin: suppression of Notch1, stabilization of beta-catenin and pre-adipocyte factor 1. Cell Microbiol. 2007;9:2485–2496. doi: 10.1111/j.1462-5822.2007.00975.x. [DOI] [PubMed] [Google Scholar]
- 19.Hirumi H, Hirumi K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- 20.Deli MA, Abraham CS, Kataoka Y, Niwa M. Permeability studies on in vitro blood-brain barrier models: physiology, pathology, and pharmacology. Cell Mol Neurobiol. 2005;25:59–127. doi: 10.1007/s10571-004-1377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tiruppathi C, Malik AB, Del Vecchio PJ, Keese CR, Giaever I. Electrical method for detection of endothelial cell shape change in real time: assessment of endothelial barrier function. Proc Natl Acad Sci U S A. 1992;89:7919–7923. doi: 10.1073/pnas.89.17.7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aschner M, Vitarella D, Allen JW, Conklin DR, Cowan KS. Methylmercury-induced astrocytic swelling is associated with activation of the Na+/H+ antiporter, and is fully reversed by amiloride. Brain Res. 1998;799:207–214. doi: 10.1016/s0006-8993(98)00399-0. [DOI] [PubMed] [Google Scholar]
- 23.Reddy L, Wang HS, Keese CR, Giaever I, Smith TJ. Assessment of rapid morphological changes associated with elevated cAMP levels in human orbital fibroblasts. Exp Cell Res. 1998;245:360–367. doi: 10.1006/excr.1998.4273. [DOI] [PubMed] [Google Scholar]
- 24.Keese C. Monitoring of cells during signal transduction. Genetic Engineering News. 2001;21:51. [Google Scholar]
- 25.Grab DJ, Perides G, Dumler JS, Kim KJ, Park J, et al. Borrelia burgdorferi, host-derived proteases, and the blood-brain barrier. Infect Immun. 2005;73:1014–1022. doi: 10.1128/IAI.73.2.1014-1022.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nyarko E, Grab DJ, Dumler JS. Anaplasma phagocytophilum-infected neutrophils enhance transmigration of Borrelia burgdorferi across the human blood brain barrier in vitro. Int J Parasitol. 2006;36:601–605. doi: 10.1016/j.ijpara.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 27.Brown A, Zhang H, Lopez P, Pardo CA, Gartner S. In vitro modeling of the HIV-macrophage reservoir. J Leukoc Biol. 2006;80:1127–1135. doi: 10.1189/jlb.0206126. [DOI] [PubMed] [Google Scholar]
- 28.Eckmann L, Smith JR, Housley MP, Dwinell MB, Kagnoff MF. Analysis by high density cDNA arrays of altered gene expression in human intestinal epithelial cells in response to infection with the invasive enteric bacteria Salmonella. J Biol Chem. 2000;275:14084–14094. doi: 10.1074/jbc.275.19.14084. [DOI] [PubMed] [Google Scholar]
- 29.Nadon NL, Mohr D, Becker KG. National Institute on Aging microarray facility--resources for gerontology research. J Gerontol A Biol Sci Med Sci. 2005;60:413–415. doi: 10.1093/gerona/60.4.413. [DOI] [PubMed] [Google Scholar]
- 30.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim SY, Volsky DJ. PAGE: parametric analysis of gene set enrichment. BMC Bioinformatics. 2005;6:144. doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaidyula VR, Rao AK. Role of Galphaq and phospholipase C-beta2 in human platelets activation by thrombin receptors PAR1 and PAR4: studies in human platelets deficient in Galphaq and phospholipase C-beta2. Br J Haematol. 2003;121:491–496. doi: 10.1046/j.1365-2141.2003.04296.x. [DOI] [PubMed] [Google Scholar]
- 34.Offermanns S, Toombs CF, Hu YH, Simon MI. Defective platelet activation in G alpha(q)-deficient mice. Nature. 1997;389:183–186. doi: 10.1038/38284. [DOI] [PubMed] [Google Scholar]
- 35.Offermanns S, Laugwitz KL, Spicher K, Schultz G. G proteins of the G12 family are activated via thromboxane A2 and thrombin receptors in human platelets. Proc Natl Acad Sci U S A. 1994;91:504–508. doi: 10.1073/pnas.91.2.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Babich M, King KL, Nissenson RA. Thrombin stimulates inositol phosphate production and intracellular free calcium by a pertussis toxin-insensitive mechanism in osteosarcoma cells. Endocrinology. 1990;126:948–954. doi: 10.1210/endo-126-2-948. [DOI] [PubMed] [Google Scholar]
- 37.Zywietz A, Gohla A, Schmelz M, Schultz G, Offermanns S. Pleiotropic effects of Pasteurella multocida toxin are mediated by Gq-dependent and -independent mechanisms. Involvement of Gq but not G11. J Biol Chem. 2001;276:3840–3845. doi: 10.1074/jbc.M007819200. [DOI] [PubMed] [Google Scholar]
- 38.Wilson BA, Ho M. Pasteurella multocida toxin as a tool for studying Gq signal transduction. Rev Physiol Biochem Pharmacol. 2004;152:93–109. doi: 10.1007/s10254-004-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aminova LR, Luo S, Bannai Y, Ho M, Wilson BA. The C3 domain of Pasteurella multocida toxin is the minimal domain responsible for activation of Gq-dependent calcium and mitogenic signaling. Protein Sci. 2008;17:945–949. doi: 10.1110/ps.083445408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orth JH, Lang S, Taniguchi M, Aktories K. Pasteurella multocida toxin-induced activation of RhoA is mediated via two families of G{alpha} proteins, G{alpha}q and G{alpha}12/13. J Biol Chem. 2005;280:36701–36707. doi: 10.1074/jbc.M507203200. [DOI] [PubMed] [Google Scholar]
- 41.Orth JH, Fester I, Preuss I, Agnoletto L, Wilson BA, et al. Activation of Galpha (i) and subsequent uncoupling of receptor-Galpha(i) signaling by Pasteurella multocida toxin. J Biol Chem. 2008;283:23288–23294. doi: 10.1074/jbc.M803435200. [DOI] [PubMed] [Google Scholar]
- 42.Mizuno N, Itoh H. Functions and regulatory mechanisms of Gq-signaling pathways. Neurosignals. 2009;17:42–54. doi: 10.1159/000186689. [DOI] [PubMed] [Google Scholar]
- 43.Seo B, Choy EW, Maudsley S, Miller WE, Wilson BA, et al. Pasteurella multocida toxin stimulates mitogen-activated protein kinase via G(q/11)-dependent transactivation of the epidermal growth factor receptor. J Biol Chem. 2000;275:2239–2245. doi: 10.1074/jbc.275.3.2239. [DOI] [PubMed] [Google Scholar]
- 44.Orth JH, Lang S, Aktories K. Action of Pasteurella multocida toxin depends on the helical domain of Galphaq. J Biol Chem. 2004;279:34150–34155. doi: 10.1074/jbc.M405353200. [DOI] [PubMed] [Google Scholar]
- 45.Wilson BA, Aminova LR, Ponferrada VG, Ho M. Differential modulation and subsequent blockade of mitogenic signaling and cell cycle progression by Pasteurella multocida toxin. Infect Immun. 2000;68:4531–4538. doi: 10.1128/iai.68.8.4531-4538.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson BA, Zhu X, Ho M, Lu L. Pasteurella multocida toxin activates the inositol triphosphate signaling pathway in Xenopus oocytes via G(q)alpha-coupled phospholipase C-beta1. J Biol Chem. 1997;272:1268–1275. doi: 10.1074/jbc.272.2.1268. [DOI] [PubMed] [Google Scholar]
- 47.Mocchetti I, Bachis A. Brain-derived neurotrophic factor activation of TrkB protects neurons from HIV-1/gp120-induced cell death. Crit Rev Neurobiol. 2004;16:51–57. doi: 10.1615/critrevneurobiol.v16.i12.50. [DOI] [PubMed] [Google Scholar]
- 48.Li F, Omori N, Jin G, Wang SJ, Sato K, et al. Cooperative expression of survival p-ERK and p-Akt signals in rat brain neurons after transient MCAO. Brain Res. 2003;962:21–26. doi: 10.1016/s0006-8993(02)03774-5. [DOI] [PubMed] [Google Scholar]
- 49.Hetman M, Xia Z. Signaling pathways mediating anti-apoptotic action of neurotrophins. Acta Neurobiol Exp (Wars) 2000;60:531–545. doi: 10.55782/ane-2000-1374. [DOI] [PubMed] [Google Scholar]
- 50.Bushell T. The emergence of proteinase-activated receptor-2 as a novel target for the treatment of inflammation-related CNS disorders. J Physiol. 2007;581:7–16. doi: 10.1113/jphysiol.2007.129577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan W, Hsuchou H, Tu H, Kastin AJ. Developmental changes of leptin receptors in cerebral microvessels: unexpected relation to leptin transport. Endocrinology. 2008;149:877–885. doi: 10.1210/en.2007-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Otvos L, Jr, Terrasi M, Cascio S, Cassone M, Abbadessa G, et al. Development of a pharmacologically improved peptide agonist of the leptin receptor. Biochim Biophys Acta. 2008;1783:1745–1754. doi: 10.1016/j.bbamcr.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 53.Farr SA, Banks WA, Morley JE. Effects of leptin on memory processing. Peptides. 2006;27:1420–1425. doi: 10.1016/j.peptides.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 54.Zieba DA, Szczesna M, Klocek-Gorka B, Molik E, Misztal T, et al. Seasonal effects of central leptin infusion on secretion of melatonin and prolactin and on SOCS-3 gene expression in ewes. J Endocrinol. 2008;198:147–155. doi: 10.1677/JOE-07-0602. [DOI] [PubMed] [Google Scholar]
- 55.Kristensson K, Claustrat B, Mhlanga JD, Moller M. African trypanosomiasis in the rat alters melatonin secretion and melatonin receptor binding in the suprachiasmatic nucleus. Brain Res Bull. 1998;47:265–269. doi: 10.1016/s0361-9230(98)00084-7. [DOI] [PubMed] [Google Scholar]
- 56.Frankel A, Yadav N, Lee J, Branscombe TL, Clarke S, et al. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J Biol Chem. 2002;277:3537–3543. doi: 10.1074/jbc.M108786200. [DOI] [PubMed] [Google Scholar]
- 57.Covic M, Hassa PO, Saccani S, Buerki C, Meier NI, et al. Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-kappaB-dependent gene expression. EMBO J. 2005;24:85–96. doi: 10.1038/sj.emboj.7600500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Girard M, Bisser S, Courtioux B, Vermot-Desroches C, Bouteille B, et al. In vitro induction of microglial and endothelial cell apoptosis by cerebrospinal fluids from patients with human African trypanosomiasis. Int J Parasitol. 2003;33:713–720. doi: 10.1016/s0020-7519(03)00033-x. [DOI] [PubMed] [Google Scholar]
- 59.Bushell TJ, Plevin R, Cobb S, Irving AJ. Characterization of proteinase-activated receptor 2 signalling and expression in rat hippocampal neurons and astrocytes. Neuropharmacology. 2006;50:714–725. doi: 10.1016/j.neuropharm.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 60.Smith-Swintosky VL, Cheo-Isaacs CT, D'Andrea MR, Santulli RJ, Darrow AL, et al. Protease-activated receptor-2 (PAR-2) is present in the rat hippocampus and is associated with neurodegeneration. J Neurochem. 1997;69:1890–1896. doi: 10.1046/j.1471-4159.1997.69051890.x. [DOI] [PubMed] [Google Scholar]
- 61.Kanke T, Macfarlane SR, Seatter MJ, Davenport E, Paul A, et al. Proteinase-activated receptor-2-mediated activation of stress-activated protein kinases and inhibitory kappa B kinases in NCTC 2544 keratinocytes. J Biol Chem. 2001;276:31657–31666. doi: 10.1074/jbc.M100377200. [DOI] [PubMed] [Google Scholar]
- 62.Belham CM, Tate RJ, Scott PH, Pemberton AD, Miller HR, et al. Trypsin stimulates proteinase-activated receptor-2-dependent and -independent activation of mitogen-activated protein kinases. Biochem J. 1996;320(Pt 3):939–946. doi: 10.1042/bj3200939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DeFea KA, Zalevsky J, Thoma MS, Dery O, Mullins RD, et al. beta-arrestin-dependent endocytosis of proteinase-activated receptor 2 is required for intracellular targeting of activated ERK1/2. J Cell Biol. 2000;148:1267–1281. doi: 10.1083/jcb.148.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin G, Hayashi T, Kawagoe J, Takizawa T, Nagata T, et al. Deficiency of PAR-2 gene increases acute focal ischemic brain injury. J Cereb Blood Flow Metab. 2005;25:302–313. doi: 10.1038/sj.jcbfm.9600021. [DOI] [PubMed] [Google Scholar]
- 65.Kennedy PG. Diagnostic and neuropathogenesis issues in human African trypanosomiasis. Int J Parasitol. 2006;36:505–512. doi: 10.1016/j.ijpara.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 66.Sternberg JM, Rodgers J, Bradley B, Maclean L, Murray M, et al. Meningoencephalitic African trypanosomiasis: Brain IL-10 and IL-6 are associated with protection from neuro-inflammatory pathology. J Neuroimmunol. 2005;167:81–89. doi: 10.1016/j.jneuroim.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 67.Masocha W, Robertson B, Rottenberg ME, Mhlanga J, Sorokin L, et al. Cerebral vessel laminins and IFN-gamma define Trypanosoma brucei brucei penetration of the blood-brain barrier. J Clin Invest. 2004;114:689–694. doi: 10.1172/JCI22104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Masocha W, Rottenberg ME, Kristensson K. Minocycline impedes African trypanosome invasion of the brain in a murine model. Antimicrob Agents Chemother. 2006;50:1798–1804. doi: 10.1128/AAC.50.5.1798-1804.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cenac N, Chin AC, Garcia-Villar R, Salvador-Cartier C, Ferrier L, et al. PAR2 activation alters colonic paracellular permeability in mice via IFN-gamma-dependent and -independent pathways. J Physiol. 2004;558:913–925. doi: 10.1113/jphysiol.2004.061721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ishikawa C, Tsuda T, Konishi H, Nakagawa N, Yamanishi K. Tetracyclines modulate protease-activated receptor 2-mediated proinflammatory reactions in epidermal keratinocytes. Antimicrob Agents Chemother. 2009;53:1760–1765. doi: 10.1128/AAC.01540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lutz S, Freichel-Blomquist A, Yang Y, Rumenapp U, Jakobs KH, et al. The guanine nucleotide exchange factor p63RhoGEF, a specific link between Gq/11-coupled receptor signaling and RhoA. J Biol Chem. 2005;280:11134–11139. doi: 10.1074/jbc.M411322200. [DOI] [PubMed] [Google Scholar]