Abstract
The gene of Cytotoxic T Lymphocyte-associated Antigen 4 (CTLA4), a negative regulator of T lymphocytes, contains a single-nucleotide polymorphism (SNP) at position +6230A->G (ct60A->G), which has been found associated with several autoimmune diseases and appears to reduce T-cell inhibitory activity. In Ghana, West Africa, we compared the frequencies of CTLA4 +6230 A/G and 6 haplotype-tagging SNPs in 2010 smear-positive, HIV-negative patients with pulmonary tuberculosis (TB) and 2346 controls matched for age, gender and ethnicity. We found no difference in allele frequencies between cases and controls. However, +6230A and a distinct CTLA4 haplotype and a diplotype comprising the +6230A allele were significantly less frequent among cases with large opacities in chest radiographs compared to those with small ones (Pcorrected [cor] = 0.002, Pcor = 0.00045, P = 0.0005, respectively). This finding suggests that an increased T-cell activity associated with the CTLA4 +6230G allele contributes to pathology rather than to protection in pulmonary TB.
Introduction
Cytotoxic T Lymphocyte-associated Antigen 4 (CTLA-4) is considered a key negative regulator of T lymphocytes [1]. Being transiently surface expressed and released by activated T cells, it inhibits T-cell functions through a variety of direct and indirect mechanisms [2]–[7]. In addition, CTLA-4 has been found to skew the activity of T-helper (Th) cells towards a Th1 phenotype [8]–[9].
A G→A nucleotide exchange at position +6230 (also addressed as CT60; according to alternate numbering located at position +6253 relative to the start codon) in the 3′-untranslated region of the human CTLA-4 gene has been found to support protection from several autoimmune diseases [10]. It was associated with an increased concentration of a splice variant of the CTLA4 transcript encoding a soluble form of the molecule, which was assumed to enhance the T-cell inhibitory function [10], [11]. The authors reasoned that the autoimmunity-supporting +6230G allele of CTLA4, which was found in the majority of members of the European study group, may have been the subject of evolutionary selection by providing enhanced cellular immune responses and, thereby, protection against particular intracellular infections [10]. The identification of this polymorphism provided an important tool to study the overall influence of T-cell responses in humans.
We studied the CTLA4 +6230 polymorphism in pulmonary tuberculosis (TB) for two reasons. First, T cells are regarded of eminent importance in controlling mycobacterial infections [12]. In addition, TB may be relevant in the evolution of the autoimmunity-associated +6230G allele because it is caused by an intracellular infection and, due to its widespread occurrence and high mortality in the past and today, it may be relevant to natural selection.
Studying an African population, we included in the analysis four single nucleotide polymorphisms (SNP) which recently have been described to characterize the relevant haplotypes of the CTLA4 gene in Africans [13] and, according to the most recent version of the HapMap database (http://www.hapmap.org/) five additional tagging SNPs providing comprehensive and appropriate coverage of the CTLA4 gene.
Materials and Methods
Study group
Study participants were enrolled in Ghana, West Africa, between September 2001 and July 2004. The study group has been described in detail elsewhere [14]. In brief, patients were recruited at the two major teaching hospitals of Accra and Kumasi, and additional hospitals or policlinics in Accra, Kumasi, Obuasi, Agona, Mampong, Agogo, Konongo, Nkawie, Nkawkaw, Atibie, Assin Fosu and Dunkwa. The characterization of patients comprised a documentation of medical histories on standardized questionnaires, two examinations for acid-fast bacilli of non-induced sputum specimens, a determination of HIV serology and confirmation of positive results by an alternative test, culturing and differentiation of mycobacterial isolates by IS6110 fingerprinting and spoligotyping as well as assessment of drug resistances, and a posterior-anterior chest radiography.
Included were patients 6 to 60 years of age, with no history of previous TB or anti-mycobacterial treatments, two sputum smears positive for acid-fast bacilli, and a negative HIV serology. Excluded were individuals with incomplete information provided by the questionnaire or evidence for alcoholism, drug addiction or other apparent generalized disease.
Unrelated personal contacts and neighbours as well as community members were recruited as controls. Inclusion criteria were no history of previous TB or anti-mycobacterial treatments and no evidence for previous TB in chest radiographs. The characterization of controls included a tuberculin skin test (Tuberculin Test PPD Mérieux, bioMérieux, Nürtingen, Germany).
Radiographic films were pseudonymized and read by an experienced radiologist (G. S.). Opacities, cavities, nodular lesions, pulmonary shrinkage (loss of lung volume as assessed by hilus dislocation or displacement of fissures), calcifications, and pleural thickening were individually assigned to the upper right, lower right, upper left, and lower left thoracic quadrants and overall assessed quantitatively being rated “0” (no lesion detectable), “1” (mild lesion), “2” (moderate lesion), and “3” (severe lesion). Enlargements of mediastinal lymph nodes, effusions, and miliary lesions were evaluated qualitatively. The evaluation presented here for the reason of simplicity was restricted to the overall scores for opacities and cavities because these are the most common signs of TB reflecting cellular infiltration and necrosis, respectively (Table 1).
Table 1. Radiographic findings in the case group1.
| Severity score | Opacities | Cavities |
| 0 | 17 (0.9%) | 103 (5.6%) |
| 1 | 340 (18.4%) | 825 (44.6%) |
| 2 | 1121 (60.6%) | 682 (36.8%) |
| 3 | 373 (20.2%) | 241 (13.0%) |
Shown are numbers of patients and, in brackets, percentages of total (n = 1851).
The final study group comprised 2010 cases and 2346 controls comprising 1211 household contacts and 1135 neighbours or community controls. They belonged to the following ethnic groups (cases/all controls/PPD-positive controls/PPD-negative controls): Akan including Ashanti, Fante, Akuapem (63.6%/59.1%/58.2%/74.8%), Ga-Adangbe (14.5%/19.8%/20.6%/7.0%), Ewe (7.1%/9.3%/9.6%/3.9%) and ethnic groups of northern Ghana, including Mole Dagbane, Gurma, Grusi (12.9%/10.4%/10.3%/12.6%). For 1.6% of study participants the ethnicity was not clearly assignable. The percentage of males among cases, all controls, PPD-positive controls and PPD-negative controls was 68.2%/59.2%/60.2%/42.5%, respectively. The mean age of study participants in these groups was 34.1±11.6/32.5±12.0/32.4±11.8/34.1±15.3.
The study protocol was approved by the Committee on Human Research, Publications and Ethics, School of Medical Sciences, Kwame Nkrumah University, Kumasi, and the Ethics Committee of the Ghana Health Service, Accra. Patients were treated according to the „Directly Observed Treatment, Short-course” (DOTS) strategy organized by the National Tuberculosis Programme. Blood samples for genetic analyses and HIV testing were taken only after a detailed explanation of the study aims and written or thumb-printed consent for participation provided, including HIV testing. Disclosure of HIV test results was dependent on the documented willingness of participants to be informed. HIV-positive patients were promptly referred to counseling and treatment as provided by the Ghanaian AIDS Control Programme.
Genetic Analysis
After DNA extraction from whole peripheral blood by a magnetic separation technology (AGOWA®, Berlin, Germany), the CTLA4 −1765 (rs11571315), −1722 (rs733618), −1661 (rs4553808), −1577 (rs11571316), −1411 (rs16840252), −443 (rs231774), +49 (rs231775), +923 (rs231777) and +6230 (rs3087243) variants were analysed by dynamic allele specific hybridization with fluorescence resonance energy transfer (FRET) in a LightTyper (Roche Diagnostics, Mannheim, Germany). Primer pair and sensor/anchor oligonucleotides for LightTyper-based genotyping are listed in Table 2.
Table 2. Primers and sensor/anchor oligonucleotides used for CTLA4 genotyping1.
| CTLA4 SNP | rs Number | Primer Oligonucleotides2 | Sensor/Anchor Oligonucleotides3 |
| −1765 A/G | rs11571315 | F–CTGCTAAGAGCATCCGC | S–TCAACTCCAGCATTGATCTCACTCTAT |
| R–TGTTGGTGTGATGCACAG | A–GCTGAACCACTGGCTTCTGCTCCTCTACATAATAC | ||
| −1722 A/G | rs733618 | F–CTGCTAAGAGCATCCGC | S–GGGTTTAGCTGCCTGTCCC |
| R–TGTTGGTGTGATGCACAG | A-GCCACTGCTGTGTGTTCCTCTTGAGG | ||
| −1661 A/G | rs4553808 | F–CTGCTAAGAGCATCCGC | S–TGGGCAACAGAGGTTTTTCAAAAAG |
| R–TGTTGGTGTGATGCACAG | A–CAATAACAACCTAATGGGCACTTCCTAATGCCAGA | ||
| −1577 C/T | rs11571316 | F–CTGCTAAGAGCATCCGC | S–CTGGGGCTTGAAGGTTTCTATAATGTGTA |
| R–TGTTGGTGTGATGCACAG | A–CAGTGTATAGAAAACAGGCAGGTCAGAAAAGGC | ||
| −1147 C/T | rs16840252 | F_AGGGAGGCATTTGGTGA | S_GGACGGACTGGAGTAGGCAA |
| R_CCGGAATTCTGTGCACTT | A_GTCATATTCCCTGTTACAACTGTCTGTTTGCATGT | ||
| −443 A/T | rs231774 | F_AGAAGGATGGTGCTTCACA | S_GCCTAGTAGTTTTGGAGATGTCAATGAAATGA |
| R_TGGAGAATTTCCTGGAGTACA | A_TGGACTGGATGGTTAAGGATGCCCAGAAG | ||
| +49 G/A | rs231775 | F–TGCCTTGGATTTCAGCG | S–CCAGGTCCTGGCAGCC |
| R–TGAAACAAATGAAACCCAGGTAG | A–AGGGATGAAGAGAAGAAAAAACAGGAGAGTGCAG | ||
| +923 C/T | rs231777 | F_TGTGGGTTTCAGATGCAG | S_TGGCTAAGAAACCATGTAGTTTGTATGAAGTAG |
| R_CTTTCCAGTATTGGGAGGG | A_CCATTGAATCTCAACCTTATCTCTCTCTAGACCTT | ||
| +6230 G/A | rs3087243 | F–TTCATTCAGTATCTGGTGGAGTCTC | S–GGGATATAACATGGGTTAACACAG |
| R–AAGGGGAGGTGAAGAACCTG | A–CATAGCAGTCCTTTATAAATCAATTGGCA |
Performed by dynamic allele specific hybridisation with fluorescence resonance energy transfer (FRET) in a LightTyper©.
F, forward primer; R, reverse primer.
S, sensor; A, anch.
Statistical Analysis
The STATA 9 software (Stata Corporation, College Station, TX, USA) was used to test for Hardy Weinberg equilibria (HWE) and odds ratios (OR) of genotype frequencies. Multivariate logistic regression analyses were applied to adjust for age, gender and ethnicities when comparing TB cases with all controls, PPD-negative and PPD-positive controls. Multivariate ordinal logistic regression analyses were performed to determine the influences of genetic variants on the severity scores of radiological signs. Calculations included adjustments for age, gender, ethnicity, recruitment centers and duration of cough, the latter because it was found correlated with the radiographic signs. Haplotype frequencies were estimated and compared using the haplo.score module of the haplo.stats package (http://cran.r-project.org/web/packages/haplo.stats/index.html).
Associations of the distinct disease phenotypes of opacities and cavities with CTLA4 diplotypes comprising the +6230A allele were determined by weighted ordinal logistic regression analyses. Weights in the regression models were the posterior probabilities of each haplotype pair (diplotype) for each individual as estimated by the haplo.score module of the haplo.stats package.
Corrections for multiple testing of single variants were made by multiplication of P values with 7, the number of genetic variants that are not in strong linkage disequilibria with each other, and the two disease phenotypes examined. The final correction factor was, thus, 14 and corrected P values are given as Pcor. To control haplotype analyses for multiple testing, 20,000 permutations were performed for global and haplotype-specific P values. The study sample provided 80% power to identify significant genotypic differences between TB cases and controls at a statistical support level of α = 10−4 with an OR of 1.4 and a minor allele frequency of 0.1 applying a multiplicative model.
Results
The study group consisted of 2010 patients with pulmonary TB and 2346 controls group-matched for age, gender and ethnicity. Controls comprised 2219 purified protein derivative (PPD)-positive and 127 PPD-negative individuals
Genotyping of 8 SNPs located in the CTLA4 gene showed allele distributions in HWE in both the patient and control groups. The minor allele frequencies (MAF) of variants was 15–46%, whereby the +6230 (ct60) A and G alleles were found in 19% and 81%, respectively, of participants. CTLA4 −1722 showed a minor deviation from HWE in the control group but as this occurred in one subgroup only it was considered fortuitous (Table 3).
Table 3. Frequencies and distributions of SNPs studied1.
| Genetic Variant | MAF2 | Cases | Controls |
| P3 | P3 | ||
| CTLA4 −1765 | 0.46 | 0.54 | 0.40 |
| CTLA4 −1722 | 0.15 | 0.81 | 0.04 |
| CTLA4 −1661 | 0.16 | 0.49 | 0.51 |
| CTLA4 −1577 | 0.19 | 0.82 | 0.47 |
| CTLA4 −1147 | 0.16 | 0.82 | 0.57 |
| CTLA4 −443 | 0.17 | 0.73 | 0.53 |
| CTLA4 +49 | 0.38 | 0.62 | 0.65 |
| CTLA4 +923 | 0.25 | 0.73 | 0.09 |
| CTLA4 +6230 | 0.19 | 0.99 | 0.64 |
The χ2 test was applied to test for deviations from Hardy-Weinberg equilibrium.
Minor allele frequency.
P value for a deviation from Hardy-Weinberg equilibrium.
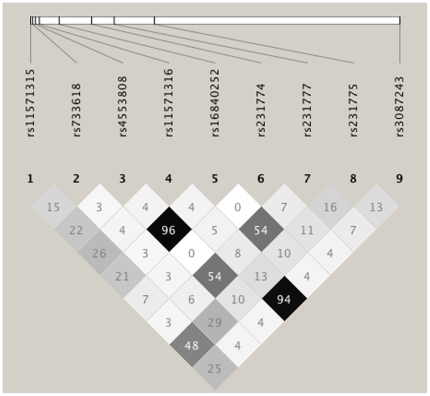
The analysis of pairwise linkage disequilibrium (LD) revealed strong LDs between the variants −1577 with +6230 (R2 = 0.94) and −1147 with −1661 (R2 = 0.96) (Figure 1). Due to these LDs, the SNPs at positions −1577 and −1147 could safely be excluded from further analyses as these variants would not add additional information.
Figure 1. Pairwise linkage disequilibrium (r2) based on 9 CTLA4 SNPs using HaploView 4.0 software.
The figure depicts the strong LD of variants −1577/+6230 and of variants −1147/−1661. The positions relative to the ATG start codon of SNPs are given in Table 2.
When comparing cases with controls, genotype frequencies were similar in all groups (Table 4) including subgroups of PPD-positive and PPD-negative controls (data not shown). Associations with disease severity were assessed by multivariate ordinal logistic regression based on quantitative estimates of radiographic signs of opacities and cavities. Age, gender, ethnicity, duration of illness, mycobacterial species (M. tuberculosis 69.1%, M. africanum 30.3%, M. bovis 0.6%), and recruitment site were included as potentially relevant co-variates. The stratifications showed that the A/A genotype of +6230 showed a significant association with low scores for opacities (Table 5) and a similar trend for cavities (data not shown). Compared to the most frequent +6230 G/G genotype, the A/A and A/G genotypes had odds ratios (ORs) of 0.39 (AA vs GG, confidence interval [CI] 0.23–0.65, Pcor = 0.005) and 1.06 (AG vs GG, CI 0.85–1.33, ns), respectively. As no difference was found between A/G and G/G and both genotypes showed similar ORs compared to A/A (GG vs AA, OR 2.57, CI 1.53–4.32, Pcor = 0.005 and AG vs AA, OR 2.72, CI 1.60–4.66, Pcor = 0.003), the disease-aggravating influence associated with the G allele appears to be dominant and the disease-mitigating effect associated with A recessive.
Table 4. CTLA4 genotype frequencies in TB cases and controls1.
| CTLA4 SNP | Cases | Controls | OR3 | 95%CI4 | p | |
| N | 1990 | 2337 | ||||
| −1765 | AA | 29.1% | 29.1% | 1 | ||
| AG | 49.0% | 50.5% | 0.97 | 0.8–1.1 | 0.623 | |
| GG | 21.8% | 20.4% | 1.04 | 0.9–1.2 | 0.639 | |
| N | 1986 | 2315 | ||||
| −1722 | AA | 72.9% | 70.2% | 1 | ||
| AG | 24.9% | 27.9% | 0.88 | 0.8–1.0 | 0.067 | |
| GG | 2.2% | 1.9% | 1.18 | 0.8–1.8 | 0.435 | |
| N | 1952 | 2269 | ||||
| −1661 | AA | 71.6% | 70.6% | 1 | ||
| AG | 25.8% | 26.6% | 0.94 | 0.8–1.1 | 0.364 | |
| GG | 2.6% | 2.8% | 0.89 | 0.6–1.3 | 0.552 | |
| N | 934 | 1057 | ||||
| −443 | AA | 68.8% | 68.0% | 1 | ||
| AT | 28.1% | 28.6% | 0.98 | 0.8–1.2 | 0.852 | |
| TT | 3.1% | 3.4% | 0.90 | 0.5–1.5 | 0.680 | |
| N | 1912 | 2278 | ||||
| +49 | GG | 40.1% | 38.7% | 1 | ||
| GA | 46.0% | 47.4% | 0.95 | 0.8–1.1 | 0.456 | |
| AA | 13.9% | 13.9% | 0.98 | 0.8–1.2 | 0.826 | |
| N | 903 | 1026 | ||||
| +923 | CC | 54.7% | 57.7% | 1 | ||
| CT | 38.2% | 35.2% | 1.15 | 1.0–1.4 | 0.146 | |
| TT | 7.1% | 7.1% | 1.07 | 0.7–1.5 | 0.724 | |
| N | 1994 | 2328 | ||||
| +6230 | GG | 64.6% | 65.7% | 1 | ||
| GA | 31.5% | 30.5% | 1.05 | 0.9–1.2 | 0.494 | |
| AA | 3.9% | 3.8% | 1.04 | 0.8–1.4 | 0.822 | |
Analysed by logistic regression.
Variations due to assay failures.
Odds ratio.
Confidence interval.
Table 5. CTLA4 genotype frequencies in TB cases stratified for the sizes of opacities in chest radiographs1.
| Opacity Score | ||||||||
| CTLA4 SNP | 0 | 1 | 2 | 3 | OR2 | 95%CI3 | P | Pcor 4 |
| N | 17 | 349 | 1075 | 364 | ||||
| −1765 AA | 0.0% | 30.1% | 29.6% | 26.4% | 1 | |||
| AG | 64.7% | 44.7% | 49.3% | 51.4% | 1.09 | 0.9–1.4 | 0.498 | |
| GG | 35.3% | 25.2% | 21.1% | 22.2% | 0.85 | 0.6–1.1 | 0.268 | |
| N | 17 | 347 | 1074 | 363 | ||||
| −1722 AA | 82.3% | 74.3% | 72.6% | 71.1% | 1 | |||
| AG | 17.6% | 23.1% | 24.9% | 27.3% | 1.14 | 0.9–1.5 | 0.277 | |
| GG | 0.0% | 2.6% | 2.4% | 1.6% | 1.09 | 0.5–2.3 | 0.822 | |
| N | 17 | 342 | 1056 | 357 | ||||
| −1661 AA | 52.9% | 73.1% | 71.9% | 68.9% | 1 | |||
| AG | 47.1% | 24.0% | 25.9% | 27.4% | 1.05 | 0.8–1.3 | 0.692 | |
| GG | 0.0% | 2.9% | 2.3% | 3.6% | 1.25 | 0.6–2.4 | 0.505 | |
| N | 3 | 183 | 565 | 183 | ||||
| −443 AA | 100.0% | 68.3% | 69.2% | 67.8% | 1 | |||
| AT | 0.0% | 29.0% | 27.1% | 30.6% | 1.03 | 0.8–1.4 | 0.838 | |
| TT | 0.0% | 2.7% | 3.7% | 1.6% | 0.83 | 0.4–1.7 | 0.618 | |
| N | 17 | 338 | 1040 | 347 | ||||
| +49 GG | 52.9% | 45.9% | 37.7% | 40.6% | 1 | |||
| GA | 47.1% | 42.6% | 47.9% | 46.1% | 1.17 | 0.9–1.5 | 0.180 | |
| GG | 0.0% | 11.5% | 14.4% | 13.3% | 1.38 | 1.0–1.9 | 0.061 | |
| N | 3 | 180 | 545 | 175 | ||||
| +923 CC | 66.7% | 51.7% | 55.2% | 56.0% | 1 | |||
| CT | 33.3% | 41.1% | 38.6% | 34.3% | 0.84 | 0.7–2.3 | 0.289 | |
| TT | 0.0% | 7.2% | 6.2% | 9.7% | 1.33 | 0.6–1.1 | 0.199 | |
| N | 17 | 350 | 1079 | 364 | ||||
| +6230 GG | 47.1% | 63.1% | 65.6% | 63.2% | 1 | |||
| GA | 47.1% | 29.4% | 30.8% | 35.2% | 1.06 | 0.8–1.3 | 0.602 | |
| AA | 5.9% | 7.4% | 3.6% | 1.6% | 0.39 | 0.2–0.7 | 0.0004 | 0.005 |
Analysed by ordinal logistic regression.
Odds ratio.
Confidence interval.
Bonferroni-corrected by the factor of 14 for testing 7 SNPs not in strong linkage disequilibria and two disease phenotypes.
The haplotype −1765G/−1722A/−1661A/−443A/+49 G/+923C/+6230A was strongly associated with high opacity scores in a recessive model (global Pcor = 0.03; haplotype specific Pcor = 0.0005) and showed the same, albeit weaker, association with cavities (global Pcor = 0.03; haplotype specific Pcor = 0.03) (Table 6). The same applied to the diplotype containing homozygously this haplotype (opacities: haplotype specific P = 0.0005; cavities: haplotype specific P = 0.05; Table 7).
Table 6. CTLA4 haplotype frequencies in TB cases stratified for the sizes of opacities and cavities in chest radiographs1.
| CTLA4 SNP | |||||||||||
| Haplotypes | −1765 | −1722 | −1661 | −443 | +49 | +923 | +6230 | Freq.4 | Score statistic5 | P6 | Pcor 7 |
| Opacities | Global p = 0.022, Global pcor = 0.033 | ||||||||||
| G | A | A | A | G | C | A | 0.19 | −3.70 | 0.00021 | 0.00045 | |
| A | A | A | T | G | C | G | 0.08 | −1.05 | 0.29 | 0.30 | |
| A | G | A | A | A | C | G | 0.15 | −0.60 | 0.55 | 0.55 | |
| A | A | A | T | G | T | G | 0.06 | −0.29 | 0.77 | 0.77 | |
| G | A | G | A | G | T | G | 0.13 | 0.39 | 0.69 | 0.70 | |
| A | A | A | A | A | C | G | 0.21 | 0.66 | 0.51 | 0.52 | |
| G | A | A | A | G | C | G | 0.11 | 0.68 | 0.50 | 0.50 | |
| Cavities | Global p = 0.0052, Global pcor = 0.0083 | ||||||||||
| A | A | A | T | G | C | G | 0.08 | −2.19 | 0.03 | 0.03 | |
| G | A | A | A | G | C | A | 0.19 | −2.11 | 0.04 | 0.03 | |
| G | A | G | A | G | T | G | 0.13 | −1.6 | 0.11 | 0.11 | |
| G | A | A | A | G | C | G | 0.11 | −1.61 | 0.11 | 0.11 | |
| A | G | A | A | A | C | G | 0.15 | −1.05 | 0.29 | 0.29 | |
| A | A | A | T | G | T | G | 0.06 | 0.15 | 0.88 | 0.88 | |
| A | A | A | A | A | C | G | 0.21 | 2.08 | 0.04 | 0.04 | |
Seven tagging SNPs reported to appropriately indicate CTLA4 haplotypes in sub-Saharan Africans (Ref. 13) analysed by the Haplo.score software assuming a recessive mode of inheritance.
P values obtained testing an overall association between haplotypes and the phenotype.
Global P values corrected for multiple testing by 20,000 permutations.
Frequency; only haplotypes with a frequency>0.01 are shown.
P values of haplotype-specific associations.
Simulated haplotype-specific P values corrected for multiple testing by 20,000 permutations.
Score statistic of the haplo.score software.
Table 7. CTLA4 diplotype analysis in TB cases stratified for the sizes of opacities and cavities in chest radiographs.
| Diplotype | Opacity Score | ||||||
| 0 | 1 | 2 | 3 | OR1 | 95% CI2 | P5 | |
| N | 17 | 305 | 961 | 331 | |||
| Other3/other | 47.1% | 60.3% | 62.8% | 61.3% | 1 | ||
| HAPct604/other | 47.0% | 31.8% | 33.2% | 36.9% | 1.03 | 0.8–1.3 | 0.826 |
| HAPct60/HAPct60 | 5.9% | 7.9% | 4.0% | 1.8% | 0.39 | 0.2–0.7 | 0.0005 |
| Cavity Score | |||||||
| N | 17 | 305 | 961 | 331 | |||
| Other/other | 64.0% | 60.9% | 63.4% | 60.4% | 1 | ||
| HAPct60/other | 31.0% | 33.7% | 33.2% | 37.2% | 1.03 | 0.8–1.3 | 0.77 |
| HAPct60/HAPct60 | 5.0% | 5.4% | 3.4% | 2.4% | 0.59 | 0.3–1.0 | 0.05 |
Odds ratios (OR).
95% confidence intervals (CI) for the weighted ordinal logistic regression model with weights being the posterior probabilities for each diplotype for each individual.
Other, haplotype not comprising the +6230A allele.
HAPct60, haplotype comprising the +6230A allele (−1765 G/−1722 A/−1661 A/−443 A/+49 G/+923 C/+6230 A).
P values given for haplotype-specific associations.
Discussion
Our findings do not provide evidence for a role of the +6230 polymorphism of human CTLA4 in protection against pulmonary TB. On the other hand, our study design was sufficiently sensitive to detect a significant influence of the +6230 variant on the more detailed phenotypes of disease severity as radiograpically assessed by the extent of opacities and size and numbers of pulmonary cavities. The +6230 A/A genotype, which had been found negatively associated with autoimmune disease, was found to also be negatively associated with severe pathology in pulmonary TB. This applied to the major forms of pathology as identified by chest X ray. Thus, the CTLA4 +6230 polymorphism was found to influence pulmonary TB only after Mycobacterium tuberculosis had overcome innate and adaptive host defence mechanisms.
Our data are partly supported by earlier studies in a murine model of TB. Using experimental aerosol infection of mice with Mycobacterium bovis bacillus Calmette-Guérin (BCG), it was shown that inhibition of CTLA-4 by antibody did not enhance protection [15]. Furthermore, in the same model, depletion of regulatory T cells, which express CTLA-4 constitutively, did not affect protection either [16]. However, the mouse model did not support a role of CTLA-4 in enhancing pathology. Although antibody mediated inhibition of CTLA-4 caused a rise in the number of T cells in lung-draining lymph nodes and an increase in the T-cell proliferative response to mycobacterial antigen, it did not result in increased lymphoid infiltration or granuloma formation in the lung assessed by careful histological examination [15]. As the radiological findings of opacities and cavities in human TB are considered to result from cellular infiltration and necrosis, respectively [17], our observations suggest that, in humans, a reduced activity of CTLA-4 causes an increased cellular infiltration of the lung, which may result from in-situ T-cell proliferation and recruitment of other cell types through cytokines. This is in agreement with the concept that pathology of TB to some extent is immune mediated, which has recently been confirmed by finding a clinical aggravation of pre-existing TB in the course of the immune reconstitution inflammatory syndrome (IRIS) seen among AIDS patients undergoing anti-retroviral therapy [18].
Collectively, the observation of the role of the CTLA4 −6230 polymorphism and the occurrence of a haplotype/diplotype bearing the CTLA4A variant in pulmonary TB indicates that a reduced CTLA-4 activity in humans does not enhance protection against pulmonary TB but enhances pathology if containment of the infection fails. Accordingly, pulmonary TB appears not to belong to those infections which may contribute to explain the high prevalence of CTLA4 +6230G in human populations. This evolutionary role may rather be with viral infections including hepatitis B, which was found to be controlled more effectively by carriers of the CTLA4 +6230G allele than by those carrying the +6230A/A genotype [19]. In that study, the +6230G allele appeared to act in a dominant fashion, which is in agreement with the results obtained in autoimmunity [20] but at variance with our data on the severity of TB. An explanation has to await further functional studies.
The higher frequency of +6230G allele in the Ghanaian study group (>80%) compared to that found in Europeans (<60%) may suggest an evolutionary influence exerted by diseases that occur at higher prevalences in Africa than in Europe.
Our finding of strong LD in a study population from West Africa between variants at position +6230 and the SNP located in the promoter region of CTLA4 (−1577) may open new avenues in the search for mechanisms which mediate the immunomodulatory effect of the +6230 variant.
Acknowledgments
The authors are grateful to the study participants. They thank hospital staff and recruitment teams as well as field workers and nurses and, furthermore, Lincoln Gankpala, Emmanuel Abbeyquaye and Gerd Ruge for technical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The study was funded by the German Ministry for Education and Research through the National Genome Research Network. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Teft WA, Kirchhof MG, Madrenas J. A molecular perspective of CTLA-4 function. Annu Rev Immunol. 2006;24:65–97. doi: 10.1146/annurev.immunol.24.021605.090535. [DOI] [PubMed] [Google Scholar]
- 2.Lee KM, Chuang E, Griffin M, Khattri R, Hong DK, et al. Molecular basis of T cell inactivation by CTLA-4. Science. 1998;282:2263–2266. doi: 10.1126/science.282.5397.2263. [DOI] [PubMed] [Google Scholar]
- 3.Sansom DM, Walker LS. The role of CD28 and cytotoxic T-lymphocyte antigen-4 (CTLA-4) in regulatory T-cell biology. Immunol Rev. 2006;212:131–148. doi: 10.1111/j.0105-2896.2006.00419.x. [DOI] [PubMed] [Google Scholar]
- 4.Darlington PJ, Baroja ML, Chau TA, Siu E, Ling V, et al. Surface cytotoxic T lymphocyte-associated antigen 4 partitions within lipid rafts and relocates to the immunological synapse under conditions of inhibition of T cell activation. J Exp Med. 2002;195:1337–1347. doi: 10.1084/jem.20011868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneider H, Valk E, da Rocha Dias S, Wei B, Rudd CE. CTLA-4 up-regulation of lymphocyte function-associated antigen 1 adhesion and clustering as an alternate basis for coreceptor function. Proc Natl Acad Sci U S A. 2005;102:12861–12866. doi: 10.1073/pnas.0505802102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneider H, Downey J, Smith A, Zinselmeyer BH, Rush C, et al. Reversal of the TCR stop signal by CTLA-4. Science. 2006;313:1972–1975. doi: 10.1126/science.1131078. [DOI] [PubMed] [Google Scholar]
- 7.Grohmann U, Orabona C, Fallarino F, Vacca C, Calcinaro F, et al. CTLA-4-Ig regulates tryptophan catabolism in vivo. Nat Immunol. 2002;3:1097–1101. doi: 10.1038/ni846. [DOI] [PubMed] [Google Scholar]
- 8.Bour-Jordan H, Grogan JL, Tang Q, Auger JA, Locksley RM, et al. CTLA-4 regulates the requirement for cytokine-induced signals in T(H)2 lineage commitment. Nat Immunol. 2003;4:182–188. doi: 10.1038/ni884. [DOI] [PubMed] [Google Scholar]
- 9.Nasta F, Ubaldi V, Pace L, Doria G, Pioli C. Cytotoxic T-lymphocyte antigen-4 inhibits GATA-3 but not T-bet mRNA expression during T helper cell differentiation. Immunology. 2006;117:358–367. doi: 10.1111/j.1365-2567.2005.02309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ueda H, Howson JM, Esposito L, Heward J, Snook H, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 11.Maier LM, Anderson DE, De Jager PL, Wicker LS, Hafler DA. Allelic variant in CTLA4 alters T cell phosphorylation patterns. Proc Natl Acad Sci U S A. 2007;104:18607–18612. doi: 10.1073/pnas.0706409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 13.Ramírez-Soriano A, Lao O, Soldevila M, Calafell F, Bertranpetit J. Haplotype tagging efficiency in worldwide populations in CTLA4 gene. Genes Immun. 2005;6:646–657. doi: 10.1038/sj.gene.6364251. [DOI] [PubMed] [Google Scholar]
- 14.Thye T, Browne EN, Chinbuah MA, Gyapong J, Osei I, et al. No associations of human pulmonary tuberculosis with Sp110 variants. J Med Genet. 2006;43:e32. doi: 10.1136/jmg.2005.037960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirman J, McCoy K, Hook S, Prout M, Delahunt B, et al. CTLA-4 blockade enhances the immune response induced by mycobacterial infection but does not lead to increased protection. Infect Immun. 1999;67:3786–3792. doi: 10.1128/iai.67.8.3786-3792.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quinn KM, McHugh RS, Rich FJ, Goldsack LM, de Lisle GW, et al. Inactivation of CD4+ CD25+ regulatory T cells during early mycobacterial infection increases cytokine production but does not affect pathogen load. Immunol Cell Biol. 2006;84:467–474. doi: 10.1111/j.1440-1711.2006.01460.x. [DOI] [PubMed] [Google Scholar]
- 17.Lee JY, Lee KS, Jung KJ, Han J, Kwon OJ, et al. Pulmonary tuberculosis: CT and pathologic correlation. J Comput Assist Tomogr. 2000;24:691–698. doi: 10.1097/00004728-200009000-00005. [DOI] [PubMed] [Google Scholar]
- 18.Lawn SD, Bekker LG, Miller RF. Immune reconstitution disease associated with mycobacterial infections in HIV-infected individuals receiving antiretrovirals. Lancet Infect Dis. 2005;5:361–373. doi: 10.1016/S1473-3099(05)70140-7. [DOI] [PubMed] [Google Scholar]
- 19.Thio CL, Mosbruger TL, Kaslow RA, Karp CL, Strathdee SA, et al. Cytotoxic T-lymphocyte antigen 4 gene and recovery from hepatitis B virus infection. J Virol. 2004;78:11258–11262. doi: 10.1128/JVI.78.20.11258-11262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kavvoura FK, Akamizu T, Awata T, Ban Y, Chistiakov DA, et al. Cytotoxic T-lymphocyte associated antigen 4 gene polymorphisms and autoimmune thyroid disease: a meta-analysis. J Clin Endocrinol Metab. 2007;92:3162–3170. doi: 10.1210/jc.2007-0147. [DOI] [PubMed] [Google Scholar]