Abstract
Previous reports have suggested that specific isoforms of the potential stem cell marker p63 may regulate corneal epithelial homeostatic renewal through control of cell proliferation. In this study, we characterized the presence of ΔNp63 isoforms in telomerase-immortalized human corneal epithelial cells (hTCEpi) in comparison to normal human corneal epithelium to validate the hTCEpi cell line as a viable model for the study of p63 isoforms. We further examined roles for ΔNp63 in proliferation and differentiation. For in vitro studies, hTCEpi cells were cultured in serum-free culture media and grown under 0.15 mM calcium or sequential 1.15 mM calcium/air-lifted culture. Fresh donor human corneal tissue was used to assess expression and localization in situ. mRNA and protein levels were assessed by real-time PCR, Immunofluorescence (IF) and Western blotting (WB). ΔNp63 expression levels throughout the cell cycle were assessed by double-labeling with ΔNp63 and Ki-67. In situ, ΔNp63 localized to nuclei throughout the human corneal epithelium and was lost only in superficial cells. WB confirmed the presence of all three ΔNp63 isoforms in the central corneal epithelium and in hTCEpi cells. ΔNp63 mRNA levels decreased when grown on collagen substrate and under increased calcium/air-lifted culture. mRNA and protein levels increased as cells approached confluence, with a significant decrease in post-confluent culture. ΔNp63 expression levels did not vary with the cell cycle, as assessed by Ki-67 labeling. Collectively, the presence of all three ΔNp63 isoforms in hTCEpi cells and in intact cornea validates the use of this cell line for the study of individual isoforms in the corneal epithelium; and these data suggest that expression of ΔNp63 isoforms are not altered as a function of the cell cycle or cell division in subcon-fluent hTCEpi cells cultured in serum-free media, but demonstrate reduced expression upon contact-inhibited growth down-regulation and differentiation. Significantly, the localization of ΔNp63 in central corneal epithelial cells with a loss of expression in superficial cells suggests that ΔNp63 may play a role in mediating desquamative events at the ocular surface.
Keywords: cornea, epithelium, ΔNp63, differentiation, desquamation
1. Introduction
The nuclear transcription factor p63 has been frequently suggested in recent years as a potential marker to identify limbal stem cells in the human cornea (Pellegrini et al., 2001). p63, a homolog of p53 and p73, contains multiple functional domains, which are isotype specific, allowing for both p53 transactivating and dominant negative signaling events (Yang et al., 1998). Separate promoters at the N terminus result in the production of two alternative transcripts; the first promoter which lies upstream of exon 1 and codes for the TA or transactivation domain, and a second cryptic promoter which codes for an alternative transcript lacking the acidic NH2 terminus, ΔN. Both isoforms TA and ΔN are subject to alternative splicing at the COOH terminus, producing α, β, and γ variants. Structurally, these proteins are composed of a proline-rich domain, a DNA binding domain, and an oligomerization domain, which all play critical roles in modulating p63 gene function (Helton et al., 2006).
ΔNp63α has been identified as the most predominant isoform in proliferating cells of stratified epithelial tissue and is thought to play critical roles in ectodermal development and tissue homeostasis (Koster et al., 2004; Parsa et al., 1999). Studies supporting a role for p63 in modulating epithelial development derive from investigations in mice lacking all p63 isoforms. Notably, mice deficient in p63 fail to form stratified epithelia, possess truncated limbs, presumably to an undeveloped apical ectodermal ridge, and die at birth (Mills et al., 1999; Yang et al., 1999). Subsequent studies have proposed roles for TA and ΔN isoforms in embryonic development and adult tissue maintenance (Koster et al., 2004). Functional analysis of conserved genomic sequences upstream of the p63 promoter have identified a long-range regulatory element which functions to modulate keratinocyte-specific expression of ΔNp63 isoforms (Antonini et al., 2006). This long-range enhancer, which contains a p53 binding element, preferentially binds p63 and may function in an auto-regulatory fashion, modulating p63 expression.
In the human cornea, ΔNp63 was first localized in the epithelium in a subpopulation of basal cells in the limbus (Pellegrini et al., 2001). Studies classifying the molecular phenotype of these cells have all cited the presence of high levels of ΔNp63 (Chen et al., 2004). Further work on ΔNp63 using primary cultured limbal epithelial cells grown on 3T3 feeder layers in the presence of serum, have attempted to quantify expression levels of p63 as a function of cell size, nuclear to cytoplasmic ratio, side population and low side scatter cells, proliferative capacity, and attachment substrate (Arpitha et al., 2005; Epstein et al., 2005; Salehi-Had et al., 2005; Wang et al., 2003, 2005). A recent study of the pattern of ΔNp63 mRNA in the human ocular surface using laser micro-dissection techniques suggests that the localization of ΔNp63 isoforms may extend beyond the previously established limbal compartment (Kawasaki et al., 2006).
Despite the growing evidence that ΔNp63 isoforms are present in both the limbal and corneal epithelium, the role of these proteins in cell cycle regulation, differentiation, and survival is still largely unknown. Further, the level of expression of these isoforms in specific cell types may modulate signaling pathways important in effecting these processes. In the present study, we characterized the expression of ΔNp63 isoforms in normal human central cornea and in telomerase-immortalized human corneal epithelial (hTCEpi) cells in serum-free culture to validate this cell line as a viable model for the study of p63 and further evaluated the role of ΔNp63 during proliferation and calcium-induced differentiation.
2. Materials and methods
2.1. Cell line and human corneal tissue
Human telomerase-immortalized corneal epithelial cells (hTCEpi) were routinely maintained in KGM-2 serum-free culture media (Cloneticse–BioWhittaker) containing 0.15 mM calcium (Robertson et al., 2005). Cells were subcultured on T75 tissue culture flasks (Falcon Labware; BD Biosciences, Bedford, MA), incubated at 37 °C in 5% CO2. and passaged every 7 – 10 days. Human corneal tissue was obtained from Tissue Transplant Services Lions’ Eye Bank (Dallas, TX). All tissue was obtained and processed fresh within 6 h of death without storage.
2.2. Immunohistochemistry
For immunofluorescence studies, hTCEpi cells were suspended in 0.15 mM calcium KGM-2 medium and plated at a density of 2 × 103 cells/cm2 on collagen-coated glass coverslips (Vitrogen, Cohesion Technologies) in a 24-well culture plate (Corning Costar, Corning, NY). For organotypic constructs, cells were plated and grown as previously described (Robertson et al., 2005). Antibody concentrations and incubation times varied for whole mount human tissue (WMH), cryostat sectioned human tissue (CSH) and cells. Tissues/ cells were fixed with RNase free 4% paraformaldehyde (Electron Microscopy Sciences, FortWashington, PA) in phosphate- buffered saline (PBS), permeabilized with cold acetone (−20 °C) for 3 min (CSH, cells) or 10 min (WMH), and washed with PBS. For WMH tissue, TD buffer (1% DMSO and 1% Triton X in PBS) was used in place of PBS. Tissue/cells were blocked with 10% donkey serum at 37 °C for 15 min for cells and 30 min for all tissues and incubated in anti-p63 rabbit and goat polyclonal antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4 °C at a concentration of 2 µg/ml for cells and 10 µg/ml for tissues. Tissues/cells were washed with PBS and stained with 30 µg/ml FITC-conjugated secondary donkey anti-mouse IgG at 37 °C for 2 h (cells, CSH) and overnight (WMH) and counterstained with a 1:50 dilution of rhodamine-conjugated Phalloidin (Invitrogen, Carlsbad, CA) during the secondary antibody incubation. A commercially available blocking peptide mapping near the N-terminus of ΔNp63α (sc-25039P, Santa Cruz Biotechnology) was used as a control. For blocking peptide experiments, the antibody was incubated in a five-fold excess of blocking peptide in 500 µl of PBS for 2 h at 4 °C with agitation. All samples were mounted on slides using a 50% v/v solution of glycerole–PBS and imaged on a Leica SP2 laser scanning confocal microscope (Leica Microsystems, Heidelberg, Germany) using a 63× water objective. A 488 nm Ar/ArKr laser was used for excitation of the FITC fluorophore and a 535 nm GreNe laser was used to excite rhodamine and PI. 512 × 512 images were collected at 400 Hz with a line average of 4. Emission collection wavelengths, gain and offset were optimized for all staining experiments. Images were processed using MetaMorph Software (Molecular Devices Corp., Downington, PA) and Photoshop (Adobe, San Jose, CA).
2.3. Protein extraction and Western blotting
Total protein was collected by lysing cells in radioimmuno-precipitation buffer (RIPA) buffer containing a protease inhibitor cocktail tablet (Complete-Mini, Roche Diagnostics, Indianapolis, IN) on ice for 10 min. Lysates were snap frozen in liquid nitrogen and vortexed. The amount of protein was determined using a DC™ Protein Assay Reagents Package (Bio-Rad Laboratories, Hercules, CA). All lysates were boiled for 5 min in 4× sample buffer, pH 6.8, containing 0.25 M Tris, 8% lauryl sulfate, 40% glycerol, 20% mercaptoethanol and 0.04% bromophenol blue, resolved on a 12% SDS–epolyacrylamide gel and subsequently transferred to a nitrocellulose membrane. Membranes were blocked in 5% non-fat milk for 1 h at room temperature and blotted using a mouse monoclonal (4A4) antibody directed against pan-ΔNp63 isoforms (1 µg/ml, Santa Cruz Biotechnology) overnight at 4 °C. Following a 1 h incubation with an anti-mouse secondary antibody (1:5000 dilution, Amersham Biosciences, Piscataway, NJ), protein was visualized using ECL Plus Detection Reagents (Amersham Biosciences) and imaged on a Typhoon Variable Mode Imager. For protein analysis, volume quantitation measuring pixel intensity was performed using Image Quant Software (Amersham Biosciences). Background correction was applied using the local average function. Independent experiments were repeated in full two or more times, as specified in the corresponding figure legends.
2.4. Real-time PCR
Real-time PCR was used to quantitate basal levels of ΔNp63 and p63α, β, and γ isoforms. hTCEpi cells were seeded onto plastic 6-well tissue culture dishes at a density of 2.2 × 105 cells/well or onto collagen-coated coverslips. To assess changes in mRNA levels during differentiation, hTCEpi cells were grown in sequential submersed/air-lifted culture, as previously described (Robertson et al., 2005). mRNA was extracted from hTCEpi cells by homogenization using the standard RNA STAT 60 protocol (TEL-TEST Inc., Friendswood, TX). Cells were lysed directly in the well using 1 ml of RNA STAT 60. Lysates were placed in sterile 1.5 ml Eppendorf tubes and incubated for 5 min on ice. 0.2 ml of chloroform was added and the sample was vortexed for 15 s and allowed to sit on ice for 3 min. The sample was then centrifuged at 12,000 × g for 15 min at 4 °C. The top colorless aqueous phase was collected and placed in a fresh Eppendorf tube. Isopropanol (0.5 ml) was added and incubated on ice for 10 min, followed by centrifugation at 12,000 × g for 10 min at 4 °C. The supernatant was removed and the remaining pellet was washed using 75% ethanol and centrifugation at 7500 × g for 5 min at 4 °C. The pellet was air-dried and resuspended in DEPC-treated water (Fisher Scientific, Burr Ridge, IL). Real-time PCR with First-Strand Synthesis using Random Primers (Invitrogen) was used to generate cDNA according to the manufacturer’s instructions. A 10 µl RNA/primer reaction mixture was made containing 2 µg RNA, 125 ng random hexamer, 1 mM dNTPs and DEPC-treated water. The reaction mixture was incubated at 65 °C for 5 min in the thermocycler, followed by incubation on ice for 1 min. A master mix was made containing 2 µl 10× RT buffer, 4 µl 24 mM MgCl2, 2 µl 0.1 M DTT and 1 µl RNase OUT Recombinant Ribonuclease Inhibitor and added to the cooled RNA/primer reaction. The total mixture was incubated for 2 min at 25 °C. One microliter of Superscript II Reverse Transcriptase was added and allowed to incubate at 25 °C for 10 min, 42 °C for 50 min, and 70 °C for 15 min (in thermocycler). The reaction was cooled to 4 °C. One microliter of RNase H was added and incubated at 37 °C for 20 min. Real-time PCR was performed with a 20-µl reaction using 10 µl of 2× Taqman Universal Master Mix (Applied Biosystems, Foster City, CA), 900 nM forward/reverse primer, 200 nM probe, and 4 µl cDNA. The reaction conditions were an initial 50 °C for 2 min and 95 °C for 10 min, followed by 95 °C for 15 s and 60 °C for 1 min for 40 cycles. Gene-specific primers were used to amplify specific p63 isoforms (outlined in Table 1). The hybridization probes were designed using Primer Express software (Table 1, Applied Biosystems). For quantitation of mRNA, a standard curve was prepared using serial dilutions of a known concentration of hTCEpi RNA. RNA concentration was determined by measuring the absorbance at 260 nm on a Beckman DU 530 spectrophotometer and converted to µg/µl. A pre-designed assay reagent kit for β-actin was used as an endogenous control (Applied Biosystems), along with a no template control. For quantitative analysis of individual isoform expression, intraprobe comparisons of different treatment groups were made using individual standard curves.
Table 1.
Primer and hybridization probe sequences for real-time PCR
| Isotype | Primers | Probe |
|---|---|---|
| ΔNp63 | Forward: 5′-GAA AAC AAT GCC CAG ACT CAA TTT-3′ | MGB-5′-TGAGCCACAGTACACG-3′ |
| Reverse: 5′-TCT GCG CGT GGT CTG TGT TAT-3′ | ||
| p63α | Forward: 5′-ATG TCG AAA TTG CTC AGG GAT TTT CAG A-3′ | MGB-5′-TGGAATGATCTGGCAAGTC-3′ |
| Reverse: 5′-TGA CCA CCA TCT ATC AGA TTG AGC ATT ACT-3′ | ||
| p63β | Forward: 5′-AGA CTT GCC AGA TCC TGA CAA TGC-3′ | MGB-5′-TCT GTG GGA TAC GGA-3′ |
| Reverse: 5′-ATC CAC CTC CCA CTG CAC A-3′ | ||
| p63γ | Forward: 5′-AGC AGC ACC AGC ACT TAC TTC A-3′ | MGB-5′-AAA CAT CTC CTT TCA GCC TG-3′ |
| Reverse: 5′-GGC TCC ACA AGC TCA TTC CTG AA-3′ |
2.5. Statistics
Statistical analysis was performed using SigmaStat 3.1 (Systat Software Inc., San Jose, CA). All data are expressed in mean ± standard deviation. Normality and equal variance assumption testing were performed using the Kolmogorov–Smirnov test and the Levene median test. For comparisons between two groups, a Mann–Whitney Rank Sum test was used to determine if a significant difference existed. A one- or two-way analysis of variance was used to determine if a significant difference existed between multiple groups. When comparing changes over time, a repeated measures analysis of variance was performed. The Student–Newman–Keuls post hoc multiple comparison test was performed to determine which groups were significantly different. All multiple comparison test results are shown on respective graphs. Statistical significance was set at p < 0.05.
3. Results
3.1. p63 expression and localization
ΔNp63 isoforms were assessed in the human corneal epithelium using whole-mount and cryostat-sectioned donor tissue and in hTCEpi cells in vitro using a rabbit polyclonal antibody directed against the N terminal region of p63 (Fig. 1). In the central human corneal epithelium, p63 localized to nuclei of all cell layers and was absent only in the most superficial layer. This pattern of expression was further confirmed using a goat polyclonal antibody recognizing amino acids 15–151 of ΔNp63 (Fig. 2). To confirm specificity of the antibody, staining was repeated following incubation of the primary antibody with a commercially available blocking peptide. Treatment with the blocking peptide abrogated all ΔNp63 staining (Fig. 2C). No staining was seen in the negative control. ΔNp63 expression was also evaluated in proliferating, subconfluent, non-polarized hTCEpi cells. All hTCEpi cells demonstrated positive staining to ΔNp63 (Fig. 3).
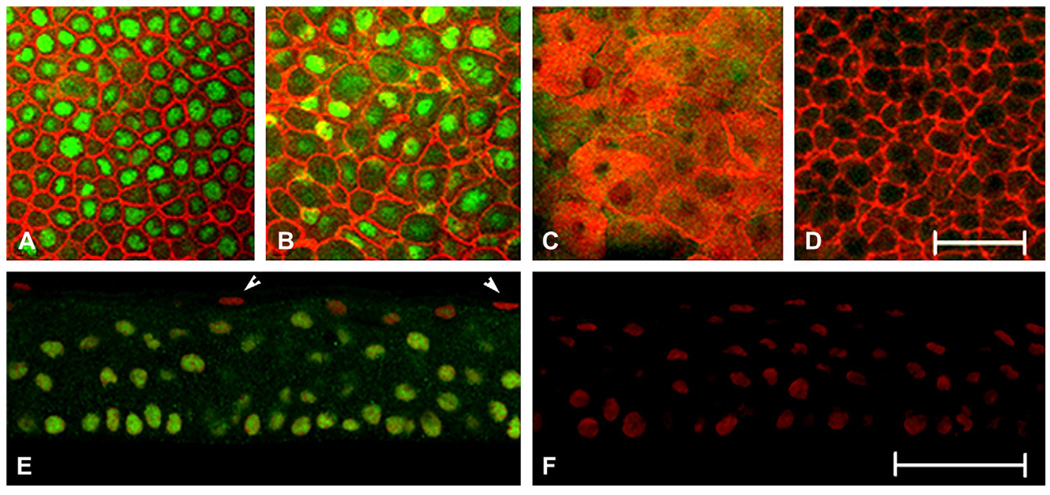
Fig. 1.
Immunostaining for ΔNp63 in normal human tissue. (A–D) Representative XY slices of whole mount human corneal epithelium double-labeled with a rabbit polyclonal antibody directed against the N terminus of ΔNp63 (green) and phalloidin (red). (A) Basal cell layer; (B) wing cell layer; (C) superficial cell layer; (D) negative control, primary antibody omitted. (E, F) 10 µm cryosections double-labeled with anti-rabbit ΔNp63 (green) and PI (red). (E) Human central corneal epithelium. Arrows indicate DNp63-negative, PI-positive cells; (F) negative control, primary antibody omitted. Whole mount experiments were repeated three independent times; cryostat sectioned experiments were repeated three independent times. Scale: 50 µm.
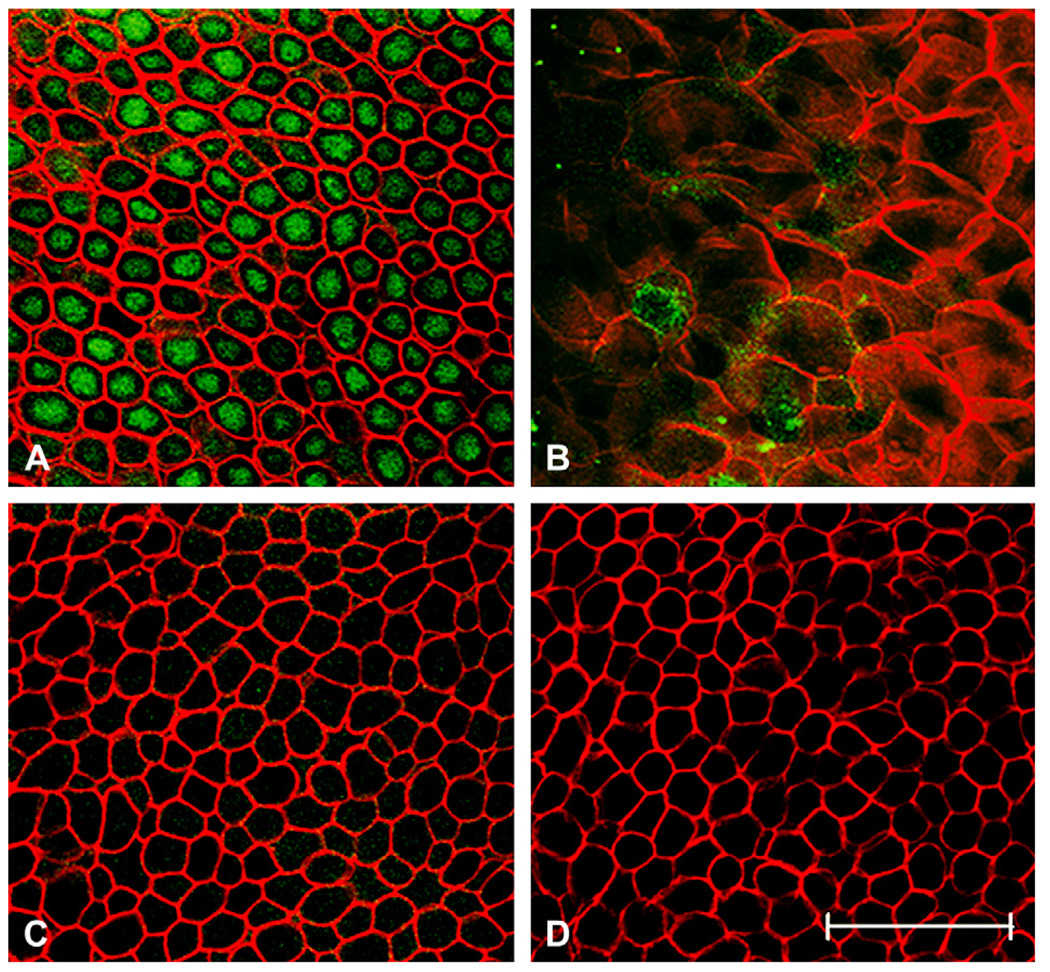
Fig. 2.
Immunostaining for ΔNp63 in normal human cornea. (A–D) Representative XY slices of whole mount human central cornea. (A) Basal cell layer doublelabeled with a goat polyclonal anti-ΔNp63 (green) and phalloidin (red); (B) superficial cells from the same tissue stack shown in (A), demonstrating loss of ΔNp63 staining in superficial cells; (C) basal cell layer following incubation of primary antibody with blocking peptide; (D) basal layer in negative control, primary antibody omitted. Experiment was repeated two independent times. Scale: 50 µm
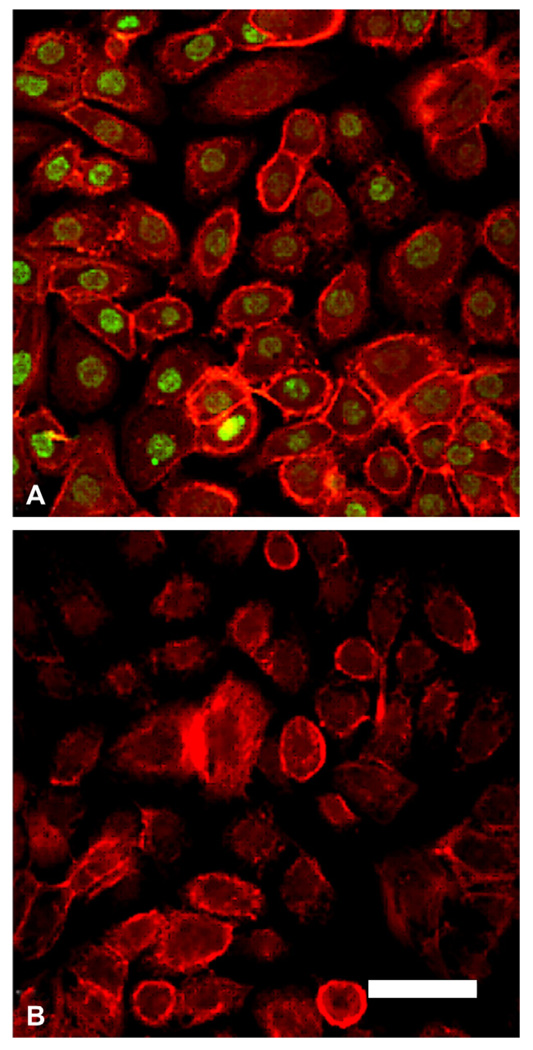
Fig. 3.
Immunostaining for ΔNp63 in hTCEpi cells. (A) hTCEpi cells grown on collagen-coated glass coverslips double-labeled with ΔNp63 (green) and rhodamine-conjugated phalloidin (red). All cells were positive for nuclear p63. (B) Negative control, primary antibody omitted, counterstained with phalloidin. Scale: 45 µm.
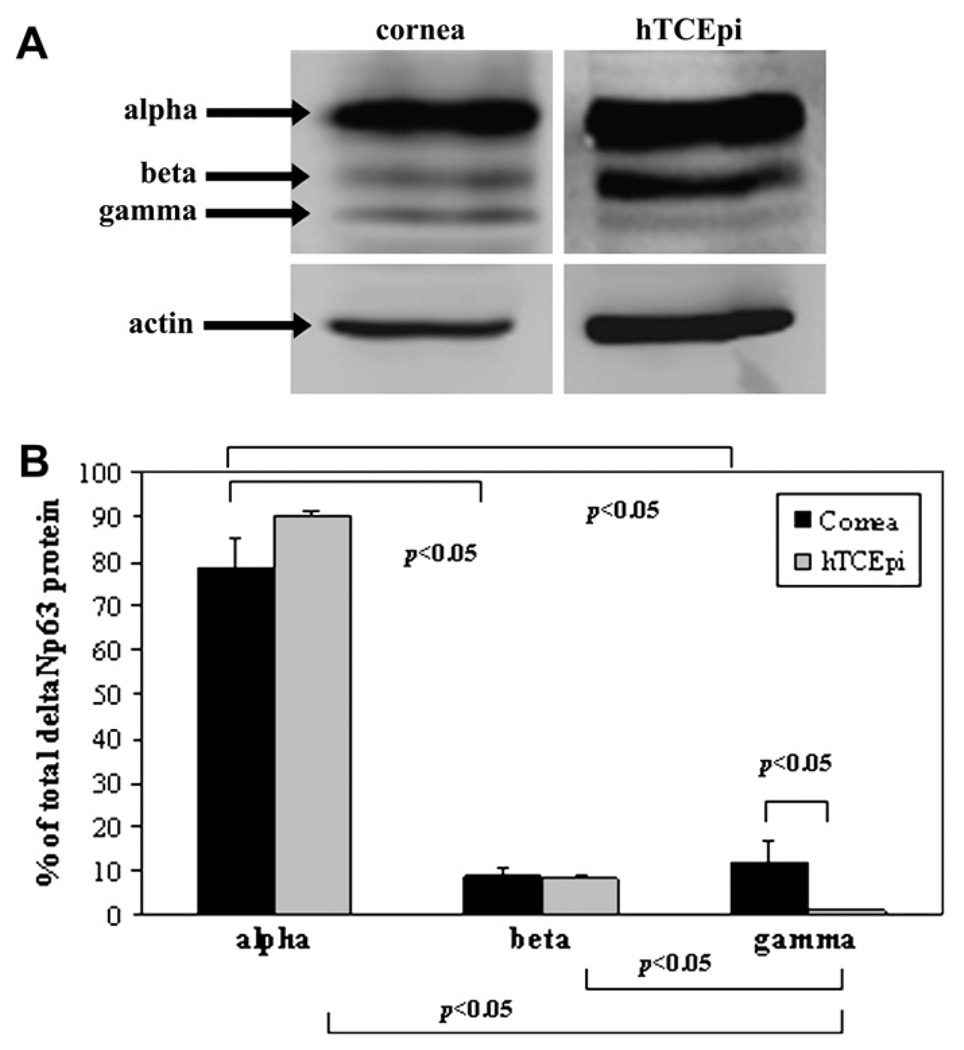
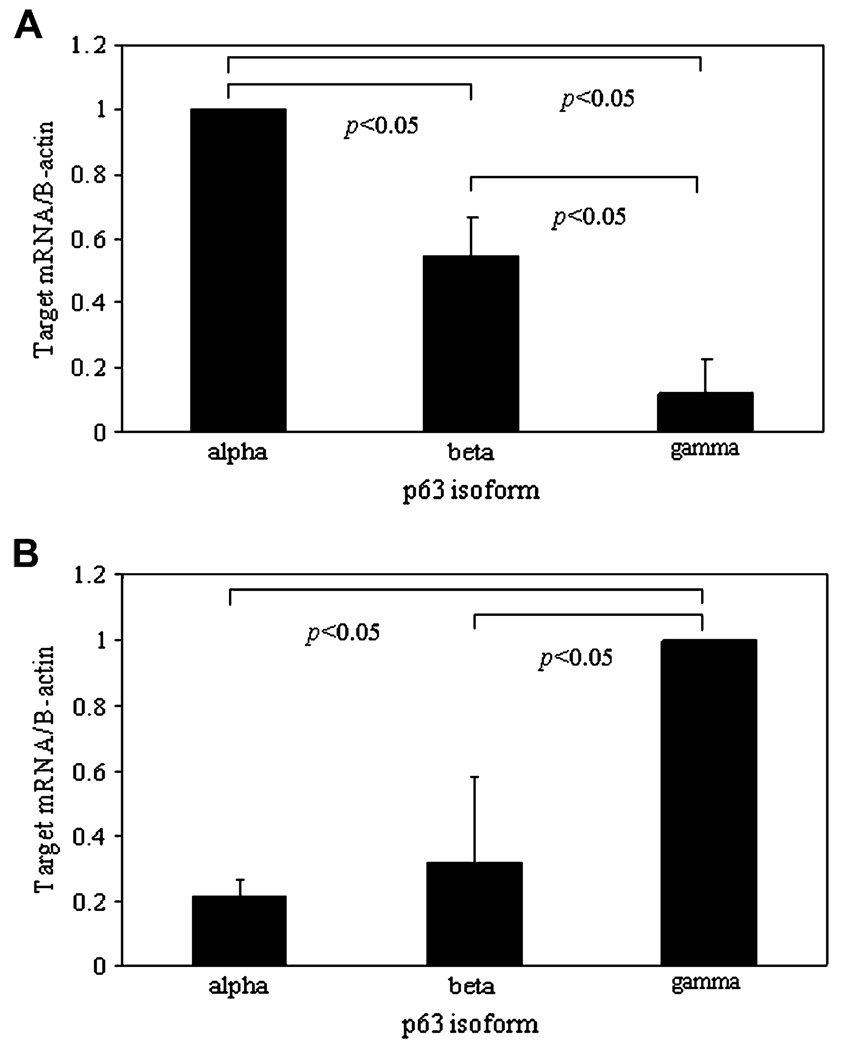
To further establish and characterize the expression of specific isoforms of ΔNp63, Western blotting was performed on lysates obtained from an epithelial scrape of human corneal epithelium and semi-confluent hTCEpi cells using a mouse monoclonal antibody clone 4A4 specific to the ΔN terminus isoforms (Fig. 4). Of all three isoforms, ΔNp63α was the most abundantly expressed isoform in human corneal epithelium and in hTCEpi cells (p < 0.05, two-way ANOVA). There was a significantly higher percentage of ΔNp63γ in human corneal epithelium compared to hTCEpi cells in vitro (p < 0.05, two-way ANOVA). In repeated experiments, levels of γ were consistently low and often difficult to detect. The distribution of p63 isoforms was further investigated by real-time PCR. Using real-time probes generated against the C-terminus of α, β, and γ, the relative concentration of mRNA for each transcript was determined. As expected from our proteomic studies, transcripts for all three isoforms were present, with a significantly higher amount of ΔNp63α compared to β or γ (Fig. 5A, p < 0.001, one-way ANOVA). Interestingly, in occasional confluent cells, expression of p63γ was significantly higher than for α or β (Fig. 5B, p = 0.015, one-way ANOVA). It is important to note that in these experiments the real-time probe was unable to differentiate between TA and ΔN isoforms.
Fig. 4.
Western blot for ΔNp63. (A) Western blotting of ΔNp63 in human corneal epithelium and hTCEpi cells demonstrated the presence of ΔNp63α, β, and γ isoforms. β-Actin was used as a loading control. (B) Quantitative analysis confirmed that α was the dominant isoform present in both cornea and cells (data represented as mean ± standard deviation of two independent experiments, p < 0.05, Two-way ANOVA) and that relative gamma levels were significantly higher in cornea compared to cells.
Fig. 5.
Real-time PCR measuring mRNA levels of p63α, β, and γs. (A) In subconfluent hTCEpi cells, all three transcripts were detected (data represented as mean ± standard deviation of four independent experiments performed in duplicate, p < 0.001, one-way ANOVA). Data were normalized to p63a. (B) In confluent samples, p63γ mRNA was significantly higher than α or.β ( p = 0.015, one-way ANOVA). Data normalized to p63γ. It is important to note that real-time probes do not differentiate between TA and ΔN isoforms.
3.2. p63, substrate effects and differentiation
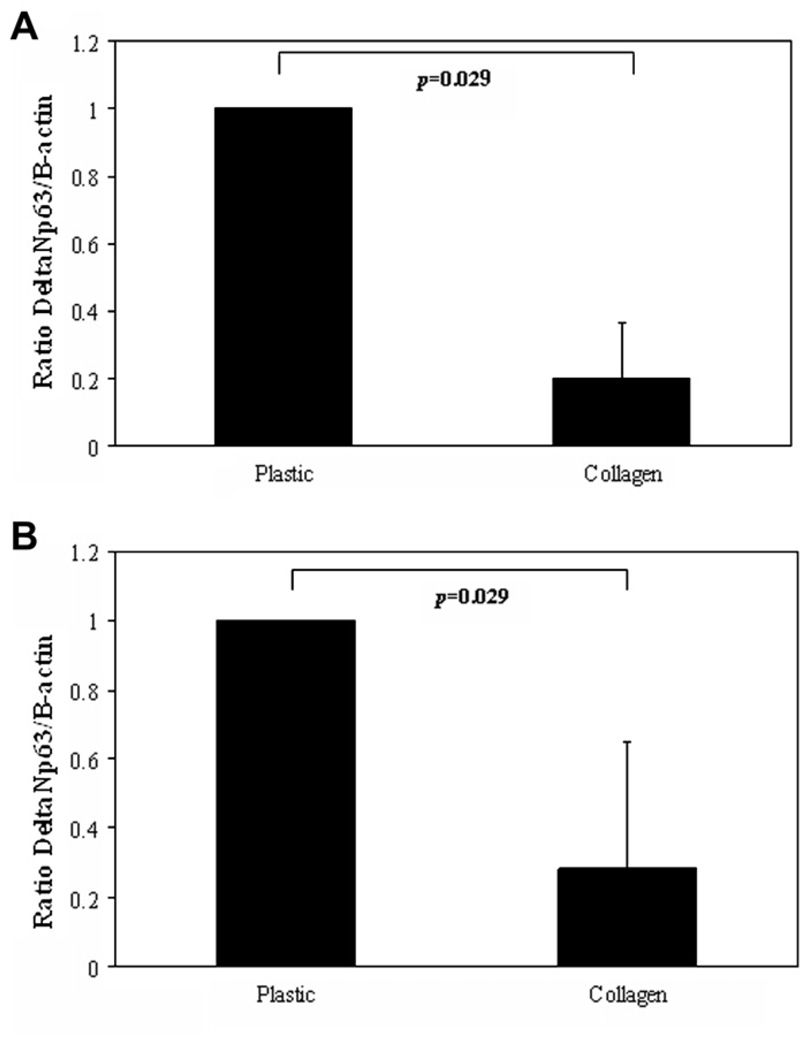
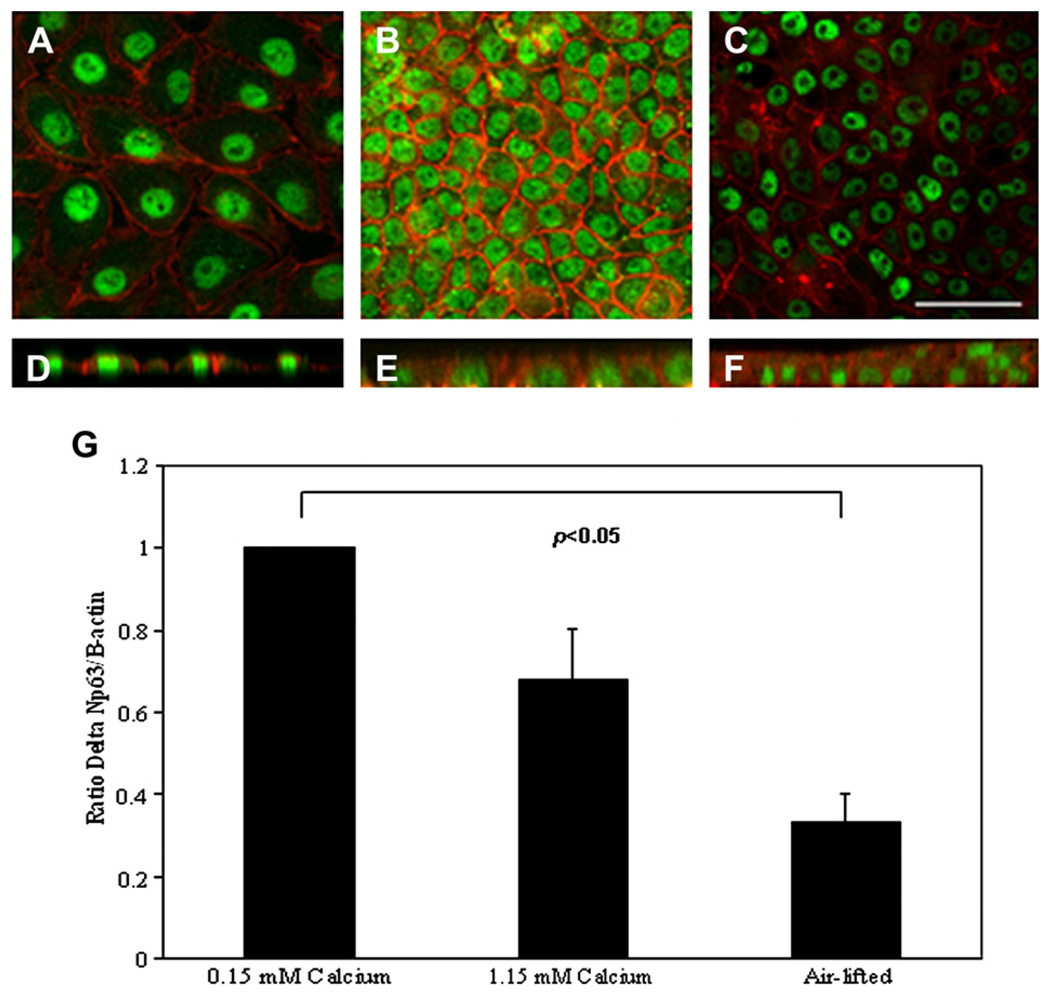
Substrate effects were evaluated by comparing mRNA and protein levels for cells grown on plastic or grown on collagen. Both mRNA and protein appeared to be significantly lower when cultured on collagen substrates compared to plastic (Fig. 6, mRNA and protein, p = 0.029, Mann–Whitney Rank Sum test). ΔNp63 expression was also evaluated as a function of differentiation. hTCEpi cells grown in 1.15 mM calcium sequential submersed/air-lifted culture were double-labeled with ΔNp63 and phalloidin. Immunofluorescence demonstrated positive ΔNp63 expression at all time points examined (Fig. 7). ΔNp63 mRNA levels were further assessed by real-time PCR (Fig. 7). mRNA expression levels were highest in 0.15 mM calcium culture. Following the addition of calcium and subsequent polarization, there was a decrease in ΔNp63 mRNA levels, which was significant following air-lifted culture (p < 0.05, one-way ANOVA).
Fig. 6.
Substrate effects on ΔNp63. (A) ΔNp63 mRNA levels were significantly reduced when grown on type I collagen substrate compared to plastic (data representative of mean ± standard deviation of four repeated experiments performed in duplicate, p = 0.029, Manne–Whitney Rank Sum Test). (B) ΔNp63 protein levels were significantly reduced when grown on type I collagen substrate compared to plastic (data representative of mean ± standard deviation of four repeated experiments, p = 0.029, Manne–Whitney Rank Sum Test). Data were normalized to the plastic condition.
Fig. 7.
ΔNp63 and differentiation. (A–C) En face XY sections of hTCEpi organotypic culture double-labeled with p63 (green) and rhodamine-conjugated phalloidin (red). (A) 0.15 mM calcium; (B) 1.15 mM calcium submersed; (C) air-lifted culture with 1.15 mM calcium media. (D–F) Vertical XZ slices of 0.15 mM calcium, 1.15 mM calcium submersed, and air-lifted culture, respectively. p63 was seen at all time points. The experiment was repeated two independent times. Scale: 50 µm. (G) Real-time PCR using a probe specific for the ΔN terminus of p63 demonstrated a decrease in pan-ΔNp63 mRNA as a function of calcium-mediated differentiation. All runs were normalized against β-actin. ΔNp63 levels following air-lifting were significantly reduced compared to ΔNp63 levels for cells grown in 0.15 mM calcium (data represented as mean ± standard deviation of three independent experiments performed in duplicate, p < 0.05, oneway ANOVA). Data normalized to the 0.15 mM calcium time point.
3.3. p63 and confluence
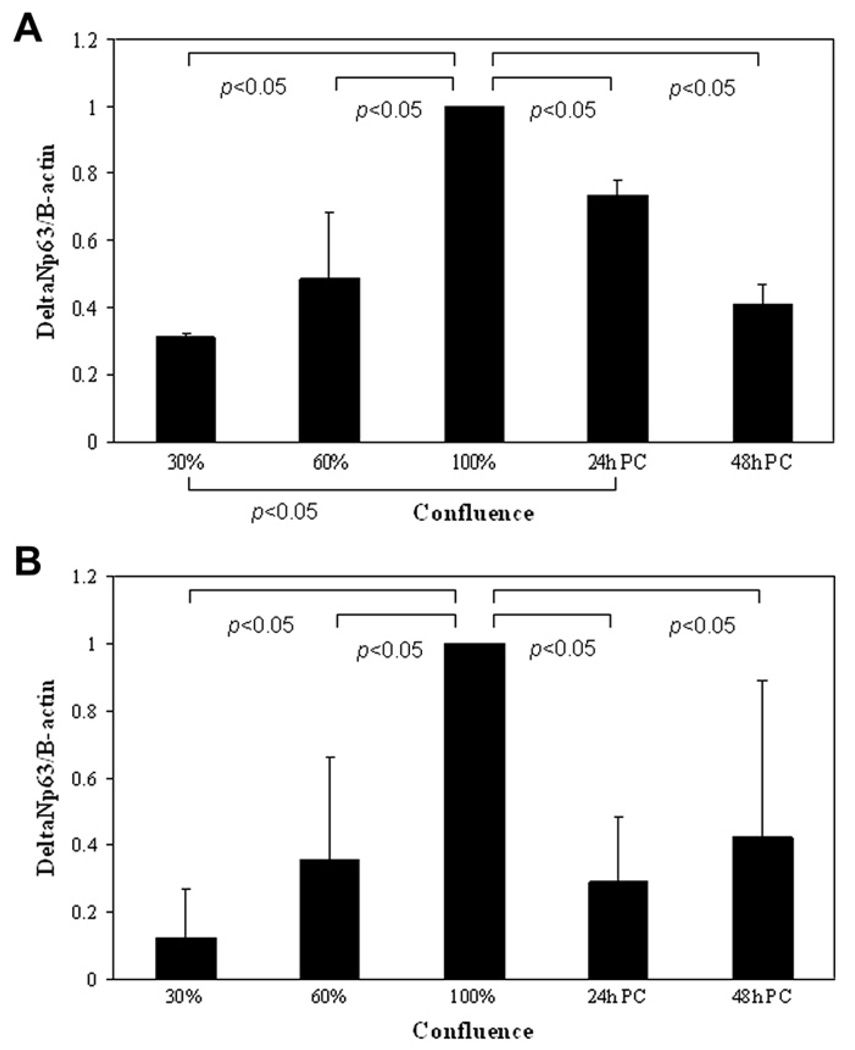
ΔNp63 levels were evaluated by real-time PCR and Western blotting as a function of confluence. For confluence studies, 5 × 104 hTCEpi cells per well were plated onto collagen-coated glass coverslips in a 24 well plate and grown for up to 7 days. ΔNp63 levels were assessed by real-time PCR and Western blotting (Fig. 8). ΔNp63 mRNA and protein levels correspondingly increased as a function of confluence, peaking at 100% confluence (mRNA, p = 0.042, one-way repeated measures ANOVA; protein, p = 0.014 Repeated Measures one-way ANOVA). Following confluence, there was a decline in ΔNp63.
Fig. 8.
ΔNp63 and confluence. (A) ΔNp63 mRNA levels increased as a function of confluence in undifferentiated cells, with a significant increase at 100% confluence compared to 30–60% confluence (data represented as mean ± standard deviation of five independent experiments performed in duplicate, p = 0.042, one-way repeated measures ANOVA). With the onset of multi-layering (24 h PC, 24 h post confluence; 48 h PC, 48 h post confluence) there was a decline in ΔNp63 mRNA levels. (B) ΔNp63α protein levels increased as a function of confluence and time post confluence in undifferentiated cells, with a significant increase at 100% confluence (data represented as mean ± standard deviation of three independent experiments, p = 0.014, oneway repeated measures ANOVA). Similar to ΔNp63 mRNA, the onset of multi-layering produced a significant decline in ΔNp63α protein levels. All data were normalized to 100% confluence.
3.4. p63 and cell cycle
To evaluate changes in ΔNp63 protein levels with phases of the cell cycle, hTCEpi cells were double-labeled with ΔNp63 and Ki-67. Using indirect immunofluorescence, all cells were positive for ΔNp63 regardless of phase (Fig. 9). Cells negative for Ki-67, indicative of having exited the cell cycle, were positive for ΔNp63. Measurements of mean pixel intensity demonstrated no difference in ΔNp63 levels throughout interphase or mitosis ( p < 0.490, one-way ANOVA) (Fig. 9).
Fig. 9.
ΔNp63 and the cell cycle. (A–G) Double labeling DNp63 (red) and Ki-67 (green) was used to identify cells in representative stages of the cell cycle. (A) Interphase; (B) prophase; (C) metaphase; (D) anaphase; (E) telophase; (F) late telophase; (G) Ki-67 negative, p63 positive cells. Scale: 25 µm. (H) Measurements of mean pixel intensity for ΔNp63 through out the cell cycle. There was no significant difference (NS) in ΔNp63 levels between phases of the cell cycle (data represented as mean ± standard deviation for one representative experiment, 10 cells per group, p < 0.490, one-way ANOVA). The entire experiment was repeated three independent times.
4. Discussion
In the human corneal epithelium, ΔNp63 has been previously postulated to be a limbal stem cell marker and which potentially functions in regulating proliferative capacity of limbal epithelial stem cells. In agreement with previous studies suggesting that ΔNp63 isoforms are not restricted to the limbal compartment, results from our in situ proteomic studies demonstrate that ΔNp63 isoforms are retained and expressed throughout the central corneal epithelium (Joseph et al., 2004; Kawasaki et al., 2006). Significantly, the loss of expression of ΔNp63 in superficial cells, which are presumably preparing to desquamate, raises new possibilities for potential function(s) for p63 in mediating epithelial differentiation and survival.
To support the use of hTCEpi cells as a viable model for elucidating the function of ΔNp63 in the human corneal epithelium in vivo, we also characterized the expression of ΔNp63 isoforms in hTCEpi cells grown in serum-free culture. Interestingly, the relative distribution of ΔNp63 isoforms varied dependent on growth conditions. In subconfluent cells, transcripts for α, β, and γ were all present, with α as the most predominant transcript. This is consistent with the presence of all three isoforms, ΔNp63 α, β, and γ, previously reported in subconfluent immortalized human keratinocyte HaCaT cells (Testoni and Mantovani, 2006). Surprisingly, upon confluence, occasional hTCEpi cells demonstrated a robust up-regulation of p63γ over α and β. This increase in p63γ was not seen in all confluent cultures, suggesting that contact inhibition alone is not enough to induce this response. While the signaling mechanism responsible for this change is still unknown, it may be due to a transient spike in p63γ mRNA levels seen at confluence and the difficulty in catching this spike due to instability of the γ transcript. Significantly, when γ is up-regulated in culture, there is a reversal in the relative mRNA distribution of α, β, and γ suggesting that potential feedback interplay between isoforms may exist. Further, the significant increase in ΔNp63γ protein in human corneal epithelium in situ compared to hTCEpi cells lends support to a potential role for ΔNp63γ in growth down-regulated tissue.
Consistent with our findings, elevated levels of p63γ have been previously reported in the corneal epithelium (Kawasaki et al., 2006). Using laser micro-dissection techniques and semi-quantitative real-time PCR, the authors localized p63γ variants in basal and suprabasal cells in the central cornea at higher levels than p63α or β. The authors further classified the three splice variants by the N-terminus region, identifying only ΔNp63 isoforms. Significantly, they failed to detect the presence of TAp63γ. While ΔNp63g mRNA levels have been reported in significant amounts in situ, the actual amount of ΔNp63γ protein in situ is quite low and is often difficult to detect. A potential mechanism for explaining and maintaining these low, steady-state protein levels may be through ubiqui-tin-mediated degradation. In skin, an E3 ubiquitin ligase which mediates TAp63α and ΔNp63α degradation through interaction with a proline-rich motif (PPxY) in the C-terminal tail has recently been identified (Rossi et al., 2006). While a specific E3 ligase for β and γ isoforms has not yet been elucidated, it is likely that both mono- and poly-ubiquitination may regulate p63 activity and proteome-mediated degradation, respectively.
The presence of ΔNp63 isoforms in the central corneal epithelium is indicative of alternate roles for p63 in addition to the previously hypothesized regulator of the cell cycle and thus, proliferative capacity. Previous studies of primary cultured limbal epithelial cells have utilized explants cultures grown on 3T3 feeder layers in the presence of serum. Using this model, expression levels of ΔNp63 have been correlated with cell size and extrapolated to define clonal and proliferative capacity (Di Iorio et al., 2005; Pellegrini et al., 1999). We evaluated ΔNp63 as a potential cell cycle regulator in corneal epithelial cells in the absence of serum by double-labeling with an antibody recognizing ΔNp63 isoforms and Ki-67. Ki-67 is a marker that can be used to identify cell cycle phases including interphase and throughout mitosis (Scholzen and Gerdes, 2000; Verheijen et al., 1989a,b). Based upon these findings, we concluded that there was no difference in fluorescence intensity throughout the cell cycle and further, that cells that had already exited the cell cycle (G0), were still positive for ΔNp63. Significantly, these findings are in agreement with previously published studies evaluating ΔNp63 expression with Ki-67 in human skin and epidermal tumors, which also concluded there was no correlation between ΔNp63 and cycling cells (Tsujita-Kyutoku et al., 2003).
It is generally thought that protein degradation is a relatively slow cellular process. Thus, it has been hypothesized that the apparent reduction in ΔNp63 levels across the limbal–ecorneal border can not be attributed to degradation alone, but is a result of protein dilution following cell division. Our findings do not support this view. Indirect immunofluorescence using double-labeling for Ki-67 and p63 did not demonstrate a detectable difference in relative protein expression regardless of the cell cycle. Further, in cultured cells, realtime PCR and Western blotting studies demonstrate increasing levels of ΔNp63 expression throughout the log growth phase, decreasing with onset of confluence. Interestingly, experiments using HaCaT cells to evaluate the association of p63 isoforms with G2/M cell cycle regulators suggest that p63 is associated with specific cell cycle-regulatory promoters containing multiple CCAAT sequences, but under normal proliferative conditions, functions as a permissive regulator of mitosis (Testoni and Mantovani, 2006). Thus, when activated, p63 is then present to exert the requisite inhibitory effect signaling cell cycle arrest and subsequent growth down-regulation. The activation or ability of p63 to induce an inhibitory effect may be due to post-translation modification and/or intramolecular modifications; however, the mechanism(s) is still unknown.
In addition to confluence, mRNA and protein levels for ΔNp63 were also shown to decrease following the addition of 1.15 mM calcium and when grown on collagen substrate. Thus culture conditions conducive to mediating early differentiative events such as calcium-induced cellular adhesion and substrate attachment may play a role in mediating changes in ΔNp63 expression and function. Further, the change in levels of ΔNp63 as a function of confluence and differentiation support a role for ΔNp63 in cellular growth control.
Taken together, these data support the use of hTCEpi cells as a viable model for the study of p63 in the corneal epithelium. Significantly, the expression of ΔNp63 isoforms in hTCEpi cells was not altered as a function of the cell cycle or cell division in subconfluent, serum-free culture, but demonstrated reduced expression upon contact inhibited growth down-regulation and differentiation. Further, the localization of ΔNp63 isoforms in differentiated cells of the corneal epithelium coupled with a loss of expression in surface epithelial cells suggests that ΔNp63 may play roles in mediating molecular signaling events at the ocular surface. Therefore, we hypothesize that the expression of ΔNp63, as previously characterized in limbal cells (Pellegrini et al., 2001), may function to block expression of target genes required for onset of differentiation and retention within the stem cell niche; whereas expression of ΔNp63 in central corneal cells mediates differentiation and surface cell desquamation. Additional studies are needed to establish these potential signaling mechanisms.
Acknowledgments
Supported in Part by NIH Grants K08 EY15713 (D.M.R.), EY10738 (H.D.C.), Infrastructure Grant EY016664, W.C. Ezell Fellowship (D.M.R.), The Contact Lens Association of Ophthalmologists, St. Paul, MN (D.M.R.), The Pearle Vision Foundation, Dallas, TX, and an unrestricted grant from Research to Prevent Blindness, Inc., New York, NY.
References
- Antonini D, Rossi B, Han R, Minichiello A, Di Palma T, Corrado M, Banfi S, Zannini M, Brissette JL, Missero C. An auto-regulatory loop directs the tissue-specific expression of p63 through a long-range evolutionary conserved enhancer. Mol. Cell. Biol. 2006;26(8):3308–3318. doi: 10.1128/MCB.26.8.3308-3318.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpitha P, Prajna NV, Srinivasan M, Muthukkaruppan V. High expression of p63 combined with a large N/C ratio defines a subset of human limbal epithelial cells: implications on epithelial stem cells. Invest. Ophthalmol. Vis. Sci. 2005;46(10):3631–3635. doi: 10.1167/iovs.05-0343. [DOI] [PubMed] [Google Scholar]
- Chen Z, De Paiva CS, Luo L, Kretzer FL, Pflugfelder SC, Li DQ. Characterization of putative stem cell phenotype in human limbal epithelia. Stem Cells. 2004;22:355–366. doi: 10.1634/stemcells.22-3-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Iorio E, Barbaro V, Ruzza A, Ponzin D, Pellegrini G, De Luca M. Isoforms of DeltaNp63 and the migration of ocular limbal cells in human corneal regeneration. Proc. Natl Acad. Sci. U.S.A. 2005;102(27):9523–9528. doi: 10.1073/pnas.0503437102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein SP, Wolosin JM, Asbell PA. P63 Expression levels in side population and low light scattering ocular surface epithelial cells. Trans. Am. Ophthalmol. Soc. 2005;103:187–199. [PMC free article] [PubMed] [Google Scholar]
- Helton ES, Zhu J, Chen X. The unique NH2-terminally deleted (deltaN) residues, the PXXP MOTIF, and the PPXY motif are required for the transcriptional activity of the DeltaN variant of p63. J. Biol. Chem. 2006;281(5):2533–2542. doi: 10.1074/jbc.M507964200. [DOI] [PubMed] [Google Scholar]
- Joseph A, Powell-Richards AOR, Shanmuganathan VA, Dua HS. Epithelial cell characteristics of cultured human limbal explants. Br. J. Ophthalmol. 2004;88:393–398. doi: 10.1136/bjo.2003.018481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki S, Tanioka H, Yamasaki K, Connon CJ, Kinoshita S. Expression and tissue distribution of p63 isoforms in human ocular surface epithelia. Exp. Eye Res. 2006;82:293–299. doi: 10.1016/j.exer.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 2004;18:126–131. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA, Zheng BH, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Parsa R, Yang A, McKeon F, Green H. Association of p63 with proliferative potential in normal and neoplastic human keratinocytes. J. Invest. Dermatol. 1999;113:1099–1105. doi: 10.1046/j.1523-1747.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- Pellegrini G, Golisano O, Paterna P, Lambiase A, Bonini S, Rama P, De Luca M. Location and clonal analysis of stem cells and their differentiated progeny in the human ocular surface. J. Cell Biol. 1999;145(4):769–782. doi: 10.1083/jcb.145.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini G, Dellambra E, Golisano O, Martinelli E, Fantozzi I, Bondanza S, Ponzin D, McKeon F, De Luca M. p63 identifies keratinocyte stem cells. Proc. Natl. Acad. Sci. U.S.A. 2001;98(6):3156–3161. doi: 10.1073/pnas.061032098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson DM, Li L, Fisher S, Pearce VP, Shay JW, Wright WE, Cavanagh HD, Jester JV. Characterization of growth and differentiation in a telomerase-immortalized human corneal epithelial cell line. Invest. Ophthalmol. Vis. Sci. 2005;46(2):470–478. doi: 10.1167/iovs.04-0528. [DOI] [PubMed] [Google Scholar]
- Rossi M, Aqeilan RI, Neale M, Candi E, Salomoni P, Knight RA, Croce CM, Melino G. The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc. Natl. Acad. Sci. U.S.A. 2006;103(34):12753–12758. doi: 10.1073/pnas.0603449103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi-Had H, Alvarenga LS, Isseroff R, Schwab IR. Factors modulating p63 expression in cultured limbal epithelial cells. Cornea. 2005;24(7):845–852. doi: 10.1097/01.ico.0000154406.76160.a5. [DOI] [PubMed] [Google Scholar]
- Scholzen T, Gerdes J. The Ki-67 Protein: from the known and the unknown. J. Cell Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Testoni B, Mantovani R. Mechanisms of transcriptional repression of cell-cycle G2/M promoters by p63. Nucleic Acids Res. 2006;34(3):928–938. doi: 10.1093/nar/gkj477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujita-Kyutoku M, Kiuchi K, Danbara N, Yuri T, Sensaki H, Tsubura A. p63 expression in normal human epidermis and epidermal appendages and their tumors. J. Cutan. Pathol. 2003;30:11–17. doi: 10.1034/j.1600-0560.2003.300102.x. [DOI] [PubMed] [Google Scholar]
- Verheijen R, Kuijpers HJH, Schlingemann RO, Boehmer ALM, van Driel R, Brakenhoff GJ, Ramaekers FCS. Ki-67 detects a nuclear matrix-associated proliferation-related antigen. I. Intracellular localization during interphase. J. Cell Sci. 1989a;92:123–130. doi: 10.1242/jcs.92.1.123. [DOI] [PubMed] [Google Scholar]
- Verheijen R, Kuijpers HJH, Van Driel R, Beck JLM, van Dierendonck JH, Brakenhoff GJ, Ramaekers FCS. Ki-67 detects a nuclear matrix-associated proliferation-related antigen. II. Localization in mitotic cells and association with chromosomes. J. Cell Sci. 1989b;92:531–540. doi: 10.1242/jcs.92.4.531. [DOI] [PubMed] [Google Scholar]
- Wang DY, Hsueh YJ, Yang VC, Chen JK. Propagation and phenotypic preservation of rabbit limbal epithelial cells on amniotic membrane. Invest. Ophthalmol. Vis. Sci. 2003;44:4698–4704. doi: 10.1167/iovs.03-0272. [DOI] [PubMed] [Google Scholar]
- Wang DY, Cheng CC, Kao MH, Hsueh YJ, Ma DHK, Chen JK. Regulation of limbal keratinocyte proliferation and differentiation by TAp63 and DeltaNp63 transcription factors. Invest. Ophthalmol. Vis. Sci. 2005;46:3102–3108. doi: 10.1167/iovs.05-0051. [DOI] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell. Biol. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun DQ, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial, and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]