Abstract
The non-enzymatic digestion of proteins by microwave D-cleavage is an effective technique for site-specific cleavage at aspartic acid (D). This specific cleavage C-terminal to D residues leads to inherently large peptides (15–25 amino acids) that are usually relatively highly charged (above +3) when ionized by electrospray ionization (ESI) due to the presence of several basic amino acids within their sequences. It is well documented that highly charged peptide ions generated by ESI are well-suited for electron transfer dissociation (ETD), which produces c- and z-type fragment ions via gas-phase ion/ion reactions. In this paper we describe the sequence analysis by ETD tandem mass spectrometry (MS/MS) of multiply charged peptides generated by microwave D-cleavage of several standard proteins. Results from ETD measurements are directly compared to CID MS/MS of the same multiply charged precursor ions. Our results demonstrate that the non-enzymatic microwave D-cleavage technique is a rapid (< 6 min) and specific alternative to enzymatic cleavage with Lys-C or Asp-N to produce highly charged peptides that are amenable to informative ETD.
Introduction
Protein identification by bottom-up proteomics involves the use of proteolytic enzymes to digest protein mixtures that are subsequently analyzed by liquid chromatography (e.g., HPLC)-tandem mass spectrometry (MS/MS) 1–3. Traditionally, digestion of proteins is performed with the enzyme trypsin (C-terminus of Lys and Arg, with some exceptions including C-terminus Pro). Collision induced dissociation (CID) of the protonated amide bonds in the peptide produces type b- and y-fragment ions, and these ions can be used to derive sequence information from the precursor ion. However, the coverage of the derived amino acid sequence depends on the extent of amide backbone cleavages reflected in either of the two complementary ion series. It is well known that the extent of the cleavages observed can be reduced by the presence of multiple basic groups (e.g., Arg) within the precursor ion sequence (as a result of trypsin missed cleavages), which inhibits proton migration along the peptide backbone that initiates the fragmentation reaction. In addition, the presence of a post translational modification (PTM) can provide a more energetically facile fragmentation than the cleavage of the amide bond. As a result, CID tandem mass spectra of peptides under these circumstances provide very limited sequence information in the form of y- and b-ion series.
The advent of electron capture dissociation (ECD) 4, 5, in which near-thermal energy electrons (0.1 eV) are captured by protonated peptides, has led to fragmentation of the peptide backbone that is less sequence dependent than CID and to the preservation of PTMs. ECD-like dissociation 6 has been observed with linear quadrupole ion traps (LIT) via gas phase ion/ion reactions 7 that involve electron transfer dissociation (ETD) of the protonated peptide.
Studies have been conducted on the effect of the precursor ion size (amino acid length), charge state and m/z on the extent of sequence coverage obtained by either ETD, CID or combined ETD/CID multistage MS. In general, ETD of triply (or higher) protonated peptides resulted in a higher degree of sequence coverage than that of doubly protonated peptides8. In this same study as well as in previous work6, a combination of electron transfer (ET) followed by CID of the charge reduction product (termed ET/CID) yielded similar data to the ETD process. Conversely, it was also established that CID of singly and doubly protonated peptides yielded more sequence information than ETD of the same precursor ions. In another large scale study that focused on the analysis of only doubly protonated tryptic peptide precursors with an analogous ET/CID approach, (in this study termed ETcaD9) noted greater sequence coverage with ETcaD than with either ETD or CID analyses alone. In a similar study using endoproteinase Lys-C enzymatic digestion (C-terminus of Lys, except for Lys-Pro) to generate large size peptides 10, doubly charged peptides yielded better fragmentation by CID than by ETD, while larger precursor ions with higher charge states were better analyzed by ETD and the ET/CID combination (termed in this latter study Charge Reduction-CID, or CRCID). From all these studies, it seems reasonable to conclude that in order to take advantage of the extensive fragmentation and information rich data obtained from ETD mass analysis, particular attention must be placed on the nature of the protonated peptide precursors being analyzed (size, charge state, sequence, etc), and that traditional tryptic peptides may not yield upon electrospray ionization (ESI) the well-suited precursors for ETD analysis. These peptide precursor properties may be controlled to a certain extent (peptide size and charge state) by a judicious choice of the protein digestion method used to generate them.
Recently, Basile and coworkers demonstrated a one-step online microwave D-cleavage of proteins, where proteins are reduced and digested in 5 minutes and products directly analyzed by LC-MS/MS11. The method is based on the specific hydrolysis of the N- and C-termini of aspartyl (D) residues by raising the temperature of a protein in an acidic (pH <2.1) solution above 108°C12, 13; however, heated by microwave radiation14. Microwave radiation has been used to rapidly raise the temperature of this reaction which dramatically increases the rate at which D is cleaved15, 16 and it is increasingly finding applications in proteomics17. Because of the relative low abundance of D residues (~5%) in proteins, the range of the average number of amino acids in peptides generated this way is around 16 (compared to 9.6 for tryptic peptides), and when missed cleavages are taken into account, the size of the peptides generated is expected to increase considerably (calculated by the in silico digestion of 250 proteins from the E. coli proteome). Accordingly, these peptides tend to have high charge states (+3 or higher), when ionized via ESI, due to their inherently large size and inclusion of one or more basic residues in their sequence. These larger peptides, when analyzed by CID tandem MS, generated tandem mass spectra with limited fragmentation information because the localization of charges within the sequence of the peptide lead to the preferential fragmentation at specific amide backbone sites rather than uniformly (randomly) across the extent of the peptide. As a result, only a fraction of the many possible fragment ions are observed yielding a poor database search match.
On the other hand, the relatively high charge states resulting from ESI of these microwave D-cleavage peptides should make them good candidate precursor ions for ETD (and ECD) analysis. Here we report the use of microwave D-cleavage digestion of proteins in conjunction with ETD mass analysis as a viable and rapid approach for “bottom-up” proteomics. Peptides were generated by microwave D-cleavage of different protein standards and analyzed by ETD and CID tandem MS. Results presented in this study demonstrate that the microwave D-cleavage method is a rapid (6 mins) and non-enzymatic alternative to protein digestion for “bottom-up” proteomics implementing ETD mass analyses.
Experimental
Chemicals
Insulin (bovine pancreas), α-lactalbumin (bovine milk), , myoglobin (horse skeletal muscle), albumin (bovine serum, BSA) and dithiothreitol (DTT) were all purchased from Sigma (St. Louis, MO). Trypsin was purchased from Promega (Madison, WI). Trifluoracetic acid (TFA) was purchased from Pierce (Rockford, IL). Methanol, formic acid, and glacial acetic acid were purchased from Mallinckrodt (Phillipsburg, NJ).
Microwave Digestion
Proteins (0.5 mg) were dissolved in 1 mL of aqueous 100 mM DTT and 12.5% formic acid solution. The solutions were transferred to a 1.5 mL eppendorf tube and heated in a CEM Focused Microwave Synthesis System (Model: Discovery; CEM, Matthews, NC). Microwave heating was achieved using 100 Watts of power to bring the temperature up to 120°C in one minute and held at that temperature for 5 minutes, for a total digestion time of 6 minutes.
Tryptic Digestion
Proteins (1 mg) were dissolved in 1 mL of deionized water. A total of 4 µg (20µL of a 0.2µg/µL solution) of trypsin was added to the protein solutions. The solutions were incubated at room temperature for ~ 12 h.
HPLC Separation
After each digestion, the peptides were separated on a reversed-phase HPLC (Agilent 1100, Palo Alto, CA) using an Aquapore RP-300 (7 µm pore size, 100 × 4.6 mm i.d.) column (Perkin-Elmer, Wellesey, MA). A linear 60 min gradient from 0 to 100% buffer B (60% ACN, 40% H20, 0.09% TFA) and buffer A consisted of 0.1% TFA aqueous solution. Fractions were collected manually and each fraction was dried in a vacuum centrifuge (Savant, Holbrook, NY). All samples were reconstituted in 100 µL of 1% acetic acid in 50% methanol/water (v/v) prior to positive ion nano-ESI.
Mass Spectrometry
All experiments were performed using a prototype version of the Q TRAP™ mass spectrometer18 (Applied Biosystems/MDS SCIEX, Concord, Ontario, Canada) modified for ion/ion reactions7. The Q TRAP electronics were modified to superpose auxiliary RF signals on the containment lenses of the Q3 quadrupole array, IQ3 and exit lenses, allowing mutual storage of oppositely charged ions in the Q3 linear ion trap (LIT). The frequency and amplitude of the auxiliary RF signals applied to the containment lenses of the Q3 LIT were optimized for the electron transfer ion/ion reaction experiments. A home-built pulsed dual nano-ESI/atmosphere pressure chemical ionization (APCI) source19 was coupled directly to the interface of the Q TRAP mass spectrometer to generate multiply charged peptide cations and azobenzene radical anions, respectively. Sequential pulsing and accumulation of the oppositely charged ions were controlled by the Daetalyst 3.14 software, a research version of software provided by MDS Sciex.
The experimental procedure for the electron transfer ion/ion reactions used here has been reported in detail elsewhere19. In brief, a typical experiment employed in this work comprised sequences as follows: (1) pulsing the high voltage (+1.0−1.5 kV) on the nano-ESI emitter and injection of positive ions into Q3 LIT; (2) switching off the high voltage on the nano-ESI emitter while the cations were cooled in Q3 for 50 ms; (3) isolating the ions of interest under RF/DC mode in Q3 cell; (4) switching on the high voltage (−2 kV) applied on the APCI needle and injection of azobenzene radical anions into Q3 LIT, selected by Q1 in the mass-resolving mode while the cations were cooled in Q3 cell; (4) mutual storage of oppositely charged ions in Q3 LIT with nitrogen as bath gas at around 2.9 × 10−5 Torr; (5) cooling the ions for 50 ms in Q3 cell and (6) mass analysis of ions in Q3 LIT via mass selective axial ejection (MSAE)20 using a supplementary RF signal at frequency 380 kHz. For the ion trap CID experiments, a dipolar radio frequency (RF) in resonance with the secular frequency of the ions was applied on the isolated ions of interest for 100 ms. Then the product ions were cooled for 50 ms in Q3 LIT and mass analyzed via MSAE using a supplementary RF signal at frequency 380 kHz. The spectra shown herein were typically the averages of 20–100 individual scans.
Results and Discussion
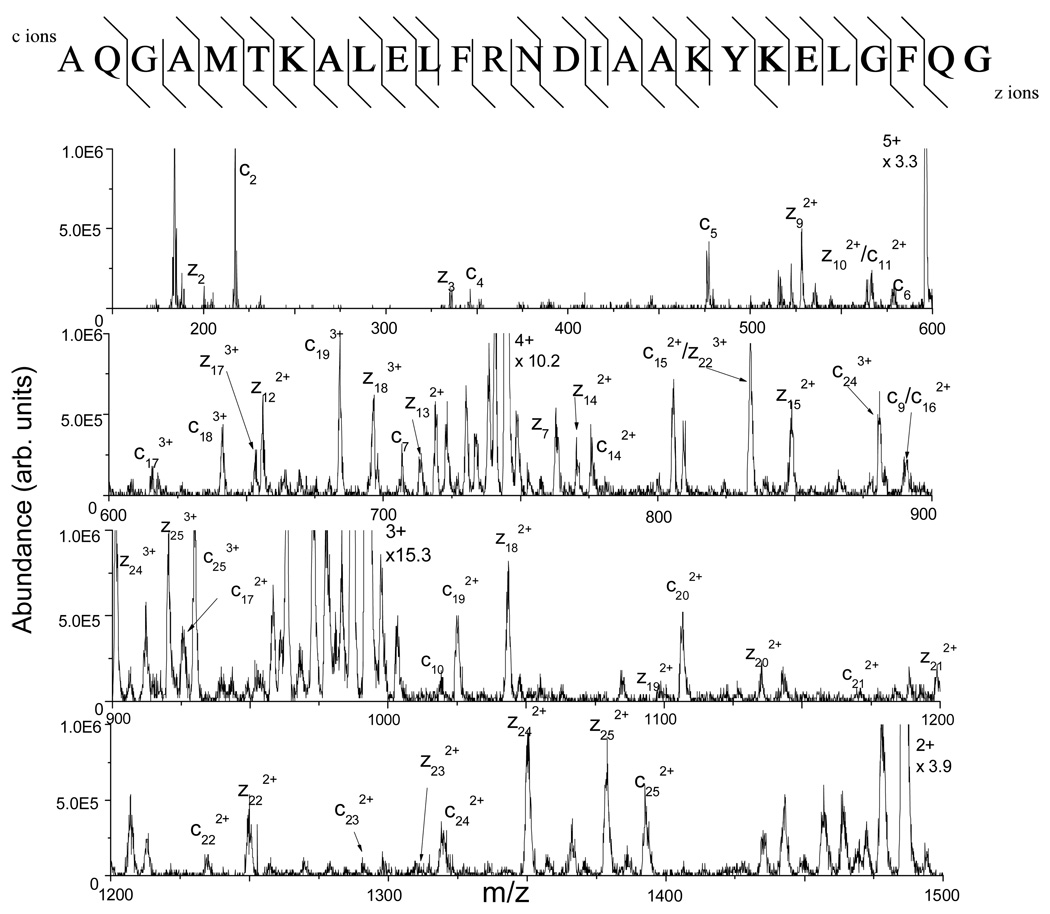
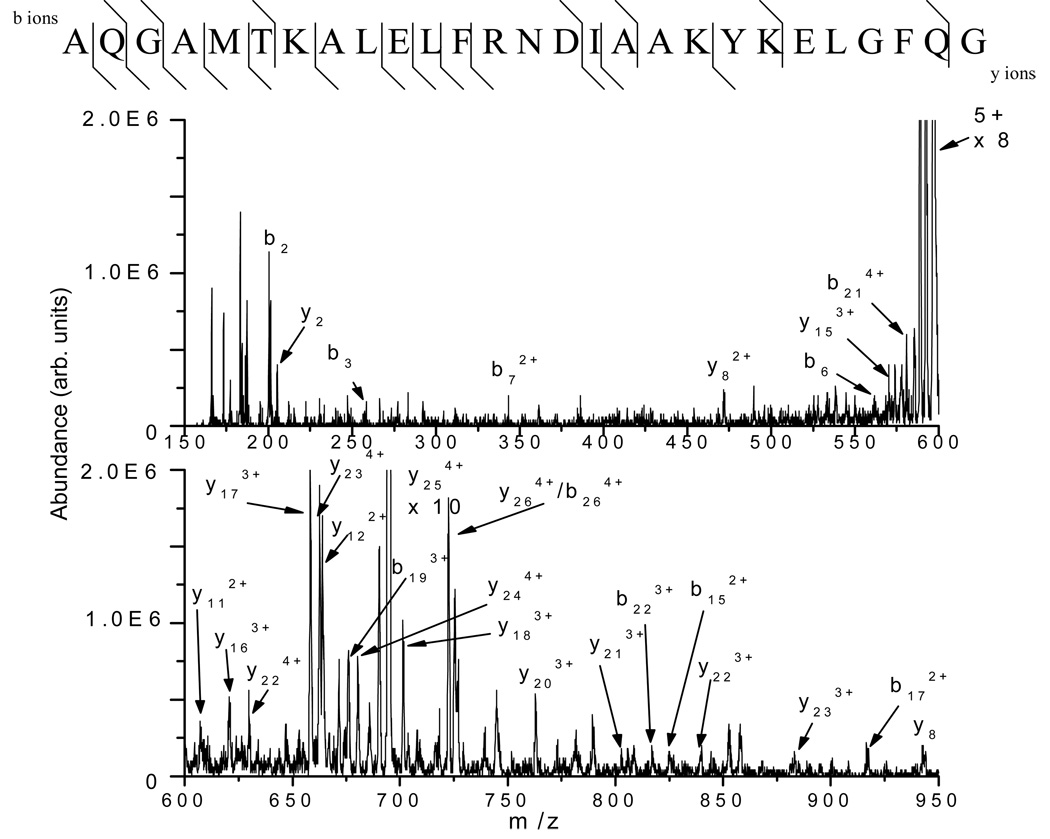
Myoglobin (MW of 16.9 kDa) has eight D residues within its sequence, and upon microwave D-cleavage, it is expected that seven of the nine possible peptides formed have at least 12 amino acids. The number of amino acids increases when partial digestion (i.e., missed cleavage) is taken into account. Three peptides ranging in amino acid length from 17–27 formed from microwave D-cleavage of myoglobin were analyzed by tandem MS (MS/MS) alternatively using ETD and CID for direct comparison of the resulting sequence information. Figure 1 shows the ETD tandem mass spectrum of the +5 charged precursor ion at m/z 595 ([M+5H]5+) obtained for the first peptide analyzed from the C-terminal cleavage at (D)126 with a missed cleavage at (D)141 (of sequence AQGAMTKALELFRNDIAAKYKELGFQG). The peptide has a molecular weight of 2971 Da and contains four basic groups (1 R and 3 K’s) within its sequence, giving rise to the observed charge states above +2. The most abundant peaks in the product ion spectrum shown in Figure 1 are from the +3 and +2 ions that can arise from either proton transfer during the ion/ion reaction or electron transfer without dissociation. From this mass spectrum, 38 out of the 52 possible c- and z-type fragment ions are observed and identified. (Note that c- and z-ions with mass-to-charge ratios lower than the low-mass cut-off used in these experiments (m/z 150), if formed, could not be observed.) Moreover, 24 of the 26 N-Cα backbone bonds are cleaved, yielding a 92% sequence coverage for this peptide. Figure 2 shows the complementary CID tandem mass spectrum obtained from the same +5 ion at m/z 595. The sequence information retrieved from this mass spectrum differs greatly from that from the ETD tandem mass spectrum. Only 20 out of the possible 52 b- and y-type ions could be identified representing cleavage of only 17 of the 26 (~65% sequence coverage) possible amide bonds (Figure 2, top).
Figure 1.
The ETD tandem mass spectrum of the ion at m/z 595, corresponding to the +5 charge state of the peptide AQGAMTKALELFRNDIAAKYKELGFQG formed from the microwave D-cleavage of myoglobin at D126 (100 single scans). The sequence at the top shows that 24 of the N-Cα backbone bonds are broken, giving nearly a full sequence coverage of the peptide.
Figure 2.
CID tandem mass spectrum of the ion at m/z 595, corresponding to the +5 charge state of the peptide AQGAMTKALELFRNDIAAKYKELGFQG formed from the microwave D-cleavage of myoglobin at D126 (20 single scans). Only 20 b- and y-type ions are observed, mostly in the +2 or +3 charge state. Top: 17 of the amide backbone bonds are cleaved.
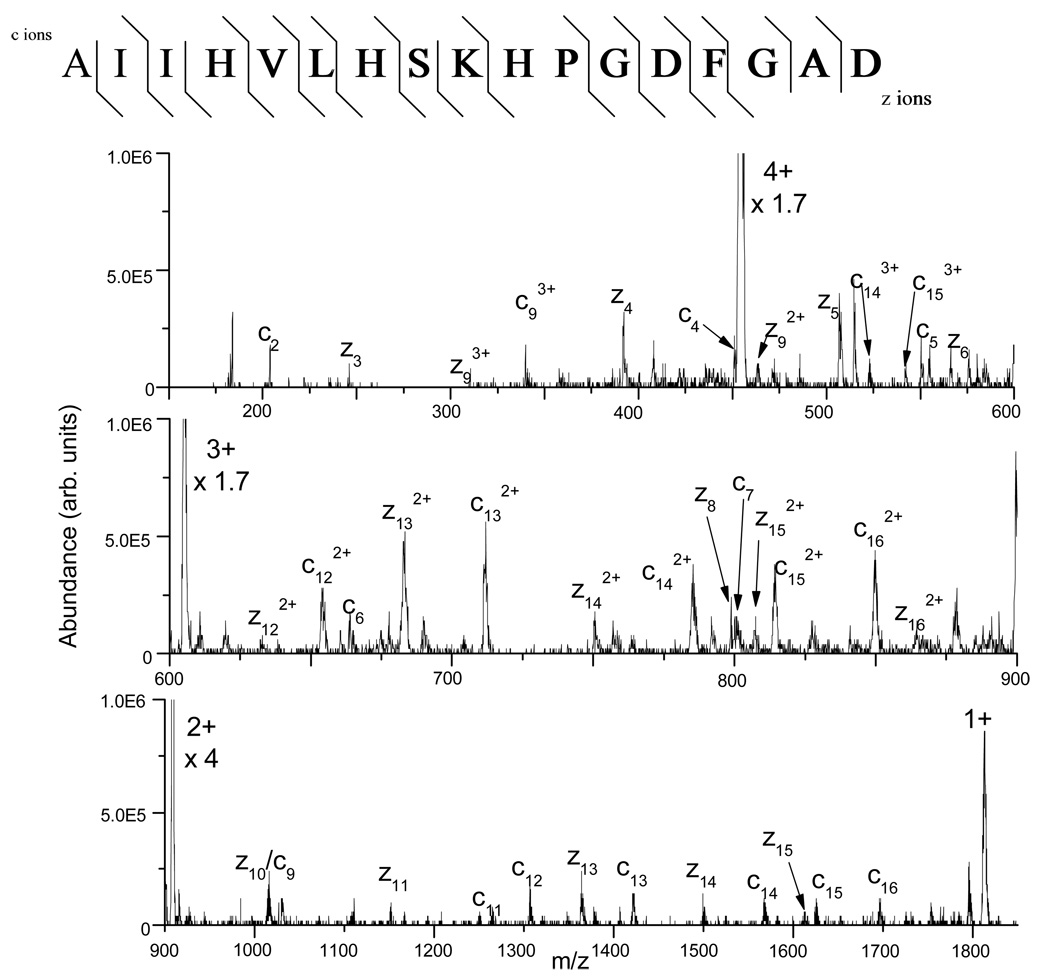
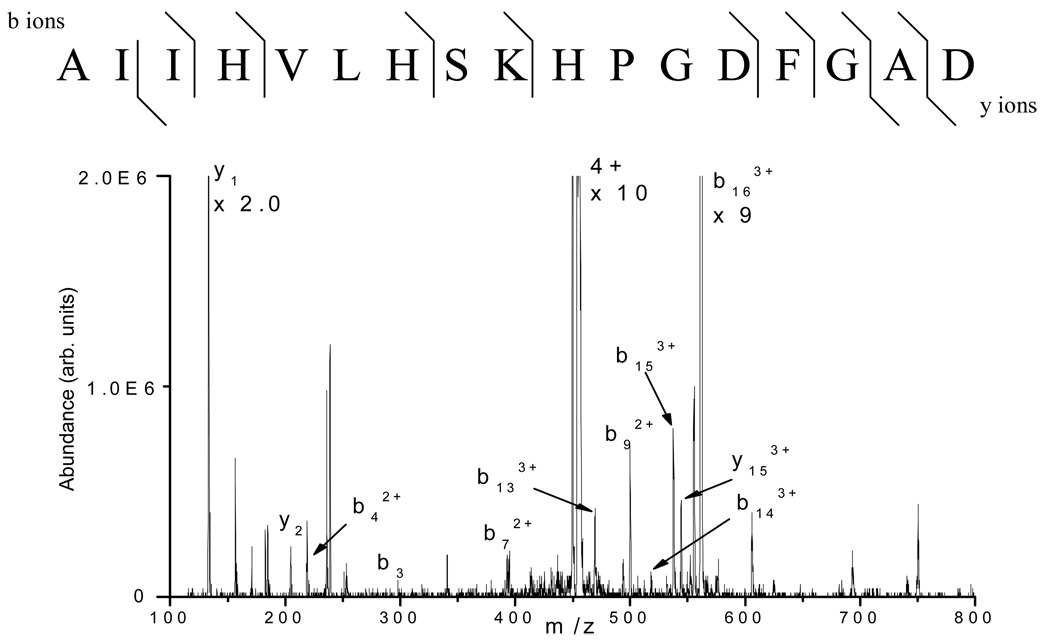
Figure 3 shows the ETD tandem mass spectrum of the +4 ion (m/z 455) from the peptide AIIHVLHSKHPGDFGAD, which is formed from the microwave D-cleavage of myoglobin at D109 and D126. Proton transfer during the ion/ion reaction or electron transfer without dissociation leads to charge state reduction of the precursor ion, the most abundant being the +2 ion. ETD of the peptide leads to 25 out of 32 possible c- and z-type fragment ions identified, including cleavage at the c-terminus of proline( P); however, N-terminal dissociation at P residues is generally not observed with ETD (analogous to ECD21). Most of the fragment ions observed are singly charged and 15 out of 16 (~94% sequence coverage) possible N-Cα backbone bond cleavages are observed. Figure 4 shows the corresponding CID tandem mass spectrum of the +4 ion from the same peptide. Limited sequence information can be derived from this spectrum as 11 out of 32 possible b- and y- ions can be identified. Additional data for myoglobin is provided in the supplementary data section.
Figure 3.
ETD tandem mass spectrum of the +4 ion at m/z 455 from the peptide AIIHVLHSKHPGDFGAD formed from the microwave D-cleavage of myoglobin at the C-termini of D109 and D126 (200 single scans). 15 out of the 16 possible backbone bonds are cleaved, leading to excellent sequence coverage.
Figure 4.
CID tandem mass spectrum of the +4 ion (m/z 455) from the peptide AIIHVLHSKHPGDFGAD formed from the microwave D-cleavage of myoglobin at the C-termini of D109 and D126 (50 single scans). Only 11 b- and y-type ions are observed and only 9 of the amide backbone bonds are cleaved.
Combined, these results demonstrate the compatibility of the microwave D-cleavage method with MS/MS using ETD. A clear advantage of this methodology is the speed of digestion (6 mins) and that no enzymes are used, decreasing the complexity of the background by eliminating possible peptides from enzyme auto-hydrolysis.
Microwave D-cleavage digestion was also performed on three other proteins including insulin, α-lactalbumin, and BSA. Table 1 presents a summary of the peptides analyzed from each protein by ETD and CID, illustrating the advantage of implementing ETD as a dissociation technique for peptides generated with the microwave D-cleavage digestion method. Insulin is a protein that contains two interchain disulfide bond linked chains, Chain A and Chain B, and does not contain D residues. Even though the disulfide bonds were reduced with the microwave D-cleavage method11, ETD is known to specifically break these bonds. The analysis of this protein serves to illustrate the specificity of the microwave D-cleavage method in that no non-specific peptides were generated and only the peptides corresponding to the Chain A and B fragments were observed (this specificity was also demonstrated in a previous study by the Basile group implementing LC-MS11). The peptide corresponding to Chain B, a 30 amino acid long peptide with a MW of 3400 Da, was analyzed with both ETD and CID (see supplementary data). The protein α-lactalbumin (14.2 kDa) with 123 amino acids, 13 of which are D residues was also analyzed using the microwave D-cleavage coupled with ETD MS/MS. Of the 14 possible peptides formed from microwave D-cleavage, 6 have at least 10 amino acids in their sequence and 4 of the peptides are comprised of a single D residue because they are found consecutively within the sequence. BSA is a large protein (66 kDa) with 583 amino acids and 40 potential D-cleavage sites. There are 20 possible peptides that contain at least 10 amino acids, and 12 of those peptides contain more than 15 amino acids. The ETD mass spectra and subsequent CID mass spectra for the peptides analyzed from these proteins can be found in the supplementary data section. Finally, it is worth noting that deamidation of asparagine (Asn, N) and glutamine (Gln, Q) are known to take place under acidic conditions; however, these modifications have not been detected in this and other studies11 under the conditions used. Also, in this study the number of peptides detected from the microwave D-cleavage digestion of these proteins may be limited by the fact that no chromatographic separation was performed on the resulting peptide mixture, and as a result some of the peptides generated may have gone undetected due to ionization suppression effects.
Table 1.
Summary of ions detected from peptides generated by microwave D-cleavage digestion and analyzed by ETD and CID tandem mass spectrometry (MS/MS).
| ETD | CID | ||||||
|---|---|---|---|---|---|---|---|
| Protein | Peptides Analyzed | MW (Da) | Charge | c ions | z ions | b ions | y ions |
| Myoglobin P68082 | |||||||
| AIIHVLHSKHPGDFGAD | 1814 | 4 | 12/16 | 13/16 | 8/16 | 3/16 | |
| AQGAMTKALELFRNDIAAKYKELGFQG | 2971 | 5 | 20/26 | 18/26 | 7/26 | 13/26 | |
| IAGHGQEVLIRLFTGHPETLEKFD | 2708 | 4 | 19/23 | 12/23 | 12/23 | 13/23 | |
| BSA CAA76847 | |||||||
| PHACYSTVFDKLKHLVD | 1973 | 4 | 13/16 | 13/16 | 5/16 | 12/16 | |
| KPLLEKSHCIAEVEKD | 1839 | 3 | 11/15 | 11/15 | 10/15 | 10/15 | |
| α-lactalbumin 1F6S_F | |||||||
| EQLTKCEVFRELKD | 1738 | 4 | 10/13 | 11/13 | 5/13 | 8/13 | |
| KVGINYWLAHKALCSEKLD | 2188 | 4 | 14/18 | 12/18 | 10/18 | 9/18 | |
| KVGINYWLAHKALCSEKLDQWLCEKL | 3090 | 4 | 15/25 | 12/25 | 10/25 | 6/25 | |
| Insulin P01317 | |||||||
| FVNQHLCGSHLVEALYLVCGERGFFYTPKA | 3400 | 5 | 21/29 | 21/29 | 22/29 | 18/29 | |
Conclusions
Multiply charged peptides that arise from the ESI of peptides produced from microwave D-cleavage were subjected to both ETD MS/MS and CID MS/MS. Results demonstrate that precursor ion fragmentation from the gas phase ETD ion/ion reaction between azobenzene anions and the protonated peptides generated by the microwave D-cleavage digestion method yielded higher sequence coverage than CID fragmentation of the same precursor ion. Our results also corroborate findings of previous studies in that: (i) CID of protonated peptides with charge states of +3 or higher yields poor sequence information, and (ii) ETD of protonated peptides with charge states of +3 or higher yields useful sequence information. Peptides generated by the microwave D-cleavage digestion method are larger in size because of the low abundance of D residues and the presence of multiple basic residues within their sequence confers them a high charge state (≥ +3) when ionized via ESI. The efficiency of ETD relies on the charge state of the precursor ion, with the efficiency decreasing with decreasing charge state, and so the peptides formed by microwave D-cleavage with higher charge states are well-suited for fragmentation by ETD. These multiply charged peptides are readily fragmented to c- and z-type ions by ETD with little reliance upon the sequence length or amino acid composition, leading to greater sequence coverage and improved protein identification. Lastly, the ability to perform the microwave D-cleavage step online with the LC-MS/MS analysis 11 will provide the means to perform rapid and high throughput “bottom-up” proteomic measurements implementing ETD, in effect making the chromatographic separation the rate limiting step in the analysis.
Supplementary Material
Acknowledgments
FB and NH acknowledge the support of the National Institutes of Health (grant R15-RR020354-01A1) and the University of Wyoming Research Office and Graduate Office. HH and SAM acknowledge support by the National Institute of General Medical Sciences under Grant GM 45372.
Contributor Information
Nicolas J. Hauser, Department of Chemistry, University of Wyoming, 1000 E. University Ave., Laramie, Wyoming, 82071.
Franco Basile, Department of Chemistry, University of Wyoming, 1000 E. University Ave., Laramie, Wyoming, 82071.
Hongling Han, Department of Chemistry, Purdue University, West Lafayette, Indiana, 47907-2084.
Scott A. McLuckey, Department of Chemistry, Purdue University, West Lafayette, Indiana, 47907-2084
References
- 1.Yates JR., III Mass spectrometry and the age of the proteome. Journal of Mass Spectrometry. 1998;33(1):1–19. doi: 10.1002/(SICI)1096-9888(199801)33:1<1::AID-JMS624>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 2.Yates JR, 3rd, Speicher S, Griffin PR, Hunkapiller T. Peptide mass maps: a highly informative approach to protein identification. Anal Biochem. 1993;214(2):397–408. doi: 10.1006/abio.1993.1514. [DOI] [PubMed] [Google Scholar]
- 3.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature (London, United Kingdom) 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 4.Zubarev RA, Kelleher NL, McLafferty FW. Electron Capture Dissociation of Multiply Charged Protein Cations. A Nonergodic Process. Journal of the American Chemical Society. 1998;120(13):3265–3266. [Google Scholar]
- 5.Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FW. Electron capture dissociation for structural characterization of multiply charged protein cations. Anal Chem. 2000;72(3):563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 6.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proceedings of the National Academy of Sciences. 2004;101(26):9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia Y, Wu J, McLuckey SA, Londry FA, Hager JW. Mutual storage mode ion/ion reactions in a hybrid linear ion trap. Journal of the American Society for Mass Spectrometry. 2005;16(1):71–81. doi: 10.1016/j.jasms.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 8.Pitteri SJ, Chrisman PA, Hogan JM, McLuckey SA. Electron Transfer Ion/Ion Reactions in a Three-Dimensional Quadrupole Ion Trap: Reactions of Doubly and Triply Protonated Peptides with SO2.- Analytical Chemistry. 2005;77(6):1831–1839. doi: 10.1021/ac0483872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swaney DL, McAlister GC, Wirtala M, Schwartz JC, Syka JEP, Coon JJ. Supplemental Activation Method for High-Efficiency Electron-Transfer Dissociation of Doubly Protonated Peptide Precursors. Analytical Chemistry. 2007;79(2):477–485. doi: 10.1021/ac061457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu S-L, Huehmer AFR, Hao Z, Karger BL. On-Line LC-MS Approach Combining Collision-Induced Dissociation (CID), Electron-Transfer Dissociation (ETD), and CID of an Isolated Charge-Reduced Species for the Trace-Level Characterization of Proteins with Post-Translational Modifications. Journal of Proteome Research. 2007 doi: 10.1021/pr070313u. Published on Web 2–2007, ACS ASAP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hauser NJ, Basile F. On-line Microwave D-Cleavage LC-ESI-MS/MS of Intact Proteins: Site-Specific Cleavages at Aspartic Acid Residues and Disulfide Bonds. Journal of Proteome Research. 2008 doi: 10.1021/pr700596e. in Press. [DOI] [PubMed] [Google Scholar]
- 12.Inglis AS. Cleavage at aspartic acid. Methods in Enzymology. 1983;91(Enzyme Struct Pt I):324–332. doi: 10.1016/s0076-6879(83)91030-3. [DOI] [PubMed] [Google Scholar]
- 13.Schultz J, Allison H, Grice M. Specificity of the cleavage of proteins by dilute acid. I. Release of aspartic acid from insulin, ribonuclease, and glucagon. Biochemistry. 1962;1:694–698. doi: 10.1021/bi00910a024. [DOI] [PubMed] [Google Scholar]
- 14.Hauser NJ, Basile F. Development of a Rapid Non-Enzymatic Cell Digestion Procedure for Proteomics Based Microorganism Identification. Site-Specific Digestion at Aspartyl Residue by Microwave Heating -Mild Acid Hydrolysis, 52nd ASMS Meeting, Nashville, TN 2004; Nashville, TN. 2004. [Google Scholar]
- 15.Zhong H, Marcus SL, Li L. Microwave-assisted acid hydrolysis of proteins combined with liquid chromatography MALDI MS/MS for protein identification. Journal of the American Society for Mass Spectrometry. 2005;16(4):471–481. doi: 10.1016/j.jasms.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 16.Swatkoski S, Russell SC, Edwards N, Fenselau C. Rapid Chemical Digestion of Small Acid-Soluble Spore Proteins for Analysis of Bacillus Spores. Analytical Chemistry. 2006;78(1):181–188. doi: 10.1021/ac051521d. [DOI] [PubMed] [Google Scholar]
- 17.Lill Jennie R, Ingle Elizabeth S, Liu Peter S, Pham V, Sandoval Wendy N. Microwave-assisted proteomics. Mass Spectrom Rev FIELD Full Journal Title:Mass spectrometry reviews. 2007;26(5):657–671. doi: 10.1002/mas.20140. [DOI] [PubMed] [Google Scholar]
- 18.Hager JW. A new linear ion trap mass spectrometer. Rapid Communications in Mass Spectrometry. 2002;16(6):512–526. doi: 10.1002/rcm.1020. [DOI] [PubMed] [Google Scholar]
- 19.Liang X, Xia Y, McLuckey SA. Alternately Pulsed Nanoelectrospray Ionization/Atmospheric Pressure Chemical Ionization for Ion/Ion Reactions in an Electrodynamic Ion Trap. Analytical Chemistry. 2006;78(9):3208–3212. doi: 10.1021/ac052288m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Londry FA, Hager JW. Mass selective axial ion ejection from a linear quadrupole ion trap. Journal of the American Society for Mass Spectrometry. 2003;14(10):1130–1147. doi: 10.1016/S1044-0305(03)00446-X. [DOI] [PubMed] [Google Scholar]
- 21.Zubarev RA. Reactions of polypeptide ions with electrons in the gas phase. Mass Spectrometry Reviews. 2003;22(1):57–77. doi: 10.1002/mas.10042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.