Abstract
Remodeling of the chromatin network plays an important role regulating embryonic development as well as differentiation. The SWI/SNF complex is an ATP dependent chromatin-remodeling complex. It consists of several proteins, including an ATPase subunit, either Brg1 or Brm. Two subunits of this complex Baf53a and Baf45 have been previously identified as being neural progenitor specific. In this study, we show that Baf60c, another important part of this large complex, acts in a similar neural-progenitor specific manner. We show that during development Baf60c is expressed in neural progenitors in human retinas as well as mouse retina, cortex and spinal cord. Baf60c expression is lost during neural differentiation and its over-expression keeps the progenitors in a proliferative state through its interaction with the Notch pathway. Finally, we show that Baf60c is re-expressed in the Müller glial cells that reenter the cell cycle after neurotoxic damage.
Keywords: Chromatin remodeling, Baf60c, retina development, retinal regeneration, neural development, Swi/Snf
Introduction
Chromatin remodeling complexes have been shown to be critical regulators of gene expression during development. One of these complexes, the SWI/SNF complex utilizes energy from ATP hydrolysis to disrupt histone–DNA interactions (Martens and Winston, 2003). Mammalian SWI/SNF complexes contain one of two catalytic ATPase subunits- Brm and Brg1- and ten core subunits known as BAFs (Brg/Brm-associated factors), which are named according to their size in kD. In the mammalian genome, 25 genes encode proteins in the BAF complexes (Wu et al., 2007). These complexes have been shown to be critical for expression of many different genes, both during development and in adult tissues; for example, a recent study of TGF-beta signaling, found that over 70% of the genes regulated by TGF-beta are dependent on Brg1 and the SWI/SNF chromatin remodeling complex (Xi et al., 2008).
The SWI/SNF complex also appears to play key developmental roles in neurogenesis. Loss of function studies of Brg1 in mice have shown that this protein is necessary to maintain normal levels of proliferation of the neural stem/progenitor cells (Lessard et al., 2007). In addition, loss of function mutations in Brg1 in zebrafish leads to defects in neural development (Seo et al., 2005). Other components of the complex also serve non-redundant functions during neurogenesis. RNAi mediated knock-down of Baf53a or Baf45a leads to a significant reduction in neural progenitor proliferation, while over-expression of these subunits stimulates neurogenesis (Lessard et al., 2007).
The specific developmental functions of BAFs correlate with changes in the composition of the core subunits of the complex. Lessard et al (2007) recently defined a specific set of subunits expressed by neural progenitors (npBAFs), which are different from those expressed in differentiated neurons (nBAFs). In their model, BAF45a and BAF53a are primarily expressed by neural progenitors and, as noted above, are necessary and sufficient to maintain them in an undifferentiated, proliferating state. Although in their model, BAF60c was presumed to be present in both the neural progenitors and the differentiated neurons, they did not specifically examine this subunit during neurogenesis. It has also been shown recently that BAF60c physically interacts with the Notch intracellular domain (ICD) and Rbpj to stabilize their interaction and potentiate Notch signaling (Takeuchi et al., 2007). Since the control of Notch signaling is critical during the transition from neural progenitor to differentiated neuron, we were motivated to investigate the expression of BAF60c during neurogenesis.
Using a combination of in vitro and in vivo analyses, we have analyzed the expression and potential function of BAF60c during retinal development and we report several novel findings concerning BAF60c in developing retina. Specifically, we have found that BAF60c is expressed in retinal progenitors, in both embryonic human and mouse, but not in differentiated neurons. We also found that like other npBAFs, over-expression of BAF60c in the progenitors of the developing mouse retina promotes their proliferation. We also found that while BAF60c is not normally present in adult mouse retina, it is re-expressed in Müller glia under conditions that stimulate their proliferation. Our results therefore indicate that BAF60c is a component of the npBAF complex and may play important roles in retinal development and regeneration.
Results
Expression of BAFs in fetal human retina
Since the antibody to BAF60c we used was raised against the human protein, we first tested its expression in human retina. We found that the expression was nuclear, and somewhat granular, similar to what has been reported previously for the various subunits of this chromatin remodeling complex. At 96 days post-conception, the human fetal retina contains layers of ganglion cells and amacrine cells (both labeled with HuC/D) and a "neuroblastic layer" that contains primarily mitotically active retinal progenitors and some post-mitotic neurons that are migrating to the ganglion cell layer from their "birth" at the ventricular surface (Figure 1 D,E). Nearly all the nuclei in the neuroblastic layer are labeled with the BAF60c antibody (Figure 1 A,C,D,E); most of those that are not are labeled with HuC/D and are likely the migrating post-mitotic newly generated ganglion cells and/or amacrine cells (Figure 1 D,E). Thus, BAF60c appears to be similar to the other npBAFs, in that it is specific to neural progenitors, at least in the developing retina.
Figure 1.
Expression of chromatin remodeling agents in developing 96 day human retina. A–C (A) BAF60c is expressed in retinal progenitors (red) and not in Hu C/D expressing inner retinal differentiated cells (green) (B) and C represents the merged view. D and E are high power view of boxed region in C. (F, G) Brg1 (red) is expressed by both retinal progenitors and Hu C/D (green) expressing differentiated retinal neurons. (H, I) BAF53a (red) is expressed by retinal progenitors as well as a subset of differentiated retinal neurons (green). (J, K) Brm (red) is only expressed by differentiated inner retinal neurons (Hu C/D, green). (NBL- neuroblastic layer, GCL- ganglion cell layer).
As noted above, the SWI/SNF complex contains one of two different ATPases, Brg1 or Brm. To determine which of these two is present in the npBAF complex in developing retina, we labeled sections with antibodies to each of them. Both antibodies show clear nuclear labeling in retinal cells; however, while the antibody to Brg1 labels both progenitors and differentiated HuC/D+ neurons, the anti-Brm antibody only labels the latter (Figure 1 F,G and J,K). As noted above, one of the previously identified npBAFs, expressed specifically in neural stem/progenitor cells in the CNS, is BAF53a. To determine whether this is true for the retina, we used an antibody to BAF53a and compared its expression to the other subunits. We found that BAF53a is highly expressed in the retinal progenitors in the NBL, and appears to be less highly expressed in the differentiated neurons, though some of the retinal ganglion cells still have some detectable expression (Figure 1 H,I). Taken together, these data indicate that Baf60c, along with BAF53a and Brg1, are part of the npBAF complex, while either Brm or Brg1, along with other npBAFs, mediate chromatin remodeling in differentiated neurons.
When we used the same BAF60c antibody to label sections of developing mouse retina, we found a similar pattern as that described for the fetal human retina. At embryonic day 14 (E14) in the mouse, there is already clear, nuclear expression by nearly all the cells in the progenitor layer (Figure 2 A–C). Co-labeling the sections with an antibody against HuC/D to label the inner retinal neurons (ganglion cells and amacrine cells) shows essentially no overlap with the anti-BAF60c labeled cells. Sox2, another transcription factor known to be expressed in progenitors is shown for comparison (Figure 2F). BAF60c continues to be expressed by the progenitors though postnatal development; by P6, it is expressed only near the retinal margin, where the remaining progenitors persist in the INL (Figure 2G). In the adult retina, there are no more progenitors, even at the margin, and there is no more BAF60c expression (Figure 2H).
Figure 2.
Expression of BAF60c during mouse retinal development. (A–C) E14 retina stained for BAF60c (red, C) and Hu C/D (green, A) with the merged view in B showing non-overlapping expression. (D, E, F) Higher power view of E14 retina similarly stained for BAF60c (red, D, E), Hu C/D (green, E) and Sox2 (green, F) showing BAF60c having a similar pattern as the neural progenitor marker. (G) P6 retinal section showing a few BAF60c positive (red) progenitor cells remaining at periphery (Hu C/D in green) with almost no remaining BAF60c remaining cell by P14 (H). At E14, HuC/D is also expressed by cells in the anterior part of the lens.
We also used quantitative PCR analysis (QPCR) to confirm the antibody expression patterns (Figure 3). We extracted mRNA from mouse retinas at embryonic day 13.5 (E13.5, n=5), when most of the retina consists of mitotically active progenitors, and postnatal day 8 (P8, n=5), when neurogenesis is complete and there are few progenitors remaining. We generated cDNA and carried out QPCR for Baf60c, Brg1, and Brm. In addition, we analyzed expression of two progenitor markers (and components of the Notch signaling pathway), Hes1 and Hes5, and a gene expressed in postmitotic rod photoreceptors, Nrl. After normalizing expression levels to GAPDH levels in each sample, we compared the dCT values for the two ages for all six genes, and the ratios of E13.5/P8 is shown in Figure 3. As expected, Hes5 and Hes1 decline as progenitors differentiate (−13.59 and −18.57 fold, respectively) while Nrl expression increases dramatically by P8 (+405 fold) as many of the progenitors differentiate into postmitotic rod photoreceptors. Similar QPCR analysis for Baf60c mRNA expression shows its down-regulation between E13.5 and P8, confirming its expression in progenitors. Although our antibody labeling showed Brg1 expression in both progenitors and differentiated neurons, the relative expression of this gene also declines with the completion of neurogenesis. By contrast, the mRNA levels of Brm are slightly higher at P8 over those observed at E13.5, consistent with our antibody labeling observations that this protein is primarily localized to differentiated neurons and not progenitors.
Figure 3.
Quantitative PCR analysis comparing postnatal day 8 retina to E13.5 retina. The PCR confirms that Baf60c, Brg, Hes1 and Hes5 decline as the retina differentiates while Brm and Nrl expression increase. Graph shows mean (+/− SD) PCR cycle difference (dCT) when mRNA expression reaches threshold of detection with the corresponding fold change in the table below. All data was normalized to GAPDH expression levels in each sample.
The above analysis in the retina shows BAF60c is localized to progenitors. To determine whether this neural progenitor expression of BAF60c is unique to the retina, or is also true for other regions of the nervous system, we assessed its expression in two other developing neuronal regions: the early embryonic cortex and the spinal cord. At E12, we found that BAF60c is expressed in the progenitor regions of both the cortex and spinal cord (Figure 4A, C). Upon co-labeling the sections with Tuj-1, a marker of differentiated neurons, BAF60c was absent in the differentiated cells in both regions (Figure 4B, D).
Figure 4.
Expression of BAF60c in the mouse embryonic cortex and spinal cord. (A) Baf60c (red) and Tuj1 (green) staining of the E12 cortex showing absence of BAF60c in differentiated cells. (B) Higher power view of a region in 4A showing absence of BAF60c in differentiated tuj1 expressing neurons. (C) BAF60c (red) and tuj1 (green) staining of the E12 spinal cord showing BAF60c in progenitor cell region. (D) Higher power view of ventral spinal cord showing lack of BAF60c expression in differentiated neurons. Tuj1-positive axons seem to instead wrap around and move between the BAF60c-expressing progenitors (arrowheads).
Baf60c over-expression promotes progenitor proliferation
A previous study characterizing npBAFs demonstrated that over-expression of either BAF45a, or the combination of BAF45a and BAF53a, stimulated proliferation and/or inhibited differentiation of neural stem/progenitor cells in embryonic mouse and chick CNS (Lessard et al., 2007). Since BAF60c also appears to be specifically expressed in neural progenitors, we asked whether over-expression in these cells would have a similar effect as that found with the other npBAFs. We made explant cultures of newborn (P0) mouse retina, electroporated an expression plasmid containing Baf60c, along with GFP, and after two days in vitro, added BrdU to label the mitotically active, S-phase progenitors. In both the control explants (electroporated with the vector expressing GFP) and the explants electroporated with GFP and the Baf60c expressing plasmid, BrdU+ cells were present in the NBL; however, cells in which Baf60c was over-expressed were more than twice as likely to incorporate BrdU as those cells transfected with the GFP-expressing vector alone (Figure 5A–G). We quantified this effect by counting all GFP expressing cells in control and Baf60c transfected explants and then counting how many of those were mitotic by looking at BrdU co-expression. Thus, these data show Baf60c behaves
Figure 5.
Baf60c over-expression maintains progenitor proliferation. (A–C) Control plasmid and GFP transfection of newborn mouse retina showing GFP (green, B) and BrdU (red, C) with merged overlap in (A). (D–F) Baf60c plasmid overexpression along with GFP (green) shows more BrdU (red) co-expressing cells (arrowheads). (G) Quantitation of the above effect showing almost twice as many proliferating transfected cells upon Baf60c overexpression.
BAF60c in proliferating Muller glia
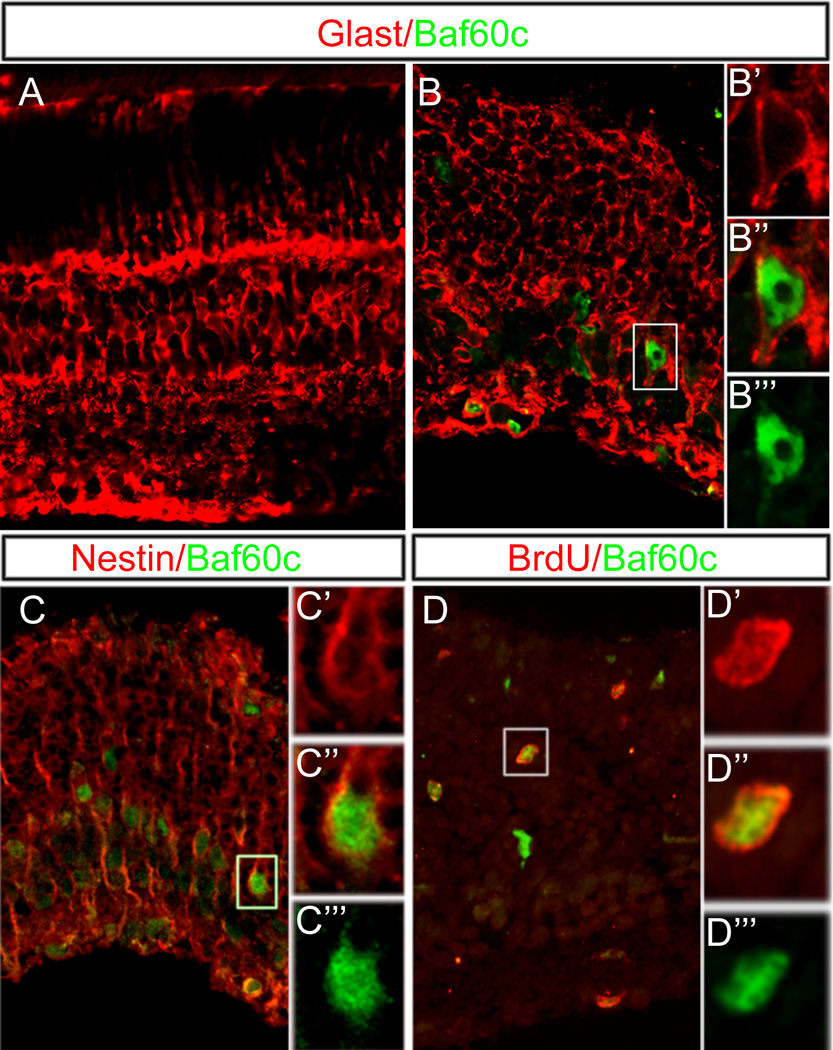
A number of reports in fish, bird and mammals have shown that the Müller glia, the resident glia in the retina, have the ability to re-enter the cell cycle following damage and adopt a progenitor identity (see Lamba et al, 2008 for review). Such a change could potentially involve remodeling of the chromatin network and the re-expression of chromatin remodeling factors normally only present in retinal progenitors. To determine whether Müller glia re-express the npBAF, BAF60c when induced to proliferate after neurotoxic damage, we carried out the following experiment. The Müller glia were induced to proliferate by causing retinal damage using intra-ocular NMDA, followed by daily intra-ocular dose of FGF-1 and insulin (Karl et al, 2008). All proliferating cells were then identified by giving the mice BrdU daily intra-peritoneally. On analyzing the retinas six days later, we found Müller glia (identified by GLAST expression) now express BAF60c (Figure 6B–B’’’). Müller glia in the control eyes which were not damaged were not labeled with the anti-BAF60c antibodies (Figure 6A). Along with the expression of BAF60c, many of the Müller glia were also immunoreactive for nestin, consistent with their de-differentiation towards a more progenitor-like state (Figure 6C–C’’’). We also labeled sections from the retinas treated with NMDA and FGF1/insulin and found BAF60c/BrdU co-expression (Figure 6D–D’’’).
Figure 6.
BAF60c is re-expressed during mouse retinal regeneration. (A, B) Retinal sections from control (A) and damaged retina (B) stained for Glast (red) and BAF60c (green). BAF60c is normally not expressed in adult retina, but is re-expressed in Muller in glia following NMDA and growth factor treatment. (B'–B''') High power view of boxed region in (B) showing BAF60c in Glast+ cell. These cells re-expressing BAF60c (green) express the progenitor marker, nestin (red, C–C''') and incorporate BrdU (red, D–D''').
Discussion
In this report, we find that one of the core subunits of the SWI/SNF complex, BAF60c, is specifically expressed in progenitors in the developing retina, in both embryonic human and mouse. We also find that of the two ATPase subunits in the complex, Brg1, but not Brm, is expressed in the retinal progenitors. We confirm and extend the findings of Lessard et al (2007) that BAF53a is expressed in neural progenitors, though in the retina we find that it is also expressed in some differentiated neurons. Consistent with its neural progenitor expression, we find that over-expression of BAF60c in the progenitors of the developing mouse retina leads to an increase in the percentage of cells that incorporate BrdU, similar to that reported for BAF53a and BAF45a over-expression (Lessard et al., 2007). Finally, we show that while BAF60c is not normally present in adult mouse retina, stimulation of proliferation by NMDA induced damage and intraocular growth factor injections leads to re-expression of BAF60c in Müller glia. These findings lend further support for a critical role of chromatin remodeling factors in retinal development and potentially regeneration.
SWI/SNF complex in retinal progenitors
Previous studies have implicated the SWI/SNF complex in retinal development. In the Zebrafish, the young mutation (which causes a premature truncation of Brg1) has a severe retinal defect; the overall organization and lamination of the retina is severely disrupted (Link et al., 2000; Gregg et al., 2003). Das et al (Das et al., 2007) have reported that Brm is also necessary for retinal development; although there are no large disruptions in retinal lamination or cell number; there is a small, but significant decline in the number of retinal ganglion cells in the Brm deficient mice. In general, Brg1 appears to be much more critical than Brm for embryogenesis; mice deficient in Brm are viable and fertile, while those deficient in Brg1 have early embryonic lethality (Bultman et al, 2000). While no study has analyzed the effects of loss of Brg1 in the mammalian retina, the effects in the rest of the CNS are dramatic. Conditional deletion of Brg1 in neural progenitor cells in mice results in reduction in proliferation throughout the CNS precocious neuronal differentiation, and a reduction in astrocyte and oligodendrocyte differentiation (Matsumoto et al., 2006). The much greater requirement for Brg1 than Brm may also relate to their expression; Brm and Brg1 are both expressed in neurons, while only Brg1 is expressed in progenitors (Randazzo et al., 1994; Schofield et al., 1999; Machida et al., 2001)(this study).
The roles of the SWI/SNF complex in neural development
Some of the first studies to implicate the SWI/SNF complex in the regulation of gene expression in the developing nervous system were those in developing Xenopus. Inhibition of Brg1 during Xenopus development inhibits neuronal differentiation from progenitor cells; proneural transcription factors require Brg1, potentially mediated through a direct physical association (Seo et al., 2005). However, more recent studies in mice have shown a very different phenotype: in mouse nervous system, loss of Brg1 leads to a premature neuronal differentiation, a loss of stem/progenitor proliferation, and a decrease in glial development (Matsumoto et al., 2006; Lessard et al., 2007). The difference in results may be due either to differences in the requirement for Brg1 in neurogenesis in mammals and frogs, or to technical differences, since the loss of function studies in frog were done primarily with dominant-negative approaches. More recently, Lessard et al (2007) reported that a specific complex of BAFs, along with Brg1, act in neural stem/progenitors to maintain their specific pattern of gene expression; they named this complex the npBAF complex, to contrast it with a similar, but distinct complex present in differentiated neurons. BAF45a over-expression in transgenic mice led to a 2–4 fold increase in mitotic cells in the ventricular zone in cerebral cortex, while overexpression of BAF45a and BAF53a in chick spinal cord both stimulated progenitor proliferation and prevented their differentiation into motor-neurons. These experiments support their hypothesis that a change in the composition of the subunits of this complex, from BAF53a/45a to BAF53b/45b, is necessary for progenitors to differentiate into neurons.
Our results are consistent with these findings, and extend this idea to include another BAF, BAF60c in the control of progenitor-specific gene expression. In this regard, over-expression of BAF45a, BAF53a (Lessard et al., 2007) and BAF60c (this study) all promote neural stem/progenitor proliferation, potentially through the Notch pathway (see below). We find that BAF60c expression is almost exclusive to the neural progenitors in the retina, and propose that its role as another member of the npBAF complex may be true of other regions of the CNS as well. Debril et al (Debril et al., 2004) reported that the embryonic brain was one of the highest regions of expression of Baf60c, though it is also expressed in other tissues. During embryogenesis in mice, the message is expressed in the proliferating ventricular zone cells. However, the same group reported expression in some types of neurons in the adult brain, and so it is possible that this BAF can be used in the neural complex as well in the mature brain, even though we find the expression highly specific to progenitors in the developing retina, cortex and spinal cord.
Notch signaling and BAF60c
The possibility that Notch signaling is regulated by the SWI/SNF complex has been examined in two previous studies. In both mice and zebrafish, Takeuchi et al (2007) found that BAF60c was necessary for the Notch mediated expression of nodal in the node. RNAi knockdown of Baf60c led to a loss in expression, while over-expression of Baf60c, along with Brg1, led to an increase in Notch signaling and a stabilization in the interaction between the Notch ICD and Rbp-J. While BAF60c and Brg1 are required for Notch signaling, Brm appears to inhibit it. Over-expression of Brm in embryonic mouse neurospheres caused a reduction in Hes1 and Hes5 expression, and a reduction in Notch-ICD stimulated Hes1 promoter driven luciferase activity in 293T cells (Das et al., 2007). We have found that Baf60c over-expression leads to an increase in Notch-ICD stimulated Hes1 and Hes5 promoter driven GFP (data not shown). Moreover, the phenotype of the conditional Brg1 knockout mouse is consistent with a reduction in Notch signaling (Matsumoto et al., 2006); reduction in progenitor markers, increased neuronal differentiation, and loss of glial differentiation are hallmarks of the loss of Notch function during CNS development (Nelson et al., 2007). Moreover, among the most significant changes in gene expression in the Brg1 conditional knock-out mouse brain were reductions in several components of the Notch pathway (Lessard et al., 2007).
Implications for regeneration
Despite the growing evidence for the importance of chromatin remodeling complexes in nervous system development, there is little known about their roles in regeneration. One of the few studies of the SWI/SNF complex during regeneration demonstrated that the decline in hepatic regeneration with aging is due a complex of cEBP-alpha and Brm that inhibits hepatocyte proliferation (Conboy et al., 2005). This increase in Brm in aged livers can be reversed through parabiosis with a younger animal, suggesting that chromatin remodeling complexes may play an important role in tissue regeneration. Studies in fish, birds and recently mammals have shown that Müller glia can de-differentiate following retinal damage to a retinal progenitor state, expressing proneural genes and components of the Notch pathway (Fischer and Reh, 2001; Fischer and Reh, 2003; Hitchcock et al., 2004; Ooto et al., 2004; Close et al., 2006; Raymond et al., 2006; Fimbel et al., 2007; Hayes et al., 2007). We therefore assayed whether BAF60c, an npBAF component was re-expressed in these cells as they de-differentiate. We found that the majority of Müller glia that had re-entered the mitotic cycle after retinal damage and growth factor stimulation also expressed BAF60c, though they did not do so with damage alone. These results thus show that at least one component of the neural progenitor BAF chromatin remodeling complex is re-expressed when mature Muller glia are stimulated to proliferate, and may provide a critical step for de-differentiation in this and other regenerating systems.
Methods
Mice
All experiments were done in accordance with approved protocols and the animals were housed and bred in the Department of Comparative Medicine at the University of Washington.
Human Fetal Eyes
Eyes from 98 day post-conception fetuses, without identifiers, were obtained from therapeutic abortions through fetal tissue bank at the University of Washington under approved protocols. Individual eyes were rinsed with sterile HBSS; retinas were then dissected from other ocular tissue and fixed in 4%PFA for 2 hours and subsequently embedded in OCT and cryosectioned.
Immunofluorescence
A standard immunofluorescence staining protocol was performed as described (Hayes et al, 2007). Antibodies used included: rabbit anti-BAF60c (1:1000, Aviva System), rabbit anti- BAF53a (1:500, ABCAM), rabbit anti-Brg1 (1:200, Santa Cruz), rabbit anti-Brm (1:500, gift from A. Imbalzano), mouse anti-Vimentin (1:50, DSHB), rat anti-BrdU (1:100, Accurate), goat anti-Sox2 (1:150, Santa Cruz), mouse anti-Tuj1 (1:750, Covance), and mouse anti-Hu C/D (1:100, Monoclonal Antibody Facility, University of Oregon). When rat anti-BrdU antibody was used, slides were treated with a 100Kunitz/mL DNAse (Sigma). Alexa-488 and Alexa-568 secondary antibodies (Invitrogen) used at 1:500. Alexa-350 (Invitrogen) secondary antibodies were used at 1:80.
Electroporation
Mouse retinal explants were prepared as described previously and electroporated with plasmids encoding GFP and either pCS2 empty vector (control) or Baf60c (Open Biosystems) driven by CMV. Retinas were collected in HBSS+, the extra-ocular tissue and pigment epithelium were removed. DNA was mixed with a glycerol-PBS/loading buffer and the DNA solution was pipetted over the retinas. Electrodes were positioned above and below the explants and the voltage applied (25 V, 5 pulses, 50 msec-interval using BTX ECM 830). The retinal explants were cultured in non-TC treated 24 well dishes in DMEM/F12 media supplemented with 0.6% glucose, 0.1125% NaHCO3, 5 mM HEPES, 1% fetal bovine serum (Gibco-BRL), penicillin (1 unit/ml) and streptomycin (1 µg/ml), and B-27 (Gibco-BRL) for 2 days. The retinal explants were then labeled with 1mg/ml BrdU for 2hours. The retinas were fixed for 30 minutes in 4% PFA and prepared for cryosectioning.
Cell Counts and Statistics
For Baf60c overexpression studies, cells were counted from > 5 sections from 3 different retinas electroporated with either Baf60c or pCS2. The sections were stained for BrdU and GFP. We counted the total number of GFP+ cells per section, and the number of the GFP+ cells which also stained for BrdU; the data were expressed as a percentage of BrdU+/GFP+ cells for each case. Data was analyzed for statistical significance using Graphpad Prism by performing a Student's T-test.
Quantitative PCR analysis
For quantitative PCR analysis, retinas from E13.5 and P0 were isolated and lysed with Trizol (Invitrogen). Genomic DNA was digested by DNAse (Invitrogen) and RNA was purified using an RNEasy column (Qiagen). cDNA was generated by oligo-dT primed reverse transcription reaction with Superscript II reverse transcriptase (Invitrogen); an RT-minus control was included for each sample. Quantative PCR was performed using an Opticon DNA Engine (Bio-Rad). The cDNAs were normalized to GAPDH levels. Primers sets: Brg Forward 5’-AACCAAAGCAACCATCGAAC, Brg Reverse 5’- TCTCCAGGGCTGTGTCTCTT, Brm Forward 5’-GGCACCCTAAAGCATTACCA, Brm Reverse 5’GCAATGGTCTGGATGGTCTT, Baf60c Forward 5’-ATGGCCGCGGACGAAGTTGCCGGA, Baf60c Reverse 5’-TCCGGTGCCACTCAACAAGG, Nrl Forward 5’-TCCCAGTCCCTTGGCTATGG, Nrl Reverse 5’-CACCGAGCTGTATGGTGTG, GAPDH Forward 5’-GGCATTGCTCTCAATGACAA, GAPDH Reverse 5’-CTTGCTCAGTGTCCTTGCTG, Hes1 Forward 5’-CCAGCCAGTGTCAACACGA, Hes1 Reverse 5’-AATGCCGGGAGCTATCTTTCT, Hes5 Forward 5’-AGTCCCAAGGAGAAAAACCGA, Hes5 Reverse 5’-GCTGTGTTTCAGGTAGCTGAC.
Müller glia proliferation
For in vivo studies, adult mice (P30) were anesthetized using 0.13mg ketamine per g body weight (BW) plus 8.8ug xylazine per g BW by an intraperitoneal injection. Proparacaine topical anesthetic was applied to the eyes prior to, and Bacitracin antibiotic ointment after injection. A 30-G needle was used to carefully make a small incision at the upper temporal ora serrata. Mice were aligned for injection using a stereotaxic instrument. Graded glass micropipettes with a fine tip aperture were used to inject the left eyes. Retinal neurotoxic damage was induced by injection of 2 µl of 0.1 M NMDA on day 0 (D0). Subsequently animals received daily (D2-5) intraocular treatment of FGF1 (100 ng /µl, R&D, bovine) and insulin (1 µg /ul, Sigma, USA, bovine) to stimulate Müller glia proliferation. Intraocular injection included 1mg /ml bromodeoxyuridine (BrdU) with a final volume of 2 µl /eye. Animals also received BrdU (50 µg /g BW) daily (D2-5) by intraperitoneal injection. Mice were sacrificed at D6 and processed for immunohistochemistry.
Acknowledgements
We would like to thank all members of the Reh Lab for their constructive comments and Paige Etter for her help during the experiments. We would like to thank Dr. A. Imbalzano (University of Massachusetts) for the Brm antibody, Dr. R. Kageyama for the Hes1 and Hes5 promoter fluorescent reporter constructs and Dr. R. Kopan for the Notch ICD construct. This work was supported by Vision Training Grant (2004–2005) to S.H., NRSA postdoctoral fellowship F32 EY016636 (2004–2008) to S.H., postdoctoral fellowship by the German Research Foundation (DFG, KA 2794/1-1) and ProRetina Travel Grants to M.O.K. and NIH grant EY13475 to T.R.
References
- Close JL, Liu J, Gumuscu B, Reh TA. Epidermal growth factor receptor expression regulates proliferation in the postnatal rat retina. Glia. 2006;54:94–104. doi: 10.1002/glia.20361. [DOI] [PubMed] [Google Scholar]
- Conboy IM, Conboy MJ, Wagers AJ, Girma ER, Weissman IL, Rando TA. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;433:760–764. doi: 10.1038/nature03260. [DOI] [PubMed] [Google Scholar]
- Das AV, James J, Bhattacharya S, Imbalzano AN, Antony ML, Hegde G, Zhao X, Mallya K, Ahmad F, Knudsen E, Ahmad I. SWI/SNF chromatin remodeling ATPase Brm regulates the differentiation of early retinal stem cells/progenitors by influencing Brn3b expression and Notch signaling. J Biol Chem. 2007;282:35187–35201. doi: 10.1074/jbc.M706742200. [DOI] [PubMed] [Google Scholar]
- Debril MB, Gelman L, Fayard E, Annicotte JS, Rocchi S, Auwerx J. Transcription factors and nuclear receptors interact with the SWI/SNF complex through the BAF60c subunit. J Biol Chem. 2004;279:16677–16686. doi: 10.1074/jbc.M312288200. [DOI] [PubMed] [Google Scholar]
- Fimbel SM, Montgomery JE, Burket CT, Hyde DR. Regeneration of inner retinal neurons after intravitreal injection of ouabain in zebrafish. J Neurosci. 2007;27:1712–1724. doi: 10.1523/JNEUROSCI.5317-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AJ, Reh TA. Muller glia are a potential source of neural regeneration in the postnatal chicken retina. Nat Neurosci. 2001;4:247–252. doi: 10.1038/85090. [DOI] [PubMed] [Google Scholar]
- Fischer AJ, Reh TA. Potential of Muller glia to become neurogenic retinal progenitor cells. Glia. 2003;43:70–76. doi: 10.1002/glia.10218. [DOI] [PubMed] [Google Scholar]
- Gregg RG, Willer GB, Fadool JM, Dowling JE, Link BA. Positional cloning of the young mutation identifies an essential role for the Brahma chromatin remodeling complex in mediating retinal cell differentiation. Proc Natl Acad Sci U S A. 2003;100:6535–6540. doi: 10.1073/pnas.0631813100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes S, Nelson BR, Buckingham B, Reh TA. Notch signaling regulates regeneration in the avian retina. Dev Biol. 2007;312:300–311. doi: 10.1016/j.ydbio.2007.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock P, Ochocinska M, Sieh A, Otteson D. Persistent and injury-induced neurogenesis in the vertebrate retina. Prog Retin Eye Res. 2004;23:183–194. doi: 10.1016/j.preteyeres.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link BA, Fadool JM, Malicki J, Dowling JE. The zebrafish young mutation acts non-cellautonomously to uncouple differentiation from specification for all retinal cells. Development. 2000;127:2177–2188. doi: 10.1242/dev.127.10.2177. [DOI] [PubMed] [Google Scholar]
- Machida Y, Murai K, Miyake K, Iijima S. Expression of chromatin remodeling factors during neural differentiation. J Biochem. 2001;129:43–49. doi: 10.1093/oxfordjournals.jbchem.a002834. [DOI] [PubMed] [Google Scholar]
- Martens JA, Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr Opin Genet Dev. 2003;13:136–142. doi: 10.1016/s0959-437x(03)00022-4. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Banine F, Struve J, Xing R, Adams C, Liu Y, Metzger D, Chambon P, Rao MS, Sherman LS. Brg1 is required for murine neural stem cell maintenance and gliogenesis. Dev Biol. 2006;289:372–383. doi: 10.1016/j.ydbio.2005.10.044. [DOI] [PubMed] [Google Scholar]
- Nelson BR, Hartman BH, Georgi SA, Lan MS, Reh TA. Transient inactivation of Notch signaling synchronizes differentiation of neural progenitor cells. Dev Biol. 2007;304:479–498. doi: 10.1016/j.ydbio.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooto S, Akagi T, Kageyama R, Akita J, Mandai M, Honda Y, Takahashi M. Potential for neural regeneration after neurotoxic injury in the adult mammalian retina. Proc Natl Acad Sci U S A. 2004;101:13654–13659. doi: 10.1073/pnas.0402129101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo FM, Khavari P, Crabtree G, Tamkun J, Rossant J. brg1: a putative murine homologue of the Drosophila brahma gene, a homeotic gene regulator. Dev Biol. 1994;161:229–242. doi: 10.1006/dbio.1994.1023. [DOI] [PubMed] [Google Scholar]
- Raymond PA, Barthel LK, Bernardos RL, Perkowski JJ. Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Dev Biol. 2006;6:36. doi: 10.1186/1471-213X-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield J, Isaac A, Golovleva I, Crawley A, Goodwin G, Tickle C, Brickell P. Expression of Drosophila trithorax-group homologues in chick embryos. Mech Dev. 1999;80:115–118. doi: 10.1016/s0925-4773(98)00207-x. [DOI] [PubMed] [Google Scholar]
- Seo S, Richardson GA, Kroll KL. The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development. 2005;132:105–115. doi: 10.1242/dev.01548. [DOI] [PubMed] [Google Scholar]
- Takeuchi JK, Lickert H, Bisgrove BW, Sun X, Yamamoto M, Chawengsaksophak K, Hamada H, Yost HJ, Rossant J, Bruneau BG. Baf60c is a nuclear Notch signaling component required for the establishment of left-right asymmetry. Proc Natl Acad Sci U S A. 2007;104:846–851. doi: 10.1073/pnas.0608118104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- Xi Q, He W, Zhang XH, Le HV, Massague J. Genome-wide impact of the BRG1 SWI/SNF chromatin remodeler on the transforming growth factor beta transcriptional program. J Biol Chem. 2008;283:1146–1155. doi: 10.1074/jbc.M707479200. [DOI] [PMC free article] [PubMed] [Google Scholar]