Abstract
Rap small GTPases regulate excitatory synaptic strength and morphological plasticity of dendritic spines. Changes in spine structure are mediated by the F-actin cytoskeleton, but the link between Rap activity and actin dynamics is unclear. Here we report a novel interaction between SPAR, a postsynaptic inhibitor of Rap, and α-actinin, a family of actin-crosslinking proteins. SPAR and α-actinin engage in bidirectional structural plasticity of dendritic spines: SPAR promotes spine head enlargement, whereas increased α-actinin2 expression favors dendritic spine elongation and thinning. Surprisingly, SPAR and α-actinin2 can function in an additive rather than antagonistic fashion at the same dendritic spine, generating combination spine/filopodia hybrids. These data identify a molecular pathway bridging the actin cytoskeleton and Rap at synapses, and suggest that formation of spines and filopodia are not necessarily opposing forms of structural plasticity.
Keywords: Rap small GTPase, actin cytoskeleton, hippocampal neurons, postsynaptic density, structural plasticity
INTRODUCTION
Dendritic spines are the primary sites of excitatory synaptic transmission in the CNS [1]. Spines are small, highly dynamic protrusions that are morphologically responsive to many types of environmental stimuli and patterns of synaptic activity, suggesting that changes in spine structure may underlie molecular encoding of information [2]. Spine morphology is developmentally regulated, progressing from thin filopodia-like structures in immature neurons to mushroom headed spines as neurons age [3], implicating filopodia as spine precursors. Functionally, spine head size is correlated with synaptic strength but inversely correlated with potential for further plasticity, and spines are dysmorphic in many dementias [4]. Thus, dendritic spine morphology may play critical roles in synaptic function and cognitive disorders.
Actin is the primary cytoskeleton in spines, and alterations in spine morphogenesis are intimately associated with regulation of actin dynamics [5]. Accordingly, several actin binding proteins have been implicated in synaptic function and plasticity [6]. SPAR (Spine-Associated Rap GTPase-activating protein (GAP)) is an attractive candidate linking synaptic activity to actin remodeling. SPAR binds to the postsynaptic density scaffold protein PSD-95 in association with NMDA receptors (NMDARs) in brain [7]. In addition, SPAR is highly associated with and induces dramatic reorganization of F-actin filaments, and promotes massive spine head growth [7]. SPAR also contains enzymatic GAP activity that inhibits Rap1 and Rap2, small GTPases involved in synaptic depression, depotentiation and spine shrinkage [7-12]. Inhibition of Rap, or remodeling of F-actin, may contribute to the ability of SPAR to cause spine enlargement.
Here we report that SPAR interacts with the actin cross-linking protein α-actinin. Actinin comprises a family of proteins that play multiple roles at synapses [13], in association with glutamate receptors [14, 15] and other excitatory synaptic proteins [16-19]. α-Actinin2 overexpression leads to elongation/thinning of dendritic protrusions and increased numbers of filopodia [20] at the expense of mature spines [21], in apparent opposition to the spine enlargement mediated by SPAR. We examined in detail the physical association of SPAR and α-actinin and determined their functional interaction in spine morphogenesis.
MATERIALS AND METHODS
Constructs
The following constructs have been described: PSD-95 PDZ1/2 in pGAD10 [22], α-actinin2 in pCDNA3 and NR1 C0-C1 tail in pBHA [14], myc-Act2 GW1 and myc-SPAR GW1 [7], and SPAR-Act2 in pBHA and SNK in pGAD10 [23]. Deletion constructs were generated by PCR and cloned into pGW1, pGAD10, or pBHA. Deletions for SPAR-Act2 domain were ΔN (aa1313-1505), ΔNC (aa1313-1415) and ΔC (aa1216-1415). All constructs were verified by DNA sequencing.
Yeast two hybrid
Two-hybrid screens and assays used the yeast strain L40 harboring β-gal and HIS3 reporters, as described [24]. 1 × 106 clones of a rat or human brain cDNA library in pGAD10 (Clontech) were screened against SPAR Act2 domain (aa1216-1505) in pBHA as bait.
Antibodies
The following antibodies have been described: rabbit-anti-SPAR [7]; rabbit Shank [25]. The following antibodies were purchased from commercial sources: myc 9E10, myc agarose conjugate, and goat anti-SPAR V20 (Santa Cruz Biotechnology); GFP monoclonal 3E6 (Quantum Biotechnologies); α-tubulin B-5-1-2 and α-actinin EA-53 monoclonals, and goat, mouse and rabbit IgG (Sigma).
COS-7 transfections, immunostaining, and immunoprecipitation
COS-7 cells were transfected using Lipofectamine (Invitrogen) according to the manufacturer’s protocol, and immunostained as described [22]. For immunoprecipitation, cells were harvested in RIPA buffer and lysates centrifuged at 16,000g for 15 minutes. Antibodies, or non-immune rabbit and mouse IgG/protein A or protein G sepharose conjugates were mixed with supernatants for 2 hr at 4°C. After washing 5x in RIPA buffer, immunoprecipitates were analyzed by immunoblotting using standard methods.
Brain immunoprecipitation
Immunoprecipitations from rat brain homogenates were performed as described [26] except that whole brain lysates were extracted by 1% sodium deoxycholate for 1 hr at 4°C. For each immunoprecipitation, clarified extract (200 μg protein) was incubated with 10 μg of desired antibodies (or nonimmune IgG) for 2 hr at 4°C. Precipitates were washed in 50 mM Tris pH 7.5, 150 mM NaCl, 0.1% Triton X-100.
Neuronal culture, transfection and immunocytochemistry
Hippocampal primary neuronal cultures prepared from embryonic day (E) 18-19 rat embryos were plated at medium density (~150 cells mm-2) as described [27]. Rats were treated in accordance with NIH and Georgetown guidelines on animal care and use. Neurons were transfected at ~14 days in vitro (DIV) using a modified calcium phosphate procedure [28]. Immunostaining was performed essentially as described [27] at 19 DIV. Secondary antibodies used were donkey-anti-rabbit AlexaFluor 555 (1:300) and donkey-anti-mouse AlexaFluor 488 (1:100) (Invitrogen).
Quantitation and image analysis
Images were acquired using a Fluoview confocal microscope (Olympus) or Axiovert 200M (Zeiss) using consistent laser intensity or camera exposure levels for each fluorophore in each experiment. Confocal z-series image stacks encompassing dendrite segments were analyzed using MetaMorph software (Universal Imaging Corporation). For morphological classification, filopodia were defined as dendritic protrusions >2 μm in length and < 0.6 μm in width. All other protrusions < 5 μm in length were classified as spines.
Statistical analyses
All data are expressed as mean ± standard error of the mean. Data were analyzed for multiple groups using one-way ANOVA with Tukey’s post hoc test or using Student’s t-test for two group comparisons. Significance was determined at p<0.05.
RESULTS AND DISCUSSION
To elucidate the mechanism by which SPAR remodels actin and spine morphology, yeast two-hybrid (Y2H) interaction screens were conducted with the SPAR Act2 domain as bait (Fig. 1A). Act2 was chosen because this region is sufficient for SPAR association with F-actin and is required for spine enlargement [7]. Two separate screens were performed, against either a rat or a human brain cDNA library. Two α-actinin family members were identified as extremely strong interactors in these screens, clone HB15 representing rat α-actinin-1 and clone HB20 encoding human α-actinin-2. The primary structure of α-actinin proteins consists of an N-terminal actin binding domain, a series of central spectrin repeats, an EF hand motif in the carboxy-terminal region, and a PDZ-domain binding ligand at the very C-terminal tail [29]. DNA sequencing revealed that HB15 and HB20 encompassed strikingly similar coding regions, spanning from spectrin repeat 2 to the C terminus (Fig. 1A). Further Y2H deletion analysis showed that the α-actinin domains required for interaction with SPAR could be narrowed down to spectrin repeats 2-4 (Fig. 1A), a region that also binds to the C0-C1 exons of the NMDAR subunit NR1 C-terminal tail [14].
Figure 1. Identification of α-actinin interaction with SPAR.
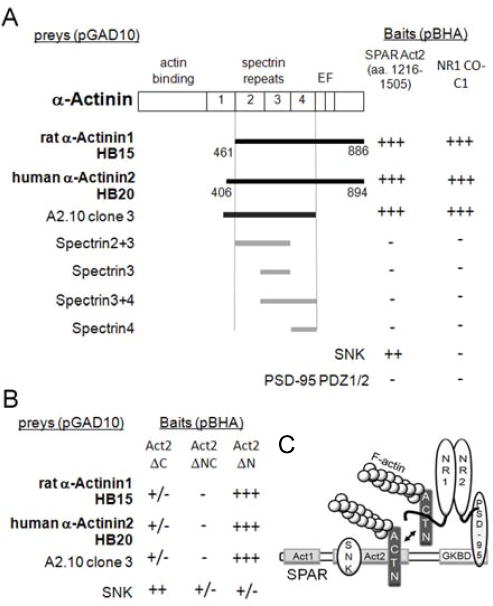
(A) Y2H screens were performed using SPAR Act2 domain as bait in vector pBHA. Interaction strength was scored by β-gal activity (+++, 0-30 min; +, 30 min-2 hrs; -, >12 hrs). Interaction time for α-actinin-SPAR was consistently under 10 minutes. Residues encompassed by positive clones (HB15 and HB20) and α-actinin deletion constructs are indicated below α-actinin schematic diagram (interacting clones shown in black). SNK in pGAD10 vector was positive control for interaction with Act2-pBHA; NR1 C0-C1 exon-containing C-terminus in pBHA was positive control for the α-actinin interactions. PSD-95 PDZ1/2 in pGAD10 was negative control for all pBHA constructs.
(B) α-Actinins bind specifically to the C-terminal portion of Act2 (Act2ΔN), whereas SNK binds primarily to the N-terminal part (Act2ΔC).
(C) Schematic of SPAR domains and interactions. Act2, actin-associated domain 2. GKBD, guanylate kinase binding domain. ACTN, α-actinin. Double headed arrow indicates SPAR binding to α-actinin may displace NR1.
Conversely, deletion mapping of SPAR showed that clones HB15 and HB20 both bound preferentially to the C-terminal part of the SPAR Act2 domain (Act2ΔN) (Fig. 1B). In contrast, the activity-inducible polo kinase SNK/Plk2, which targets SPAR for degradation via phosphorylation-dependent ubiquitination [23, 30, 31] and also interacts with the Act2 domain [23], bound preferentially to the N-terminal portion (Act2ΔC). Such adjacent Act2 binding sites suggest that SNK and α-actinin could bind SPAR simultaneously and independently (Fig. 1B; see Fig. 1C for schematic network of proposed interactions).
Coimmunoprecipitation and colocalization of actinin and SPAR
To confirm the Y2H results, lysates from adult rat whole brain were detergent extracted using 1% deoxycholate and subjected to immunoprecipitation. Native SPAR and α-actinin (detected using pan-α-actinin antiserum that is nonselective for α-actinin family members) were robustly co-immunoprecipitated from brain lysates using a goat SPAR antibody, but not by nonimmune goat IgG (Fig. 2A). As another specificity control, immunoblots were stripped and reprobed for tubulin, but no association with SPAR was observed (Fig. 2A). Thus, the co-immunoprecipitation of α-actinin and SPAR was unlikely to be due to nonspecific cytoskeletal aggregation. We also performed the reciprocal coimmunoprecipitation, using monoclonal α-actinin antibodies. These antibodies efficiently precipitated brain α-actinin as well as a substantial amount of SPAR (Fig. 2B). Furthermore, COS-7 cells were cotransfected with myc epitope-tagged SPAR and α-actinin-2. The full-length recombinant proteins could be readily co-immunoprecipitated in the absence of neuron-specific proteins (Fig. 2C).
Figure 2. Biochemical and immunocytochemical association of SPAR and α-actinin.
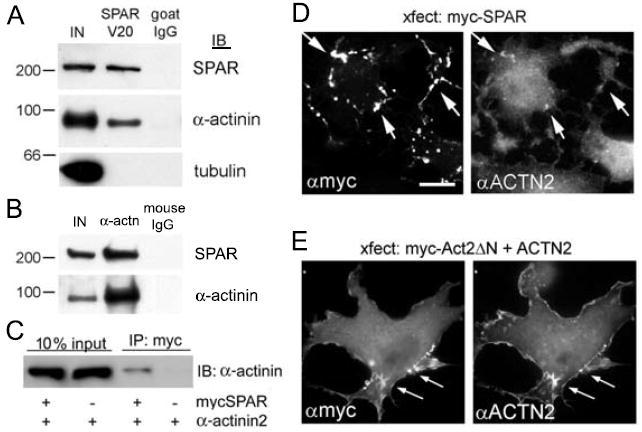
(A) Rat brain lysates were immunoprecipitated with SPAR V20 goat antibodies or with nonimmune goat IgG control, and precipitates were immunoblotted for SPAR, α-actinin and tubulin as indicated.
(B) Rat brain lysates were immunoprecipitated with mouse monoclonal antibodies to α-actinin or with nonimmune mouse IgG control, and precipitates were immunoblotted for SPAR and α-actinin. IN represents 10% of input used in each immunoprecipitation reaction in (A, B).
(C) COS-7 cells transfected with myc-tagged SPAR or α-actinin2 as indicated were immunoprecipitated with myc agarose and immunoblotted for α-actinin. 10% of input is shown as expression control. (D-E) COS-7 cells transfected with myc-tagged SPAR alone (D) or with mycAct2ΔN domain of SPAR together with α-actinin2 (E). Cells were immunostained with myc and α-actinin antibodies, as indicated. Arrows show examples of colocalization. Scale, 5 μm.
Next, we performed immunocytochemistry of heterologous cells. As reported previously, recombinant SPAR overexpressed in COS-7 cells formed self-aggregating clusters (Fig. 2D) that reorganize and recruit F-actin [7]. Endogenous α-actinin also became recruited to these SPAR clusters (Fig. 2D, arrows). When the minimal α-actinin binding Act2ΔN portion of SPAR was cotransfected with α-actinin2, the exogenous proteins colocalized even more robustly (Fig. 2E).
We then determined the degree of colocalization of native proteins using immunocytochemistry of primary cultured hippocampal neurons. Extensive colocalization of α-actinin with SPAR was observed in a punctate pattern and at the tips of dendritic spines (Fig. 3A-C and G, quantified in I, J). However, many SPAR puncta did not overlap with α-actinin (Fig. 3C, arrowhead), and α-actinin puncta often did not colocalize with SPAR (Fig. 3C, arrow). A similar partial colocalization existed between α-actinin and the excitatory synaptic marker Shank (Fig. 3D-F and H, quantified in I, J). Taken together, these data have identified a novel protein complex between SPAR and α-actinin that was readily observed in a variety of biochemical, genetic, and immunocytochemical assays, consistent with a robust, high affinity interaction.
Figure 3. Colocalization of SPAR and α-actinin in neurons.
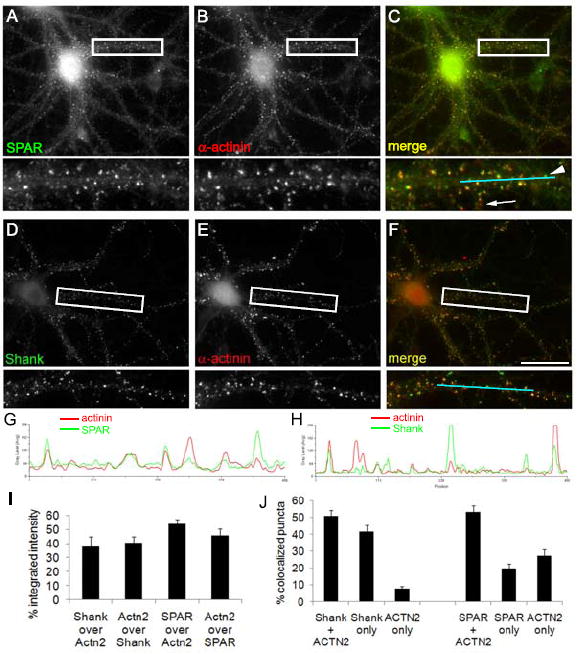
(A-F) Cultured hippocampal neurons at 19 DIV were double immunostained for proteins as indicated, with merged images shown at right (C, F). Boxed regions are shown below each image at higher magnification. Arrow, example of α-actinin cluster lacking SPAR colocalization. Arrowhead, example SPAR cluster lacking α-actinin colocalization. Scale, 20 μm. (G, H) Linescans of cyan segments shown in (C, F) demonstrating partial colocalization of α-actinin with SPAR (G) and with Shank (H). (I) Pixel colocalization between Shank/α-actinin or SPAR/α-actinin, calculated as the percentage of the integrated intensity of the first marker overlapping with the integrated intensity of the second marker. (J) Percentage of puncta colocalized between Shank/α-actinin and SPAR/α-actinin. ‘Marker only’ represents puncta that contain only the marker indicated out of the pair examined.
Combinatorial regulation of spine morphology
Previously, both α-actinin2 and SPAR were implicated in shaping dendritic spine morphology, albeit in different ways. α-Actinin promotes thinning and elongation of spines [21]. In contrast, SPAR causes spine head enlargement, with increased abundance of glutamate receptors and other synaptic markers [7]. We initially confirmed both of these findings (Fig. 4B-C, quantified in F-G). These properties implied that SPAR and α-actinin2 may drive opposing morphological changes, the former toward more stable and larger spines and synapses, and the latter toward more dynamic and immature filopodia with smaller and weaker synapses. This potential antagonism was also suggested by the similarity between boosting α-actinin-2 levels and blocking SPAR with dominant negative constructs [7].
Figure 4. SPAR and α-actinin2 regulation of dendritic spine morphogenesis.
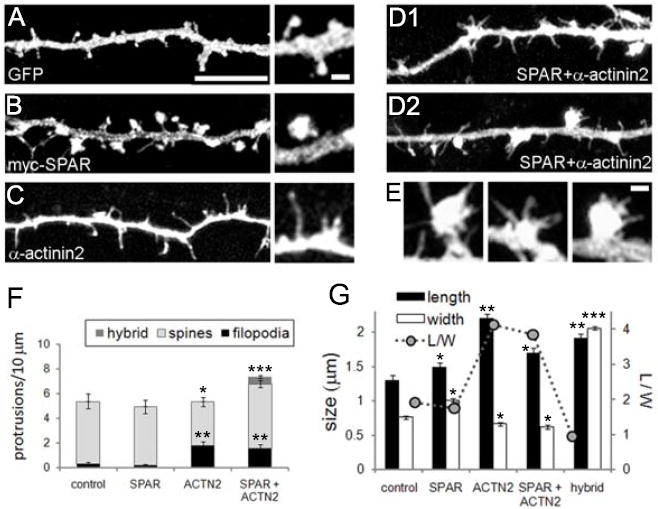
(A-E) Cultured hippocampal neurons (15 DIV) were transfected with DNA constructs as indicated and with pEGFP to visualize neuronal morphology. Insets show higher magnification views of representative dendritic protrusions. (E) Representative hybrid spine/filopodia from transfected neurons in (D), shown at same scale as higher magnification insets in (A-C). Scale bars: 5 μm for dendrite images and 1 μm for all higher magnification images. (F) Quantification of dendritic spine morphological classifications as indicated. Hybrid refers to combination spine/filopodia protrusions. (G) Dendritic protrusion length, width, and length-to-width ratio (L/W) for transfected neurons as indicated. *p<0.05, **p<0.01, ***p<0.001, compared to control.
Thus, we cotransfected hippocampal neurons with both SPAR and α-actinin-2 to test whether their actions were mutually antagonistic, and predicted that their morphogenetic effects would cancel out, or that one would predominate. Instead, we found that the coexpression of α-actinin-2 and SPAR resulted in a mixed phenotype, with highly enlarged spines formed in addition to overly abundant filopodia (Fig. 4D, quantified in F-G). This result could represent a situation in which SPAR ‘won’ at certain synapses, yielding large spines, but ‘lost’ to actinin at other synapses, resulting in filopodial transformation. However, it is not known what factors influence one outcome over the other.
Unexpectedly, we observed that many of the greatly enlarged dendritic spines sprouted multiple filopodia directly from the head itself (Fig. 4E), a ‘hybrid’ phenomenon that was rarely observed in control neurons (Fig. 4F). Thus, spine and filopodia morphogenetic pathways controlled by SPAR and α-actinin are not mutually exclusive, even at the same locus. It is presently unclear whether such hybrid spine/filopodia (‘spinopodia’) are physiological. These structures could be artifacts of misregulated overexpression of α-actinin and SPAR; perhaps mechanisms exist normally to prevent both proteins from being active simultaneously at the same synapse. On the other hand, filopodia atop spines are reminiscent of spinules [32, 33], thin protrusions on spines observed in vivo that penetrate into presynaptic terminals and may play a role in trans-synaptic retrograde signaling [34]. Thus, the extraordinary spine plasticity we observed here could be exaggerated versions of physiologically relevant processes. Regardless, our data extend the traditional view of the relationship between spines and filopodia, and argue that filopodia cannot simply be precursors of spines or products of spine shrinkage.
In addition to SPAR, α-actinins interact with several other synaptic proteins and likely play a wide variety of roles at synapses [16-19, 35]. As an actin-crosslinking protein, α-actinin may directly link SPAR to actin to enable the Act2 domain to remodel F-actin cytoskeleton. α-Actinin-2 also binds to NR1 via the same region as it binds to SPAR (spectrin repeats 2-4)[14]. Because of the known role of α-actinin in calcium-dependent inactivation of NMDARs [36, 37], it will be interesting to examine whether SPAR regulates this process by competitive interference with the α-actinin-NR1 interaction.
A final intriguing possibility is suggested by the observation that α-actinin is associated with numerous receptors that activate Rap in different contexts, including integrins [38, 39], NMDARs [12, 14, 37], cadherins [40, 41], insulin receptors [42, 43], and mGluRs [44, 45]. It is thus tempting to speculate that the α-actinin-SPAR complex may act as a widespread convergence point and general tethering mechanism between cell surface receptors, intracellular Rap signaling, and actin remodeling.
Acknowledgments
This work was supported by NIH/NINDS grant NS048085 (D.P.). We thank members of the Pak laboratory for critical reading of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bourne JN, Harris KM. Balancing structure and function at hippocampal dendritic spines. Annu Rev Neurosci. 2008;31:47–67. doi: 10.1146/annurev.neuro.31.060407.125646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Roo M, Klauser P, Garcia PM, Poglia L, Muller D. Spine dynamics and synapse remodeling during LTP and memory processes. Prog Brain Res. 2008;169:199–207. doi: 10.1016/S0079-6123(07)00011-8. [DOI] [PubMed] [Google Scholar]
- 3.Ethell IM, Pasquale EB. Molecular mechanisms of dendritic spine development and remodeling. Prog Neurobiol. 2005;75:161–205. doi: 10.1016/j.pneurobio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Bourne J, Harris KM. Do thin spines learn to be mushroom spines that remember? Curr Opin Neurobiol. 2007;17:381–386. doi: 10.1016/j.conb.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Cingolani LA, Goda Y. Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nat Rev Neurosci. 2008;9:344–356. doi: 10.1038/nrn2373. [DOI] [PubMed] [Google Scholar]
- 6.Tada T, Sheng M. Molecular mechanisms of dendritic spine morphogenesis. Curr Opin Neurobiol. 2006;16:95–101. doi: 10.1016/j.conb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Pak DT, Yang S, Rudolph-Correia S, Kim E, Sheng M. Regulation of dendritic spine morphology by SPAR, a PSD-95-associated RapGAP. Neuron. 2001;31:289–303. doi: 10.1016/s0896-6273(01)00355-5. [DOI] [PubMed] [Google Scholar]
- 8.Zhu JJ, Qin Y, Zhao M, Van Aelst L, Malinow R. Ras and Rap control AMPA receptor trafficking during synaptic plasticity. Cell. 2002;110:443–455. doi: 10.1016/s0092-8674(02)00897-8. [DOI] [PubMed] [Google Scholar]
- 9.Zhu Y, Pak D, Qin Y, McCormack SG, Kim MJ, Baumgart JP, Velamoor V, Auberson YP, Osten P, van Aelst L, Sheng M, Zhu JJ. Rap2-JNK removes synaptic AMPA receptors during depotentiation. Neuron. 2005;46:905–916. doi: 10.1016/j.neuron.2005.04.037. [DOI] [PubMed] [Google Scholar]
- 10.Fu Z, Lee SH, Simonetta A, Hansen J, Sheng M, Pak DT. Differential roles of Rap1 and Rap2 small GTPases in neurite retraction and synapse elimination in hippocampal spiny neurons. J Neurochem. 2007;100:118–131. doi: 10.1111/j.1471-4159.2006.04195.x. [DOI] [PubMed] [Google Scholar]
- 11.Ryu J, Futai K, Feliu M, Weinberg R, Sheng M. Constitutively active Rap2 transgenic mice display fewer dendritic spines, reduced extracellular signal-regulated kinase signaling, enhanced long-term depression, and impaired spatial learning and fear extinction. J Neurosci. 2008;28:8178–8188. doi: 10.1523/JNEUROSCI.1944-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie Z, Huganir RL, Penzes P. Activity-dependent dendritic spine structural plasticity is regulated by small GTPase Rap1 and its target AF-6. Neuron. 2005;48:605–618. doi: 10.1016/j.neuron.2005.09.027. [DOI] [PubMed] [Google Scholar]
- 13.Shirao T, Sekino Y. Clustering and anchoring mechanisms of molecular constituents of postsynaptic scaffolds in dendritic spines. Neurosci Res. 2001;40:1–7. doi: 10.1016/s0168-0102(01)00209-7. [DOI] [PubMed] [Google Scholar]
- 14.Wyszynski M, Lin J, Rao A, Nigh E, Beggs AH, Craig AM, Sheng M. Competitive binding of alpha-actinin and calmodulin to the NMDA receptor. Nature. 1997;385:439–442. doi: 10.1038/385439a0. [DOI] [PubMed] [Google Scholar]
- 15.Nuriya M, Oh S, Huganir RL. Phosphorylation-dependent interactions of alpha-Actinin-1/IQGAP1 with the AMPA receptor subunit GluR4. J Neurochem. 2005;95:544–552. doi: 10.1111/j.1471-4159.2005.03410.x. [DOI] [PubMed] [Google Scholar]
- 16.Walikonis RS, Oguni A, Khorosheva EM, Jeng CJ, Asuncion FJ, Kennedy MB. Densin-180 forms a ternary complex with the (alpha)-subunit of Ca2+/calmodulin-dependent protein kinase II and (alpha)-actinin. J Neurosci. 2001;21:423–433. doi: 10.1523/JNEUROSCI.21-02-00423.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robison AJ, Bass MA, Jiao Y, MacMillan LB, Carmody LC, Bartlett RK, Colbran RJ. Multivalent interactions of calcium/calmodulin-dependent protein kinase II with the postsynaptic density proteins NR2B, densin-180, and alpha-actinin-2. J Biol Chem. 2005;280:35329–35336. doi: 10.1074/jbc.M502191200. [DOI] [PubMed] [Google Scholar]
- 18.Schulz TW, Nakagawa T, Licznerski P, Pawlak V, Kolleker A, Rozov A, Kim J, Dittgen T, Kohr G, Sheng M, Seeburg PH, Osten P. Actin/alpha-actinin-dependent transport of AMPA receptors in dendritic spines: role of the PDZ-LIM protein RIL. J Neurosci. 2004;24:8584–8594. doi: 10.1523/JNEUROSCI.2100-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia H, Winokur ST, Kuo WL, Altherr MR, Bredt DS. Actinin-associated LIM protein: identification of a domain interaction between PDZ and spectrin-like repeat motifs. J Cell Biol. 1997;139:507–515. doi: 10.1083/jcb.139.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- 21.Nakagawa T, Engler JA, Sheng M. The dynamic turnover and functional roles of alpha-actinin in dendritic spines. Neuropharmacology. 2004;47:734–745. doi: 10.1016/j.neuropharm.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 22.Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 23.Pak DT, Sheng M. Targeted protein degradation and synapse remodeling by an inducible protein kinase. Science. 2003;302:1368–1373. doi: 10.1126/science.1082475. [DOI] [PubMed] [Google Scholar]
- 24.Niethammer M, Sheng M. Identification of ion channel-associated proteins using the yeast two-hybrid system. Methods Enzymol. 1998;293:104–122. doi: 10.1016/s0076-6879(98)93010-5. [DOI] [PubMed] [Google Scholar]
- 25.Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, Weinberg RJ, Worley PF, Sheng M. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- 26.Dunah AW, Luo J, Wang YH, Yasuda RP, Wolfe BB. Subunit composition of N-methyl-D-aspartate receptors in the central nervous system that contain the NR2D subunit. Mol Pharmacol. 1998;53:429–437. doi: 10.1124/mol.53.3.429. [DOI] [PubMed] [Google Scholar]
- 27.Sala C, Rudolph-Correia S, Sheng M. Developmentally regulated NMDA receptor-dependent dephosphorylation of cAMP response element-binding protein (CREB) in hippocampal neurons. J Neurosci. 2000;20:3529–3536. doi: 10.1523/JNEUROSCI.20-10-03529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang M, Chen G. High Ca2+-phosphate transfection efficiency in low-density neuronal cultures. Nat Protoc. 2006;1:695–700. doi: 10.1038/nprot.2006.86. [DOI] [PubMed] [Google Scholar]
- 29.Otey CA, Carpen O. Alpha-actinin revisited: a fresh look at an old player. Cell Motil Cytoskeleton. 2004;58:104–111. doi: 10.1002/cm.20007. [DOI] [PubMed] [Google Scholar]
- 30.Seeburg DP, Feliu-Mojer M, Gaiottino J, Pak DT, Sheng M. Critical role of CDK5 and Polo-like kinase 2 in homeostatic synaptic plasticity during elevated activity. Neuron. 2008;58:571–583. doi: 10.1016/j.neuron.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ang XL, Seeburg DP, Sheng M, Harper JW. Regulation of postsynaptic RapGAP SPAR by Polo-like kinase 2 and the SCFbeta-TRCP ubiquitin ligase in hippocampal neurons. J Biol Chem. 2008;283:29424–29432. doi: 10.1074/jbc.M802475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner HJ, Djamgoz MB. Spinules: a case for retinal synaptic plasticity. Trends Neurosci. 1993;16:201–206. doi: 10.1016/0166-2236(93)90155-f. [DOI] [PubMed] [Google Scholar]
- 33.Sorra KE, Fiala JC, Harris KM. Critical assessment of the involvement of perforations, spinules, and spine branching in hippocampal synapse formation. J Comp Neurol. 1998;398:225–240. [PubMed] [Google Scholar]
- 34.Spacek J, Harris KM. Trans-endocytosis via spinules in adult rat hippocampus. J Neurosci. 2004;24:4233–4241. doi: 10.1523/JNEUROSCI.0287-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakagawa N, Hoshijima M, Oyasu M, Saito N, Tanizawa K, Kuroda S. ENH, containing PDZ and LIM domains, heart/skeletal muscle-specific protein, associates with cytoskeletal proteins through the PDZ domain. Biochem Biophys Res Commun. 2000;272:505–512. doi: 10.1006/bbrc.2000.2787. [DOI] [PubMed] [Google Scholar]
- 36.Zhang S, Ehlers MD, Bernhardt JP, Su CT, Huganir RL. Calmodulin mediates calcium-dependent inactivation of N-methyl-D-aspartate receptors. Neuron. 1998;21:443–453. doi: 10.1016/s0896-6273(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 37.Krupp JJ, Vissel B, Thomas CG, Heinemann SF, Westbrook GL. Interactions of calmodulin and alpha-actinin with the NR1 subunit modulate Ca2+-dependent inactivation of NMDA receptors. J Neurosci. 1999;19:1165–1178. doi: 10.1523/JNEUROSCI.19-04-01165.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otey CA, Pavalko FM, Burridge K. An interaction between alpha-actinin and the beta 1 integrin subunit in vitro. J Cell Biol. 1990;111:721–729. doi: 10.1083/jcb.111.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cingolani LA, Thalhammer A, Yu LM, Catalano M, Ramos T, Colicos MA, Goda Y. Activity-dependent regulation of synaptic AMPA receptor composition and abundance by beta3 integrins. Neuron. 2008;58:749–762. doi: 10.1016/j.neuron.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knudsen KA, Soler AP, Johnson KR, Wheelock MJ. Interaction of alpha-actinin with the cadherin/catenin cell-cell adhesion complex via alpha-catenin. J Cell Biol. 1995;130:67–77. doi: 10.1083/jcb.130.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asuri S, Yan J, Paranavitana NC, Quilliam LA. E-cadherin dis-engagement activates the Rap1 GTPase. J Cell Biochem. 2008 doi: 10.1002/jcb.21902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Talior-Volodarsky I, Randhawa VK, Zaid H, Klip A. Alpha-actinin-4 is selectively required for insulin-induced GLUT4 translocation. J Biol Chem. 2008;283:25115–25123. doi: 10.1074/jbc.M801750200. [DOI] [PubMed] [Google Scholar]
- 43.Okada S, Matsuda M, Anafi M, Pawson T, Pessin JE. Insulin regulates the dynamic balance between Ras and Rap1 signaling by coordinating the assembly states of the Grb2-SOS and CrkII-C3G complexes. Embo J. 1998;17:2554–2565. doi: 10.1093/emboj/17.9.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cabello N, Remelli R, Canela L, Soriguera A, Mallol J, Canela EI, Robbins MJ, Lluis C, Franco R, McIlhinney RA, Ciruela F. Actin-binding protein alpha-actinin-1 interacts with the metabotropic glutamate receptor type 5b and modulates the cell surface expression and function of the receptor. J Biol Chem. 2007;282:12143–12153. doi: 10.1074/jbc.M608880200. [DOI] [PubMed] [Google Scholar]
- 45.Huang CC, You JL, Wu MY, Hsu KS. Rap1-induced p38 mitogen-activated protein kinase activation facilitates AMPA receptor trafficking via the GDI.Rab5 complex. Potential role in (S)-3,5-dihydroxyphenylglycene-induced long term depression. J Biol Chem. 2004;279:12286–12292. doi: 10.1074/jbc.M312868200. [DOI] [PubMed] [Google Scholar]


