Abstract
Background and Aims
Land-use changes and associated extinction/colonization dynamics can have a large impact on population genetic diversity of plant species. The aim of this study was to investigate genetic diversity in a founding population of the self-incompatible forest herb Primula elatior and to elucidate the processes that affect genetic diversity shortly after colonization.
Methods
AFLP markers were used to analyse genetic diversity across three age classes and spatial genetic structure within a founding population of P. elatior in a recently established stand in central Belgium. Parentage analyses were used to assess the amount of gene flow from outside the population and to investigate the contribution of mother plants to future generations.
Results
The genetic diversity of second and third generation plants was significantly reduced compared with that of first generation plants. Significant spatial genetic structure was observed. Parentage analyses showed that <20 % of the youngest individuals originated from parents outside the study population and that >50 % of first and second generation plants did not contribute to seedling recruitment.
Conclusions
These results suggest that a small effective population size and genetic drift can lead to rapid decline of genetic diversity of offspring in founding populations shortly after colonization. This multigenerational study also highlights that considerable amounts of gene flow seem to be required to counterbalance genetic drift and to sustain high levels of genetic diversity after colonization in recently established stands.
Key words: AFLP, colonization, forest regeneration, genetic diversity, genetic drift, parentage analysis, spatial genetic structure
INTRODUCTION
In fragmented landscapes, the degree of genetic differentiation among populations and the genetic diversity maintained within local populations depend on the history of colonization/extinction dynamics within the landscape (Pannell and Dorken, 2006), the amount of gene flow between populations (Sork and Smouse, 2006) and the effective size of individual populations. Whereas the effects of population turnover on inter-population genetic differentiation have been relatively well studied empirically (e.g. Whitlock, 1992; McCauley et al., 1995; Giles and Goudet, 1997; Ingvarsson et al., 1997; Jacquemyn et al., 2004, 2006; Vandepitte et al., 2007), there are few studies that have focused on estimating the effects of population turnover on the distribution of neutral genetic variation within single populations (Pannell and Charlesworth, 2000).
Modelling studies have shown that population turnover is expected to result in a decline of within-population genetic diversity, due to genetic bottlenecks or founder events (Slatkin, 1977; Pannell and Charlesworth, 1999). Additional factors that can affect the dynamics of genetic variation after colonization are the amount of gene flow among populations (Pannell and Charlesworth, 1999; Sork and Smouse, 2006), the effective size of founding populations and associated mating patterns within these populations. Sork and Smouse (2006) showed that the risk of loss of genetic variation after colonization for a given fragment depends on both the degree of spatial isolation of that fragment and the diversity of pollen and seed sources that come into the fragment. Fragments which are highly isolated and subject to little incoming gene flow, will have the highest loss of genetic variation, whereas highly connected patches with high numbers of seed and pollen sources will have the lowest loss of genetic variation. Intermediate situations are found when the fragment has high spatial isolation, but still receives a high number of incoming pollen and seed sources or when a fragment has low spatial isolation, but receives high gene flow from just a few sources.
Forest herbs represent ideal study species with which to investigate the effects of population turnover on genetic diversity and differentiation as, in most European and North-American landscapes, forests have shown dramatic changes in distribution due to forest clearance and post-agricultural recovery. The associated colonization/extinction dynamics allow testing of specific hypotheses about the effects of population turnover on the genetic diversity of forest herbs (Flinn and Vellend, 2005). For example, colonization of recently established forests allows assessment of the impact of land-use changes on within-population genetic diversity by comparing genetic diversity of forest herbs between populations in recently established and ancient forests. Although this has only rarely been done (Jacquemyn et al., 2004; Vellend, 2004; Vandepitte et al., 2007), results differ between species. Vellend (2004) found decreased genetic diversity in populations of Trillium grandiflorum located in secondary forests compared with populations located in ancient stands. In contrast, Vandepitte et al. (2007) did not find decreased genetic diversity in Geum urbanum, suggesting that the processes that affect genetic diversity in populations located in recently established forests might vary from one system to the next.
A different, more direct approach to studying the effects of population turnover on genetic diversity would be to investigate temporal changes in genetic diversity within founding populations shortly after colonization. Ideally, this can be achieved by sampling over several years, resulting in a time series of genetic diversity estimates (Berg et al., 2002), yet this approach is logistically difficult. Alternatively, when plants are long-lived and an age-structure can be delineated within these populations, one could investigate genetic diversity across different generations (e.g. Hossaert-McKey et al., 1996; Kalisz et al., 2001). Combined with detailed studies of the fine-scale genetic structure and gene flow patterns within recently founded populations, this approach can reveal insights into the processes that affect variation in genetic diversity shortly after founding.
In this study, the genetic diversity and the fine-scale spatial genetic structure were investigated within a founding population of the forest herb Primula elatior. Based on a detailed reconstruction of the land use within a 42-km2 study area and data on the distribution of P. elatior within this area, a secondary forest was selected that was <20 years old and that had only very recently been colonized by the species. Because in P. elatior the age of individuals can be related to the number of rosettes produced, an age structure within populations can be easily delineated, allowing assessment of genetic diversity across different age classes. More specifically, genetic diversity parameters across three different age classes and the extent of fine-scale spatial genetic structure within the population were estimated. Parentage analyses were used to study the amount of gene flow from outside the population and the differential contribution of mother plants to future generations. It is hypothesized that, if gene flow from outside the population is high after colonization, no or little differences in genetic diversity across age classes should be observed. If on the other hand, gene flow after colonization is limited, differences in genetic diversity across generations and significant fine-scale spatial genetic structure may arise if only a few individuals contribute to future generations and pollen and/or seed dispersal within populations is limited.
MATERIALS AND METHODS
Study species
Primula elatior Hill (oxlip) is a small, herbaceous, diploid (2n = 22) perennial, growing in mixed deciduous temperate forests. It is widely distributed in western and central Europe and extends its distribution range to Denmark and further eastwards to central Asia (Woodell, 1969). It appears as a late successional species typically occurring in ancient forests, although colonization of recent forest patches has frequently been observed (Jacquemyn et al., 2002, 2004). In early spring, on average 5·25 (±4·20) inflorescences per plant that carry on average 7·81 (±2·91) flowers per inflorescence are formed (Jacquemyn et al., 2002). Like most other species of the genus Primula, P. elatior is distylous and self-incompatible (Richards, 1997). Flowers are mostly pollinated by Hymenoptera (mostly bumble-bees) and Diptera, although other insects (e.g. Lepidotera) may be involved in the pollination process (Schou, 1983). Only cross-pollination between thrum and pin morphs results in seed set. In late May or early June, on average 29·18 (±26·30) fruits per plant are produced (Jacquemyn et al., 2002). The number of seeds per fruit can vary considerably, ranging from 1 to 61 (average 34·72 ± 17·29; Jacquemyn et al., 2002).
Seeds of P. elatior lack specific dispersal mechanisms and are generally dispersed in the immediate vicinity of the mother plant (i.e. barochory). However, as fruit predation by roe deer was frequently observed in the study area, occasional long-distance seed dispersal may occur through seed transfer by roe deer (i.e. endozoochory). Seeds germinate in spring. The next year they grow to be juveniles, from which they develop into small flowering plants within 1 or 2 years. Young plants form a rhizome by their third summer, on which additional leaf rosettes can arise in subsequent years. Primula elatior is long-lived: Rackham (2003) has suggested from photographic evidence that the half-life of P. elatior is approx. 50 years. Demographic monitoring has shown that it takes several years for plants to progress from small flowering adults to become flowering plants with multiple rosettes (Jacquemyn and Brys, 2008). Plants with more than five rosettes are therefore most likely to be >10 years old, so they are almost certainly the founding plants of this population.
Study population, data collection and AFLP protocol
The study was conducted in the central part of Belgium (Vlaams-Brabant), between Diest, Tienen and Leuven. This area is characterized by considerable turnover of forest habitat within the last 200 years (for more details, see Jacquemyn et al. 2003). The annual rate of change in forest cover, calculated as Rt = [(Ae – As)/As] × (t–1) × 100, where Ae is the area of forest at the end of the period between subsequent maps, As is he area of forest cover at the start of that period and t is the number of years between observations, varied between –2·58 % and 0·55 % (mean –0·34 %). As a result, forest area declined from 1947 ha in 1775 to 464 ha in 1970. Only in the last 30 years have reforestation rates exceeded deforestation rates, resulting in a final forest area of 525 ha in 2007. Since P. elatior occasionally colonizes recent forests, populations differ in age. Populations establish most likely from a small number of founders, which may come from several forests (Jacquemyn et al., 2004).
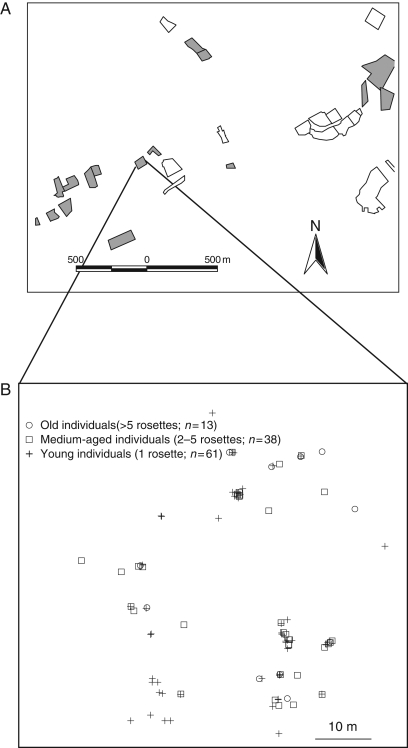
For this study, a recent forest fragment (50°50′50″N; 4°53′26″E) was selected (age: 20 years; area: 0·4 ha) that that had been recently colonized by the species. The forest occurred isolated within the landscape (mean distance to the five nearest forests >400 m) and was surrounded by meadows (Fig. 1A). Average size of neighbouring populations was 366 (range 8–1235) flowering individuals. At the time of sampling (April 2002), the population consisted of 112 flowering individuals in total. When the population had been visited at peak flowering 2 years earlier in 2000 (Jacquemyn et al., 2002), it only contained 39 flowering individuals, indicating rapid population expansion after colonization. Within this population, each flowering individual was mapped to the nearest centimetre (Fig. 1B), its size was determined and from each individual young leaf material was collected and immediately frozen in liquid nitrogen. Based on previous research investigating the demography of the species (Jacquemyn and Brys, 2008), plants were subdivided in three age classes: young individuals (1 rosette), medium-aged individuals (2–5 rosettes) and old individuals (>5 rosettes). The frequency distribution of these age classes showed a typical J-shaped curve, with a large proportion of very young individuals (54 %, n = 61) and a small proportion of old adults (12 %, n = 13). Medium-aged individuals made up the remaining 34 % (n = 38) of the population.
Fig. 1.
Map of the study area and location of the sampled population within the landscape. (A) Overview of the landscape. Each polygon represents a single forest with a unique land-use history. Shaded polygons represent forest fragments occupied by Primula elatior. (B) Detailed map of the investigated population of Primula elatior with clear depiction of the different age classes within the studied population.
Before DNA extraction, leaf material was freeze dried for 48 h and homogenized with a mill (Retsch MM 200) to fine powder. Total DNA was extracted from 30 mg of freeze-dried leaf material using methods described in Lefort and Douglas (1999). DNA concentrations were estimated on 1·5 % (w/v) agarose gels. AFLP analysis was carried out according to Vos et al. (1995), using commercial kits and following standard protocols. The enzymes EcoRI and MseI were used for DNA digestion. Each individual plant was assayed with three primer combinations. The primer extensions used were EcoACC/MseCAA, EcoACC/MseCAT and EcoACT/MseCAT. Fragment separation and detection took place on an ABI Prism 377 DNA sequencer on 36-cm denaturing gels using 4·25 % polyacrylamide (4·25 % acrylamide/bisacrylamide 19/1, 6 m urea in 1× TBE). GeneScan 500 Rox labelled size standard (Perkin Elmer) was loaded in each lane.
The fluorescent AFLP patterns were scored using Genotyper (Perkin Elmer 1996). To assess reproducibility of the markers retained, three independent DNA extractions were carried out for ten individuals and AFLP fingerprints were generated for each replicate and primer combination (a total of 120 AFLP fingerprints). The replicated samples used to estimate the error rate were fully randomized with all other samples in order to avoid biases in this estimation. Only once the scoring had been completed were the replications compared to estimate the reproducibility of the results. In total, 41 differences were observed among a total of 3660 comparisons, yielding an error rate of 1·12 % (Bonin et al., 2004).
Data analysis
Two measures of within-population genetic diversity were estimated: Nei's gene diversity (Hj) and band richness (Br). Nei's gene diversity was estimated using AFLPsurv V.1·0 (Vekemans et al., 2002). Estimates of allelic frequencies at AFLP loci were calculated using the Bayesian method with a non-uniform prior distribution of allele frequencies following Zhivotovsky (1999), assuming either no, or some deviation (FIS = 0·1) from Hardy–Weinberg genotypic proportions according to the outcrossing nature of the species. After estimating allele frequencies, Nei's gene diversity (Hj) was computed according to Lynch and Milligan (1994). Band richness was calculated for a standardized sample size according to the rarefaction method of Petit et al. (1998). This measure of genetic diversity represents the number of phenotypes expected at each locus (i.e. each scored AFLP fragment) and can be interpreted as an analogue of allelic richness (Coart et al., 2005).
The software program FaMoZ was used for reconstruction of parent–offspring relationships (Gerber et al., 2000). A likelihood-based approach was used to detect the most-likely parents and parent pairs using the logarithm of the likelihood ratio (LOD score) for dominant markers (Meagher and Thompson, 1986; Gerber et al., 2000). LOD-score thresholds for assignment of parents/parent pairs were set using the cross-section of the LOD distributions of simulated offspring from inside versus outside the stand, using 10 000 synthetic offspring and 0·1 % as error rate for simulation and calculation (see Gerber et al. (2000, 2003) for a detailed discussion of the simulation procedures). Only highly reliable parent–offspring matches (LOD single parent >4·5, parent pair >7·5) were used. Offspring were assigned to the most likely parental pair or parent if any were found using the set thresholds and the following procedure: (a) offspring were assigned to the parent pair with the highest LOD value if this value exceeded the set threshold for pairs and both adults scored higher than the set threshold for single parent assignment; (b) if not, offspring were assigned to the most likely parent if exceeding the single parent threshold (see Gerber et al., 2003). Following assignment, tests of the gene flow parameters were done using 10 000 simulations. Using the derived observed gene flow and set thresholds, true gene flow, cryptic gene flow and alpha errors were assessed using methods outlined in Gerber et al. (2003). Analyses were only conducted for the youngest individuals with both old and medium-aged individuals as potential parents. Using the physical distances between parents and offspring, estimates of pollen and seed dispersal were obtained. Because AFLPs are nuclear markers, it is not possible to distinguish between seed and pollen parents. However, because the species lacks any specific adaptations for long-distance seed dispersal and seeds in most cases fall next to the mother plant (i.e. barochory), a more conservative approach to estimating seed dispersal distances was followed and the nearest parent was designated as the seed parent (Sezen et al., 2007).
In order to determine the extent of fine-scale spatial genetic structure, spatial autocorrelation analyses were used. The multilocus estimator of the pairwise relatedness coefficient between individuals specifically adapted to dominant genetic markers (AFLP and RAPD) was used (Hardy, 2003). Under isolation by distance processes, theory predicts that the multilocus kinship coefficient F decreases approximately linearly with the natural logarithm of the physical distance between individuals. Average multilocus kinship coefficients per distance interval were then computed for the following distance classes: 1, 3, 6, 10, 15, 20, 25, 30, 40 and 55 m and were plotted against distance. To test the hypothesis that there was significant spatial genetic structure, the observed regression slope of Fdij on ln(rij), 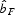 , was compared with those obtained after 10 000 random permutations of individuals among positions. In this formula ln(rij) is the natural logarithm of the physical distance between samples i and j. This procedure has the advantage that all the information is contained in one single test statistic, and the results are independent of arbitrarily set distance intervals (Vekemans and Hardy, 2004). Spatial genetic structure was also quantified by the ‘Sp’ statistic, which is calculated as
, was compared with those obtained after 10 000 random permutations of individuals among positions. In this formula ln(rij) is the natural logarithm of the physical distance between samples i and j. This procedure has the advantage that all the information is contained in one single test statistic, and the results are independent of arbitrarily set distance intervals (Vekemans and Hardy, 2004). Spatial genetic structure was also quantified by the ‘Sp’ statistic, which is calculated as 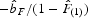 , where
, where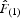 is the mean
is the mean 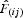 between individuals belonging to the first distance interval and
between individuals belonging to the first distance interval and 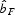 is the regression slope of
is the regression slope of 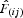 on rij (Vekemans and Hardy, 2004).
on rij (Vekemans and Hardy, 2004). 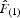 can be considered a good estimate of the kinship between pairs of neighbours, on the condition that the first distance interval contains enough pairs of individuals to obtain reasonably precise
can be considered a good estimate of the kinship between pairs of neighbours, on the condition that the first distance interval contains enough pairs of individuals to obtain reasonably precise 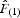 values, even if these pairs are not necessarily related (Vekemans and Hardy, 2004). All analyses were performed using SPAGeDi v.1·2 (Hardy and Vekemans, 2002).
values, even if these pairs are not necessarily related (Vekemans and Hardy, 2004). All analyses were performed using SPAGeDi v.1·2 (Hardy and Vekemans, 2002).
RESULTS
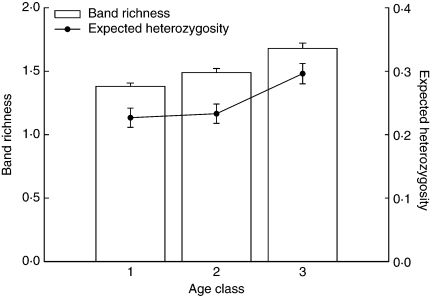
The three AFLP primer pairs detected a total of 122 scorable bands. No monomorphic bands were scored. In a few cases, some individuals failed to amplify for a certain primer combination and these individuals were removed from further analysis. Expected heterozygosity and band richness decreased across age classes in P. elatior (Fig. 2). Expected heterozygosity (Hj) was approx. 20 % lower in the youngest age class compared with that in oldest individuals. Similarly, band richness decreased across age classes and was lowest in the youngest individuals.
Fig. 2.
Band richness and genetic diversity (± s.e.) within different age classes in a founding population of Primula elatior: 1, young individuals (1 rosette; n = 57); 2, medium-aged individuals (2–5 rosettes; n = 34); 3, old individuals (>5 rosettes; n = 10).
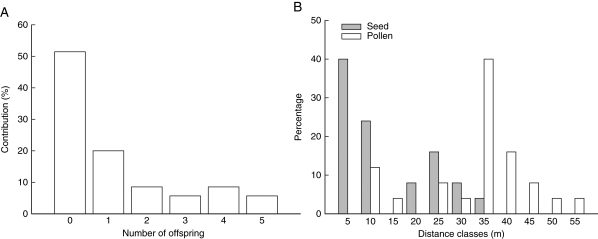
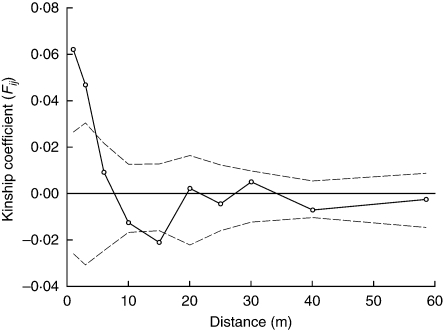
Cumulative exclusion probabilities calculated on the 122 AFLP markers were high (paternity exclusion was 0·97761 and parent pair exclusion 0·99892). Of the sampled young individuals that amplified successfully (n = 57), 82 % could be unambiguously assigned to a parent [17 (36·2 %) individuals] or parent pair [30 (63·8 %) individuals] from within the stand, indicating that most plants originated from parents within the forest stand and that gene flow from outside the stand was relatively limited. Out of 44 possible parents, only 21 (48 %) were in the parental pool (Fig. 3A). In all cases where both parents were found, they always consisted of a pin and thrum individual, confirming the self-incompatibility system of the species and validating the parentage assignment. Pollination and minimum dispersal distances imply a relatively large genetic neighbourhood (seed dispersal: median 7·6 m, range 0·2–30·8 m; pollen dispersal: median 31·4 m, range 6·2–50·1 m; Fig. 3B). Finally, permutation tests showed that the population as a whole showed weak, but significant (P < 0·0001) spatial genetic structure, with the slope of the regression coefficient bF = –0·0094 (± 0·0022; Fig. 4). Significant positive kinship coefficients were found for distances up to 3 m. The corresponding Sp statistic was 0·010.
Fig. 3.
(A) Differential contribution (%) of parent plants (n = 44) to future generations in a founder population of Primula elatior. (B) Pollen and seed dispersal distribution inferred from the parentage analysis.
Fig. 4.
Spatial correlogram of mean pairwise kinship coefficients (Fij) for a founder population of Primula elatior in central Belgium. The dashed lines represent the upper and lower 95 % confidence envelopes, respectively, around the null hypothesis of Fij = 0.
DISCUSSION
Within-population genetic diversity
Theory predicts that population turnover results in decreased genetic diversity within populations (Pannell and Charlesworth, 2000), due to founder events and genetic bottlenecks. Although there is mounting evidence that some loss of within-population genetic diversity is common in introduced populations of exotic plant species [e.g. about half of 80 species investigated by Dlugosch and Parker (2008) showed losses of allelic richness >20 %], few studies have investigated effects of population turnover on genetic diversity in fragmented landscapes. In addition, studies that have provided insights into the processes that affect within-population genetic diversity across the early generations in founding populations are also scant. The present multigenerational study in a founding population of P. elatior demonstrated that genetic diversity of plant populations might decrease substantially shortly after colonization. Genetic diversity of founding individuals was high (Hj = 0·291) and comparable to that of plants in populations located in old forests (range 0·262–0·320, mean 0·288) (Jacquemyn et al., 2004). These results suggest that founding individuals were from several source populations in the landscape and confirm earlier findings of Jacquemyn et al. (2004) who compared FST values between populations in recently established forests and old forests and concluded that colonizing individuals were most likely to originate from a few, nearby populations. Roe deer, which are supposed to be the main long-distance dispersal agents of P. elatior seeds, may consume and deposit seeds from a large number of populations. Losses of >50 % of the fruits due to predation are not unusual in this species. Given that roe deer have large home ranges, seeds from a wide range of populations may be simultaneously deposited within a given forest patch.
Across the three age classes delineated, however, genetic diversity had decreased by approx. 20 %, which may be partly explained by restricted gene flow from outside the population (Sork and Smouse, 2006). Parentage analyses indeed showed that the majority of young individuals (82 %) was assigned to parent plants from within the stand. Although this may suggest some influx of pollen and seeds from outside the stand or that some of the parents had died in the period between seed dispersal and sampling, the possibility that some plants could not be assigned to parents might be related to the fact that some individuals were removed from the analysis because they failed to amplify. Gene flow from outside the stand is therefore probably even lower than estimated here. Nonetheless, these results thus suggest that considerable amounts of gene flow are needed to counterbalance genetic drift after founding.
Decreased genetic diversity in secondary stands was also reported for the long-lived perennial Trillium grandiflorum (Vellend, 2004). Both T. grandiflorum and P. elatior lack specific adaptations facilitating long-distance seed dispersal, which may impede efficient gene flow through the landscape. Vandepitte et al. (2007), on the other hand, found no differences in genetic diversity between populations of the forest herb Geum urbanum in secondary and primary stands, and attributed this to high levels of gene flow preventing founding events from being reflected in the present genetic structure of the species. Because G. urbanum has hairy seeds containing a 5–7 mm long hook, facilitating exozoochorous seed dispersal, higher levels of gene flow can indeed be expected for this species than for P. elatior. Moreover, since this species is also an early successional species, it is less likely to show decreased genetic diversity due to its more widespread dispersal pattern.
Parentage analyses further showed that only one half of the possible parents contributed to recruitment, suggesting that a small effective population size and genetic drift are the principal factors explaining the decreased genetic diversity in this population (Nei et al., 1975). Although the observed reduction in genetic diversity might also be the result of inbreeding (selfing or biparental inbreeding) leading to inbreeding depression and the subsequent loss of more inbred individuals over time, several lines of evidence suggest that this is unlikely to cause the decreased genetic diversity in the youngest age classes. First, inbreeding through selfing is implausible given the strict self-incompatibility of the species (Schou, 1983). Secondly, parentage analyses clearly showed that in the case where both parents were found, they always consisted of a pin and thrum individual, reducing the possibility of selfing to contribute to decreased genetic diversity. Thirdly, the possibility that genetic diversity decreased due to biparental inbreeding (i.e. mating among close relatives), is also unlikely because, in this case, pollen dispersal distances should be small, which was also not the case (see below). On the other hand, the observed decline in genetic diversity might also reflect loss of variation due to selection. Although recently established forest stands have similar soil characteristics to older forests, mortality rates of especially seedlings can be high due to strong competition with nitrophilous species such Urtica dioica. However, more research is needed to show whether selection indeed explains the reduced diversity in later generations.
Differential contribution of mother plants
The differential contribution of parent plants to future generations can be best explained by the ecology of the species. Predation by roe deer or snails may lead to losses of >50 % of the seed crop (H. Jacquemyn, unpubl. res.). Thus, as many plants fail to reproduce, they obviously do not contribute to seedling recruitment. Besides, as mentioned before, mortality rates of seedlings can be quite high in these recent stands due to competition with nitrophilous species such as Urtica dioica. Therefore, seedling survival often occurs at particular micro-sites that are not too heavily affected by competition, suggesting that genetically related seedlings germinate and establish in small micro-sites that allow survival of seedlings. Inspection of Fig. 1B indeed shows a non-random distribution of medium-aged and, in particular, young individuals, which is consistent with the importance of safe sites for establishment of this species in secondary stands. Combined with limited seed dispersal distances, this leads to highly clustered distribution patterns within the population (Fig. 1B).
Spatial genetic structure
Limited seed dispersal and kin-structured germination and establishment are expected to result in significant spatial genetic structure (Ingvarsson and Giles, 1999; Vekemans and Hardy, 2004; Torimaru et al., 2007). The present observation of fine-scale spatial genetic structure was consistent with these expectations. Moreover, the magnitude of spatial genetic structure observed in P. elatior was similar to expectations based on previous studies of outcrossing plants. Comparing the present findings with a recent survey of 47 plant species (Vekemans and Hardy, 2004) revealed that the Sp statistic for P. elatior (0·010) was very similar to that of other self-incompatible plant species (0·013 ± 0·008). Because only pollination from long-styled individuals by pollen of short-styled individuals (or vice versa) results in seed set, relatively large pollen distances are to be expected. In addition, the low population density might also have contributed to the observed extent of spatial genetic structure, as low densities tend to increase flight distances of pollinators and thus pollen dispersal distances, resulting in increased neighbourhood areas and thus lower values of the Sp statistic (Vekemans and Hardy, 2004).
CONCLUSIONS
The results from this study combined with previous studies on the effects of land use changes on genetic diversity of forest herbs (e.g. Jacquemyn et al., 2004; Vellend, 2004) indicate that land use changes may lead to decreased genetic diversity within populations shortly after colonization of secondary stands. They are also consistent with the general expectation that a metapopulation that includes many new patches experiencing transitory effects of founding events (where populations may grow quickly from the contributions of a few individuals) will have reduced diversity overall. Although the variation surveyed is most likely selectively neutral, its loss may indicate the correlated loss of potentially adaptive genetic variation (Young et al. 1996). Low genetic diversity may also be associated with decreased fitness through decreasing heterozygosity and the expression of deleterious alleles (Fischer and Matthies, 1998; Keller and Waller, 2002). However, the observed losses of genetic variation were still fairly small in absolute terms, suggesting that no effects on population viability are to be expected in the short term. Moreover, studies on introduced species have shown that over longer timescales (>100 years) allelic variation might increase again due to continuing gene flow after founding (Novak and Mack, 2005; Dlugosch and Parker, 2008). Given that the species is also very long-lived, the older cohorts can also be expected to continue to contribute to the diversity of the patch.
ACKNOWLEDGEMENTS
This research was supported by the Flemish Fund for Scientific Research (FWO) to H.J. Two anonymous reviewers provided very useful comments that improved the quality of this manuscript.
LITERATURE CITED
- Berg DJ, Garton DW, MacIsaac HJ, Panov VE, Telesh IV. Changes in genetic structure of North American Bythotrephes populations following invasion from Lake Ladoga, Russia. Freshwater Biology. 2002;47:275–282. [Google Scholar]
- Bonin A, Bellemain E, Eidesen B, Pompanon F, Brochmann C, Taberlet P. How to track and assess genotyping errors in population genetics studies. Molecular Ecology. 2004;13:3261–3273. doi: 10.1111/j.1365-294X.2004.02346.x. [DOI] [PubMed] [Google Scholar]
- Coart E, Van Glabeke S, Petit RJ, Van Bockstaele E, Roldán-Ruiz I. Range wide versus local patterns of genetic diversity in hornbeam (Carpinus betulus L.) Conservation Genetics. 2005;6:259–273. [Google Scholar]
- Dlugosch KM, Parker IM. Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Molecular Ecology. 2008;17:431–449. doi: 10.1111/j.1365-294X.2007.03538.x. [DOI] [PubMed] [Google Scholar]
- Fischer M, Matthies D. RAPD variation in relation to population size and plant fitness in the rare Gentianella germanica (Gentianaceae) American Journal of Botany. 1998;85:811–819. [PubMed] [Google Scholar]
- Flinn KM, Vellend M. Recovery of forest plant communities in post-agricultural landscapes. Frontiers in Ecology and the Environment. 2005;3:243–250. [Google Scholar]
- Gerber S, Mariette S, Streiff R, Bodénès C, Kremer A. Comparison of micro-satellites and amplified fragment length polymorphism markers for parentage analysis. Molecular Ecology. 2000;9:1037–1048. doi: 10.1046/j.1365-294x.2000.00961.x. [DOI] [PubMed] [Google Scholar]
- Gerber S, Chabrier P, Kremer A. FAMOZ: a software for parentage analysis using dominant, codominant and uniparentally inherited markers. Molecular Ecology Notes. 2003;3:479–481. [Google Scholar]
- Giles BE, Goudet J. Genetic differentiation in Silene dioica metapopulations: estimation of spatiotemporal effects in a successional plant species. American Naturalist. 1997;149:507–526. [Google Scholar]
- Hardy OJ. Estimation of pairwise relatedness between individuals and characterization of isolation-by-distance processes using dominant genetic markers. Molecular Ecology. 2003;12:1577–1588. doi: 10.1046/j.1365-294x.2003.01835.x. [DOI] [PubMed] [Google Scholar]
- Hardy OJ, Vekemans X. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes. 2002;2:618–620. [Google Scholar]
- Hossaert-McKey M, Valero M, Magda D, Jarry M, Cuguen J, Vernet P. The evolving genetic history of a population of Lathyrus sylvestris: evidence from temporal and spatial genetic structure. Evolution. 1996;50:1808–1821. doi: 10.1111/j.1558-5646.1996.tb03567.x. [DOI] [PubMed] [Google Scholar]
- Ingvarsson PK, Giles BE. Kin-structured colonization and small-scale genetic differentiation in Silene dioica. Evolution. 1999;53:605–611. doi: 10.1111/j.1558-5646.1999.tb03795.x. [DOI] [PubMed] [Google Scholar]
- Ingvarsson PK, Olsson K, Ericson L. Extinction-recolonization dynamics in the mycophagous beetle Phalacrus substriatus. Evolution. 1997;51:187–195. doi: 10.1111/j.1558-5646.1997.tb02400.x. [DOI] [PubMed] [Google Scholar]
- Jacquemyn H, Brys R. Effects of stand age on the demography of a temperate forest herb in post-agricultural landscapes. Ecology. 2008;89:3480–3489. doi: 10.1890/07-1908.1. [DOI] [PubMed] [Google Scholar]
- Jacquemyn H, Brys R, Hermy M. Patch occupancy, population size and reproductive success of a forest herb (Primula elatior) in a fragmented landscape. Oecologia. 2002;130:617–625. doi: 10.1007/s00442-001-0833-0. [DOI] [PubMed] [Google Scholar]
- Jacquemyn H, Butaye J, Hermy M. Influence of environmental and spatial variables on regional distribution of forest plant species in a fragmented and changing landscape. Ecography. 2003;26:768–776. [Google Scholar]
- Jacquemyn H, Honnay O, Galbusera P, Roldán-Ruiz I. Genetic structure of the forest herb Primula elatior in a changing landscape. Molecular Ecology. 2004;13:211–219. doi: 10.1046/j.1365-294x.2003.02033.x. [DOI] [PubMed] [Google Scholar]
- Jacquemyn H, Honnay O, Van Looy K, Breyne P. Spatiotemporal structure of genetic variation of a spreading plant metapopulation on dynamic riverbanks along the Meuse River. Heredity. 2006;96:471–478. doi: 10.1038/sj.hdy.6800825. [DOI] [PubMed] [Google Scholar]
- Kalisz S, Nason JD, Hanzawa FM, Tonsor SJ. Spatial population genetic structure in Trillium grandiflorum: the roles of dispersal, mating, history, and selection. Evolution. 2001;55:1560–1568. doi: 10.1111/j.0014-3820.2001.tb00675.x. [DOI] [PubMed] [Google Scholar]
- Keller LF, Waller DM. Inbreeding effects in wild populations. Trends in Ecology and Evolution. 2002;17:230–241. [Google Scholar]
- Lefort F, Douglas GC. An efficient micro-method of DNA isolation from mature leaves of four hardwood tree species Acer, Fraxinus, Prunus and Quercus. Annals of Science. 1999;56:259–263. [Google Scholar]
- Lynch M, Milligan BG. Analysis of population genetic structure with RAPD markers. Molecular Ecology. 1994;3:91–99. doi: 10.1111/j.1365-294x.1994.tb00109.x. [DOI] [PubMed] [Google Scholar]
- McCauley DE, Raveill J, Antonovics J. Local founding events as determinants of genetic structure in a plant metapopulation. Heredity. 1995;75:630–636. [Google Scholar]
- Meagher TR, Thompson E. The relationship between single point and point pair genetic likelihoods in genealogy reconstruction. Theoretical Population Biology. 1986;29:87–106. [Google Scholar]
- Nei M, Maruyama T, Chakraborty R. The bottleneck effect and genetic variability in populations. Evolution. 1975;29:1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x. [DOI] [PubMed] [Google Scholar]
- Novak SJ, Mack RN. Genetic bottlenecks in alien plant species: influence of mating systems and introduction dynamics. In: Sax DF, Stachowicz JJ, Gaines SD, editors. Species invasions: insights into ecology, evolution, and biogeography. Sunderland, MA: Sinauer Associates; 2005. pp. 201–228. [Google Scholar]
- Pannell JR, Charlesworth B. Neutral genetic diversity in a metapopulation with recurrent local extinction and recolonization. Evolution. 1999;53:664–676. doi: 10.1111/j.1558-5646.1999.tb05362.x. [DOI] [PubMed] [Google Scholar]
- Pannell JR, Charlesworth B. Effects of metapopulation processes on measures of genetic diversity. Philosophical Transactions of the Royal Society London B. 2000;355:1851–1864. doi: 10.1098/rstb.2000.0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannell JR, Dorken ME. Colonisation as a common denominator in plant metapopulations and range expansions: effects on genetic diversity and sexual systems. Landscape Ecology. 2006;21:837–848. [Google Scholar]
- Petit RJ, El Mousadik A, Pons O. Identifying populations for conservation on the basis of genetic markers. Conservation Biology. 1998;12:844–855. [Google Scholar]
- Rackham O. Ancient woodland: its history, vegetation and uses in England. 2nd edn. Colvend: Castlepoint Press; 2003. [Google Scholar]
- Richards AJ. Plant breeding systems. London: Allen & Unwyn; 1997. [Google Scholar]
- Schou O. The distyly in Primula elatior (L.) Hill (Primulaceae), with a study of flowering phenology and pollen flow. Botanical Journal of the Linnean Society. 1983;86:1679–1690. [Google Scholar]
- Sezen UU, Chazdon RL, Holsinger KE. Multigenerational genetic analysis of tropical secondary regeneration in a canopy palm. Ecology. 2007;88:3065–3075. doi: 10.1890/06-1084.1. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Gene flow and genetic drift in a species subject to frequent local extinction. Theoretical Population Biology. 1977;12:253–262. doi: 10.1016/0040-5809(77)90045-4. [DOI] [PubMed] [Google Scholar]
- Sork VL, Smouse PE. Genetic analysis of landscape connectivity in tree populations. Landscape Ecology. 2006;21:821–836. [Google Scholar]
- Torimaru T, Tani N, Tsumura Y, Nishimura N, Tomaru N. Effects of kin-structured seed dispersal on the genetic structure of the clonal dioecious shrub Ilex leucoclada. Evolution. 2007;61:1289–1300. doi: 10.1111/j.1558-5646.2007.00108.x. [DOI] [PubMed] [Google Scholar]
- Vandepitte K, Jacquemyn H, Roldán-Ruiz I, Honnay O. Landscape genetics of the self-compatible forest herb Geum urbanum: effects of habitat age, fragmentation and local environment. Molecular Ecology. 2007;16:4171–4179. doi: 10.1111/j.1365-294X.2007.03473.x. [DOI] [PubMed] [Google Scholar]
- Vekemans X, Hardy OJ. New insights from fine-scale structure analyses in plant populations. Molecular Ecology. 2004;12:921–935. doi: 10.1046/j.1365-294x.2004.02076.x. [DOI] [PubMed] [Google Scholar]
- Vekemans X, Beauwens T, Lemaire M, Roldán-Ruiz I. Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Molecular Ecology. 2002;11:139–151. doi: 10.1046/j.0962-1083.2001.01415.x. [DOI] [PubMed] [Google Scholar]
- Vellend M. Parallel effects of land-use history on species diversity and genetic diversity of forest herbs. Ecology. 2004;85:3043–3055. [Google Scholar]
- Vos P, Hogers R, Bleeker M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock MC. Nonequilibrium population structure in forked fungus beetles: extinction, colonization, and the genetic variance among populations. American Naturalist. 1992;139:952–970. [Google Scholar]
- Woodell SRJ. Natural hybridization in Britain between Primula vulgaris Huds (the primrose) and P. elatior (L.) Hill (the oxlip) Watsonia. 1969;7:115–127. [Google Scholar]
- Young A, Boyle T, Brown T. The population genetic consequences of habitat fragmentation for plants. Trends in Ecology and Evolution. 1996;11:413–418. doi: 10.1016/0169-5347(96)10045-8. [DOI] [PubMed] [Google Scholar]
- Zhivotovsky LA. Estimating population structure in diploids with multilocus dominant DNA markers. Molecular Ecology. 1999;8:907–913. doi: 10.1046/j.1365-294x.1999.00620.x. [DOI] [PubMed] [Google Scholar]