Abstract
Background and Aims
Detailed knowledge of variations in ploidy levels and their geographic distributions is one of the key tasks faced in polyploid research in natural systems. Flow cytometry has greatly facilitated the field of cytogeography by allowing characterization of ploidy levels at both the regional and population scale, and at multiple stages of the life cycle. In the present study, flow cytometry was employed to investigate the patterns and dynamics of ploidy variation in the taxonomically challenging complex Knautia arvensis (Dipsacaceae) and some of its allies (K. dipsacifolia, K. slovaca) in Central Europe.
Methods
DNA ploidy levels were estimated by DAPI flow cytometry in 5205 adult plants, 228 seedlings and 400 seeds collected from 292 Knautia populations in seven European countries. The flow cytometric data were supplemented with conventional chromosome counts. A subset of 79 accessions was subjected to estimation of the absolute genome size using propidium iodide flow cytometry.
Key Results and Conclusions
Five different ploidy levels (from 2x to 6x) were found, with triploids of K. arvensis being recorded for the first time. The species also exhibited variation in the monoploid genome size, corresponding to the types of habitats occupied (grassland diploid populations had larger genome sizes than relict and subalpine diploid populations). Disregarding relict populations, the distribution of 2x and 4x cytotypes was largely parapatric, with a diffuse secondary contact zone running along the north-west margin of the Pannonian basin. Spatial segregation of the cytotypes was also observed on regional and microgeographic scales. The newly detected sympatric growth of diploids and tetraploids in isolated relict habitats most likely represents the primary zone of cytotype contact. Ploidy level was found to be a major determinant of the strength of inter-cytotype reproductive barriers. While mixed 2x + 4x populations virtually lacked the intermediate ploidy level at any ontogenetic stage, pentaploid hybrids were common in 4x +6x populations, despite the cytotypes representing different taxonomic entities.
Key words: Contact zone, cytogeography, flow cytometry, genome size, hybridization, Knautia arvensis, ploidy mixture, polyploidy, relict, reproductive isolation, serpentine
INTRODUCTION
Polyploidy, the presence of more than two complete genomes per cell, has long been recognized as an important force in plant evolution. Recent estimates suggest that about 70 % of angiosperms and up to 95 % of pteridophytes underwent one or more rounds of genome duplication in their evolutionary history (Soltis and Soltis, 1999), and genomic data even assumes near ubiquity of polyploidy (Soltis, 2005). In fact, it has been suggested that at least 2–4 % of all speciation events in angiosperms involve polyploidization (Otto and Whitton, 2000). Because polyploidization potentially confers immediate reproductive isolation, it is widely recognized as the major mode of sympatric speciation in plants (Coyne and Orr, 2004). While different ploidy levels may correspond to different taxa (e.g. Rosenbaumová et al., 2004; see also Soltis et al., 2007), intraspecific ploidy variation is not a rare phenomenon (Lihová et al., 2003; Suda et al., 2007b). When several cytotypes occur within the same species, zones of ploidy overlap are often formed. These contact zones are of particular interest to evolutionary biologists because they allow for the study of the mechanisms involved in early stages of polyploid speciation and/or the assessment of selective forces operating in mixed-ploidy populations. Two types of contact zone are recognized according to their evolutionary history (Petit et al., 1999): primary, where a new cytotype originates in situ, and secondary, where two formerly allopatric cytotypes meet.
One of the prerequisites for polyploid research in natural systems is knowledge of the geographical distribution of cytotypes or closely related taxa exhibiting ploidy heterogeneity (Hodálová et al., 2007; Suda et al., 2007b; Rivero-Guerra, 2008). Distributional data provide useful insights into the evolutionary history of diploid–polyploid groups and can aid in interpretation of phylogenetic relationships and/or experimental results (Baack, 2004, 2005; Baack and Stanton, 2005). In addition, distributional data serve as a foundation for exploring the ecological preferences of individual cytotypes and the degree of cytotype interactions (e.g. patterns of mating, competition) and allow for the assessment of the historical development of modern distribution patterns and/or evolutionary forces governing ploidy coexistence (Petit et al., 1999). Thus, the unravelling of variation in ploidy levels and their distributions represents one of the key tasks faced by polyploid research (Favarger, 1984; Perný et al., 2008).
Knautia arvensis agg. (Dipsacaceae) is an intricate polyploid complex of perennial outcrossing herbs inhabiting dry and mesophilous, often human-influenced, grasslands, shrublands, forest margins and open woodlands (Ehrendorfer, 1962b; Štěpánek, 1982, 1997). Two species are recognized in Central Europe. While the exclusively tetraploid (2n = 4x = 40) K. kitaibelii (Schult.) Borbás is restricted to the Western Carpathians (Ehrendorfer, 1962b; Štěpánek, 1982), K. arvensis (L.) J. M. Coult. s.s. is widely distributed throughout Europe. The latter taxon harbours two ploidy levels, usually classified as separate subspecies or varieties [i.e. diploid subsp. pannonica (Heuff.) O. Schwarz and tetraploid subsp. arvensis] with more-or-less parapatric distribution. Diploids and tetraploids of K. arvensis occur in south-eastern and north-western parts of Europe, respectively (Ehrendorfer, 1962b); the contact zone runs through Central Europe, and most authors localize it to the northern margin of the Pannonian basin (Ehrendorfer, 1962b; Štěpánek, 1982; Kaplan, 1998). In addition, a few diploid populations were discovered in Bohemia and Northern Bavaria in areas otherwise occupied by tetraploids. These populations inhabit relict habitats of serpentine outcrops (sometimes treated as a separate taxon, subsp. serpentinicola Smejkal ined.) and a subalpine glacial cirque [= subsp. pseudolongifolia (Szabó) O. Schwarz] (Štěpánek, 1982, 1989; Kaplan, 1998). The serpentine localities are rather small (no more than several square kilometres), geographically isolated rocky outcrops with distinct vegetation (usually a mosaic of rocks and open pine forest). Most likely, they have never been covered by compact deciduous forest and are noted for the occurrence of several other relic and/or endemic taxa confined to ultramafic substrates (see, for example, Novák, 1960; Dvořáková, 1988). The subalpine population is restricted to a small outcrop of carbonate rocks in the Krkonoše Mountains (Štěpánek, 1989). Other peculiar diploid populations formerly recognized as K. arvensis agg. are known to occur in relict stands (open pine forests on limestone outcrops) in Central–Eastern Slovakia. Based on several unique morphological characters (e.g. type of indumentum and leaf shape), these populations were described as a separate taxon, K. slovaca Štěpánek, and assigned to a mainly (sub)Mediterranean group of K. velutina agg. (Štěpánek, 1983).
In areas of sympatry, the nominate tetraploid subspecies of K. arvensis hybridizes freely with other homoploid species [with both the closely related K. kitaibelii, forming the introgressive hybrid K. × posoniensis Degen, and with the rather distantly related K. drymeja Heuffel and 4x Carpathian cytotypes of K. dipsacifolia (Schrank) Kreutzer]. In fact, the virtual lack of interspecific reproductive barriers at the same ploidy level leads to the high incidence or even predominance of introgressants in some regions (Ehrendorfer, 1964; Breton Sintes, 1974). In addition to homoploid hybridization, some heteroploid crosses between polyploids (e.g. between 4x K. arvensis and 6x cytotypes of K. dipsacifolia) have also been documented based on both morphology (Štěpánek, 1997) and chromosome counts (Ehrendorfer, 1962a; Štěpánek, 1982). In contrast, there are no records of triploid Knautia arvensis, despite the assumed close relationship between putative diploid and tetraploid parents and their occasional sympatric growth (Ehrendorfer, 1962a, b). The only documented case of interploid 2x × 4x hybridization [involving diploid K. drymeja Heuffel var. tergestina (Beck) Briq. and tetraploid K. illyrica Beck] resulted in a putative tetraploid hybrid (Ehrendorfer 1962a). Strong reproductive barriers between 2x and 4x Knautia plants have also been confirmed by several pollination experiments (Breton Sintes, 1974, 1975; Ehrendorfer, 1962a).
Although previous karyological studies utilizing conventional chromosome counts have provided a rough picture of the ploidy distribution of K. arvensis in Central Europe (Ehrendorfer, 1962a, b; Štěpánek, 1982; Kaplan, 1998), they have been unable to describe the pattern in sufficient detail due to limited sampling. Therefore, in order to obtain robust and representative results for ploidy variation on various spatial scales, we employed DNA flow cytometry (FCM). This high-throughput technique generates much larger sample sizes that can (1) provide broader geographical information, (2) uncover rare and previously unrecognized cytotypes (e.g. heteroploid crosses), and (3) characterize ploidy variation during different life stages (e.g. seeds, seedlings, and mature plants) (Husband and Schemske, 1998; Suda et al., 2007a). In addition, FCM offers the possibility of detecting differences in nuclear DNA content, even in organisms with the same number of chromosomes (e.g. Dimitrova et al., 1999; Mahelka et al., 2005; see Kron et al., 2007, for a review). In many plant groups, genome size has been found to be a useful taxonomic marker that may guide taxonomic decisions and allow for detection of cryptic diversity or incipient speciation (Murray, 2005).
Using FCM, we addressed the following questions. (1) What is the cytotype distribution of K. arvensis in Central Europe? What is the structure of the zone of ploidy overlap? Does the cytotype contact correspond to the primary or the secondary zone? (2) How frequent are mixed-ploidy populations? What is the fine-scale distribution of cytotypes in such populations? Do ploidy proportions in mixed populations differ between the various life stages? (3) Is the pattern observed in 2x + 4x populations of K. arvensis different from the situation present in populations formed by K. arvensis (4x) and K. dipsacifolia (6x)? (4) Is there any variation in genome size within the K. arvensis agg. complex? If so, is this variation systematically, geographically and/or ecologically structured?
MATERIALS AND METHODS
Field sampling
Plant material was sampled from 2005 to 2008 in the Czech Republic, Slovakia, Austria, Hungary, Poland, Ukraine and Germany. In total, 283 populations of Knautia arvensis agg. (i.e. K. arvensis s.s., K. kitaibelii and their introgressive hybrid K. × posoniensis) were analysed. For comparative purposes, four populations of presumably related K. slovaca and five mixed populations consisting of K. arvensis and K. dipsacifolia were also included in the study. GPS co-ordinates and basic environmental characteristics (e.g. type of habitat, vegetation cover and irradiation) were recorded at each locality. Leaves from approx. ten plants per population were collected, placed in plastic bags and stored at cold temperatures (no more than 1 week) until FCM analyses; to avoid collecting the same genet, the distance between sampled individuals was at least 1 m. The locality details and numbers of plants analysed is available online in the Supplementary Data. Herbarium vouchers are deposited in the CBFS. For mapping the cytotype distribution on a coarse spatial scale, the following literature sources were also considered: Ehrendorfer (1962a) – Austria, Hungary; Frey (1969) – Poland; Májovský (1976) – Slovakia; Štěpánek (1982) – the Czech Republic, Slovakia and Poland; and Kaplan (1998) – the Czech Republic.
Three mixed-ploidy populations (see Supplementary Data, available online) were subjected to detailed microspatial screening. The populations were selected to encompass (1) different ploidy combinations (2x + 4x vs. 4x + 6x); (2) different taxonomic compositions (subspecies of K. arvensis vs. mixture of K. arvensis and K. dipsacifolia); and (3) different habitat types (semi-natural vs. relict habitats). At each locality, initial ploidy screening was performed to locate representative mixed-ploidy plots. Subsequently, every adult individual (both flowering plants and sterile rosettes) in the plot was labelled, its position recorded in a rectangular co-ordinate system, and the DNA ploidy level determined by FCM. Moreover, all seedlings (young individuals with less than three pairs of basal leaves, presumably germinated the same year) and achenes from all fructiferous plants were collected. One additional tetraploid and hexaploid mixed population (Želnava, no. 290; see Supplementary Data) was screened in a different way. Approximately 50 % of the individuals were sampled in a priori defined patches (circles of 1 m radius) along a 50-m transect.
Estimation of DNA ploidy levels and genome sizes
DNA ploidy levels (Suda et al., 2006) and genome sizes (C- and Cx-values; Greilhuber et al., 2005) were determined by flow cytometry using Partec PA II and CyFlow instruments (Partec GmbH., Münster, Germany) equipped with a HBO mercury arc lamp and a green (532 nm) solid-state laser, respectively. Sample preparation generally followed the two-step procedure using Otto buffers (Doležel et al., 2007). Intact leaf tissue of analysed Knautia plant(s) with an appropriate volume of the internal reference standard (Pisum sativum ‘Ctirad’, Doležel et al., 1998; 2C value set to 8·84 pg following Greilhuber et al., 2007) was chopped with a sharp razor blade in a Petri dish containing 0·5 mL of ice-cold Otto I buffer (0·1 m citric acid, 0·5 % Tween-20; Otto, 1990). The suspension was filtered through a 42-μm nylon mesh and incubated for approx. 30 min at room temperature. The staining solution consisted of 1 mL of Otto II buffer (0·4 m Na2HPO4.12 H2O), β-mercaptoethanol (final concentration of 2 µL mL−1), and a fluorochrome. Propidium iodide (PI) and RNase IIA (both at final concentrations of 50 µL mL−1) were used to determine the genome size in absolute values (pg of DNA), while AT-selective DAPI (4′,6-diamidino-2-phenylindole) at a final concentration of 4 µL mL−1 was preferred for the ploidy analyses. Samples were stained for 10 min at room temperature and run on the flow cytometer. Isolated nuclei were excited using either a laser (for PI staining) or a mercury arc lamp (for DAPI staining), and the fluorescence intensity of 3000–5000 particles was recorded. Only histograms with coefficients of variation (CVs) for the G0/G1 peak of the analysed Knautia sample below 3·0 % were considered. For ploidy screening, pooled samples (up to ten individuals) were often used. Our previous experiments showed that such practice enables reliable detection of a minority cytotype present even at a low proportion (<10 %). Moreover, the lack of endopolyploidy and virtual absence of mitotic activity in the selected leaf tissue guaranteed unbiased DNA ploidy level estimates. Nevertheless, each plant was separately re-analysed if mixed samples were suspected, peaks were asymmetrical, or the CV of the Knautia peak exceeded the above-set threshold. In fact, short-term storage of plant tissues in the cold (4 °C) did not negatively influence the quality of analyses, and high-resolution histograms were obtained even after 1 week of storage. More stringent criteria were applied to analyses aimed at genome size determination: (1) peaks for both the sample and the internal standard had to be of approximately the same height; (2) each sample was measured at least three times on different days to minimize potential random instrumental drift; and (3) the between-day variation was defined to not exceed a 2 % threshold; otherwise the most remote value was discarded and the sample was re-analysed. The reliability of FCM measurements (i.e. between-plant differences) was repeatedly confirmed in simultaneous runs of Knautia accessions having distinct genome sizes.
DNA ploidy levels in seeds were analysed using the same protocol as for leaf tissues, with the following modifications: (1) only single-seed analyses were performed; and (2) Bellis perennis L. (2C = 3·38 pg; Schönswetter et al., 2007) was selected as an internal reference standard. Altogether, 400 seeds with well-developed embryo/endosperm obtained from 103 mother plants were examined. The criteria for seed analyses were more relaxed – the CVs of embryo peaks varied from 2·2 % to 7·3%.
Chromosome counts
To confirm the FCM results, 14 individuals representing different DNA ploidy levels and genome size variants (i.e. ten K. arvensis individuals and one individual each of K. kitaibelii, K. slovaca, K. dipsacifolia and a putative hybrid between K. arvensis and K. dipsacifolia; see Supplementary Data, available online) were subjected to conventional karyological analysis using rapid squash methods. The apical shoot meristems of mature plants were pre-treated with p-dichlorbenzene for 3 h, fixed in ice-cold 3 : 1 ethanol : acetic acid for 12–14 h, macerated for 1 min in 1 : 1 ethanol : hydrochloric acid, and stained with lacto-propionic-orcein (Dyer, 1963).
Statistical analyses
Differences in the altitudinal distribution between diploid and tetraploid cytotypes of K. arvensis were tested using a two-tailed t-test available in the Statistica package, ver. 7 (StatSoft, 2005; mixed-ploidy populations were excluded from the analysis). The GLM procedure available in SAS 8.1 (SAS Institute, 2000) was used to assess differences in genome sizes (C-values), and Tukey's procedure was applied to compare mean values of particular groups. The spatial segregation of cytotypes in mixed-ploidy plots was analysed using the Mantel test (Manly, 1991) implemented in the zt software (Bonnet and van de Peer, 2002). A correlation coefficient (rM) was calculated for (1) the matrix of mutual distances among individuals, and (2) the binary matrix of ploidies, and compared to the distribution of coefficients obtained from matrices generated by random rearrangements of the original matrices (999 permutations were performed). Only majority ploidies were considered (i.e. rare odd cytotypes were omitted).
RESULTS
Variation in DNA ploidy levels, chromosome numbers and genome sizes
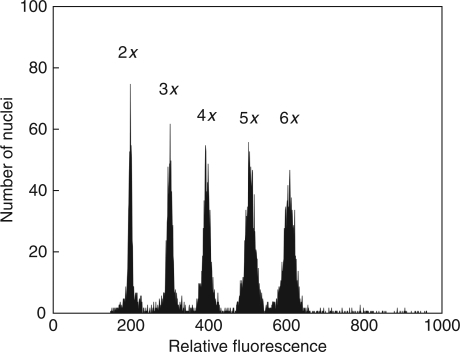
Five different DNA ploidy levels (from 2x to 6x) were detected during FCM analyses of 5205 adult individuals from 292 Knautia populations (Fig. 1). While K. arvensis s.s. showed ploidy heterogeneity (2x + 4x cytotypes), other species were karyologically uniform (i.e. K. slovaca, 2x; K. kitaibelii, 4x; and K. dipsacifolia, 6x). Two minority odd ploidy levels were detected: (1) DNA pentaploids, which corresponded to crosses between 4x K. arvensis and 6x K. dipsacifolia (24 plants from four populations); and (2) DNA triploids (two plants from two populations of K. arvensis otherwise formed by diploids; see Supplementary Data, available online). In fact, the latter cytotype represents one of the few triploid records for the entire Knautia genus and the very first record for K. arvensis agg.
Fig. 1.
Fluorescence histogram of propidium iodide-stained nuclei isolated from fresh leaf tissues of diploid, triploid and tetraploid Knautia arvensis plants, pentaploid hybrid K. arvensis × K. dipsacifolia, and hexaploid K. dipsacifolia. Nuclei from all samples were isolated, stained and analysed simultaneously.
Chromosome counts confirmed the estimated ploidy levels and revealed 2n = 2x = 20 in five accessions of K. arvensis and one accession of K. slovaca, 2n = 3x ≈ 30 in one accession of K. arvensis, and 2n = 4x = 40 in four accessions of K. arvensis and one accession of K. kitaibelii (see Supplementary Data). No aneuploidy/accessory chromosomes were recorded. Exact chromosome numbers could not be unambiguously determined in a DNA pentaploid and a DNA hexaploid; nevertheless, more than 40 chromosomes were observed in their somatic cells.
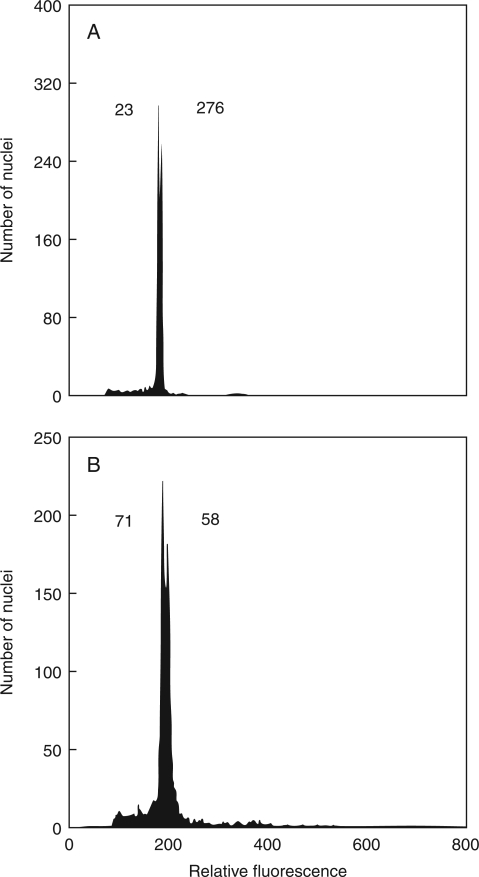
A subset of Knautia accessions was subjected to genome size estimation, including 68 K. arvensis individuals, six K. kitaibelii individuals and five K. slovaca individuals (Table 1; see also Supplementary Data). To assess the genome size variation, the K. arvensis accessions were further divided into five groups corresponding to ploidy levels and habitat types. The intra-group variation in genome size was always small (≤4·3 %; Table 1) but genuine, as confirmed by simultaneous analyses of individuals with distinct nuclear DNA amounts (Fig. 2A). No geographical or ecological structuring was observed within the groups (data not shown). Analysis of between-group differences revealed five more-or-less distinct clusters of holoploid genome size (F = 8879·94, P < 0·0001; Table 1). The first, most-distant cluster (mean 2C-value = 7·23 pg) corresponded to non-relict diploids of K. arvensis, which were widely distributed in the eastern part of the area studied (i.e. subsp. pannonica). Geographically vicariant diploids of K. arvensis from relict habitats in the western region (serpentine outcrops and a subalpine glacial cirque) formed, together with K. slovaca, the second cluster (mean 2C-values = 6·86–6·95 pg). Once again, the divergence in estimated nuclear DNA content between these two groups was confirmed in simultaneous FCM analyses (Fig. 2B). Genome size differentiation at the tetraploid level was less apparent. Despite 2C-values of non-serpentine 4x K. arvensis (mean = 14·08 pg) differing significantly from those of K. kitaibelii (mean = 13·75 pg), both values were quite similar (average difference of only 2·4 %), and the situation was further complicated by relict serpentine populations of K. arvensis with mean 2C-values of 13·94 pg.
Table 1.
Genome sizes (C- and Cx-values) obtained for different ploidy levels and ecological types of K. arvensis agg. and K. slovaca
| No. of populations/no. of individuals analysed | 2C-value (mean ± s.d.; pg of DNA)* | 2C-value range (min–max; pg of DNA) | Variation (%) | Mean Cx-value (pg of DNA) | |
|---|---|---|---|---|---|
| K. arvensis (2x): non-relict | 14/23 | 7·23 ± 0·09d | 7·08–7·37 | 4·1 | 3·62 |
| K. arvensis (2x): relict serpentine | 5/18 | 6·86 ± 0·09e | 6·72–7·00 | 4·2 | 3·43 |
| K. arvensis (2x): relict subalpine | 1/5 | 6·95 ± 0·04e | 6·91–7·00 | 1·3 | 3·48 |
| K. slovaca (2x) | 4/5 | 6·87 ± 0·11e | 6·70–6·96 | 3·9 | 3·43 |
| K. arvensis (3x): relict serpentine | 1/1 | 10·51c | – | – | 3·50 |
| K. arvensis (4x): non-relict | 4/11 | 14·08 ± 0·14a | 13·92–14·34 | 3·0 | 3·52 |
| K. arvensis (4x): relict serpentine | 3/10 | 13·94 ± 0·18ab | 13·62–14·16 | 4·0 | 3·48 |
| K. kitaibelii (4x) | 4/6 | 13·75 ± 0·25b | 13·38–13·96 | 4·3 | 3·44 |
* Different letters indicate groups of taxa that are significantly different at α = 0·05.
Fig. 2.
Flow cytometric histograms documenting variation in genome size between selected diploid accessions of K. arvensis. (A) Simultaneous analysis of non-relict populations nos. 23 (2C = 7·21 pg) and 276 (2C = 7·37 pg). (B) Simultaneous analysis of a relict serpentine population, no. 71 (2C = 6·90 pg), and non-relict population, no. 58 (2C = 7·26 pg); see Supplementary Data (available online) for population details. Nuclei from both samples were isolated, stained with DAPI and analysed simultaneously.
Cytotype distribution on a coarse spatial scale
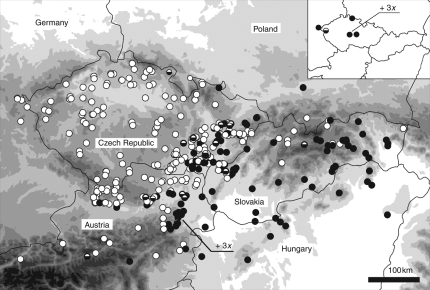
The geographic distribution of cytotypes of K. arvensis (including introgressants with K. kitaibelii) in Central Europe is shown in Fig. 3 (both our FCM results and selected literature records were used in constructing the map). The two majority cytotypes exhibit a clear spatial segregation along a NW–SE gradient. Disregarding the relict populations, diploids were restricted to the SE part of the area investigated, while tetraploids predominated in the NW region. For diploid plants, the centre of distribution was located in Pannonia, from where they extended in several directions, mainly along large river valleys (e.g. the Váh River in Slovakia and the Danube River in Austria). A few scattered diploid populations having distinct genome sizes occurred in relict habitats, such as serpentine outcrops and a glacial subalpine cirque in the Bohemian massif (see inset in Fig. 3). Tetraploids occupied the entire Bohemian massif, the outer ranges of the Western Carpathians, and the northern portions of the Eastern Alps. In the zone of overlap with K. kitaibelii (e.g. in the Carpathians and eastern parts of the Bohemian massif), tetraploids were almost exclusively represented by the introgressive hybrid K. × posoniensis; this taxon can be easily recognized by the intermediate colour of its flower parts (i.e. whitish-to-pale-violet corolla with violet anthers).
Fig. 3.
Distribution of the cytotypes of Knautia arvensis s.s. (including an introgressive hybrid K. × posoniensis) in Central Europe. The overall distribution of cytotypes is shown: note the largely parapatric distribution. The inset shows the distribution of relict populations (these are not depicted in the main figure). Solid circles: diploid; open circles: tetraploid; half-filled circles: mixed 2x + 4x population. Two triploids are designated by ‘3x’. The diploid population from west Ukraine (close to Lviv) is not displayed on the map. While the majority of the populations mapped were analysed in the present study, the figure also includes some populations extracted from selected literature sources (see Materials and Methods for details).
Diploids and tetraploids of K. arvensis coexist along the NW margin of the Pannonian basin (eastern foothills of the Eastern Alps, eastern part of Lower Austria, South and Central Moravia) and in the Polonian lowlands of north Moravia and south Poland. The contact zone then continues through Slovakia, where the situation is more complex because diploids penetrate along river valleys far into the Carpathian Mountains. Generally, the zone of ploidy contact is quite wide (several dozens of kilometres) and diffuse.
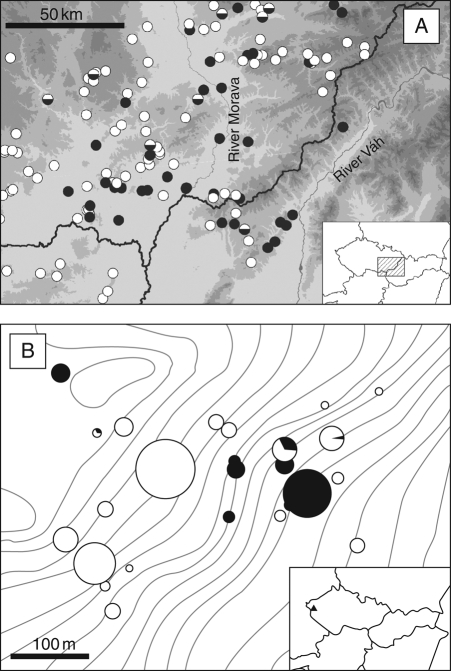
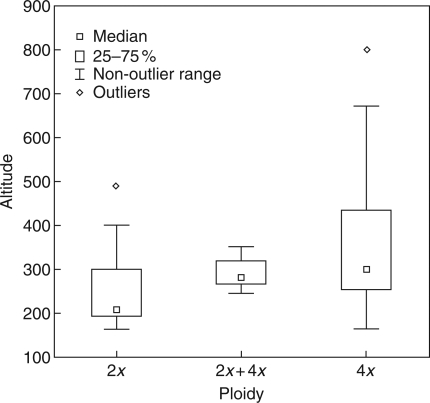
Single-cytotype populations of K. arvensis clearly prevailed, even in areas where both ploidies were recorded (Fig. 4A). In the two contact zones where the most representative sampling was performed (south and central Moravia, and north and north-east Austria), elevational differences among the sites of 2x and 4x cytotypes were tested using a two-tailed t-test. Significant differences were found in both cases (t = –4·51, d.f. = 68, P < 0·0001 and t = –2·47, d.f. = 36, P = 0·018 for the Moravian and Austrian localities, respectively). In each region, diploids generally occupied lower altitudes than their tetraploid counterparts, with an average difference of >100 m (Fig. 5).
Fig. 4.
Distribution of cytotypes of Knautia arvensis s.s. (including an introgressive hybrid K. × posoniensis) on regional and microgeographic scales. (A) Detailed distribution in the zone of ploidy overlap in South and Central Moravia, SW Slovakia and NE Austria. The inset shows the location of the region in within Central Europe. (B) Cytotype composition in the mixed-ploidy relict serpentine population Planý vrch, no. 278 (see Supplementary Data, available online, for population details). Solid circles: diploid; open circles: tetraploid; partially-filled circles: mixed 2x + 4x patch. The size of the symbols in (B) corresponds to the number of individuals analysed and the relative proportions of cytotypes are visualized as pie charts. The inset shows the position of the locality within the Czech Republic.
Fig. 5.
Box-and-whisker plots demonstrating altitudinal ranges of the diploid, tetraploid and mixed (2x + 4x) populations in the contact zone in South and Central Moravia.
Cytotype distribution on fine spatial scales
A detailed study of cytotype distribution at the level of the whole locality and/or selected plots was performed for several populations exhibiting ploidy heterogeneity. Mixed-ploidy populations occurred both in relict serpentine (three populations) and semi-natural grassland (20 populations) habitats. The following types of ploidy mixtures were detected: 2x + 4x (16 populations), 2x + 3x (two populations), 4x + 6x (one population), and 4x + 5x + 6x (four populations). There was a striking difference in ploidy composition between populations formed by diploids + tetraploids vs. tetraploids + hexaploids. All of the former populations lacked the intermediate (3x) ploidy level, suggesting the existence of efficient breeding barriers. In contrast, pentaploids of a putative hybrid origin were detected in all but one population comprising 4x K. arvensis and 6x K. dipsacifolia, and constituted a considerable proportion of the individuals analysed (3·9–9·5 %).
In Southern and Central Moravia, visual assessment of the spatial distribution of cytotypes in localities was easy due to the differences in flower colour between diploids (violet) and tetraploids (whitish due to the introgression of K. kitaibelii). All but one population showed clear cytotype segregation, with different ploidy levels growing in more-or-less aggregated patches (at least several metres apart) or even occupying different parts of the same locality. The only exception applied to the locality of Ždánice (no. 267, see Supplementary Data), where a plot with a genuine ploidy mixture was detected and further examined (see below). Cytotype aggregation also occurred in two 2x + 4x populations of K. arvensis growing in serpentine outcrops (Vlčí hřbet, no. 277, and Planý vrch, no. 278). The majority of the patches analysed (94 % and 92 %, respectively) were cytologically uniform and with prevailing tetraploids. Figure 4B shows the spatial arrangement and ploidy composition of the patches at the latter locality, Planý vrch (a very similar pattern was also found at the other one, Vlčí hřbet; data not shown). Because serpentine tetraploids are phenotypically more similar to coexisting diploids than to their widespread non-serpentine counterparts, we considered this zone as a primary contact.
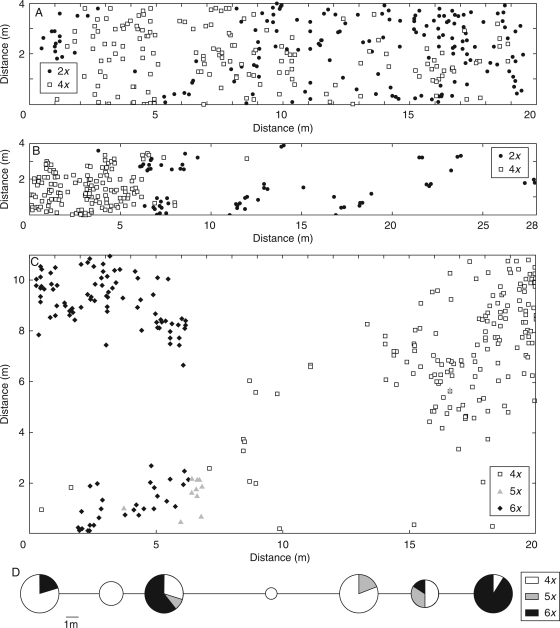
A microspatial survey was undertaken for selected mixed-ploidy plots in each of the three localities (Ždánice, no. 267, non-serpentine 2x + 4x; Planý vrch, no. 278, relict serpentine 2x + 4x; and Horní Planá, no. 288, 4x + 5x + 6x; see Supplementary Data) to ascertain whether or not the cytotypes were randomly distributed. Figure 6 shows the spatial arrangement of adult individuals. While clear ploidy grouping is apparent in the latter two localities (Fig. 6B, C), the cytotypes seem to be more intermingled in the first locality (Fig. 6A). Nevertheless, the Mantel test strongly supported the non-randomness of ploidy distribution in all plots (Table 2). In addition to the above three plots, the proportions of cytotypes were also assessed along a transect through a mixed population of K. arvensis and K. dipsacifolia near Želnava (no. 290). The complex distribution patterns of 4x, 5x and 6x ploidies are depicted in Fig. 6D.
Fig. 6.
Detailed distributions of cytotypes in selected mixed-ploidy plots (all adult plants were mapped). (A) Ždánice, no. 267: non-relict 2x + 4x population of K. arvensis. (B) Planý vrch, no. 278: relict serpentine 2x + 4x population of K. arvensis. (C) Horní Planá, no. 288: mixed population of 4x K. arvensis + 6x K. dipsacifolia (+ 5x hybrids). (D) Cytotype composition along a transect through a mixed population of 4x K. arvensis, 6x K. dipsacifolia and 5x hybrids near Želnava, no. 290. The size of the symbols in (D) corresponds to the number of individuals analysed and the relative proportions of cytotypes are visualised as pie charts. See Supplementary Data (available online) for population details.
Table 2.
Cytotype composition of adult plants and seedlings in three mixed-ploidy Knautia populations subjected to detailed ploidy screening (see Supplementary Data, available online, for population details). The spatial structure of adult individuals was evaluated using the Mantel test.
| Population | Ontogenetic stage | Total number of samples analysed | Proportions of ploidy levels (%) |
Mantel statistics |
||||
|---|---|---|---|---|---|---|---|---|
| 2x | 4x | 5x | 6x | rM | P-value | |||
| Ždánice, no. 267 (non-relict 2x + 4x K. arvensis) | Adult plants | 324 | 49·7 | 50·3 | – | – | –0·154 | 0·001 |
| Seedlings | 93 | 77·4 | 22·6 | – | – | |||
| Planý vrch, no. 278 (relict serpentine 2x + 4x K. arvensis) | Adult plants | 209 | 31·6 | 68·4 | – | – | –0·519 | 0·001 |
| Seedlings | 37 | 48·6 | 51·4 | – | – | |||
| Horní Planá, no. 288 (4x K. arvensis + 6x K. dipsacifolia + 5x hybrid) | Adult plants | 278 | – | 56·0 | 4·0 | 40·0 | –0·824* | 0·001* |
| Seedlings | 98 | – | 42·8 | 3·1 | 54·1 | |||
* The minority pentaploid cytotype was omitted from the statistical analysis.
Life stage-specific variation in ploidy composition in mixed populations
To acquire a holistic picture of ploidy composition and dynamics in the three mixed populations, data on adult plants were supplemented with ploidy estimates obtained for seedlings and seeds (Tables 2 and 3). For seedlings, both the ploidy variation and distribution pattern clearly reflected the conditions found in adult plants, i.e. seedlings of different ploidy levels were largely restricted to sectors of the plot where cytotype-mixing in adult plants had also been recorded. Interestingly, a few DNA pentaploid seedlings were found at the locality of Horní Planá (no. 288) in a sector primarily occupied by mature tetraploids (see Fig. 6C). In both 2x + 4x populations, the proportion of diploids in seedlings was higher than in mature plants. These differences could either reflect differential survival of the two cytotypes or could arise due to a ploidy-specific supply of seeds from outside the experimental plot.
Table 3.
Ploidy variation in seeds obtained from three mixed-ploidy Knautia populations with different cytotype compositions (see Supplementary Data, available online, for population details)
| Population | Ploidy level of mother plants | No. of analysed seeds/no. of mother plants | Numbers and proportions (%) of seeds of different ploidy levels |
||||
|---|---|---|---|---|---|---|---|
| 2x | 3x | 4x | 5x | 6x | |||
| Ždánice, no. 267 (non-relict K. arvensis) | 2x | 80 / 25 | 80 (100) | – | – | – | – |
| 4x | 64 / 20 | – | – | 64 (100) | – | – | |
| Planý vrch, no. 278 (relict serpentine K. arvensis) | 2x | 12 / 4 | 12 (100) | – | – | – | – |
| 4x | 75 / 21 | – | 1 (1·3) | 74 (98·7) | – | – | |
| Horní Planá, no. 288 (4x K. arvensis + 6x K. dipsacifolia + 5x hybrid) | 4x | 100 / 17 | – | – | 99 (99·0) | 1 (1·0) | – |
| 5x | 10 / 1 | – | – | 1 (10·0) | 9 (90·0)* | – | |
| 6x | 59 / 15 | – | – | – | – | 59 (100·0) | |
* Aneuploids are likely to be included due to large variations in fluorescence intensity.
A more complex pattern of ploidy variation emerged when ripe seeds were analysed (Table 3). Notably, a single triploid seed produced by a tetraploid maternal plant was detected in a relict serpentine 2x + 4x population in Planý vrch (no. 278). In addition, a pentaploid seed was recorded among a seed set of tetraploid plants from a 4x + 6x population in Horní Planá (no. 288). Pentaploid hybrids in the latter population produced mostly 5x seeds, although fluorescence intensity corresponding to the tetraploid cytotype was also recorded, along with some putative aneuploids (see Table 3).
DISCUSSION
Cytogeography of Knautia arvensis
The results obtained by ploidy screening of a representative set of Knautia samples support previous theories regarding the parapatric distribution of diploid and tetraploid cytotypes of K. arvensis s.s. in Central Europe (Ehrendorfer, 1962b; Štěpánek, 1982; Kaplan, 1998). While the centre of the distribution of diploids in Pannonia was also confirmed, marked extensions of the distributional range both to the west (the Danube valley in Upper Austria) and north (Central and North Moravia, and South Poland) were newly detected. Of particular significance is the occurrence of diploids in the southern Polonian lowlands (several localities stretching from North Moravia up to West Ukraine), which indicates the existence of a previously largely unnoticed area (but see Frey, 1969) separated from the Pannonian basin by the mountain ranges of the Western Carpathians (Fig. 3). The Pannonian and Polonian populations seem to be connected via the Moravská brána depression on the western border of the Carpathians. Additional possible connections along the eastern margin of the Carpathians (through Ukraine, Moldova and Romania) remain to be determined. In addition, there are several deep penetrations of diploids into the Western Carpathian mountain systems, mainly along the flat valleys of rivers flowing to Pannonia (e.g. the Bečva and the Váh rivers).
A group of diploid K. arvensis plants displaying significantly different genome sizes was found in relict habitats (serpentine outcrops and a subalpine glacial cirque) in the western part of the area, otherwise occupied solely by tetraploids. The same genome size was shared by the Slovakian endemic K. slovaca, which also grows exclusively in relict habitats, such as open pine forests on limestone outcrops (Štěpánek, 1983, 1985). These habitat preferences sharply contrast with the semi-natural/semi-ruderal character of stands occupied by the widespread (i.e. Pannonian) diploids and non-serpentine tetraploids. The genome size difference between diploids growing in relict (subsp. serpentinicola and subsp. pseudolongifolia) vs. semi-natural (subsp. pannonica) habitats provides support for the different evolutionary history of both types of plants. It is plausible that relict diploids originated in the early Holocene, and subsequently were forced into serpentine, limestone or alpine refugia by the expanding forest vegetation. Non-relict diploids and tetraploids could have migrated to Central Europe later, perhaps as a consequence of human-induced changes in the landscape, such as deforestation, grazing and/or meadow agriculture (Štěpánek, 1989; Kaplan, 1998).
Although caution should be taken when small differences in genome size are interpreted (Bennett et al., 2008), we are convinced that our results are not negatively influenced by instrumental or methodological errors, including the presence of disturbing secondary metabolites. First, low coefficients of variation were achieved, which are not compatible with the presence of interfering metabolites. It should also be noted that several plants from each group were cultivated in homogeneous conditions (for up to 1 year) before flow analyses. Moreover, between-plant differences remained stable in analyses with either intercalating propidium iodide or AT-selective DAPI (a fluorochrome less sensitive to secondary metabolites). In addition, co-processed samples with different genome sizes always gave two distinct peaks, which is considered the most convincing evidence for genuine differences in nuclear DNA content (Greilhuber, 2005).
Primary and secondary contact zones
If more chromosomal races exist within a species, zones of ploidy overlap are often formed (Husband and Schemske, 1998; Weiss et al., 2002; Suda et al., 2007b). Primary contact zones differ from secondary contact zones in their evolutionary history (Petit et al., 1999). The former zones arise as a direct consequence of the emergence of a (higher) polyploid within a diploid/lower polyploid population, while the latter results from a secondary contact between previously allopatric cytotypes. The close genetic proximity between coexisting individuals of different ploidy levels characterizes primary zones; in contrast, secondary contact zones are usually composed of distantly related individuals because they combine genetically distinct parental gene pools. While genetic variation is the most robust criterion for discrimination between the two types of contact zones, additional evidence can be obtained from cytological, phenotypic and/or distributional data. Our FCM results support the existence of both primary and secondary contact zones in K. arvensis. A distinct geographical pattern in non-relict diploids and tetraploids indicates a secondary contact zone, which stretches from northern Austria through Moravia to southern Poland. An approximate westward continuation of the zone of sympatry is outlined by Ehrendorfer (1962b, 1976) and Breton Sintes (1969); the cytotype contact zone runs from SE Austria along the southern flanks of the Alps, and through southern France up to the Pyrenees. The situation in east and SE Europe is, however, unknown and, together with the sparse information available from west Europe, calls for further investigation. In general, the transient zone of K. arvensis is diffuse and quite wide (several dozens of kilometres). Similar conditions (mixed populations scattered over large geographical areas) have been observed, for instance, for the grass Pennisetum sect. Brevivalvula, Poaceae (Renno et al., 1995). Nonetheless, narrow sympatric zones usually seem to prevail in diploid–polyploid plant groups (e.g. van Dijk et al., 1992; Liebenberg et al., 1993; Husband and Schemske, 1998).
Relict Knautia populations demonstrate a very different pattern. The coexistence of 2x and 4x plants in two serpentine populations in Western Bohemia (Vlčí hřbet, no. 277, and Planý vrch, no. 278; see Supplementary Data) suggests the independent autopolyploid origin of serpentine tetraploids from their diploid counterparts, and thus primary cytotype contact. There are a few more plant groups for which both primary and secondary contacts have been suggested, including Dianthus, Caryophyllaceae (Weiss et al., 2002) and Melampodium, Asteraceae (Stuessy et al., 2004). The hypothesis stating the independent evolutionary history of serpentine Knautia populations was first postulated by Kaplan (1998), on the basis of phenotypic variation and ecological observations. The data herein demonstrating novel ploidies and genome sizes support this hypothesis. Initially, a minority of diploid cytotypes was detected in two serpentine localities where only tetraploids had been previously known to exist. Furthermore, the slightly smaller genome size of serpentine tetraploids in comparison with their non-serpentine counterparts may be regarded as additional, supplementary evidence for in situ polyploid evolution in relict serpentine habitats. Independent evolution of serpentine populations was also proposed by Westerbergh and Saura (1992) when explaining genetic differences between ruderal and serpentine plants of Silene dioica (Caryophyllaceae) in central Sweden. Nevertheless, the possibility of independent colonization of serpentine outcrops by neighbouring grassland tetraploids of Knautia can not be rejected at this stage of investigation and molecular data are required to reach a final conclusion. Provided that the independent in situ origin of serpentine tetraploids is confirmed, the K. arvensis agg. renders a unique opportunity for a comparative study of patterns and processes in primary vs. secondary diploid–polyploid contact zones.
Spatial segregation of cytotypes in contact zones
Special attention was paid to the distribution of different ploidy levels in contact zones in order to (1) evaluate the level of among-cytotype interactions and (2) assess potential ecological and/or phenological differences among cytotypes. On both the regional and microgeographic scales, the two majority cytotypes (2x, 4x) were more-or-less spatially segregated. This segregation is well documented by a low number of mixed-ploidy populations (see Figs 3 and 4A). One reason for ploidy segregation at the regional scale seems to be certain cytotype preferences for different altitudes and/or geomorphology of the terrain. Whereas diploids in the contact zone mostly grew in flat lowlands or in valleys of large rivers, tetraploids were more often found in more rolling landscapes. Altitudinal separation of different cytotypes does not seem to be a rare phenomenon in plants exhibiting ploidy heterogeneity, as previously documented, for instance, for Anthoxanthum alpinum, Poaceae (Felber-Girard et al., 1996), Senecio carniolicus, Asteraceae (Schönswetter et al., 2007) and Allium przewalskianum, Alliaceae (Xie-Kui et al., 2008).
The spatial segregation observed at the regional scale was mirrored in selected mixed-ploidy plots within localities. The cytotype distribution observed in the two diploid–tetraploid plots (i.e. non-serpentine Ždánice, no. 267, and serpentine Planý vrch, no. 278) was more-or-less non-random. Because the vegetation cover at both plots was quite homogeneous (F. Kolář et al., pers. obs.), we do not consider microhabitat differentiation as the major driving force behind the spatial segregation. Rather, our non-exclusive explanation takes into account a founder effect and limited distribution capability of Knautia seeds. The seeds are dry, rather heavy, and lack any surface structures that facilitate exozoochory. Ants are sometimes discussed as dispersing agents (Ehrendorfer, 1962a; Štěpánek, 1997), yet they usually carry seeds for short distances only (Gómez and Espadaler, 1998). A combination of the above-mentioned factors can lead to the formation of single-cytotype patches, even in populations exhibiting ploidy heterogeneity. Indeed, clusters of seedlings were often observed in close proximity to the putative mother plant. An alternative hypothesis assuming that the observed distribution pattern in fact represents a transitional stage in the gradual competitive exclusion of one cytotype can not be ruled out at this stage of investigation.
On the other hand, cytotype sorting along an environmental gradient was observed in the locality of Horní Planá (no. 288) where 4x K. arvensis coexisted with 6x K. dipsacifolia (plus some 5x hybrids). The former species preferred open grassy parts of the plot, while the latter taxon grew mostly in shady parts of the plot close to the forest margin. DNA-pentaploids occupied an ecologically transitional semi-shaded zone of the plot.
Differential reproductive barriers among different cytotypes
In recent years, an increasing number of studies have investigated the role of different pre- and post-zygotic reproductive barriers in the evolutionary dynamics of mixed-ploidy populations (see Husband and Sabara, 2004, and references therein). A cumulative effect of all isolating mechanisms is apparent in the frequency of heteroploid crosses in a population.
In the present study, we observed dramatic differences in the frequency of heteroploid crosses between different cytotype combinations, irrespective of the taxonomic identity of the plant material. Only two triploids were encountered among more than 5400 Knautia plants (adults + seedlings) from 292 populations cytotyped in the present study (<0·04 %). They both occurred in otherwise diploid populations (Haidhof bei Baden, no. 262, and a serpentine locality Bernartice, no. 263; see Supplementary Data), suggesting that a fusion of unreduced and reduced gametes is the most plausible mechanism giving rise to triploidy. No triploids have been detected among adult plants or seedlings in mixed 2x+ 4x populations, despite intensive ploidy screening. A virtual lack of adult triploids in our study is consistent with previous karyological surveys because, within the entire genus, triploidy has been reported only twice: (1) in the artificial interspecific hybrid K. basaltica Chassagne & Szabó × K. arvernensis (Briq.) Szabó in France (Breton Sitens and Cauderon, 1969), and (2) in a natural population of K. tatarica (L.) Szabó in Russia (Plaksina, 1999). Additional support for the evolution of strong reproductive barriers between diploids and polyploids is provided by a series of experimental crosses performed by Ehrendorfer (1962a) and Breton Sintes (1974, 1975). Certain ecological segregation of cytotypes in the zone of sympatry (e.g. sorting according to altitude or geomorphology), as observed in our study, may serve as an additional pre-zygotic barrier. Moreover, pollinator foraging behaviour as a consequence of different flower colours in some regions with ploidy mixing (violet in diploids vs. whitish in tetraploids due to the introgression of K. kitaibelii) may further contribute to reproductive isolation. However, the mating barrier between 2x and 4x Knautia cytotypes is not absolute, as revealed by the presence of a triploid seed recorded in a serpentine population exhibiting ploidy heterogeneity (Planý vrch, no. 278). It is therefore possible that interploidy crosses resulting in triploid seeds may occasionally occur in mixed 2x + 4x populations; however, the offspring are either non-viable or are later outcompeted due to their reduced fitness. It should also be noted that we cannot at present reject an alternative hybridization scenario involving unidirectional gene flow from 2x to 4x cytotypes via the formation of unreduced gametes from diploid parents and the production of tetraploid hybrids.
The increase in the genome copy number of Knautia seems to be associated with the relaxation of interploidy reproductive barriers. In the present study, we recorded individuals having intermediate ploidy levels in four out of five mixed 4x + 6x populations, even though these cytotypes corresponded to different parental species (K. arvensis and K. dipsacifolia, respectively). In fact, pentaploid crosses were present in all three stages examined (i.e. adult plants, seedlings and seeds). Although both parental species have distinct ecological preferences (Štěpánek, 1997), individuals can coexist at ecotones such as forest margins, where they are highly susceptible to hybridization. Unlike Štěpánek (1997), we are convinced that pentaploid hybrids are fertile and may further influence the evolutionary dynamics of mixed-ploidy populations. Vigorous and regularly flowering plants that set well-developed seeds were observed in population Horní Planá (no. 288). Pentaploids often seem to form aneuploid seeds, as indicated by the pronounced variation in fluorescence intensity observed for such samples. In addition, gametes having a euploid number of chromosomes may be involved in back-crossing with parental species. The incidence of a presumably tetraploid seed on a 5x maternal plant (Table 3) and the discovery of an adult hexaploid with divided leaves (i.e. a characteristic otherwise restricted to tetraploids) in a mixed-ploidy population both support this hypothesis. The observed ploidy-specific reproductive behaviour of Knautia (strong mating barriers between 2x and 4x cytotypes vs. certain compatibility between different polyploids) closely matches the situation reported about five decades ago for Achillea, Asteraceae (Schneider, 1958).
Conclusions
In the present study, it has been demonstrated that Knautia arvensis is an intriguing taxon that shows variation in both genome copy number and monoploid genome size. Diploid and tetraploid cytotypes exhibit spatial segregation at all the geographical scales examined, including the Central European portion of the distribution range, the zone of ploidy overlap, the mixed-ploidy populations, and the selected mixed-ploidy plots within such populations. Distributional and phenotypic data support the existence of both primary and secondary zones of cytotype contact. Irrespective of taxonomic affinity, ploidy level seems to be the major determinant of the strength of interploidy reproductive isolation. In addition, our study also highlights the importance of involving different ontogenetic stages when assessing evolutionary processes in mixed-ploidy populations. Collectively, Knautia arvensis provides a unique model system for studying the evolutionary dynamics of populations exhibiting ploidy heterogeneity, and for examining ecological and genetic circumstances that govern the interactions between different cytotypes in mixed populations.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank J. Vojta for aiding with the map construction, V. Jarolímová for advice regarding chromosome counting, J. Štěpánek, Z. Kaplan and F. Ehrendorfer for fruitful discussions, and J. Loureiro for comments on an earlier draft of the manuscript. We are also indebted to our colleagues who collected some samples and/or recommended suitable localities, namely P. Koutecký, M. Dortová, R. Sudová, J. Hurtová, J. Těšitel, T. Malinová, Z. Mruzíková, T. Tyml, T. Bodnár, E. Patáčová, A. Matoušů, K. Zimmerman, D. Novotný, P. Janšta, J. Straka, P. Kúr, M. Slovák, K. Prach, A. Jírová, J. Kochánková, P. Lepší, M. Lepší, P. Petřík, K. Boublík, J. Douda, R. Hlaváček, L. Mořkovský and M. Weiser. This work was supported by the Grant Agency of the Academy of Sciences of the Czech Republic (grant number B601110627). Additional support was supplied by the Ministry of Education, Youth and Sports of the Czech Republic (MSM 0021620828 and MSM6007665801), and the Academy of Sciences of the Czech Republic (AV0Z60050516).
LITERATURE CITED
- Baack EJ. Cytotype segregation on regional and microgeographic scales in snow buttercups (Ranunculus adoneus: Ranunculaceae) American Journal of Botany. 2004;91:1783–1788. doi: 10.3732/ajb.91.11.1783. [DOI] [PubMed] [Google Scholar]
- Baack EJ. Ecological factors influencing tetraploid, establishment in snow buttercups (Ranunculus adoneus, Ranunculaceae): minority cytotype exclusion and barriers to triploid formation. American Journal of Botany. 2005;92:1827–1835. doi: 10.3732/ajb.92.11.1827. [DOI] [PubMed] [Google Scholar]
- Baack EJ, Stanton ML. Ecological factors influencing tetraploid speciation in snow buttercups (Ranunculus adoneus): Niche differentiation and tetraploid establishment. Evolution. 2005;59:1936–1944. [PubMed] [Google Scholar]
- Bennett MD, Price HJ, Johnston JS. Anthocyanin inhibits propidium iodide DNA fluorescence in Euphorbia pulcherrima: implications for genome size variation and flow cytometry. Annals of Botany. 2008;101:777–790. doi: 10.1093/aob/mcm303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet E, van de Peer Y. zt: a software tool for simple and partial Mantel tests. Journal of Statistical Software. 2002;7:1–12. [Google Scholar]
- Breton-Sintes S. Étude cytotaxonomique du Knautia arvensis (L.) Coult. diploïde (2n = 20). Sa répartition en Auvergne. Bulletin de la Société botanique de France. 1969;116:189–193. [Google Scholar]
- Breton-Sintes S. Etude biosystematique du genre Knautia (Dipsacaceae) dans le Massif Central français. I. Analyse morphologique et cytogenetique d'hybrides experimentaux. Annales des Sciences Naturelles, Botanique. 1974;12:277–320. [Google Scholar]
- Breton-Sintes S. Cytogenetic contribution to the study of Knautia (Dipsacaceae) of the central French plateau. Annee Biologique. 1975;14:45–68. [Google Scholar]
- Breton-Sintes S, Cauderon Y. Analyse morphologique et cytogénétique d'un hybride triploïde (Knautia balsatica × K. arvernensis) Bulletin de la Société botanique de France. 1969;116:125–136. [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- van Dijk P, Hartog M, Vandelden W. Single cytotype areas in autopolyploid Plantago media L. Biological Journal of the Linnean Society. 1992;46:315–331. [Google Scholar]
- Dimitrova D, Ebert I, Greilhuber J, Kozhuharov S. Karyotype constancy and genome size variation in Bulgarian Crepis foetida s. l. (Asteraceae) Plant Systematics and Evolution. 1999;217:245–257. [Google Scholar]
- Doležel J, Greilhuber J, Lucretti S, et al. Plant genome size estimation by flow cytometry: inter-laboratory comparison. Annals of Botany. 1998;82(Suppl. A):17–26. [Google Scholar]
- Doležel J, Greilhuber J, Suda J. Estimation of nuclear DNA content in plants using flow cytometry. Nature Protocols. 2007;2:2233–2244. doi: 10.1038/nprot.2007.310. [DOI] [PubMed] [Google Scholar]
- Dvořáková M. Minuartia smejkalii, eine neue Art aus der Minuartia gerardii-Gruppe (Caryophyllaceae) Preslia. 1988;60:1–9. [Google Scholar]
- Dyer AF. The use of lacto-propionic orcein in rapid squash methods for chromosome preparations. Stain Technology. 1963;38:85–90. [Google Scholar]
- Ehrendorfer F. Beiträge zur Phylogenie der Gattung Knautia (Dipsacaceae), I. Cytologische Grundlagen und allgemeine Hinweise. Österreichische Botanische Zeitschrift. 1962;a 109:276–343. [Google Scholar]
- Ehrendorfer F. Cytotaxonomische Beiträge zur Genese der mitteleuropäischen Flora und Vegetation. Berichte der Deutschen Botanischen Gesellschaft. 1962;b 75:137–152. [Google Scholar]
- Ehrendorfer F. Evolution and karyotype differentiation in a family of flowering plants: Dipsacaceae. In: Geerts SJ, editor. Genetics today (Proceedings of the XI International Congress of Genetics) Oxford: Symposium Publications Division, Pergamon Press; 1964. pp. 399–407. [Google Scholar]
- Ehrendorfer F. Knautia L. In: Tutin T, editor. Flora Europaea. Vol. 4. Cambridge University Press; 1976. pp. 60–67. [Google Scholar]
- Favarger C. Cytogeography and biosystematics. In: Grant WF, editor. Plant biosystematics. Toronto, New York: Academic Press; 1984. pp. 453–476. [Google Scholar]
- Felber-Girard M, Felber F, Buttler A. Habitat differentiation in a narrow hybrid zone between diploid and tetraploid Anthoxanthum alpinum. New Phytologist. 1996;133:531–540. [Google Scholar]
- Frey L. Karyological studies in some flowering plants in Poland. Fragmenta Floristica et Geobotanica. 1969;15:261–267. [Google Scholar]
- Gómez C, Espadaler X. Seed dispersal curve of a Mediterranean myrmecochore: influence of ant size and the distance to nests. Ecological Research. 1998;13:347–354. [Google Scholar]
- Greilhuber J. Intraspecific variation in genome size in angiosperms: identifying its existence. Annals of Botany. 2005;95:91–98. doi: 10.1093/aob/mci004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greilhuber J, Doležel J, Lysák MA, Bennett MD. The origin, evolution and proposed stabilization of the terms ‘genome size’ and ‘C-value’ to describe nuclear DNA contents. Annals of Botany. 2005;95:255–260. doi: 10.1093/aob/mci019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodálová I, Grulich V, Horová L, Valachovič M, Marhold K. Occurrence of tetraploid and octoploid cytotypes in Senecio jacobaea ssp. jacobaea (Asteraceae) in Pannonia and the Carpathians. Botanical Journal of the Linnean Society. 2007;153:231–242. [Google Scholar]
- Husband BC, Sabara HA. Reproductive isolation between autotetraploids and their diploid progenitors in fireweed, Chamerion angustifolium (Onagraceae) New Phytologist. 2004;161:703–713. doi: 10.1046/j.1469-8137.2004.00998.x. [DOI] [PubMed] [Google Scholar]
- Husband BC, Schemske DW. Cytotype distribution at a diploid–tetraploid contact zone in Chamerion (Epilobium) angustifolium (Onagraceae) American Journal of Botany. 1998;85:1688–1694. [PubMed] [Google Scholar]
- Kaplan Z. Relict serpentine populations of Knautia arvensis s. l. (Dipsacaceae) in the Czech Republic and an adjacent area of Germany. Preslia. 1998;70:21–31. [Google Scholar]
- Kron P, Suda J, Husband BC. Applications of flow cytometry to evolutionary and population biology. Annual Review of Ecology Evolution and Systematics. 2007;38:847–876. [Google Scholar]
- Liebenberg H, Lubbinge J, Fossey A. Cytotaxonomic studies in Themeda triandra Forssk.: IV. A population cytogenetic study of a contact zone between tetraploid and hexaploid populations. South African Journal of Botany. 1993;59:305–310. [Google Scholar]
- Lihová J, Tribsch A, Marhold K. The Cardamine pratensis (Brassicaceae) group in the Iberian Peninsula: taxonomy, polyploidy and distribution. Taxon. 2003;52:783–802. [Google Scholar]
- Mahelka V, Suda J, Jarolímová V, Trávníček P, Krahulec F. Genome size discriminates between closely related taxa Elytrigia repens and E. intermedia (Poaceae: Triticeae) and their hybrid. Folia Geobotanica. 2005;40:367–384. [Google Scholar]
- Májovský J. Index of chromosome numbers of Slovakian flora. Part 5. Acta Facultatis Rerum Naturalium Universitatis Comeniae. 1976;25:1–18. [Google Scholar]
- Manly BJF. Randomization, bootstrap and Monte Carlo methods in biology. London: Chapman and Hall; 1991. [Google Scholar]
- Murray BG. When does intraspecific C-value variation become taxonomically significant? Annals of Botany. 2005;95:119–125. doi: 10.1093/aob/mci007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák FA. Fylogenese serpentinových typů. Preslia. 1960;32:1–8. [Google Scholar]
- Otto F. DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. In: Crissman HA, Darzynkiewicz Z, editors. Methods in cell biology. vol. 33. New York: Academic Press; 1990. pp. 105–110. [DOI] [PubMed] [Google Scholar]
- Otto SP, Whitton J. Polyploid incidence and evolution. Annual Review of Genetics. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Perný M, Kolarčik V, Majeský L, Mártonfi P. Cytogeography of the Phleum pratense group (Poaceae) in the Carpathians and Pannonia. Botanical Journal of the Linnean Society. 2008;157:475–485. [Google Scholar]
- Petit C, Bretagnolle F, Felber F. Evolutionary consequences of diploid–polyploid hybrid zones in wild species. Trends in Ecology & Evolution. 1999;14:306–311. doi: 10.1016/s0169-5347(99)01608-0. [DOI] [PubMed] [Google Scholar]
- Plaksina TI. Samarskaya luka as a natural phenomenon of the middle Povolzhje. Vestnik Samarskogo Gosudarstvennogo Universiteta. Matematika, Mekhanika, Fizika, Khimiya. 1999;2:158–171. [Google Scholar]
- Renno JF, Schmelzer GH, deJong JH. Variation and geographical distribution of ploidy levels in Pennisetum section Brevivalvula (Poaceae) in Burkina Faso, Benin and southern Niger. Plant Systematics and Evolution. 1995;198:89–100. [Google Scholar]
- Rivero-Guerra AO. Cytogenetics, geographical distribution, and pollen fertility of diploid and tetraploid cytotypes of Santolina pectinata Lag. (Asteraceae: Anthemideae) Botanical Journal of the Linnean Society. 2008;156:657–667. [Google Scholar]
- Rosenbaumová R, Plačková I, Suda J. Variation in Lamium subg. Galeobdolon (Lamiaceae) – insights from ploidy levels, morphology and isozymes. Plant Systematics and Evolution. 2004;244:219–244. [Google Scholar]
- SAS Institute. SAS OnlineDoc®, v.8. Cary, NC: SAS Institute; 2000. (available online) [Google Scholar]
- Schneider I. Zytogenetische Untersuchungen an Sippen des Polyploid-Komplexes Achillea millefolium L. s. lat. (Zur Phylogenie der Gattung Achillea, I) Österreichische Botanische Zeitschrift. 1958;105:111–158. [Google Scholar]
- Schönswetter P, Suda J, Popp M, Weiss-Schneeweiss H, Brochmann C. Circumpolar phylogeography of Juncus biglumis (Juncaceae) inferred from AFLP fingerprints, cpDNA sequences, nuclear DNA content and chromosome numbers. Molecular Phylogenetics and Evolution. 2007;42:92–103. doi: 10.1016/j.ympev.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS. Polyploidy: recurrent formation and genome evolution. Trends in Ecology & Evolution. 1999;14:348–352. doi: 10.1016/s0169-5347(99)01638-9. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Schemske DW, et al. Autopolyploidy in angiosperms: have we grossly underestimated the number of species? Taxon. 2007;56:13–30. [Google Scholar]
- Soltis PS. Ancient and recent polyploidy in angiosperms. New Phytologist. 2005;166:5–8. doi: 10.1111/j.1469-8137.2005.01379.x. [DOI] [PubMed] [Google Scholar]
- StatSoft. STATISTICA (data analysis software system), version 7.1. Tulsa, OK, USA: StatSoft Inc; 2005. [Google Scholar]
- Stuessy TF, Weiss-Schneeweiss H, Keil DJ. Diploid and polyploid cytotype distribution in Melampodium cinereum and M. leucanthum (Asteraceae, Heliantheae) American Journal of Botany. 2004;91:889–898. doi: 10.3732/ajb.91.6.889. [DOI] [PubMed] [Google Scholar]
- Štěpánek J. Die Chromosomenzahlen von tschechoslowakischen Arten der Gattung Knautia L. (Dipsacaceae) Folia Geobotanica et Phytotaxonomica. 1982;17:359–386. [Google Scholar]
- Štěpánek J. Eine neue Art der Gattung Knautia (Dipsacaceae) aus Westkarpaten. Preslia. 1983;55:1–8. [Google Scholar]
- Štěpánek J. Knautia L. In: Bertová L, editor. Flóra Slovenska, Vol. IV/2. Bratislava: Veda; 1985. pp. 154–177. [Google Scholar]
- Štěpánek J. Chrastavec rolní krkonošský – Knautia arvensis (L.) Coulter subsp. pseudolongifolia (Szabó) O. Schwarz. In: Slavík B, et al., editors. Studie ČSAV 10: Vybrané ohrožené druhy flóry ČSR. Praha: Academia; 1989. pp. 25–36. [Google Scholar]
- Štěpánek J. Knautia L. – chrastavec. In: Slavík B, editor. Květena České republiky. Vol. 6. Praha: Academia; 1997. pp. 543–554. [Google Scholar]
- Suda J, Krahulcová A, Trávníček P, Krahulec F. Ploidy level versus DNA ploidy level: an appeal for consistent terminology. Taxon. 2006;55:447–450. [Google Scholar]
- Suda J, Kron P, Husband BC, Trávníček P. Flow cytometry and ploidy: applications in plant systematics, ecology and evolutionary biology. In: Doležel J, Greilhuber J, Suda J, editors. Flow cytometry with plant cells. Analysis of genes, chromosomes and genomes. a. Weinheim: Wiley-VCH; 2007. pp. 103–130. [Google Scholar]
- Suda J, Weiβ-Schneeweiβ H, Tribsch A, Schneeweiβ G, Trávníček P, Schönswetter P. Complex distribution patterns of di-, tetra- and hexaploid cytotypes in the European high mountain plant Senecio carniolicus Willd. (Asteraceae) American Journal of Botany. 2007;b 94:1391–1401. doi: 10.3732/ajb.94.8.1391. [DOI] [PubMed] [Google Scholar]
- Weiss H, Dobes C, Schneeweiss GM, Greimler J. Occurrence of tetraploid and hexaploid cytotypes between and within populations in Dianthus sect. Plumaria (Caryophyllaceae) New Phytologist. 2002;156:85–94. [Google Scholar]
- Westerbergh A, Saura A. The effect of serpentine on the population structure of Silene dioica (Caryophyllaceae) Evolution. 1992;46:1537–1548. doi: 10.1111/j.1558-5646.1992.tb01143.x. [DOI] [PubMed] [Google Scholar]
- Xie-Kui C, Ao CQ, Zhang Q, Chen LT, Liu JQ. Diploid and tetraploid distribution of Allium przewalskianum Regel. (Liliaceae) in the Qinghai–Tibetan Plateau and adjacent regions. Caryologia. 2008;61:190–198. [Google Scholar]