Abstract
Background and Aims
Section Acrocentron of the genus Centaurea is one of the largest sections of Centaurea with approx. 100 species. The geographic distribution, centred in the Mediterranean, makes it an excellent example for studies of the biogeographic history of this biodiversity-rich region.
Methods
Plastid (trnH-psbA) and nuclear (ITS and ETS) DNA sequence analysis was used for phylogenetic reconstruction. Ancestral biogeographic patterns were inferred by dispersal-vicariance analysis (DIVA).
Key Results
The resulting phylogeny has implications for the sectional classification of Acrocentron and confirms merging sect. Chamaecyanus into Acrocentron as a subsection. Previous suggestions of an eastern Mediterranean origin of the group are confirmed. The main centres of diversification established in previous studies are now strongly supported. Expansion of the group in two different radiations that followed patently diverse paths is inferred.
Conclusions
Radiation followed two waves, widely separated in time scale. The oldest one, from Turkey to Greece and the northern Balkans and then to North Africa and Iberia, should be dated at the end of the Miocene in the Messinian period. It reached the Iberian Peninsula from the south, following a route that is landmarked by several relictic taxa in Sicily and North Africa. A later radiation during the Holocene interglacial periods followed, involving species from the north of the Balkan Peninsula, along a Eurasian pathway running from Central Iberia to the steppes of Kazakhstan. A generalized pattern of reticulation is also evident from the results, indicating past contacts between presently separated species. Molecular data also confirmed the extent of hybridization within Acrocentron and were successful in reconstructing the paleogeography of the section.
Key words: Centaurea sect. Acrocentron, Cardueae, dispersal-vicariance, ETS, ITS, Mediterranean, phylogeny, psbA-trnH, radiation, reticulation
INTRODUCTION
The tribe Cardueae (Compositae) has been deeply reviewed and widely modified in the past 10 years. Most of the many systematic problems within the tribe, from the subtribal classification to the delineation of genera, are now more clear (Susanna et al., 1995, 2006; Garcia-Jacas et al., 2001). This has been a complicated task because the tribe includes some of the largest genera of the whole family (Centaurea L., Cirsium Mill., Cousinia Cass. and Saussurea DC.). Our present research centres in one of these large genera, Centaurea.
In its present delimitation as adopted in the latest revision of the Cardueae (Susanna and Garcia-Jacas, 2007), Centaurea comprises approx. 250 species (from 400 in earlier classifications; e.g. Dittrich, 1977). Centaurea encompasses only the large Jacea group (revised by Garcia-Jacas et al., 2006), the section Cyanus and the Acrocentron group.
The Acrocentron group is formed by Centaurea sect. Acrocentron (Cass.) DC. with the addition of two much smaller sections: Centaurea sect. Chamaecyanus Willk. and C. sect. Stephanochilus Coss. & Dur. A previous proposal by Wagenitz and Hellwig (1996) also included Centaurea sect. Crocodylium DC. and Centaurea sect. Aegialophila (Boiss. & Heldr.) O. Hoffm., but these two sections should be excluded and merged in a different genus, Crocodylium Cass. (Garcia-Jacas et al., 2001; Vilatersana et al., 2001; Font et al., 2002). Among other characters, the Acrocentron group is well defined by having one of the four pollen types described in the genus Centaurea, the Centaurea scabiosa pollen type (Wagenitz, 1955). A previous molecular survey of the group using sequences of the ITS region (Font et al., 2002) only confirmed the inclusion of sect. Stephanochilus and the exclusion of the restored genus Crocodylium, and suggested merging C. sect. Chamaecyanus in C. sect. Acrocentron. However, for the rest, the resulting trees consisted largely of unsupported polytomies. The largest part of the Acrocentron group is Centaurea sect. Acrocentron, a huge section with approx. 100 species mainly in the Mediterranean region.
An aspect of special interest to the present contribution is the history of expansion of Centaurea sect. Acrocentron through the Mediterranean. Reconstruction of the paleogeographic history of plants is a relatively novel focus in botany, and the present distribution of C. sect. Acrocentron in the Mediterranean region makes the group an excellent object of study. In our previous molecular survey, the existence of three main centres of speciation (Anatolian–Aegean, North Balkanic–Eurasian, and Iberian–North African) was suggested. However, as previously noted, the resulting phylogeny was flawed by an extremely low support for most of the clades. A detailed phylogeny of the related Centaurea group Jacea (Garcia-Jacas et al., 2006) confirmed the existence of the three centres of speciation hinted at in Font et al. (2002). A recent paper dealing with the related Acrolophus–Phalolepis complex of Centaurea has contributed to our knowledge of the expansion of Centaurea in the Iberian Peninsula and North Africa (Suárez-Santiago et al., 2007). It is tempting to verify whether Acrocentron follows similar patterns of colonization of the Mediterranean.
The general lack of support in our previous report was considered a result of intense historical gene flow (Font et al., 2002). Hybridization in the Acrocentron group is well documented (Kummer, 1977; Fernández Casas and Susanna, 1986; Garcia-Jacas and Susanna, 1993, 1994; Garcia-Jacas, 1998; Hellwig et al., 1994). Hybrids are often homoploid (Kummer, 1977; Fernández Casas and Susanna, 1986), which favours gene flow because homoploid hybrids act as a bridge between the parental species (McKinnon, 2005).
Based on these previous results we decided to carry out a new study with the addition of the ETS region of the nuclear ribosomal DNA and the non-coding intergenic spacer trnH-psbA of plastid DNA. Incongruence between nuclear and plastid phylogenies is an important indicator of historical interspecific gene flow (Wendel and Doyle, 1998). The ETS region has proved to be a good tool for the phylogenetic reconstruction of closely related species in many genera from the Compositae (Calycadenia, Baldwin and Markos, 1998; Helianthus, Linder et al., 2000; Lessingia, Markos and Baldwin, 2001; Xylothamia and Gundlachia, Urbatsch et al., 2003; Montanoa, Plovanich and Panero, 2004) and has already been very useful in the Cardueae–Carduinae (Kelch and Baldwin, 2003) and Cardueae–Centaureinae (Hidalgo et al., 2006). Moreover, the ETS region has demonstrated great potential for unraveling the extent of gene flow in Centaurea sect. Willkommia (Suárez-Santiago et al., 2007). Our goals with this combined approach are as follows: (1) to examine whether a phylogenetic subsectional classification of Centaurea sect. Acrocentron is supported by molecular data and to confirm the inclusion of C. sect. Chamaecyanus in sect. Acrocentron with subsectional rank; (2) to seek for molecular evidence of hybridization and gene flow among C. sect. Acrocentron; (3) to verify the existence of the suggested centres of speciation of the group; and (4) to explore and reconstruct the history of the expansion of C. sect. Acrocentron through the Mediterranean and establish the centre of origin of the group.
MATERIAL AND METHODS
Plant material
The ETS (external transcribed spacer) and ITS (internal transcribed spacer) regions of the nrDNA of 45 taxa of Centaurea sect. Acrocentron and C. sect. Chamaecyanus were sequenced. In some cases, more than one population was sequenced. Our ITS1 and ITS2 sequences from previous work (Font et al., 2002) were complemented by sequencing the 5·8S region. Sampling included all the geographical areas of distribution of the section: eastern Mediterranean (Balkan Peninsula and Anatolia), Irano-Turanian, western Mediterranean (Iberian and Italian Peninsulas, and North Africa) and some species of wide distribution, such as Centaurea scabiosa L.
A plastid non-coding region, the intergenic spacer trnH-psbA, was also sequenced. In view of the very low number of informative characters found in this region, only a representative subset of 31 taxa was sequenced (Table 1).
Table 1.
Species, origin of the material, herbaria and GenBank accession numbers
| Species | Voucher | ITS accession | ETS accession | trnH-psbA accession |
|---|---|---|---|---|
| Centaurea aetholica Phytos & Georgadis | Greece, Achaia: Antirion-Gavrolimni, Damboldt s. n. (B). | FJ459646 | FJ459583 | – |
| Centaurea amblensis Graells | Spain, Ávila: El Barco de Ávila, sine col. (BC). | FJ459647 | FJ459584 | FJ459697 |
| Centaurea amourensis Pomel | Morocco, Zagora: north-east side of the Atlas, M. Staudinger & M. Finckh (BC).* | FJ459648 |
FJ459585 FJ459586 |
FJ459698 |
| Centaurea argecillensis Gredilla | Spain, Guadalajara: Argecilla, mountain slopes on the road to Ledanca, M. Font & Susanna 1811 (BC).* | FJ459649 |
FJ459587 FJ459588 FJ459589 |
FJ459699 |
| Centaurea borjae Valdés-Berm. & Rivas Goday | Spain, La Coruña: near El Ferrol, cabo Prior, S side, Garcia-Jacas & Susanna 2074 (BC). | FJ459650 | FJ459590 | FJ459700 |
| Centaurea carduiformis DC | Armenia, Talin: between Pokr Arthik and Bagravan, Susanna 1527 & al. (BC). | FJ459651 | FJ459591 | – |
| Centaurea carolipauana Fern. Casas & Susanna | Morocco, Tetuan: Djebel Tasaot, 14·5 km from the cross to Talembote, Garcia-Jacas & Susanna 1437 (BC). | AY826253 | FJ459592 | FJ459701 |
| Centaurea cephalariifolia Willk. | Spain, Huesca: between Capdesaso and San Lorenzo del Flumen, Vilatersana 20 (BC). | FJ459652 | FJ459593 | FJ459702 |
| Centaurea clementei Boiss. | Spain, Botanical Garden of Barcelona (BC).* | FJ459653 | FJ459594 | FJ459703 |
| FJ459595 | ||||
| Centaurea collina L. | Spain, Alacant: between Muro de Alcoi and Beniarrés, Susanna 1481 & al. (BC). | FJ459654 | FJ459596 | FJ459704 |
| Centaurea crocata Franco | Portugal, Algarve: 5 km N of Monchique on the road to Odemira, Roché & Susanna 1917 (BC). | FJ459655 | FJ459597 | FJ459705 |
| Centaurea ebenoides Heldr. | Greece, Euboea island: near Limni, on the coast road, Phytos & Kamari 20427 (B). | FJ459656 | FJ459598 | – |
| Centaurea euboica Rech. f. | Greece, Euboea island: Kandili mountains, between Achmet Aga and Hagios, Rechinger 18215 (B). | FJ459657 | FJ459599 | – |
| Centaurea gabrielis-blancae Fern. Casas | Spain, Navarra: Lumbier, Foz de Lumbier, Garcia-Jacas & Susanna 1592 (BC). | FJ459658 | FJ459600 | FJ459706 |
| Centaurea galianoi Fern. Casas & Susanna | Spain, Huelva: Sierra de Aracena, between Linares de la Sierra and Alaján, Garnatje 55 & Vilatersana (BC). | FJ459659 | FJ459601 | FJ459707 |
| Centaurea glehnii Trautv. | Armenia, Peninsula of Artanish, Susanna 1527 & al. (BC).* | FJ459660 | FJ459602 | FJ459708 |
| FJ459661 | ||||
| Centaurea granatensis Boiss. | Spain, Granada: Sierra Guillimona, Gorgas de Torilla, Valdés s. n. & al. (SEV). | FJ459662 | FJ459603 | FJ459709 |
| Centaurea grbavacensis (Rohlena) Stoj. & Acht. | Greece, Macedonia: Kozuf mountains, south side of mt. Tzena, Greuter 14108 (B). | FJ459663 | FJ459604 | – |
| Centaurea haenseleri Boiss. | Spain, Málaga: 10 km to Jubrique, Garcia-Jacas & Susanna 1888 (BC). | FJ459664 | FJ459605 | FJ459710 |
| Centaurea jankae Brandza | Romania: Dobrogea, Badarau s. n. (CLCB). | FJ459665 | FJ459606 | – |
| Centaurea josiae Humbert | Morocco, Ksar el Souk: Tizi'n Talrhemt, Susanna 1792 & al. (BC).* | FJ459666 | FJ459607 | – |
| FJ459608 | ||||
| Centaurea kandavanensis Wagenitz | Iran, Mazandaran: Chalus road, 2 km from the cross to Baladeh, Susanna 1621 & al. (BC). | FJ459667 | FJ459609 | FJ459711 |
| Centaurea kotschyana Heuff. | Romania, Botanical Garden ‘Al. Borza’, Cluj-Napoca (BC). | FJ459668 | FJ459610 | – |
| Centaurea kunkelii N. Garcia | Spain, Almería: road AL-411 between Roquetas and Canjáyar, Susanna 1612 & al. (BC).* | FJ459669 |
FJ459611 FJ459612 |
FJ459712 |
| Centaurea lagascana Graells | Spain, Palencia: 500 m from Alba de los Cardaños, M. Font & Susanna 1822 (BC).* | FJ459670 |
FJ459613 FJ459614 |
FJ459713 |
| Centaurea lainzii Fern. Casas | Spain, Sierra Bermeja: 11 Km from Estepona on the road to Igualeja, Garcia-Jacas & Susanna 1330 (BC). | FJ459671 | FJ459615 | FJ459714 |
| Centaurea legionis-septimae Fern. Casas & Susanna | Spain, León: pr. vicum Crémenes, Fernández Casas 5600 & Susanna (SEV). | FJ459672 | FJ459616 | FJ459715 |
| Centaurea litardierei Jahand. & Maire | Morocco, Ksar es Souk: south side of Col du Zad, Susanna 1797 & al. (BC). | FJ459673 | FJ459617 | FJ459716 |
| Centaurea luristanica Rech. f. | Iran, Khuzistan: ca. 15 km from Eizeh on the way to Dehdez, Mozaffarian s. n. (TARI). | FJ459674 | FJ459618 | – |
| Centaurea lydia Boiss. | Turkey, Konya: Seydisehir, Mortas, R. Ilarsan 4312 (ANK). | FJ459675 | FJ459619 | FJ459717 |
| Centaurea mariana Nyman | Spain, Almería: north side of the Sierra María, Garcia-Jacas & Susanna 1342 (BC). | FJ459676 | FJ459620 | FJ459718 |
| Centaurea nana Desf. | Morocco, Ksar es Souk: north side of Col du Zad, 1 km from Aïn Leuh on the road to Azrou, Susanna 1799 & al. (BC). | FJ459677 | FJ459621 | – |
| Centaurea ochrocephala Wagenitz | Iran, Azarbayjan-e-Gharbi: Orumiyeh, between Maranah and Haki, Susanna 1693 & al. (BC). | FJ459678 | FJ459622 | – |
| Centaurea ornata Willd. | Spain, Soria: near San Esteban de Gormaz, Garcia-Jacas & Susanna 1823 (BC), population 1. | FJ459679 | FJ459623 | FJ459719 |
| Spain, Huesca: road A-132 between Salinas and reservoir la Peña, Vilatersana 58 (BC), population 2. | FJ459680 | |||
| Centaurea podospermifolia Losc. & Pard. | Spain, Tarragona: Tortosa, 5 km from Monte Caro, Susanna 2072 & al. (BC). | FJ459681 | FJ459624 | FJ459720 |
| Centaurea prolongi Boiss. | Spain, Málaga: north side of the Sierra de Mijas, Garcia-Jacas & Susanna 1335 (BC), population 1. | FJ459682 | FJ459625 | FJ459721 |
| Spain, Málaga: Sierra of Almijara, Frigiliana, Galbany & Arrabal, s. n. (BC), population 2.* | FJ459683 |
FJ459627 FJ459628 |
– | |
| Portugal, Algarve: near Loulé, Cerro Botello over Fonte da Murta, Garcia-Jacas & Susanna 1353 (BC), population 3.* |
FJ459684 FJ459685 |
FJ459629 | FJ459722 | |
| Centaurea pseudoscabiosa Boiss. & Buhse | Turkey, Erzurum: between Erzurum and Varto, 13 km from Varto, Uysal 893 (KON). | FJ459686 | FJ459630 | – |
| Centaurea pubescens Wiilld. | Morocco, Ksar es Souk: 2 km from Oued Amesheguir, Garnatje, Susanna 1795 & Vilatersana (BC).* | FJ459687 | FJ459631 | FJ459723 |
| Centaurea raphanina Sibth. & Sm. | Greece, Chanion: Kydonias, mounts Levka, Strid 15093 & Papanicolau (B). | FJ459688 | FJ459632 | – |
| Centaurea rupestris L. | Greece, Macedonia: road E-90 between Véria and Kozáni, Roché & Susanna 1967 (BC). | FJ459689 | FJ459633 | – |
| Centaurea salonitana De Vis. | Greece, Macedonia: W from Efkarpia, Roché & Susanna 2005 (BC). | FJ459690 | FJ459634 | FJ459724 |
| Centaurea saxicola Lag. | Spain, Murcia: La Azohía, near the watchtower, Garcia-Jacas, Susanna 1616 & Vilatersana (BC). | FJ459691 | FJ459635 | FJ459725 |
| Centaurea scabiosa L. | France, Lozère: between Le Sec and L'Aumède, near Chanac, Vilatersana 52 (BC). | FJ459692 | FJ459636 | FJ459726 |
| Centaurea tauromenitana Guss. | Italy, Sicily: Botanical Garden of Catania (BC). | FJ459693 | FJ459637 | – |
| Centaurea toletana Boiss. & Reut. (2x) | Spain, Toledo: mountains above San Pablo on the track to Baños del Robledillo, M. Font & Susanna 1818 (BC), population 1. | FJ459694 | FJ459638 | FJ459727 |
| Spain, Toledo: Risco de las Paradas, M. Font & Susanna 1817 (BC), population 2. | FJ459695 | FJ459639 | FJ459728 | |
| Centaurea toletana Boiss. & Reut. (4x)Outgroup | Spain, Madrid: Redueña, road N-320, 4 km to Torrelaguna, M. Font & Susanna 1819 (BC).* | FJ459696 |
FJ459640 FJ459641 |
FJ459729 |
| Carduncellus duvauxii Batt. & Trab. | Morocco, Al-Hoceima: 8 km S to Tafraoute, Gómiz, s. n. (BC). | AY826239 | FJ459642 | – |
| Centaurea ensiformis P. H. Davis | Turkey, Muğla: Köycegiz district, Sandras Dağ range 13 km from Agla, Susanna 2251 & al. (BC). | DQ319112 | FJ459643 | – |
| Centaurea mollis Waldst. & Kit. | Ukraine, Podolia: Lysa Hora, 2 km E of Vilshanitsa near Zolochiv, Boratyñski & Romo 0506D (BC). | DQ319133 | FJ459644 | – |
| Phonus riphaeus (Font Quer & Pau) G. López | Morocco, Al-Hoceima: Tleta oued Laou between Tarerha and Azenti, J. M. Montserrat 4360 & al. (BC). | AY826310 | FJ459645 | – |
* Samples that have been cloned (for these taxa, each different accession number correspond to one cloned sequence).
Outgroups were chosen among the genera of the ‘derived clade’ of the subtribe Centaureinae (Garcia-Jacas et al., 2001), where the Acrocentron group belongs, and included one species each from the Cyanus and Jacea groups of Centaurea and the genera Carduncellus Adans. and Phonus Hill.
Voucher data, source and GenBank sequence accession numbers are given in Table 1.
DNA extraction, amplification and sequencing
Total genomic DNA was extracted following the CTAB method of Doyle and Doyle (1987) and Cullings (1992) from silica-gel-dried leaves collected in the field. In some cases, herbarium material was used.
nrDNA ITS and ETS region and cpDNA amplification strategies
Double-stranded DNA of the ITS region was amplified using ITS1 (White et al., 1990) and 1406F (Nickrent et al., 1994) as the forward primer and ITS4 as the reverse primer (White et al., 1990). The profile used for PCR amplification follows the protocol described in Susanna et al. (2006). The ETS region was amplified with ETS1F as the forward primer and as reverse primer either 18S-2L (Linder et al., 2000) or 18SETS (Baldwin and Markos, 1998). The PCR was executed with the following conditions: 5 min denaturing at 95 °C, followed by 30 cycles of 94 °C denaturing for 45 s, 48 °C annealing for 45 s and 72 °C extension for 40 s, with an additional 7 min at 72 °C. PCR products of the species with more than one band were cloned using a TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA) following the manufacturer's instructions, except that only half of the reaction volume was used. When possible, from eight-to-fifteen positive colonies from each reaction were screened with direct PCR using T7 and M13R universal primers following the protocol of Vilatersana et al. (2007). Five-to-eight PCR products were selected for sequencing in both directions using the same primers.
The intergenic spacer trnH-psbA was amplified using the primers trnH(GUG)F and psbAR (Hamilton, 1999). The profile used for PCR amplification followed the protocol of Vilatersana et al. (2007).
nrDNA and cpDNA sequencing strategies
Plastid and nuclear PCR products were purified with the QIAquick PCR Purification Kit (Qiagen Inc., Valencia, CA). Direct sequencing of the amplified DNA segments was performed using a BigDye Terminator Cycle Sequencing v3.1 (Applied Biosystems, Foster City, CA), following the manufacturer's protocol. Nucleotide sequencing was performed on an ABI PRISM 3100 DNA Analyzer (Applied Biosystems). When cloned sequences from one accession presented only a single substitution with regard to the others, they were excluded. Consensus sequences were generated for each accession and region, condensing the single base-pair differences among cloned sequences into a degenerated base. This strategy reduced the size of the matrices as well as the impact of potential PCR artifacts (chimerical sequences and Taq errors; Cline et al., 1996; Popp and Oxelman, 2001).
Phylogenetic analyses
Sequences were aligned visually by sequential pairwise comparison (Swofford and Olsen, 1990). The data matrices are available on request from the corresponding author. In view of the practical impossibility of unambiguously aligning the most variable 5′ end of the ETS region of the ingroup and the outgroup, two analyses were carried out. First, we analysed the more conserved 3′ETS region plus the ITS region including the outgroups, with the aim of identifying the earliest-branching clade of Acrocentron. Second, we carried out an analysis of the complete-region ETS plus ITS, including only the species of the Acrocentron group, using as outgroup the clade identified as earliest-branching in the first analysis. For the analysis of the trnH-psbA plastid region, we used as outgroup some representatives from the clade identified as earliest-branching in the first analysis.
Phylogenetic analyses were conducted using both parsimony and Bayesian inference optimality criteria. The plastid dataset was analysed independently. Both nrDNA regions (ETS plus ITS) were combined and analysed in one matrix because both datasets resulted in fully compatible topologies. A partition homogenity test was impossible to complete because the heuristic search collapsed after reaching the tree limits of PAUP in the first replicate.
Parsimony analysis involved heuristic searches conducted with PAUP ver. 4.0b10 (Swofford, 2002) using tree-bisection-reconnection (TBR) and branch swapping with character states specified as unordered and unweighted. The indels were treated as missing data in all the analyses. All most-parsimonious trees were saved. To locate other potential islands of most-parsimonious trees (Maddison, 1991) 100 replications were performed with random taxon addition, also with TBR branch swapping. Consistency index (CI) and retention index (RI) are always given excluding uninformative characters. Bootstrap analysis (BS) was performed in order to assess the confidence of the branches (Felsenstein, 1985). For the combined matrices (ETS plus ITS), bootstraping followed the approach of Lidén et al. (1997) using 1000 replicates, random taxon addition with ten replicates, and no branch swapping.
For the Bayesian analysis, the data sets were analysed using MrModeltest ver. 2.2 (Nylander, 2004) to determine the sequence evolution model that best described the data. This model was used to perform a Bayesian analysis using the program MrBayes ver. 3.1.2 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003). Four Markov chains were run simultaneously for 1 000 000 generations and sampled every 100 generations. Data from the first 1000 generations were discarded as the ‘burn-in’ period, after confirming that likelihood values had stabilized prior to the 1000th generation. The 50 % majority rule consensus phylogeny and posterior probability of nodes (PP) were calculated from the remaining sample.
Dispersal-vicariance analysis (DIVA)
Ancestral biogeographic reconstruction was inferred with the dispersal-vicariance analysis implemented in DIVA (Ronquist, 1996, 1997). Eight areas of endemism were defined by the presence of one or more endemic taxons: A = Iberian; B = North African; C = Balkanic; D = Aegean; E = Anatolian + Iranian; F = Eurasian; G = Caucasian; and H = Sicilian. Because DIVA needs a dichotomic tree, we used as input data the tree with the highest posterior probability, obtained with the first analysis (3'ETS plus ITS data; see Fig. 1). Outgroup taxa were omitted when running the DIVA because these taxa were not biogeographically informative. The analysis was carried out constraining the number of ancestral areas to a maximum of two, which is the maximum geographic range of the most widespread species.
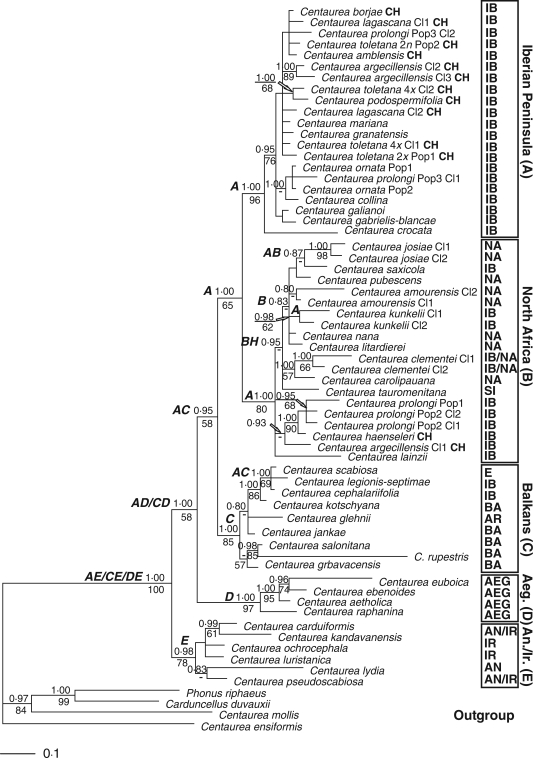
Fig. 1.
Consensus phylogram obtained from 9000 Bayesian trees with higher posterior probability (PP) from the combined 3′ETS and ITS data with an extended outgroup. Numbers above branches indicate Bayesian-credibility values (PP); numbers below branches indicate parsimony bootstrap values. The lettering of the nodes indicates the areas defined for the DIVA: A = Iberian Peninsula; B = North Africa; C = Balkans; D = Aegean; E = Anatolia + Iran; H = Sicily. Biogeographic regions on the right of the figure are: AEG = Aegean; AN = Anatolia; AR = Armenia; BA = Balkans; E = Eurasia; IB = Iberian Peninsula; IR = Iran; NA = North Africa; SI = Sicily. Other abbreviations: CH = section Chamaecyanus; Cl = clone; Pop = population.
RESULTS
Numerical results of the analyses are shown in Table 2. The analysis of the 3'ETS plus ITS regions is shown in Fig. 1. Centaurea sect. Acrocentron group is monophyletic with high support (BS = 100 %, PP = 1·0) only if we include C. sect. Chamaecyanus, which is not confirmed as a natural group. The earliest-branching group of Acrocentron is the clade of the Anatolian and Irano-Turanian taxa (clade ANAT), without parsimony support but with very high Bayesian suport (BS = 58 %, PP = 1·0). Next, there comes the Aegean taxa (clade AEG) almost without support (BS = 58 %, PP = 0·95). Some species of wide distribution (the Centaurea scabiosa group) ranging from the Iberian Peninsula to Central Asia, together with a group of taxa from the north Balkans and north Italy (clade Balkans), are sister to the rest (BS = 65 %, PP = 1·0). Finally, there is a dichotomy comprising two clades. The first subclade is a well-supported group (BS = 80 %, PP = 1·0) with all the species from North Africa and Sicily and some others from the Iberian Peninsula (clade North Africa). The second subclade (BS = 96 %, PP = 1·0) includes only species from the Iberian Peninsula.
Table 2.
Numeric results of the phylogenetic analyses
| Data set | 3′ETS + ITS | 5′ and 3′ ETS + ITS | trnH-psbA |
|---|---|---|---|
| Taxa | 49 | 45 | 31 |
| Number of sequences | 64 | 60 | 33 |
| Total characters | 1859 | 1142 | 443 |
| Informative characters | 225 | 173 | 8 |
| Number MPTs | 815 072 | 34 446 | 16 |
| Number of steps | 494 | 399 | 13 |
| Consistency index (CI) | 0·5423 | 0·5396 | 0·3913 |
| Retention index (RI) | 0·8582 | 0·8090 | 0·6410 |
| Range of divergence, ingroup (%) | 0–0·1193 | 0·0008–0·091 | 0–0·875 |
| Bayesian model of evolution | GTR + I + G | GTR + I + G | GTR + I + G |
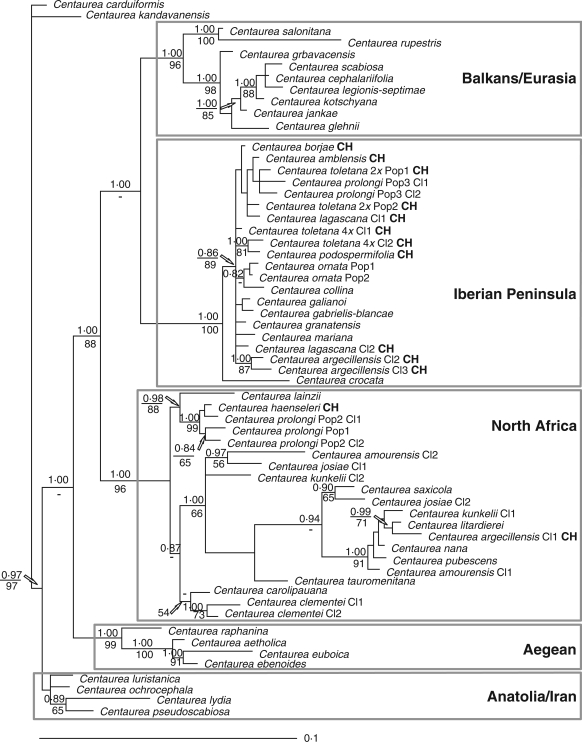
The analysis of the complete ETS plus ITS region (Fig. 2) shows an important difference with the partial analysis: the Iberian Peninsula and North African clades, which formed a well-supported clade in the first analysis (Fig. 1), are not grouped together in the complete analysis. In this second analysis, the Balkans–Eurasia clade is sister to the Iberian Peninsula clade, and this joint clade is sister to the North African clade (Fig. 2). Besides this divergent result, a few additional differences become apparent. The cloned sequences of C. josiae Humbert form a monophyletic clade in the partial analysis (Fig. 1), but one of the cloned sequences is associated with C. amourensis Pomel in the complete analysis (Fig. 2). One of the cloned sequence of C. argecillensis Gredilla (species from Central Iberia), which in the partial analyses occupies an unresolved position at the base of the North African clade (Fig. 1), is grouped together with C. litardierei Jahand. & Maire in the complete analysis (Fig. 2). There is also a difference in the group formed by the cloned sequences of Portuguese C. prolongi Boiss. (Pop. 3), which appear on different clades in the partial analysis (clade Iberian Peninsula, Fig. 1) and unsupportedly united in the complete analysis (clade Iberian Peninsula, Fig. 2). Finally, C. lainzii Fern. Casas, which appears as an isolated taxon in the partial analysis (clade North Africa, Fig. 1), is united with C. prolongi (Pop. 1 and 2) and C. haenseleri Boiss. in the complete analysis (clade North Africa, Fig. 2). Differences in the topology resulting from the two analyses (ITS + ETS 3′, Fig. 1; ITS + ETS 5′ and 3′, Fig. 2) are caused by the number of informative characters yielded by the markers used in each case – much higher for the ETS 5′ end.
Fig. 2.
Consensus phylogram obtained from 9000 Bayesian trees with higher posterior probability (PP) from the combined 5′-3′ ETS and ITS data with the Anatolian–Iranian clade as outgroup. Numbers above branches indicate Bayesian-credibility values (PP); numbers below branches indicate parsimony bootstrap values. CH = section Chamaecyanus; Cl = clone; Pop = population.
The complete analysis (Fig. 2) suggests that the species from the Aegean are closer to the outgroup formed by the Anatolian and Irano-Turanian species. Next, there come two clades, one with species from the Balkans and some others of wide distribution (clade Balkans–Eurasia), and the taxa from the Iberian Peninsula and a clade with species of North Africa, Sicily and the rest of the Iberian taxa (clade North Africa) with high support both from BS and PP.
The low number of informative characters in the plastid dataset (Table 2) explains the lack of resolution in the phylogenetic analysis (tree not shown). Table 3 shows the changes of the sequences in the plastid dataset.
Table 3.
Informative characters in the chloroplast trnH-psbA sequences. Variable characters with respect to the first sequence are in bold type
| Positions in data set |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 1 | 1 | 1 | 3 | 3 | 3 | 3 | |||||||
| 6 | 7 | 7 | 7 | 8 | 5 | 6 | 6 | 7 | 7 | 3 | 6 | 8 | 9 | ||
| Species and population | Origin* | 5 | 5 | 7 | 8 | 0 | 1 | 8 | 9 | 5 | 9 | 9 | 2 | 8 | 8 |
| C. glehnii | AR | C | T | T | T | T | A | – | – | A | G | C | A | A | A |
| C. lydia | ANT | C | T | T | T | T | A | – | – | G | G | C | A | A | A |
| C. salonitana | BAL | C | T | T | T | T | A | – | – | A | G | C | A | C | A |
| C. amblensis | IB | C | T | T | T | T | A | – | – | A | G | A | A | A | A |
| C. argecillensis | IB | C | T | T | T | T | A | – | – | A | G | A | A | A | A |
| C. borjae | IB | C | A | A | A | A | A | – | – | A | G | C | A | A | A |
| C. cephalariifolia | IB | C | T | T | T | T | A | – | – | A | G | A | A | A | A |
| C. clementei | IB | C | T | T | T | T | A | – | – | A | G | A | A | A | A |
| C. crocata | IB | C | A | A | A | A | A | – | – | A | G | A | A | A | A |
| C. collina | IB | C | T | T | T | T | A | – | – | A | G | C | A | A | A |
| C. gabrielis-blancae | IB | C | T | T | T | T | C | – | – | A | G | A | A | A | A |
| C. galianoi | IB | C | T | T | T | T | A | – | – | A | G | C | A | A | A |
| C. granatensis | IB | C | T | T | T | T | A | – | – | A | G | A | A | A | A |
| C. haenseleri | IB | C | T | T | T | T | A | – | – | A | G | A | A | A | A |
| C. kunkelii | IB | C | T | T | T | T | A | – | – | A | G | A | A | A | C |
| C. lagascana | IB | C | T | T | T | T | A | – | – | A | G | A | G | A | A |
| C. lainzii | IB | C | T | T | T | T | C | – | – | A | G | C | A | A | A |
| C. legionis-septimae | IB | C | T | T | T | T | A | – | – | A | G | C | A | A | A |
| C. mariana | IB | C | T | T | T | T | A | A | A | A | G | C | T | A | A |
| C. ornata Pop 1 | IB | C | T | T | T | T | A | – | – | A | G | A | A | A | A |
| C. podospermifolia | IB | C | T | T | T | T | A | – | – | A | G | A | A | A | A |
| C. prolongi Pop 1 | IB | C | T | T | T | T | A | – | – | A | G | C | A | A | A |
| C. prolongi Pop 3 | IB | C | T | T | T | T | A | – | – | A | G | A | A | A | A |
| C. saxicola | IB | C | T | T | T | T | A | – | – | A | G | A | A | A | A |
| C. toletana 2x Pop 1 | IB | C | T | T | T | T | A | – | – | A | G | A | A | A | A |
| C. toletana 2x Pop 2 | IB | C | T | T | T | T | A | – | – | A | G | A | A | A | A |
| C. toletana 4x | IB | C | T | T | T | T | A | – | – | A | G | A | A | A | A |
| C. kandavanensis | IR | C | A | A | A | A | A | – | – | A | G | A | A | A | A |
| C. amourensis | NA | C | A | A | A | A | A | A | A | A | A | A | A | A | A |
| C. carolipauana | NA | C | A | A | A | A | A | A | A | A | G | C | G | A | A |
| C. litardierei | NA | C | A | A | A | A | A | – | – | A | G | C | G | A | A |
| C. pubescens | NA | C | T | T | T | T | A | A | A | A | A | C | T | A | A |
| C. scabiosa | E | T | T | T | T | T | A | – | – | A | G | A | A | A | A |
* AR = Armenia; AN = Anatolia, BA = Balkans; E = Euro-Siberian; IB = Iberian; IR = Iran; NA = North Africa; – indicates gap.
The output of the DIVA is the most-parsimonious ancestral area for each node, and the results are reflected in Fig. 1. Three equally parsimonious solutions for the ancestral area are suggested for the deepest node (Iberian/Anatolian, Balkanic/Anatolian and Aegean/Anatolian). This ancestor would have then dispersed to the Iberian/Aegean or Balkanic/Aegean areas. The ancestral area suggested for the third deepest node is Iberian/Balkanic. Beyond this node, the DIVA results suggest predominance of vicariance.
DISCUSSION
Concerted evolution in the ETS–ITS regions
The presence of different rybotypes in both regions (ETS and ITS) confirms that in some species of the group concerted evolution has not occurred or has been incomplete. This is an acute contrast with the related Centaurea sect. Willkommia in which biased concerted evolution was extensive (Suárez-Santiago et al., 2007).
Taxonomic implications: sectional classification
The classic taxonomic classification in two sections, Acrocentron and Chamaecyanus, is untenable after our results. The segregation of Centaurea sect. Chamaecyanus (species of this section are marked CH in Figs 1 and 2) would leave C. sect. Acrocentron paraphyletic. The most appropriate status for C. sect. Chamaecyanus is subsectional within C. sect. Acrocentron, confirming previous suggestions (Font et al., 2002). In the Iberian clade (Figs 1 and 2), species of both sections are part of a polytomy, and C. sect. Chamaecyanus is not resolved as a natural group. However, both sections are morphologically very well defined (Fernández Casas and Susanna, 1986), and we suspect that the lack of confirmation of the monophyly of C. sect. Chamaecyanus is caused by introgression. Intermediates between them are very frequent (Fernández Casas and Susanna, 1986), a fact that is probably the cause of this entanglement and the reason for the low support for the clade that comprises many of the species of C. sect. Chamaecyanus (Figs 1 and 2). The position of Centaurea crocata Franco as sister to the Iberian clade is in this sense very significant: on the basis of morphology, C. crocata is suspected as a hybrid between taxa of both sections (Garcia-Jacas and Susanna, 1994), which would explain this basal position.
Regarding the possibility of establishing a subsectional classification within C. sect. Acrocentron as suggested by previous workers (Hayek, 1901; Routsi and Georgiadis, 1994), molecular data are of little help because intense gene flow hinders molecular reconstruction of phylogenetic affinities, and the extreme homogeneity of morphological characters makes any suggestion of infrasectional classification a very difficult task. As is evident from Figs 1 and 2, molecular data group the taxa on a geographical basis.
Hybridization and reticulation
In our previous study based on the ITS region (Font et al., 2002), we suggested that the very low definition within Centaurea sect. Acrocentron was probably caused by the high levels of hybridization and reticulation within the group. Our new results could support this view, for example the presence of an ETS cloned sequence of C. argecillensis from central and north-east Iberia within the North African clade, whilst the rest of the cloned sequences of this species are placed in the Iberian clade (Figs 1 and 2). There are some other examples, perhaps not so extreme but also indicating intense gene flow: Centaurea toletana Boiss. & Reut. (2n = 4x) has a cloned sequence united to C. podospermifolia Loscos & J. Pardo with high PP support, and another one ranged within the Iberian polytomy (Figs 1 and 2). Another case is a population of C. prolongi from Portugal, firstly cited as C. collina L., later on identified as C. prolongi (Garcia-Jacas and Susanna, 1994) and even considered a new species, C. occasus Fern. Casas (Fernández Casas, 1997). The two cloned sequences of this population of C. prolongi (Pop. 3) appear in the same clade, but in different positions (C. prolongi 3, clade Iberian Peninsula; Fig. 1).
Gene flow could also explain the differences within C. prolongi: the Portuguese population is associated with the taxa from central and north Iberia, whilst populations from the extreme south of Spain are placed in the African clade (C. prolongi Pop. 1 and Pop. 2, clade North Africa; Figs 1 and 2). This segregation shows that they have different ribotypes, a fact that could reinforce the hypothesis that they are different species. However, morphological differences alleged by Fernández Casas (1997) are only quantitative (namely, the size of the leaves and florets), which could easily be explained by the different climatic conditions of both localities (milder and wetter in Portugal). A prefereable explanation for the positions of populations of C. prolongi in opposite clades is gene flow: populations of C. prolongi in south Spain are very close to the African coast (Fig. 3), and other species of C. sect. Acrocentron growing nearby (C. haenseleri and C. lainzii) also appear in the African group (Figs 1 and 2). In contrast, the studied Portuguese locality was far apart from the waves of African taxa.
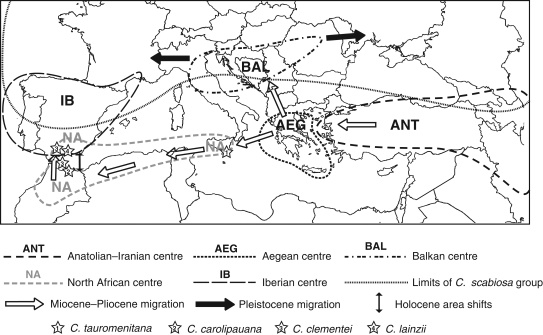
Fig. 3.
Geographic representation of the two migration pathways of sect. Acrocentron in the Mediterranean, and the landmark species in Sicily and North Africa.
In our analyses, C. legionis-septimae Fern. Casas & Susanna, a narrow endemic from the mountains of northern Spain, is united with C. cephalariifolia Willk., a species from the C. scabiosa widespread group. This position contradicts morphologic evidence, which relates C. legionis-septimae to C. ornata Willd. (Garcia-Jacas and Susanna, 1992), and could be considered another indication of hybridization, as suggested by the existence of homoploid hybrid populations between both taxa described as C. ×cephalariiseptimae Fern. Casas & Susanna (Fernández Casas and Susanna, 1985).
The position of the Balkan clade, which is united to the Iberian clade in the phylogenetic analysis (Fig. 2), could be explained in terms of hybridization and introgression. This relationship could be caused by gene flow of the widely distributed taxa from the clade Balkans–Eurasia and the Iberian species. The culprit is certainly C. cephalariifolia, a tetraploid race of the widespread diploid C. scabiosa. To date, up to six naturally occurring hybrids have been reported between C. cephalariifolia and other Iberian taxa (we have discussed one of them above, C. ×cephalariiseptimae). In four cases, the crosses involve tetraploid parentals and the hybrids are also tetraploid (Kummer, 1977; Fernández Casas and Susanna, 1986) – thereafter they are not genetically isolated from their progenitors.
A final reason for considering high levels of introgression is the result of the plastid sequencing (Table 3). The few changes observed in the sequences of the intergenic spacer trnH-psbA do not agree with any known relationship, either taxonomic or biogeographic. Cytoplasmic gene flow could be one of the main causes of low resolution in phylogenies (Rieseberg and Soltis, 1991) or, alternately, C. sect. Acrocentron shows high levels of homoplasy in the trnH-psbA (Hamilton et al., 2003).
Biogeography
The combined ITS plus ETS phylogeny (Figs 1 and 2) shows that there are five speciation centres: Anatolia–Iran, the Aegean (these two were united into one by Font et al., 2002), the Balkans (including some species of wide Eurasian distribution), the Iberian Peninsula, and North Africa. These geographic denominations for the clades are general and do not imply that all the taxa within the clade grow in this precise geographic area, but they are accurate enough for the purpose of briefly discussing each centre and the history of the geographic distribution of C. sect. Acrocentron (Fig. 3).
Distribution of Acrocentron has been presented as an east-to-west migration (Garcia-Jacas and Susanna, 1992). As a confirmation of this hypothesis, the Anatolian–Iranian centre is the earliest-branching clade in the first of our analyses, 3'ETS plus ITS (Fig. 1). This is one of the three hypotheses suggested by the DIVA, and indeed the most likely because the two areas are contiguous. The alternate solutions suggest either an unlikely Iberian–Anatolian origin for the group or a Balkanic–Anatolian origin that would obviate the Aegean milestones. The origin of Acrocentron should be placed in the eastern Mediterranean, much like the rest of the subtribe according to the greater richness of genera of the Centaureinae in this area (Susanna and Garcia-Jacas, 2007). The hypothesis of the south of the Irano-Turanian region (Iranian plateau) as the place of origin of the group was rejected on karyologic grounds, and Iranian species of C. sect. Acrocentron were considered a later expansion from Anatolian populations (Garcia-Jacas et al., 1998).
From Anatolia, species of C. sect. Acrocentron radiated to the Aegean islands, the species of which (C. aetholica Phitos & T. Georgiadis, C. ebenoides Heldr., C. euboica Rech. f. and C. raphanina Sibth. & Sm.) form a well-supported clade (AEG in Fig. 1 and Aegean in Fig. 2) that is sister to the next branches. This radiation is supported by the DIVA results, which suggests two equally possible solutions for this dispersal: an unlikely (not contiguous) Iberian–Aegean ancestral area, or the Balkanic–Aegean hypothesis (Fig. 1). The Aegean islands and south Greece constitute the easiest way of penetration from Anatolia to the western Mediterranean, which is consistent with the intermediate position of the Aegean clade in the phylogeny (Figs 1 and 2).
The Aegean clade is at the basis of the two main radiations of C. sect. Acrocentron, as reflected in our phylogeny (Fig. 1). In the north of its range, it originated the Balkanic clade, and in the south it migrated to the extreme south of the Italian Peninsula (Fig. 3). A dual radiation is supported by the DIVA results (Fig. 1), which suggest an Iberian–Balkanic ancestral area for this node. Predominance of Iberian species in the upper part of the tree could be the reason for DIVA assigning an Iberian origin to this branch, which should be better considered as North African–Balkanic.
The Messinian salinity crisis (between 6 and 5·2 MYA) offers a good opportunity for hypothesizing a date for the migration of the Acrocentron group. In the east, a pass between Anatolia and the Aegean was already open by the middle Miocene (Çağatay et al., 2006). The lowering of the sea level opened the way from the Aegean and Greece to the extreme south of Italy and Sicily, and then to North Africa and the Iberian Peninsula (Fig. 3). The main migration of C. sect. Acrocentron through this southern pathway probably would have terminated in the Pliocene because species of C. sect. Acrocentron are absent in all the western Mediterranean archipelagos, which during the Messinian were connected to the mainland. This dating has also been suggested for the expansion of the related complex Acrolophus–Phalolepis of the genus Centaurea (Suárez-Santiago et al., 2007), a group highly speciated in Iberia but absent from the Balearic Islands.
Besides paleogeographic arguments, a series of landmarks illustrate this hypothetical path. Four species closely related on morphological grounds mark the southern path of the migration: C. tauromenitana Guss. from Sicily, C. carolipauana Fern. Casas & Susanna from northern Morocco, C. clementei Boiss. from northern Morocco and south Iberia and C. lainzii from south Iberia (Fig. 3). All of them are relictic mountain plants and, with the exception of C. clementei, are known from a single locality. They are chasmophytes sharing some unusual characters in C. sect. Acrocentron, such as very wide involucral bracts ending in an unarmed black appendage (Fernández Casas and Fernández Morales, 1979). Figure 3 summarizes the hypothesized pathes of expansion of the group, together with the distribution of these landmark species. The expansion of the group to the rest of the Iberian Peninsula and North Africa was a natural consequence, favoured by the development of a Mediterranean climate. The poverty in species of C. sect. Acrocentron in the North African lowlands stretching between Algeria and Libya reflects the deep changes in the vegetation of this area in recent times. Only one species of the group survived in the desert, but at the price of a dramatic adaptation: Centaurea omphalodes (Coss. & Dur.) O. Hoffm. from the oasis of Algeria is the only annual species of the Acrocentron group.
As a final argument for the predominance of the southern path of migration, in northern and middle Italy some species of C. sect. Acrocentron can be found, but they are related to the Balkan clade and did not cross the huge barrier of the Alps into France (Fig. 3). The only species of C. sect. Acrocentron present in France belongs to the widespread C. scabiosa group, with the exception of only one species shared with east Iberia (C. collina).
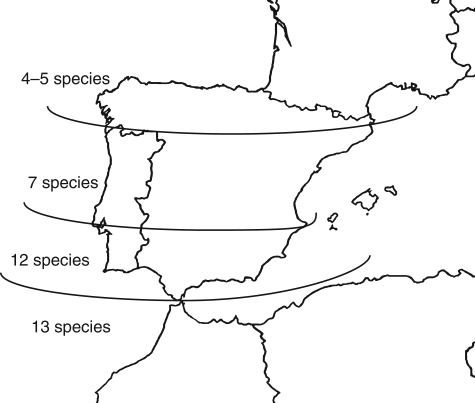
Migration via the southern path would also explain the pattern of distribution of C. sect. Acrocentron in Iberia. Figure 4 shows the richness in number of species in the Iberian Peninsula and Morocco. The number of taxa diminishes northwards, and in northern Iberia species of C. sect. Acrocentron are scarce and most of them (three out of four) are relictic paleopolyploids (Garcia-Jacas and Susanna, 1992).
Fig. 4.
Species density gradient for the Acrocentron group in the Iberian Peninsula and North Africa.
In contrast, DIVA reconstruction suggests the Iberian Peninsula as the ancestral area for the clade comprising Iberian, North African and Sicilian species (Fig. 1). This solution seems unlikely because it contradicts biogeographic evidence: if taxa of Acrocentron migrated from Anatolia to the Aegean and the south Balkans, the easiest way to the Iberian Peninsula is the southern route via Sicily and North Africa, where landmark species exist (Fig. 3). When interpreting the DIVA results, we have to be aware that the DIVA method does not take in account geographic connections between the areas defined for the analysis (Ronquist, 1996, 1997).
A later entry of C. sect. Acrocentron following a northern pathway can be tracked in the Iberian Peninsula, originating among the species of the Balkan clade. The continuous distribution of species of this group in Europe and Middle Asia, from the Iberian Peninsula and Great Britain to Kazakhstan, should be considered a recent radiation based on a single species, C. scabiosa. Many local variants of this species have been described as new taxa on insufficient morphological basis, which confirms that they are of very recent age. Even those taxa from the C. scabiosa complex that are usually accepted by synantherologists, C. cephalariifolia from Spain (a tetraploid), C. glehnii Trautv. from Armenia and C. kotschyana Heuff. from Poland, have been diversely considered at subspecific rank within C. scabiosa. This extreme geographic radiation (Fig. 3) was favoured by the occurrence of a cold steppe continuum from Iberia to Asia during the repeated interglacial periods in the Pleistocene–Holocene (Prentice et al., 2000).
In our molecular reconstruction based on the ITS plus ETS regions, the phylogeny of the Iberian and African taxa is obscure and unresolved because the present distribution of ribotypes in Iberia is significantly mediated by the troublesome climatic history of the Iberian Peninsula and North Africa during the Holocene. The succession of glaciations forced mountain species of Acrocentron to migrate to lower latitudes (Peñalba et al., 1997), and species that were isolated became sympatric. Iberian species could migrate south to Africa during glacial maxima, and African taxa moved to the north during the warmer interglacial periods (Fig. 3). The resulting pattern of generalized introgression is reflected in the nrDNA phylogeny, with even north-Iberian cloned sequences deeply nested in the North African clade. Perhaps the most striking result, which confirms the impact of glaciation-mediated migrations and hybridization in C. sect. Acrocentron, is the otherwise implausible group formed by one cloned sequence of C. argecillensis from central Spain and another of C. litardierei from the High Atlas mountain range (Figs 1 and 2).
CONCLUSIONS
Our study demonstrates the usefulness of the analysis of the ETS region for phylogenetic and biogeographic investigation of highly hybridized groups. The unravelling of the pathways followed by the Acrocentron group offers a possible model for many other groups with a suggested eastern Mediterranean origin and secondary centres of speciation in the Iberian Peninsula and North Africa.
ACKNOWLEDGEMENTS
Financial support from the Spanish Ministery of Education and Science (Projects CGL2004–04563-C02-01/BOS and CGL2006–01765/BOS) and the Generalitat de Catalunya (‘Ajuts a Grups de Recerca Consolidats’ 2005/SGR/00344) is gratefuly acknowledged. The authors thanks the curators of the institutions and the collectors listed in Table 1 for kindly supplying material of some species. Suggestions by two anonymous referees for improving the manuscript are gratefully acknowledged.
LITERATURE CITED
- Baldwin BG, Markos S. Phylogenetic utility of the external transcribed spacer (ETS) of 18S-26S rDNA: congruence of ETS and ITS trees of Calycadenia (Compositae) Molecular Phylogenetics and Evolution. 1998;10:449–463. doi: 10.1006/mpev.1998.0545. [DOI] [PubMed] [Google Scholar]
- Cline J, Braman JC, Hogrefe HH. PCR fidelity of pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Research. 1996;24:3546–3551. doi: 10.1093/nar/24.18.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullings KW. Design and testing of a plant-specific PCR primer for ecological and evolutionary studies. Molecular Ecology. 1992;1:233–240. [Google Scholar]
- Çağatay MN, Görür N, Flecker R, et al. Paratethyan–Mediterranean connectivity in the Sea of Marmara region (NW Turkey) during the Messinian. Sedimentary Geology. 2006;188–189:171–187. [Google Scholar]
- Dittrich M. Cynareae – systematic review. In: Heywood VH, Harborne JB, Turner BL, editors. The biology and chemistry of the Compositae. London, New York, San Francisco: Academic Press; 1977. pp. 999–1015. [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin, Botanical Society of America. 1987;19:11–15. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fernández Casas FJ. De Centaureis occidentalibus notulae sparsae X. Fontqueria. 1997;48:203–222. [Google Scholar]
- Fernández Casas FJ, Fernández Morales MJ. Centaurea lainzii, un triploide natural. Mémoires de la Société Botanique de Genève. 1979;1:115–122. [Google Scholar]
- Fernández Casas FJ, Susanna A. De Centaureis occidentalibus notulae sparsae, VII, Fontqueria. 1985;9:13–16. [Google Scholar]
- Fernández Casas FJ, Susanna A. Monografía de la sección Chamaecyanus Willkomm del género Centaurea L. Treballs de l'Institut Botànic de Barcelona. 1986;10:5–174. [Google Scholar]
- Font M, Garnatje T, Garcia-Jacas N, Susanna A. Delineation and phylogeny of Centaurea sect. Acrocentron based on DNA sequences: a restoration of the genus Crocodylium and indirect evidence of introgression. Plant Systematics and Evolution. 2002;234:15–26. [Google Scholar]
- Garcia-Jacas N. Centaurea kunkelii, a new hybridogenic endecaploid species of sect. Acrocentron from Spain. Annales Botanici Fennici. 1998;35:159–167. [Google Scholar]
- Garcia-Jacas N, Susanna A. Karyological notes on Centaurea sect. Acrocentron. Plant Systematics and Evolution. 1992;179:1–18. [Google Scholar]
- Garcia-Jacas N, Susanna A. Centaurea ×polymorpha Lagasca: los problemas de un híbrido. Fontqueria. 1993;36:65–66. [Google Scholar]
- Garcia-Jacas N, Susanna A. Centaurea prolongi and Centaurea crocata in Portugal: an old confusion. Nordic Journal of Botany. 1994;14:31–38. [Google Scholar]
- Garcia-Jacas N, Susanna A, Mozaffarian V. New chromosome counts in the subtribe Centaureinae (Asteraceae, Cardueae) from West Asia, III. Botanical Journal of the Linnean Society. 1998;128:413–422. [Google Scholar]
- Garcia-Jacas N, Susanna A, Garnatje T, Vilatersana R. Generic delimitation of phylogeny of the subtribe Centaureinae (Asteraceae): a combined nuclear and chloroplastic DNA analysis. Annals of Botany. 2001;87:503–515. [Google Scholar]
- Garcia-Jacas N, Uysal T, Romashchenko K, Suárez-Santiago VN, Ertuğrul K, Susanna A. Centaurea revisited: A molecular survey of the Jacea group. Annals of Botany. 2006;98:741–753. doi: 10.1093/aob/mcl157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton MB. Four primers pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Molecular Ecology. 1999;8:521–523. [PubMed] [Google Scholar]
- Hamilton MB, Braverman JM, Soria-Hernanz DV. Patterns and relative rates of nucleotide and insertion/deletion evolution at six chloroplast intergenic regions in New World species of the Lecythidaceae. Molecular Biology and Evolution. 2003;20:1710–1721. doi: 10.1093/molbev/msg190. [DOI] [PubMed] [Google Scholar]
- Hayek AV. Die Centaurea Arten Oesterreich-Ungarns. Denkschriften der Kaiserlichen Akademie der Wissenschaften, Mathematisch-Naturwissenschaftliche Klasse. 1901;70:585–773. [Google Scholar]
- Hellwig FH, Oberprieler C, Vogt R, Wagenitz G. Chromosome numbers of North African phanerogams. III. Some counts in Centaurea (Compositae) Willdenowia. 1994;24:249–254. [Google Scholar]
- Hidalgo O, Garcia-Jacas N, Garnatje T, Susanna A. Phylogeny of Rhaponticum (Asteraceae, Cardueae–Centaureinae) and related genera inferred from nuclear and chloroplast DNA sequence data: taxonomic and biogeographic implications. Annals of Botany. 2006;97:704–714. doi: 10.1093/aob/mcl029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Kelch DG, Baldwin BG. Phylogeny and ecological radiation of New World thistles (Cirsium, Cardueae–Compositae) based on ITS and ETS rDNA sequence data. Molecular Ecology. 2003;12:141–151. doi: 10.1046/j.1365-294x.2003.01710.x. [DOI] [PubMed] [Google Scholar]
- Kummer C. Untersuchungen von natürlichen Bastarden zwischen Centaurea cephalariifolia Willkomm und Centaurea ornata Willdenow. Mitteilungen der Botanischen Staatssammlung München. 1977;13:129–202. [Google Scholar]
- Lidén M, Fukuhara T, Rylander J, Oxelman B. Phylogeny and classification of Fumariaceae, with emphasis on Dicentra s. l. based on the plastid gene rps16 intron. Plant Systematics and Evolution. 1997;206:411–420. [Google Scholar]
- Linder CR, Goertzen LR, Vanden Heuvel B, Francisco-Ortega J, Jansen RK. The external transcribed spacer of the rDNA repeat: a new nuclear region for low-level taxonomic analysis of the Asteraceae and closely allied families. Molecular Phylogenetics and Evolution. 2000;14:285–303. doi: 10.1006/mpev.1999.0706. [DOI] [PubMed] [Google Scholar]
- Maddison DR. The discovery and importance of multiple islands of most-parsimonious trees. Systematic Zoology. 1991;40:315–328. [Google Scholar]
- Markos S, Baldwin BG. Higher-level relationships and major lineages of Lessingia (Compositae, Astereae) based on nuclear rDNA internal and external transcribed spacers (ITS and ETS) sequences. Systematic Botany. 2001;26:168–183. [Google Scholar]
- McKinnon GE. Reticulate evolution in higher plants. In: Henry RJ, editor. Plant diversity and evolution: genotypic and phenotypic variation in higher plants. Wallingford, UK: CABI Publishing; 2005. pp. 81–96. [Google Scholar]
- Nickrent DL, Schuette KP, Starr EM. A molecular phylogeny of Arceuthobium (Viscaceae) based on nuclear ribosomal DNA internal transcribed spacer sequences. American Journal of Botany. 1994;81:1149–1160. [Google Scholar]
- Nylander JAA. Uppsala, Sweden: Evolutionary Biology Centre, Uppsala University; 2004. MrModeltest ver. 2. Program distributed by the author. [Google Scholar]
- Peñalba MC, Arnold M, Guiot J, Duplessy JC, Beaulieu JL. Termination of the last glaciation in the Iberian Peninsula inferred from the pollen sequence of Quintanar de la Sierra. Quaternary Research. 1997;48:205–214. [Google Scholar]
- Plovanich AE, Panero JL. A phylogeny of the ITS and ETS for Montanoa (Asteraceae: Heliantheae) Molecular Phylogenetics and Evolution. 2004;31:815–821. doi: 10.1016/j.ympev.2003.10.021. [DOI] [PubMed] [Google Scholar]
- Popp M, Oxelman B. Inferring the history of the polyploid Silene aegaea (Caryophyllaceae) using plastic and homoeologous nuclear DNA sequences. Molecular Phylogenetics and Evolution. 2001;20:474–481. doi: 10.1006/mpev.2001.0977. [DOI] [PubMed] [Google Scholar]
- Prentice IC, Jolly D BIOME 6000 participants. Mid-Holocene and glacial-maximum vegetation geography of the northern continents and Africa. Journal of Biogeography. 2000;27:507–519. [Google Scholar]
- Rieseberg LH, Soltis DE. Phylogenetic consequences of cytoplasmic gene flow in plants. Evolutionary Trends in Plants. 1991;5:65–84. [Google Scholar]
- Ronquist F. DIVA ver. 1.1. Computer manual. Uppsala, Sweden: Evolutionary Biology Centre, Uppsala University; 1996. Available from http://www.ebc.uu.se/systzoo/research/diva/diva.html . [Google Scholar]
- Ronquist F. Dispersal-vicariance analysis: a new approach to quantification of historical biogeography. Systematic Biology. 1997;46:195–203. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Routsi E, Georgiadis T. Contribution to the systematics of the genus Centaurea section Acrocentron (Asteraceae) in Greece. Nordic Journal of Botany. 1994;14:369–378. [Google Scholar]
- Suárez-Santiago VN, Salinas MJ, Garcia-Jacas N, Soltis PS, Soltis DE, Blanca G. Evolution by reticulation of the Acrolophus subgroup (Centaurea L., Compositae) in the occidental Mediterranean: origin and diversification of the section Willkommia Blanca. Molecular Phylogenetics and Evolution. 2007;43:156–172. doi: 10.1016/j.ympev.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Susanna A, Garcia-Jacas N. Tribe Cardueae. In: Kadereit JW, Jeffrey C, editors. Flowering plants. Eudicots. Asterales. Berlin, Heidelberg: Springer Verlag; 2007. pp. 123–146. (vol. 8 in series The families and genera of vascular plants, Kadereit JW, ed.) [Google Scholar]
- Susanna A, Garcia-Jacas N, Soltis DE, Soltis PS. Phylogenetic relationships in tribe Cardueae (Asteraceae) based on ITS sequences. American Journal of Botany. 1995;82:1056–1068. [Google Scholar]
- Susanna A, Garcia-Jacas N, Hidalgo O, Vilatersana R, Garnatje T. The Cardueae (Compositae) revisited: insights from ITS, trnL-trnF, and matK nuclear and chloroplast DNA analysis. Annals of the Missouri Botanical Garden. 2006;93:150–171. [Google Scholar]
- Swofford DL. ver. 4.0b10. Sunderland: Sinauer Associates; 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods) [Google Scholar]
- Swofford DL, Olsen GJ. Phylogeny reconstruction. In: Hillis D, Moritz C, editors. Molecular systematics. Sunderland: Sinauer Associates; 1990. pp. 411–501. [Google Scholar]
- Urbatsch LE, Roberts RP, Karaman V. Phylogenetic evaluation of Xylothamia, Gundlachia, and related genera (Asteraceae, Astereae) based on ETS and ITS nrDNA sequence data. American Journal of Botany. 2003;90:634–649. doi: 10.3732/ajb.90.4.634. [DOI] [PubMed] [Google Scholar]
- Vilatersana R, Brysting AK, Brochmann C. Molecular evidence for hybrid origins of the invasive polyploids Carthamus creticus and C. turkestanicus (Cardueae, Asteraceae) Molecular Phylogenetics and Evolution. 2007;44:610–621. doi: 10.1016/j.ympev.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Vilatersana R, Martín J, Susanna A, Garcia-Jacas N, Garnatje T. Pollen studies in subtribe Centaureinae (Asteraceae): the Carthamus complex and the genus Aegialophila analyzed with electron microscopy. Plant Biology. 2001;3:607–615. [Google Scholar]
- Wagenitz G. Pollenmorphologie und Systematik in der Gattung Centaurea L. s. l. Flora. 1955;142:213–279. [Google Scholar]
- Wagenitz G, Hellwig FH. Evolution of characters and phylogeny of the Centaureinae. In: Hind DJN, Beentje HG, editors. Compositae: systematics. Proceedings of the International Compositae Conference, Kew, 1994. Kew, London: Royal Botanic Gardens; 1996. pp. 491–510. [Google Scholar]
- Wendel JF, Doyle JJ. Phylogenetic incongruence: window into genome history and molecular evolution. In: Soltis PS, Soltis DE, Doyle JJ, editors. Molecular systematics of plants II. Dordrecht: Kluwer; 1998. pp. 265–296. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. pp. 315–322. [Google Scholar]