Abstract
Background and Aims
Life form, mating system and seed dispersal are important adaptive traits of plants. In the first effort to characterize in detail the population genetic structure and dynamics of wild Medicago species in China, a population genetic study of two closely related Medicago species, M. lupulina and M. ruthenica, that are distinct in these traits, are reported. These species are valuable germplasm resources for the improvement of Medicago forage crops but are under threat of habitat destruction.
Methods
Three hundred and twenty-eight individuals from 16 populations of the annual species, M. lupulina, and 447 individuals from 15 populations of the perennial species, M. ruthenica, were studied using 15 and 17 microsatellite loci, respectively. Conventional and Bayesian-clustering analyses were utilized to estimate population genetic structure, mating system and gene flow.
Key Results
Genetic diversity of M. lupulina (mean HE = 0·246) was lower than that of M. ruthenica (mean HE = 0·677). Populations of M. lupulina were more highly differentiated (FST = 0·535) than those of M. ruthenica (FST = 0·130). For M. lupulina, 55·5 % of the genetic variation was partitioned among populations, whereas 76·6 % of the variation existed within populations of M. ruthenica. Based on the genetic data, the selfing rates of M. lupulina and M. ruthenica were estimated at 95·8 % and 29·5 %, respectively. The genetic differentiation among populations of both species was positively correlated with geographical distance.
Conclusions
The mating system differentiation estimated from the genetic data is consistent with floral morphology and observed pollinator visitation. There was a much higher historical gene flow in M. ruthenica than in M. lupulina, despite more effective seed dispersal mechanisms in M. lupulina. The population genetic structure and geographical distribution of the two Medicago species have been shaped by life form, mating systems and seed dispersal mechanisms.
Key words: Medicago lupulina, Medicago ruthenica, microsatellite, genetic diversity, gene flow, forage legume
INTRODUCTION
Life form, mating systems and seed dispersal are important adaptive traits shaping genetic structure and geographical distribution of plant populations (Levin, 1981; Loveless and Hamrick, 1984; Ennos, 1994; Hamrick and Godt, 1996; Bohonak, 1999; Clauss and Mitchell-Olds, 2006; Song et al., 2006; Mable and Adam, 2007). Analyses of phenotypic variation of these traits together with population genetic variation should provide insights into the evolutionary history and processes of plant species (Barrett et al., 1996; Juan et al., 2004), which in turn will help determine evolutionary potentials and conservation strategies for natural populations.
Here, a population genetic study of two wild Medicago (Fabaceae) species, Medicago lupulina and Medicago ruthenica, which differ markedly in life form, mating systems, seed dispersal mechanisms and distribution ranges, is reported. The genus Medicago is distributed worldwide and consists of approx. 83 species, including two forage crops, M. sativa and M. truncatula (Small and Jomphe, 1989). Thirteen wild species of Medicago are found in China (Wei and Huang, 1998). They are adapted to a diverse range of habitats located in different geographical regions of China, from cold northern desert to warm and humid southern and central China, and from near the sea level in eastern China to high mountains in the Himalayas. These wild species hold a rich source of natural variation for the better understanding of plant population dynamics and for the improvement of Medicago cultivars. With rapid urbanization and overgrazing in China, however, these wild Medicago populations are threatened by severe reductions in number and size (J. Yan and H.-J. Chu, pers. obs.). Thus, there is an urgent need to investigate the population genetics and evolutionary dynamics of wild relatives of important forage crops.
Medicago truncatula has been chosen as one of the two representatives of the Fabaceae to have its entire genome sequenced. Taking advantage of the availability of genomic information for M. truncatula (http://medicago.org/), the population genetic structures of M. lupulina and M. ruthenica were studied using microsatellite markers. Medicago lupulina is annual, biennial or occasionally short-lived perennial, predominantly self-fertilizing, and widely distributed, whereas M. ruthenica is long-lived perennial, outcrossing, and much more narrowly distributed (Wei and Huang, 1998). Medicago lupulina has small indehiscent pods that facilitate long-distance seed dispersal by biotic and abiotic agents, whereas M. ruthenica has dehiscent pods and lacks effective mechanisms for seed dispersal.
Medicago lupulina, although never cultivated, has been grown as a green fodder or manure (Turkington and Cavers, 1979). The species is characterized as a water-saving and easily-maintained turf legume that contains a high level of protein and a low level of fibre suitable for grazing (Cao et al., 2003). However, the biomass of the forage legume is relatively low. Medicago ruthenica is adapted to dry, stony habitats or desert with extremely low snowfall and very cold winters (Campbell et al., 1999). It was considered to be superior to the cultivated species M. sativa in soil nutrient-use efficiency and thus might be more suitable for low-input systems (Campbell et al., 1999). Both wild species are adapted to a much wider range of habitats than the cultivated species and are valuable genetic resources for developing better grazing legumes especially in drier and colder regions. Although the morphology, physiology, phenology, chromosomal variation and in-breeding of the two species had been studied previously (Lammerink, 1968; Sidhu, 1971; Hébert et al., 1994; Mariani et al., 1996; Qi, 1996; van Berkum et al., 1998; Campbell et al., 1999; Wilson, 2005; Li and Shi, 2006), their population genetic structures had not been characterized.
In the present paper, the genetic variation was investigated in a sample consisting of 16 M. lupulina populations and 15 M. ruthenica populations using 15 and 17 microsatellite markers, respectively. These included five populations of each species occurring sympatrically with those of the other species, and five microsatellite markers shared between the two species. This made it possible to evaluate to a certain extent the genetic differentiation between M. lupulina and M. ruthenica in the common environments and at the same loci. In characterizing the population genetic structures of the two species, the aim was to (a) quantify genetic variability; (b) estimate gene flow; (c) infer the correlation between the genetic relationships and geographical distributions; and (d) integrate the genetic information with phenotypic variation in mating system, life form and seed dispersal to understand the population dynamics and evolutionary processes of the two Medicago species.
MATERIALS AND METHODS
Plant materials
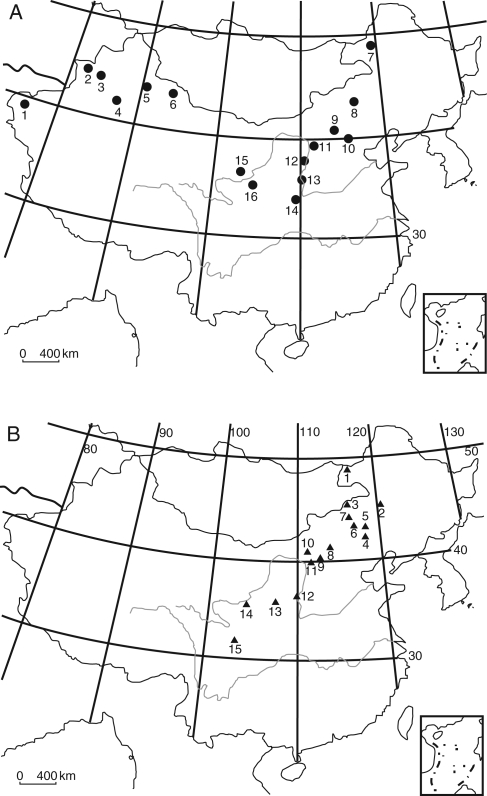
Medicago ruthenica Trautv. is distributed from central China to Mongolia and Siberia (Small and Jomphe, 1989). Our population sampling covers its entire distributional range in China (Fig. 1). Medicago lupulina L. has a broader distribution, occurring natively in temperate and subtropical Eurasia and North Africa (Dunbier, 1972). To compare its population genetic structure with M. ruthenica, populations of M. lupulina were sampled from the distributional range of M. ruthenica, including five sympatric populations of each species. In addition, populations of M. lupulina were sampled from north-western China. A total of 328 individuals were sampled from 16 populations of Medicago lupulina and 447 individuals were sampled from 15 populations of Medicago ruthenica between August and October in 2004 and 2005 (Table 1). Populations were recorded by GPS co-ordinates. Leaf samples were collected from randomly selected individuals in each population and immediately dried using silica gel for DNA isolation.
Fig. 1.
Distribution of the populations sampled in this study: (A) Medicago lupulina (numbers correspond to abbreviations ML-1 to ML-16 in Table 1); (B) Medicago ruthenica (numbers correspond to MR-1 to MR-15 in Table 1).
Table 1.
Collection localities of Medicago lupulina (ML) and Medicago ruthenica (MR) populations
| Population | Latitude | Longitude | N | na | ne | HO | HE | FIS | s | Private allele |
|---|---|---|---|---|---|---|---|---|---|---|
| ML-1 | 39·44 | 75·988 | 23 | 2·6 | 1·7 | 0·014 | 0·334 | 0·959 | 0·979 | 0 |
| ML-2 | 43·88 | 81·305 | 20 | 3·7 | 1·9 | 0·040 | 0·378 | 0·899 | 0·947 | 8 |
| ML-3 | 43·45 | 83·283 | 20 | 2·9 | 2·1 | 0·030 | 0·397 | 0·928 | 0·963 | 1 |
| ML-4 | 41·75 | 86·128 | 17 | 2·8 | 2·1 | 0·012 | 0·382 | 0·970 | 0·985 | 2 |
| ML-5 | 43·70 | 89·604 | 23 | 2·7 | 1·7 | 0·012 | 0·292 | 0·961 | 0·980 | 1 |
| ML-6 | 43·58 | 93·008 | 20 | 2·0 | 1·3 | 0·027 | 0·174 | 0·852 | 0·920 | 0 |
| ML-7 | 48·90 | 119·837 | 18 | 2·4 | 1·3 | 0·015 | 0·175 | 0·920 | 0·958 | 4 |
| ML-8 | 43·53 | 117·236 | 21 | 2·0 | 1·2 | 0·006 | 0·160 | 0·962 | 0·981 | 0 |
| ML-9 | 40·89 | 113·885 | 21 | 1·9 | 1·3 | 0·006 | 0·183 | 0·965 | 0·982 | 0 |
| ML-10 | 39·96 | 115·439 | 12 | 2·1 | 1·5 | 0·006 | 0·263 | 0·981 | 0·990 | 0 |
| ML-11 | 39·44 | 111·461 | 22 | 2·0 | 1·2 | 0·024 | 0·130 | 0·820 | 0·901 | 1 |
| ML-12 | 37·98 | 109·853 | 23 | 2·4 | 1·5 | 0·021 | 0·263 | 0·925 | 0·961 | 1 |
| ML-13 | 36·32 | 109·651 | 20 | 1·9 | 1·3 | 0·000 | 0·168 | 1·000 | 1·000 | 3 |
| ML-14 | 34·28 | 108·958 | 27 | 1·5 | 1·1 | 0·015 | 0·055 | 0·741 | 0·851 | 0 |
| ML-15 | 36·94 | 102·589 | 21 | 3·3 | 1·5 | 0·044 | 0·265 | 0·840 | 0·913 | 9 |
| ML-16 | 35·78 | 104·048 | 20 | 2·4 | 1·7 | 0·000 | 0·311 | 1·000 | 1·000 | 0 |
| ML mean | 20·5 | 2·4 | 1·5 | 0·017 | 0·246 | 0·920 | 0·958 | 1·875 | ||
| MR-1 | 49·56 | 117·45 | 51 | 8·824 | 4·840 | 0·627 | 0·715 | 0·133 | 0·235 | 3 |
| MR-2 | 45·13 | 112·49 | 46 | 9·000 | 5·449 | 0·636 | 0·729 | 0·139 | 0·244 | 4 |
| MR-3 | 45·56 | 117·01 | 51 | 8·941 | 5·141 | 0·564 | 0·706 | 0·210 | 0·347 | 2 |
| MR-4 | 42·29 | 119·02 | 20 | 6·941 | 3·927 | 0·619 | 0·684 | 0·121 | 0·216 | 3 |
| MR-5 | 42·97 | 119·01 | 21 | 7·412 | 4·221 | 0·584 | 0·686 | 0·173 | 0·295 | 1 |
| MR-6 | 43·26 | 117·53 | 35 | 8·000 | 4·359 | 0·599 | 0·692 | 0·149 | 0·259 | 6 |
| MR-7 | 43·53 | 117·24 | 36 | 8·176 | 4·768 | 0·557 | 0·706 | 0·225 | 0·367 | 4 |
| MR-8 | 40·89 | 113·87 | 20 | 6·941 | 4·003 | 0·581 | 0·660 | 0·145 | 0·253 | 5 |
| MR-9 | 40·37 | 113·24 | 20 | 7·706 | 4·829 | 0·641 | 0·722 | 0·138 | 0·243 | 1 |
| MR-10 | 40·91 | 111·69 | 35 | 7·941 | 4·728 | 0·603 | 0·722 | 0·179 | 0·304 | 2 |
| MR-11 | 39·44 | 111·46 | 23 | 6·059 | 3·866 | 0·594 | 0·677 | 0·146 | 0·255 | 3 |
| MR-12 | 36·31 | 109·65 | 20 | 6·000 | 3·772 | 0·551 | 0·617 | 0·135 | 0·238 | 1 |
| MR-13 | 35·75 | 107·98 | 21 | 7·647 | 4·547 | 0·591 | 0·664 | 0·135 | 0·238 | 4 |
| MR-14 | 35·80 | 104·06 | 20 | 5·647 | 3·664 | 0·457 | 0·608 | 0·273 | 0·429 | 3 |
| MR-15 | 31·68 | 103·84 | 28 | 6·000 | 3·220 | 0·413 | 0·569 | 0·292 | 0·452 | 3 |
| MR mean | 30 | 7·416 | 4·356 | 0·574 | 0·677 | 0·173 | 0·295 | 3 |
N, number of individual plants; na, observed alleles number; ne, effective allele number; HE, expected heterozygosity; HO, observed heterozygosity; FIS, inbreeding coefficient; s, selfing rate.
Microsatellite analysis
DNA was isolated from approx. 0·5 g dried leaves using a modified cetyltrimethy lammonium bromide (CTAB) protocol (Doyle and Doyle, 1987). DNA quality was tested on 0·8 % agarose gels. After measuring DNA concentration with an Eppendorf BioPhotometer, samples were diluted to 5 ng μL−1.
For microsatellite analysis, 92 pairs of primers were selected from previous publications (Diwan et al., 2000; Baquerizo-Audiot et al., 2001; Julier et al., 2003; Eujayl et al., 2004) and the Medicago genome website (http://medicago.org/genome/downloads.php). These primers were tested using 32 individuals from each of M. lupulina and M. ruthenica. Fifteen and 17 primers that detected a suitable level of polymorphism in M. lupulina and M. ruthenica, respectively, were identified (Table 2). The two species shared five microsatellite loci, MTIC14, MTIC188, MTIC189, MTIC432 and AFct45.
Table 2.
Diversity statistics and summary of F-statistics of microsatellite loci for Medicago lupulina and Medicago ruthenica
| Locus | A | HE | HO | FST | Nm |
|---|---|---|---|---|---|
| M. lupulina | |||||
| Mtgsp_005g08.ag.21-1 | 10 | 0·395 | 0·006 | 0·491 | 0·259 |
| Mtgsp_001b05.ag.17-1 | 7 | 0·384 | 0·031 | 0·476 | 0·276 |
| MAA660870 | 5 | 0·162 | 0·003 | 0·604 | 0·164 |
| MtSSRNFAL05 | 7 | 0·278 | 0·018 | 0·638 | 0·142 |
| MTR58 | 16 | 0·524 | 0·012 | 0·411 | 0·358 |
| MTIC210 | 9 | 0·275 | 0·003 | 0·564 | 0·193 |
| MTIC251 | 3 | 0·063 | 0·002 | 0·762 | 0·078 |
| MTIC339 | 3 | 0·037 | 0·000 | 0·062 | 3·774 |
| MTIC345 | 8 | 0·323 | 0·090 | 0·421 | 0·344 |
| MTIC451 | 8 | 0·222 | 0·020 | 0·670 | 0·123 |
| MTIC14 | 6 | 0·187 | 0·000 | 0·604 | 0·164 |
| MTIC188 | 11 | 0·258 | 0·015 | 0·585 | 0·177 |
| MTIC189 | 9 | 0·267 | 0·016 | 0·551 | 0·203 |
| MTIC432 | 5 | 0·184 | 0·037 | 0·612 | 0·158 |
| AFct45 | 5 | 0·127 | 0·003 | 0·567 | 0·191 |
| ML mean | 7·5 | 0·246 | 0·017 | 0·535 | 0·218 |
| M. ruthenica | |||||
| Mtgsp_005e04.taa.9-1 | 13 | 0·681 | 0·617 | 0·186 | 1·097 |
| Mtgsp_002c04.ac.29-1 | 39 | 0·858 | 0·641 | 0·082 | 2·800 |
| MtSSRNFAA02 | 7 | 0·445 | 0·484 | 0·082 | 2·788 |
| MtSSRNFAL45 | 20 | 0·830 | 0·698 | 0·071 | 3·292 |
| MAL369471 | 13 | 0·741 | 0·665 | 0·074 | 3·126 |
| AI974357 | 16 | 0·668 | 0·572 | 0·154 | 1·377 |
| MTIC48 | 24 | 0·735 | 0·573 | 0·139 | 1·547 |
| MTIC134 | 9 | 0·574 | 0·450 | 0·141 | 1·527 |
| MTIC249 | 9 | 0·534 | 0·299 | 0·155 | 1·367 |
| MTIC343 | 17 | 0·808 | 0·671 | 0·068 | 3·422 |
| MTIC354 | 11 | 0·453 | 0·489 | 0·057 | 4·175 |
| MTIC471 | 13 | 0·674 | 0·493 | 0·174 | 1·189 |
| MTIC14 | 6 | 0·475 | 0·302 | 0·244 | 0·776 |
| MTIC188 | 22 | 0·859 | 0·835 | 0·072 | 3·214 |
| MTIC189 | 22 | 0·885 | 0·834 | 0·053 | 4·425 |
| MTIC432 | 28 | 0·710 | 0·603 | 0·211 | 0·936 |
| AFct45 | 9 | 0·582 | 0·539 | 0·251 | 0·747 |
| MR mean | 16·4 | 0·677 | 0·577 | 0·130 | 1·672 |
A, Average number of alleles per locus; HE, expected heterozygosity; HO, observed heterozygosity; FST, coefficient of genetic differentiation; Nm, gene flow.
The five loci shared between the two species are indicated in bold.
Microsatellites were amplified by polymerase chain reaction (PCR). A 10-μL reaction contained 10–50 ng of template DNA, 1 × PCR reaction buffer, 1·5 mm of MgCl2, 0·2 mm of dNTP mix, 0·5 µm of each primer, and 0·5 U of Taq polymerase (Invitrogen). Reactions were performed in a Gene Amp PCR system 9700 (PE Applied Biosystems) with the following programme: 5 min at 95 °C, then 30 cycles of 45 s at 95 °C, primer-specific annealing temperature for 30 s at 50–60 °C, and 72 °C for 1 s, followed by a final extension of 7 min at 72 °C. PCR products were denatured for 5 min at 95 °C and run on a 6 % polyacrylamide denaturing gels that were made 0·4 mm thick. A 25-bp DNA ladder (Promega, Madison, WI, USA) was used as the size standard. The gels were treated with silver stained to visualize DNA bands and score microsatellite alleles. The gel could reliably resolve alleles with 2-bp length difference.
Genetic diversity and mating system analyses
For each population, genetic diversity was estimated across all loci using the observed number of alleles (na), effective number of alleles (ne), HE, HO, FIS, and the number of private alleles. For each microsatellite locus, genetic polymorphism was assessed by calculating the total number of alleles (A, allelic diversity), the expected and observed heterozygosity (HE and HO). The inbreeding coefficient, FIS, was calculated by FSTAT 2·9·3 (Goudet, 2001). GenAlEx 6 software was used to estimate ne, na, A, HO, HE (Peakall and Smouse, 2006). Selfing rate was estimated as s = 2 FIS/(1 + FIS) (Ritland, 1990). Deviation from the Hardy–Weinberg equilibrium and linkage disequilibrium were tested using the FSTAT program (Goudet, 2001). The significant values for the linkage disequilibrium were corrected for multiple comparisons by Bonferroni correction (Rice, 1989)
Genetic structure and genetic differentiation
STRUCTURE 2·2 (Pritchard et al., 2000; Falush et al., 2003) was used to test whether M. lupulina and M. ruthenica were genetically differentiated without a priori classification of individuals. The program STRUCTURE implements a model-based clustering method to demonstrate the presence of population structure, assign individuals to populations and identify migrants and admixed individuals (Pritchard et al., 2000), by assuming that the markers are unlinked and at linkage equilibrium within populations. The program calculates an estimate of the posterior probability of the data for a given K, Pr(X/K) (Pritchard et al., 2000). The range of possible K was from 1 or 2 to the true number of populations plus 3 (Evanno et al., 2005). Therefore, the number of populations (clusters), K, was set from 1 to 18. Under the assumption that admixture model and allele frequencies correlated, each K was replicated 3–5 times with different probability (v) for 100 000 iterations after a burn-in period of 50 000 without prior information on the population of origin. Additionally, population divergence was quantified using θ, and an unbiased estimator of FST (Weir and Cockerham, 1984; Slatkin, 1995) was estimated using FSTAT version 2·9·3 (Goudet, 2001). Genetic differentiation within and among populations was further measured by analysis of molecular variance (AMOVA) using ARLEQUIN 3·1 (Excoffier et al., 2005).
The POPULATION 1·2 software (Langella, 2000) was used to calculate the Nei's genetic distance (DA) (Nei, 1983)among individuals within each species. Unrooted neighbor-joining trees based on Nei's genetic distance were constructed and visualized with the TREEVIEW software (Page, 1996). To investigate spatial genetic structure, the relationship between the matrix of pairwise genetic distance [FST/(1 – FST)] and the matrix of the logarithm of geographical distances was analysed via a Mantel's test (Mantel, 1967) with 100 000 random permutations using the program IBD (Bohonak, 2002). Geographic distances between pairs of populations were calculated from linear distances between latitude and longitude positions (http://jan.ucc.nau.edu/~cvm/latlongdist.html).
Gene flow
Individual-based assignment tests have been used to estimate contemporary rates of gene flow and dispersal (Berry et al., 2004; Mix et al., 2006). A partial exclusion Bayesian-based individual assignment test (Rannala and Mountain, 1997), implemented in the GeneClass 2·0 (Piry et al., 2004), was used to assess the recent gene flow between pairs of populations. Assignment probabilities are computed based on the resampling method of Paetkau et al. (2004). A total number of 1000 individuals was simulated and a threshold of 0·01 was used. The limitation of this method is that the migration rate observed during a short study might not accurately reflect long-term patterns of gene flow (Manel et al., 2005). Therefore, FST was used to estimate historical rates of gene flow (Nm) according to Wright's island model of population genetic structure, where FST ≈ 1/(1 + 4Nm) (Wright, 1951; Slatkin and Barton, 1989; Gaggiotti et al., 1999; Sork et al., 1999).
RESULTS
Genetic diversity and mating system
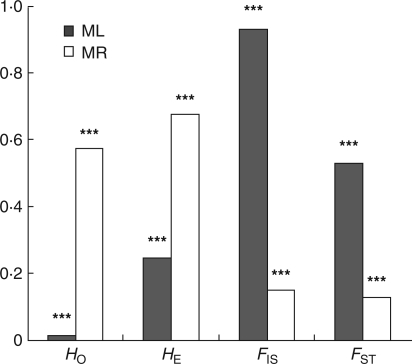
The level and pattern of estimated genetic variation differed substantially between populations of M. lupulina and M. ruthenica (Table 1 and Fig. 2). For M. lupulina, a total of 328 individuals from 16 populations was surveyed, in which 112 alleles were identified at 15 microsatellite loci. The average of observed heterozygosity (HO) within populations was 0·017, ranging from 0 to 0·044. The values are considerably lower than expected heterozygosity (HE) assuming the Hardy–Weinberg equilibrium, which averaged 0·246.
Fig. 2.
Comparison of microsatellite diversity between Medicago lupulina (ML) and Medicago ruthenica (MR); ***P < 0·001.
In comparison to M. lupulina, M. ruthenica has a higher level of genetic variation. In 447 individuals from 15 sampled populations of M. ruthenica, 278 alleles were found at 17 loci. The average HO of each populations is 0·574, ranging from 0·413 to 0·641. This is slightly lower than the average HE of 0·677. Medicago ruthenica populations also possessed a larger average number of private alleles than M. lupulina populations (3 versus 1·9). However, the variation in the number of private alleles is greater among M. lupulina populations (range 0–9) than that of the M. ruthenica populations (range 1–5).
When the genetic variation was compared between the two species at the five shared microsatellite loci, M. ruthenica accessions also showed a higher allelic diversity than M. lupulina (Table 2). At these five loci, 17·4 alleles per locus were found for M. ruthenica and 7·2 alleles per locus for M. lupulina. This is similar to the averages of all sampled loci, which was 16·4 and 7·5 alleles per locus for M. ruthenica and M. lupulina, respectively.
The test for the Hardy–Weinberg equilibrium found that of 240 locus–population combinations in M. lupulina, 165, 128 and 109 or 68·8 %, 53·3 % and 45·4 % showed significant deviation at P = 0·05, 0·01 and 0·001, respectively. For M. ruthenica, of 255 locus-population combinations, 112, 79 and 48 or 43·9 %, 30·9 % and 18·8 % showed significant deviation at P = 0·05, 0·01 and 0·001, respectively. The test for the genotypic disequilibrium in all samples within each species found that 80 of 105 locus pairs in M. lupulina and 36 of 136 locus pairs in M. ruthenica showed significant deviation at the P = 5 %, but none of locus pairs was found to be in significant genotypic disequilibrium after the Bonferroni-type correction.
The average FIS values were 0·920 (range 0·741–1·000) and 0·173 (range 0·121–0·292) for M. lupulina and M. ruthenica, respectively. From these values, the rates of self-fertilization of M. lupulina and M. ruthenica are calculated at 95·8 % and 29·5 %, respectively.
Population genetic structure and gene flow
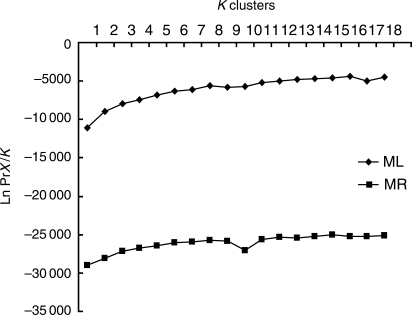
The analysis of molecular variance (AMOVA, Table 3) revealed that the majority of genetic variation occurred among populations in M. lupulina (55·5 %) and within individuals in M. ruthenica (76·06 %). In contrast, the minimum partitions of genetic variation resided within individuals in M. lupulina (3·07 %) and among populations in M. ruthenica (10·81 %). Bayesian clustering without prior information about geographical origin of populations showed that the highest likelihood value (Ln PrX/K) occurred at K = 16 in M. lupulina and K = 15 in M. ruthenica (Fig. 3), where the number of clusters (K) was consistent with the natural populations sampled in this study. The result held for different values of v (the probability that an individual was an immigrant to a given population; v = 0·01, 0·05 and 0·1).
Table 3.
The analysis of molecular variance (AMOVA) for 16 Medicago lupulina populations and 15 Medicago ruthenica populations
| Source of variation | d. f. | Variance components | Percentage of variation | P |
|---|---|---|---|---|
| M. lupulina | ||||
| Among populations | 15 | 2·39 | 55·50 | <0·001 |
| Among individuals within populations | 312 | 1·83 | 41·43 | <0·001 |
| Within individuals | 328 | 0·13 | 3·07 | <0·001 |
| Total | 655 | 4·22 | ||
| M. ruthenica | ||||
| Among populations | 14 | 0·68 | 10·81 | <0·001 |
| Among individuals within populations | 432 | 0·83 | 13·13 | <0·001 |
| Within individuals | 447 | 4·79 | 76·06 | <0·001 |
| Total | 893 | 6·30 | ||
Fig. 3.
Estimated posterior probability of K (1–18) for Medicago lupulina (n = 328) and Medicago ruthenica (n = 447) averaged over five runs, where K represents the number of clusters and n represent the number of samples.
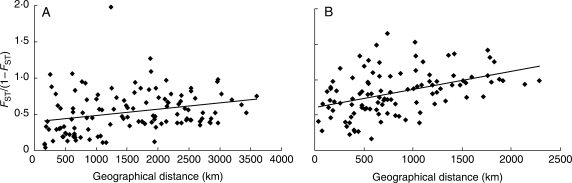
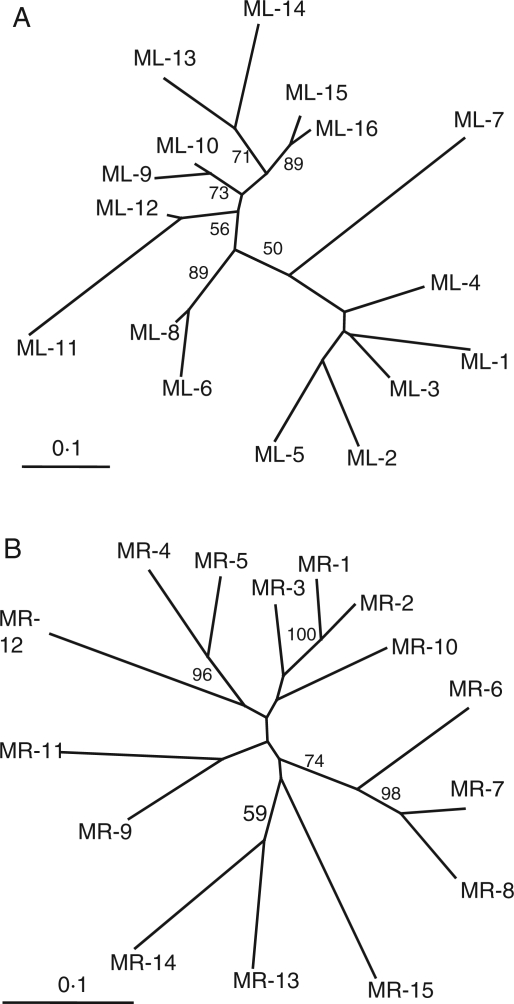
Substantial genetic differentiation was found among populations for both M. lupulina (FST = 0·535 P < 0·001) and M. ruthenica (FST = 0·130, P < 0·001; Table 2). The Mantel's test showed that the genetic distance [FST/(1 – FST)] and the geographical distance of the populations of each species are positively correlated (M. lupulina: r = 0·27, P = 0·0094; M. ruthenica: r = 0·41, P = 0·0018; Fig. 4). The cluster analyses of population relationships based on genetic distance showed the populations from closely situated regions were grouped together (Fig. 5). Nevertheless, some exceptions are noteworthy. Populations MR-10 and MR-12 from central China were clustered with populations from north-eastern China. ML-7 in north-eastern China was clustered with populations from north-western China, ML1-5, rather than clustered with the nearest population ML-8. Similarly, populations ML-8 was clustered with ML-6 in western China.
Fig. 4.
Scatter plots of pairwise genetic distance [FST/(1 – FST)] versus geographical distance (km) of all sampled populations of (A) M. lupulina and (B) M. ruthenica.
Fig. 5.
Unrooted neighbor-joining tree showing relationships among the (A) 16 M. lupulina and (B) 15 M. ruthenica populations. Numbers associated with branches indicate bootstrap values higher than 50 %, based on 1000 replications. Refer to Table 1 for abbreviations of populations.
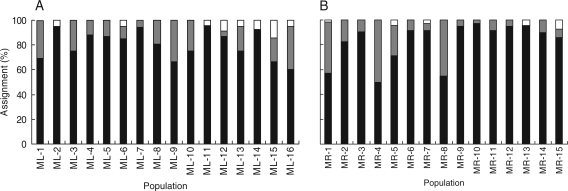
Based on estimated FST, historical gene flow among the sampled populations (Nm) was calculated at 0·218 in M. lupulina and 1·672 in M. ruthenica (Table 2). With regard to the contemporary gene flow, the assignment tests showed that an average of 80·8 % (P < 0·01) and 82·6 % (P < 0·01) of individuals were correctly assigned to their own source populations in M. lupulina and M. ruthenica, respectively (Fig. 6). For the remaining individuals, 3·4 % of M. lupulina and 1·4 % of M. ruthenica individuals did not belong to any of the populations sampled, and 15·7 % of M. lupulina and 16 % of M. ruthenica individuals were assigned to a population different from which they were collected.
Fig. 6.
Results of the assignment test for (A) Medicago lupulina and (B) Medicago ruthenica. Columns indicate the percentages of individuals per population correctly assigned to the source population (black), assigned to other sampled populations (grey) and not assigned to any sampled population (white).
DISCUSSION
Genetic variation
This study provides the genetic estimate of the mating systems of the two Medicago species. From FIS values, selfing rates were estimated to be higher than 95 % for M. lupulina but lower than 30 % for M. ruthenica. Together with previous studies (Mulligan, 1972; Bena et al. 1998; Campbell et al., 1999) and the observed heterozygosity (HO), this suggests that M. lupulina is predominantly selfing whereas M. ruthenica is highly outcrossing.
The estimated mating system differentiation is consistent with differences in floral morphology, similar to the correlation between floral morphology and pollinator attraction found in other plant groups (e.g. Juan et al., 2004; Gómez et al., 2008). Medicago lupulina has relatively small flowers with yellow papilionaceous corollas of 2–4 mm long (Fig. 7A). Medicago ruthenica has larger and more showy flowers with yellow corollas of approx. 8 mm long tinged with dark purple on the outside of the petals and on the inside toward the base (Fig. 7C). It was observed that M. ruthenica was able to attract substantially more insect pollinators such as bees, bumblebees and butterflies.
Fig. 7.
Morphology of flowers and fruits of M. lupulina and M. ruthenica: (A, B) an inflorescence and an indehiscent fruit of M. lupulina; (C, D) a flower and a dehiscent fruit with an exposed seed of M. ruthenica.
Mating system and life form play important roles in shaping population genetic structure and distribution of plants (e.g. Loveless and Hamrick, 1984; Hamrick and Godt, 1989, 1996; Stenøien et al., 2005; Clauss and Mitchell-Olds, 2006; Drummond and Hamilton, 2007; Mable and Adam, 2007; Michalski and Durka, 2007). The substantially higher selfing rate in M. lupulina could have contributed to a lower overall level of estimated heterozygosity (HE = 0·246) than those from M. ruthenica (HE = 0·677). A low level of heterozygosity (HE = 0·348–0·476) was also found for the selfing species M. truncatula in the French Mediterranean region (Bonnin et al., 2001; Ellwood et al., 2006), whereas a higher level of heterozygosity (HE = 0·665–0·717) was observed for an outcrossing species Medicago sativa (Flajoulot et al., 2005). These values are also comparable to the genetic diversity previously reported for other plant species with the similar mating system and life form. For instance, Arabidopsis thaliana, a selfing annual plant, had a much lower level of the genetic diversity (HE = 0·01, 0·06) than the self-incompatible perennial relatives A. halleri (HE = 0·31) and A. petraea (HE = 0·41) (Clauss et al., 2002; Stenøien et al., 2005). The present results are generally consistent with the trends of genetic variation observed in many flowering plant groups based on microsatellite data, with average HE values being 0·41 for inbreeding populations versus 0·65 for outcrossing populations and 0·46 for annuals versus 0·68 for perennials (Nybom, 2004).
Population differentiation and gene flow
The estimate of population differentiation showed a much higher population genetic differentiation in M. lupulina (FST = 0·535) than in M. ruthenica (FST = 0·130). This fits well with the general estimates of approx. 5-fold higher FST in selfing, annual species than outcrossing, perennial species for flower plants (Hamrick and Godt, 1996).
The significantly positive correlation between genetic and geographical distances detected in M. lupulina (r = 0·2703, P = 0·0094) and M. ruthenica (r = 0·4113, P = 0·0018) indicates that spatial separation has played a role in shaping the population genetic structure of the species. There is a general tendency that closely situated populations are genetically more similar. However, a close relationship between two disjunct populations, ML-6 and ML-8, suggests that there exists a dispersal or gene flow corridor connecting these regions across the Mongolian grasslands. Although it was not possible to sample M. lupulina populations from Mongolia, finding genetically similar populations flanking the width of Mongolia implies that the gene pool from this broad region may be represented in the present samples.
While geographical isolation has played a role in population genetic differentiation, there is evidence for gene flow between populations of each species. Among individuals of M. lupulina and M. ruthenica, 3·4 % and 1·4 %, respectively could not be assigned to sampled populations, suggesting that they were immigrants from outside the areas sampled (Fig. 6). In addition, the assignment of 15·7 % and 16 % of individuals to populations different to their sampled populations may be a result of gene flow between these populations.
Mating system, seed dispersal and population structure and distribution
Plants and their genes migrate through seed and pollen dispersal. The study of the interplay between seed dispersal and pollination is essential for understanding plant population structure and distribution (Govindaraju, 1988; Ennos, 1994; Bohonak, 1999; McCauley, 1997; Heuertz et al., 2003; Juan et al., 2004; Otero-Arnaiz, 2005). Seed dispersal of M. lupulina and M. ruthenica may be substantially affected by the distinct fruit morphologies (e.g. Janson, 1983; Gautier-Hion et al., 1985; Willson and Traveset, 2000). Pods of M. lupulina are rough-ridged, indehiscent, up to 3 mm long and 1 mm wide and each contains one seed (Fig. 7B). Rough-surfaced, indehiscent pods are effectively dispersed by birds and animals through adhesion or ingestion (Lammerink, 1968; Dunbier, 1972; Sorensen 1986). In addition, indehiscent pods of M. lupulina can float in water for up to 12 d (Turkington and Cavers, 1979).
Medicago ruthenica, however, lacks an effective mechanism of seed dispersal. Its pod is up to 15 mm long and 5 mm wide with an oblong-falcate suture and contains three to five seeds (Fig. 7D). The dorsal suture is strongly convex and the ventral suture weakly convex to straight, facilitating dehiscence when pods mature. Pods of M. ruthenica dehisce and release seeds near the mother plant. Seeds have no wings and are dispersed mainly by gravity. This is similar to wild soybean which was reported to disperse seeds within 4·5 m of the mother plants (Oka, 1983; Kuroda et al., 2006).
The difference in current distributional ranges of the two species could be attributed at least partly to seed dispersal abilities. Medicago lupulina, with more effective seed dispersal mechanisms, occurs in a much broader geographical range than M. ruthenica. The narrower distributional range of M. ruthenica does not seem to have been a result of population extinction. Medicago ruthenica populations that were studied in the field appeared healthy, and the level of genetic variation currently maintained within and among populations does not provide any indication that this outcrossing species has experienced a severe reduction in genetic variation.
Despite more effective seed dispersal of M. lupulina, the present genetic data showed that there has been a much higher level of historical gene flow between M. ruthenica populations. This is not surprising, given a greater pollen dispersal capacity of the outcrosser, M. ruthenica than the selfer, M. lupulina. In predominantly selfing species, individuals migrating into other populations may not effectively incorporate their private alleles into the local populations through cross-pollination. As a result, these alleles may easily get lost through drift if they are not favoured by selection. New alleles from other populations are likely to be less fit than the alleles of the native populations that have been selected by local ecological factors. Furthermore, migrating alleles are especially susceptible to loss through drift in annual species. For an out-crossing species, on the other hand, migration into new populations via seed dispersal allows alleles to be much more easily integrated into the gene pool of the recipient populations through cross-pollination. Less-fit alleles may be maintained in recipient populations at the heterozygous loci as long as they are at least partially recessive, and may persist for a relatively long period of time in a perennial species even though they are not postively selected into the local gene pool.
Taken together, this study between the two Medicago species with a combination of several distinct biological features has allowed us to gain a better understanding of population processes of plant evolution. As a perennial species, M. ruthenica benefits from an outcrossing mating system for the maintenance of genetic variation within populations. As a result, the populations can be more stable and less susceptible to pathogen and environmental changes. Consequently, there might have been relatively little pressure for the development of effective seed-dispersal mechanisms. In M. lupulina, on the other hand, effective seed-dispersal mechanisms are essential for a predominantly annual species. Indeed, effective seed-dispersal mechanisms could have broadened its distribution. For an annual species possessing effective seed dispersal, self-pollination provides reproductive assurance so that a single or a few seeds could potentially establish a new population in a new locality (Holsinger, 1996). Characterization of the population genetic structure of M. lupulina and M. ruthenica has provided an understanding of historical population dynamics of the two species and their current distribution.
ACKNOWLEDGEMENTS
We thank M. Kang and X.-H. Yao for technical assistance. This study was partially supported by the Chinese Academy of Sciences.
LITERATURE CITED
- Baquerizo-Audiot E, Desplanque B, Prosperi JM, Santoni S. Characterization of microsatellite loci in the diploid legume Medicago truncatula (barrel medic) Molecular Ecology Notes. 2001;1:1–3. [Google Scholar]
- Barrett SC, Harder LD, Worley AC. The comparative biology of pollination and mating in flowering plants. Philosophical Transactions of the Royal Society of London, Series B. 1996;351:1271–1280. [Google Scholar]
- Bena G, Lejeune B, Prosperi JM, Olivieri I. Molecular phylogenetic approach for studying life-history evolution: the ambiguous example of the genus Medicago L. Proceedings of the Royal Society of London, Series B. 1998;265:1141–1151. doi: 10.1098/rspb.1998.0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berkum P, Beyene D, Bao G, Campbell AT, Eardly BD. Rhizobium mongolense sp. nov., is one of three rhizobial genotypes identified which nodulate and form nitrogen-fixing symbioses with Medicago ruthenica. International Journal of Systematic Bacteriology. 1998;48:13–22. doi: 10.1099/00207713-48-1-13. [DOI] [PubMed] [Google Scholar]
- Berry O, Tocher MD, Sarre SD. Can assignment tests measure dispersal? Molecular Ecology. 2004;13:551–561. doi: 10.1046/j.1365-294x.2004.2081.x. [DOI] [PubMed] [Google Scholar]
- Bohonak AJ. Dispersal, gene flow, and population structure. The Quarterly Review of Biology. 1999;74:21–45. doi: 10.1086/392950. [DOI] [PubMed] [Google Scholar]
- Bohonak AJ. IBD (Isolation by Distance): a program for analyses of isolation by distance. Journal of Heredity. 2002;93:153–154. doi: 10.1093/jhered/93.2.153. [DOI] [PubMed] [Google Scholar]
- Bonnin I, Ronfort J, Wozniak F, Olivier I. Spatial effects and rare outcrossing events in Medicago truncatula (Fabaceae) Molecular Ecology. 2001;10:1371–1384. doi: 10.1046/j.1365-294x.2001.01278.x. [DOI] [PubMed] [Google Scholar]
- Campbell TA, Bao G, Xia ZL. Completion of the agronomic evaluations of Medicago ruthenica [(L.) Ledebour] germplasm collected in Inner Mongolia. Genetic Resources and Crop Evolution. 1999;46:477–484. [Google Scholar]
- Cao ZhZh, Feng YQ, Ma HL, Liu XN, Zhou YL, Xu ZM. Medicago lupulina – beautiful water-saving and easily-maintained turf legume. Pratacultural Science. 2003;4:58–60. [Google Scholar]
- Clauss MJ, Mitchell-Olds T. Population genetic structure of Arabidopsis lyrata in Europe. Molecular Ecology. 2006;15:2753–2766. doi: 10.1111/j.1365-294X.2006.02973.x. [DOI] [PubMed] [Google Scholar]
- Clauss MJ, Cobban H, Mitchell-Olds T. Cross-species microsatellite markers for elucidating population genetic structure in Arabidopsis and Arabis (Brassicaeae) Molecular Ecology. 2002;11:591–601. doi: 10.1046/j.0962-1083.2002.01465.x. [DOI] [PubMed] [Google Scholar]
- Diwan N, Bouton JH, Kochert G, Cregan Mapping of simple sequence repeat (SSR) DNA markers in diploid and tetraploid alfalfa. Theoretical and Applied Genetics. 2000;101:165–172. [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987;19:11–15. [Google Scholar]
- Drummond CS, Hamilton MB. Hierarchical components of genetic variation at a species boundary: population structure in two sympatric varieties of Lupinus microcarpus (Leguminosae) Molecular Ecology. 2007;16:753–769. doi: 10.1111/j.1365-294X.2006.03186.x. [DOI] [PubMed] [Google Scholar]
- Dunbier MW. Genetic variability in Medicago lupulina L. across a valley in the Weka Pass, New Zealand. New Zealand Journal of Botany. 1972;10:48–58. [Google Scholar]
- Ellwood SR, Souza NKD, Kamphuis LG, Burgess TI, Nair RM, Oliver RP. SSR analysis of the Medicago truncatula SARDI core collection reveals substantial diversity and unusual genotype dispersal throughout the Mediterranean basin. Theoretical and Applied Genetics. 2006;112:977–983. doi: 10.1007/s00122-005-0202-1. [DOI] [PubMed] [Google Scholar]
- Ennos RA. Estimating the relative rates of pollen and seed migration among plant populations. Heredity. 1994;72:250–259. [Google Scholar]
- Eujayl I, Sledge MK, Wang L, et al. Medicago truncatula EST-SSRs reveal cross-species genetic markers for Medicago spp. Theoretical and Applied Genetics. 2004;108:414–422. doi: 10.1007/s00122-003-1450-6. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin version 3·0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajoulot S, Ronfort J, Baudouin P, et al. Genetic diversity among alfalfa (Medicago sativa) cultivars coming from a breeding program, using SSR markers. Theoretical and Applied Genetics. 2005;111:1420–1429. doi: 10.1007/s00122-005-0074-4. [DOI] [PubMed] [Google Scholar]
- Gaggiotti OE, Lange O, Rassmann K, Gliddon C. A comparison of two indirect methods for estimating average levels of gene flow using microsatellite data. Molecular Ecology. 1999;8:1513–1520. doi: 10.1046/j.1365-294x.1999.00730.x. [DOI] [PubMed] [Google Scholar]
- Gautier-Hion A, Duplantier JM, Quris R, et al. Fruit characters as a basis of fruit choice and seed dispersal in a tropical forest vertebrate community. Oecologia. 1985;65:324–337. doi: 10.1007/BF00378906. [DOI] [PubMed] [Google Scholar]
- Goudet J. FSTAT, a program to estimate and test gene diversities and fixation indices. 2001. version 2.9.3. http://www2.unil.ch/popgen/softwares/fstat.htm .
- Govindaraju DR. Relationship between dispersal ability and levels of gene flow in plants. Oikos. 1988;52:31–35. [Google Scholar]
- Gómez J, Bosch J, Perfectti F, Fernández JD, Abdelaziz M, Camacho JPM. Association between floral traits and rewards in Erysimum mediohispanicum (Brassicaceae) Annals of Botany. 2008;101:1413–1420. doi: 10.1093/aob/mcn053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamrick JL, Godt MJW. Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS, editors. Plant population genetics, breeding and germplasm resources. Sunderland, MA: Sinauer; 1989. pp. 43–63. [Google Scholar]
- Hamrick JL, Godt MJW. Effects of life-history traits on genetic diversity in plant species. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 1996;351:1291–1298. [Google Scholar]
- Hébert D, Fauré S, Olivieri I. Genetic, phenotypic, and environmental correlations in black medic, Medicago lupulina L., grown in three different environments. Theoretical and Applied Genetics. 1994;88:604–613. doi: 10.1007/BF01240925. [DOI] [PubMed] [Google Scholar]
- Heuertz M, Vekemans X, Hausman JF, Palada M, Hardy OJ. Estimating seed vs. pollen dispersal from spatial genetic structure in the common ash. Molecular Ecology. 2003;12:2483–2495. doi: 10.1046/j.1365-294x.2003.01923.x. [DOI] [PubMed] [Google Scholar]
- Holsinger KE. Pollination biology and the evolution of mating systems in flowering plants. Evolutionary Biology. 1996;29:107–149. [Google Scholar]
- Janson CH. Adaptation of fruit morphology to dispersal agents in a Neotropical forest. Science. 1983;219:187–189. doi: 10.1126/science.219.4581.187. [DOI] [PubMed] [Google Scholar]
- Juan A, Crespo MB, Cowan RS, Lexer C, Fay MF. Patterns of variability and gene flow in Medicago citrina, an endangered endemic of islands in the western Mediterranean, as revealed by amplified fragment length polymorphism (AFLP) Molecular Ecology. 2004;13:2679–2690. doi: 10.1111/j.1365-294X.2004.02289.x. [DOI] [PubMed] [Google Scholar]
- Julier B, Flajoulot S, Barre P, et al. Construction of two genetic linkage maps in cultivated tetraploid alfalfa (Medicago sativa) using microsatellite and AFLP markers. BMC Plant Biology. 2003;3:1471–1490. doi: 10.1186/1471-2229-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda Y, Kaga A, Tomooka N, Vaughan DA. Population genetic structure of Japanese wild soybean (Glycine soja) based on microsatellite variation. Molecular Ecology. 2006;15:959–974. doi: 10.1111/j.1365-294X.2006.02854.x. [DOI] [PubMed] [Google Scholar]
- Lammerink J. Genetic variability in commencement of flowering in Medicago lupulina L. in the south island of New Zealand. New Zealand Journal of Botany. 1968;6:33–42. [Google Scholar]
- Langella O. POPULATIONS 1·2: population genetic software, individuals or population distance, phylogenetic trees. 2000. http://bioinformatics.org/~tryphon/populations/
- Levin DA. Dispersal versus gene flow in plants. Annals of the Missouri Botanical Garden. 1981;68:233–253. [Google Scholar]
- Li HX, Shi FL. Current situation of yield improvement and seed production of Melilotoides ruthenica in China. Grassland and Turf. 2006;3:14–16. [Google Scholar]
- Loveless MD, Hamrick JL. Ecological determinants of genetic structure in plant populations. Annual Review of Ecology and Systematics. 1984;15:65–95. [Google Scholar]
- McCauley DE. The relative contributions of seed and pollen movement to the local genetic structure of Silene alba. Journal of Heredity. 1997;88:257–263. [Google Scholar]
- Mable BK, Adam A. Patterns of genetic diversity in outcrossing and selfing populations of Arabidopsis lyrata. Molecular Ecology. 2007;16:3565–3580. doi: 10.1111/j.1365-294X.2007.03416.x. [DOI] [PubMed] [Google Scholar]
- Manel S, Gaggiotti OE, Waples RS. Assignment methods: matching biological questions with appropriate techniques. Trends in Ecology and Evolution. 2005;20:136–142. doi: 10.1016/j.tree.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Research. 1967;27:209–220. [PubMed] [Google Scholar]
- Mariani A, Pupilli F, Calderini O. Cytological and molecular analysis of annual species of the genus Medicago. Canadian Journal of Botany. 1996;74:299–307. [Google Scholar]
- Michalski SG, Durka W. High selfing and high inbreeding depression in peripheral populations of Juncus atratus. Molecular Ecology. 2007;16:4715–4727. doi: 10.1111/j.1365-294X.2007.03547.x. [DOI] [PubMed] [Google Scholar]
- Mix C, Arens PFP, Rengelink R, Smulders MJM, van Groenendael JM, Ouborg NJ. Regional gene flow and population structure of the wind-dispersal plant species Hypochaeris radicata (Asteraceae) in an agricultural landscape. Molecular Ecology. 2006;15:1749–1758. doi: 10.1111/j.1365-294X.2006.02887.x. [DOI] [PubMed] [Google Scholar]
- Mulligan GA. Autogamy, allogamy, and pollination in some Canadian weeds. Canadian Journal of Botany. 1972;50:1767–1771. [Google Scholar]
- Nei M, Tajima F, Tateno Y. Accuracy of estimated phylogenetic trees from molecular data. Journal of Molecular Evolution. 1983;19:153–170. doi: 10.1007/BF02300753. [DOI] [PubMed] [Google Scholar]
- Nybom H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molecular Ecology. 2004;13:1143–1155. doi: 10.1111/j.1365-294X.2004.02141.x. [DOI] [PubMed] [Google Scholar]
- Oka HI. Genetic control of regenerating success in semi-natural conditions observed among lines derived from a cultivated × wild soybean hybrid. Journal of Applied Ecology. 1983;20:937–947. [Google Scholar]
- Otero-Arnaiz A, Casas A, Hamrick JL. Direct and indirect estimates of gene flow among wild and managed populations of Polaskia chichipe, an endemic columnar cactus in Centro Mexico. Molecular Ecology. 2005;14:4313–4322. doi: 10.1111/j.1365-294X.2005.02762.x. [DOI] [PubMed] [Google Scholar]
- Paetkau D, Slade R, Burden M, Estoup A. Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation-based exploration of accuracy and power. Molecular Ecology. 2004;13:55–65. doi: 10.1046/j.1365-294x.2004.02008.x. [DOI] [PubMed] [Google Scholar]
- Page RDM. TREEVIEW: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. GENALE6: Genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piry S, Alapetite A, Cornuet JM, Paetkau D, Baudouin L, Estoup A. GeneCalss2: a software for genetic assignment and first generation migrants detection. Journal of Heredity. 2004;95:536–539. doi: 10.1093/jhered/esh074. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephans M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi QH. Studies on properties of photosynthetic ecology in Melilotides ruthenica. Acta Agrestia Sinica. 1996;2:110–115. [Google Scholar]
- Rannala B, Mountain JL. Detecting immigration by using multilocus genotypes. Proceedings of the National Academy of Sciences of the USA. 1997;94:9197–9221. doi: 10.1073/pnas.94.17.9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- Ritland K. Inferences about inbreeding depression based on changes of the inbreeding coefficient. Evolution. 1990;44:1230–1241. doi: 10.1111/j.1558-5646.1990.tb05227.x. [DOI] [PubMed] [Google Scholar]
- Sidhu SS. Some aspects of the ecology of black medick (Medicago lupulina L.) Canada: University of Western Ontario; 1971. PhD Dissertation. [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M, Barton NH. A comparison of three indirect methods for estimating average levels of gene flow. Evolution. 1989;43:1349–1368. doi: 10.1111/j.1558-5646.1989.tb02587.x. [DOI] [PubMed] [Google Scholar]
- Small E, Jomphe M. A synopsis of the genus Medicago (Leguminosae) Canadian Journal of Botany. 1989;67:3260–3294. [Google Scholar]
- Song BH, Clauss MJ, Pepper A, Mitchell-Olds T. Geographic patterns of microsatellite variation in Boechera stricta, a close relative of Arabidopsis. Molecular Ecology. 2006;15:357–369. doi: 10.1111/j.1365-294X.2005.02817.x. [DOI] [PubMed] [Google Scholar]
- Sorensen AE. Seed dispersal by adhesion. Annual Reviews in Ecology and Systematics. 1986;17:443–463. [Google Scholar]
- Sork VL, Nason J, Campbell DR, Fernández JF. Landscape approaches to historical and contemporary gene flow in plants. Trends in Ecology and Evolution. 1999;14:219–224. doi: 10.1016/s0169-5347(98)01585-7. [DOI] [PubMed] [Google Scholar]
- Stenøien HK, Fenster CB, Tonteri A, Savolainen O. Genetic variability in natural populations of Arabidopsis thaliana in northern Europe. Molecular Ecology. 2005;14:137–148. doi: 10.1111/j.1365-294X.2004.02359.x. [DOI] [PubMed] [Google Scholar]
- Turkington R, Cavers PB. The biology of Canadian weeds. 33. Medicago lupulina L. Canadian Journal of Plant Science. 1979;59:99–110. [Google Scholar]
- Wei Z, Huang YL. 2. Vol. 42. Beijing: Science Press; 1998. Flora Reipublicae Popularis Sinicae, Tomus; pp. 314–316. [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Willson MF, Traveset A. The ecology of seed dispersal. In: Fenner M, editor. Seeds: the ecology of regeneration in plant communities. 2nd edn. Wallingford: CAB International; 2000. pp. 85–110. [Google Scholar]
- Wilson LC. Characteristics of black medic (Medicago lupulina L.) seed dormancy loss in western Canada. Canada: University of Manitoba; 2005. Master Thesis. [Google Scholar]
- Wright S. The genetic structure of population. Annals of Eugenics. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]