Abstract
C1q is a versatile recognition protein which binds to a variety of targets and consequently triggers the classical pathway of complement. C1q is a hetero-trimer composed of three chains (A, B and C) arranged in three domains, a short N-terminal region, followed by a collagenous repeat domain that gives rise to the formation of (ABC) triple helices, each ending in a C-terminal hetero-trimeric globular domain, called gC1q, which is responsible for the recognition properties of C1q. The mechanism of the trimeric assembly of C1q and in particular the role of each domain in the process is unknown. Here, we have investigated if the gC1q domain was able to assemble into functional trimers, in vitro, in the absence of the collagenous domain, a motif known to promote obligatory trimers in other proteins. Acid-mediated gC1q protomers reassembled into functional trimers, once neutralized, indicating that it is the gC1q domain which possesses the information for trimerization. However, reassembly occurred after neutralization, only if the gC1q protomers had preserved a residual tertiary structure at the end of the acidic treatment. Thus, the collagenous domain of C1q might initialize the folding of the gC1q domain so that subsequent assembly of the entire molecule can occur.
Keywords: C1q, Assembly mechanism, Trimer, Folding, Function
1. Introduction
The classical pathway of complement is one of the routes of innate immunity, and is triggered by C1, a multi-molecular complex which combines the ability to bind pathogens and to convert a recognition signal into specific proteolytic activities [1]. As the recognition subunit of C1, C1q is the first player in the complement cascade and is able to recognize a plethora of different targets, including viruses, bacteria and even misfolded proteins such as amyloid and prions [2–7]. Though, the mechanism of such versatile recognition is unknown, it is clearly related to the structure of C1q which is a heterotrimer comprising three chains, A, B and C of similar lengths (~220 residues) and of homologous amino acid sequences. Each chain comprises a short N-terminal region, involved in the formation of A–B and C–C inter chain disulfide bonds, followed by a repeating collagen-like sequence which assembles into six ABC triple helices. These six triple helices first associate to form a “stalk” and then, due to interruptions in the repeating Gly-Xaa-Yaa collagen-like sequence, diverge to form six “arms” [8,9]. Each arm merges at its C-terminal end into a globular “head” region consisting of ABC heterotrimers, known as gC1q and responsible for the recognition properties of C1q [10–12]. The C-terminal domains assembled from gC1q modules are also found at the C-terminal end of various other collagen-containing proteins such as types VIII and X collagens and adipocyte complement-related protein-30 [12,13]. These proteins share similar macromolecular organization with C1q, i.e. a short N-terminal domain followed by collagenous repeats forming triple helices and ending with a trimeric globular C-terminal domain. The folding and the assembly processes leading to the formation of such a complex structure has not been addressed on the full-length protein, but for some members of the family, the C-terminal domain expressed in bacterial system or synthetic peptides from the C-terminal domain have been shown to trimerize in vitro [14,15]. In those cases, the sequence of the C-terminal domain contains the information required for trimerization and the domain constitutes an independent folding unit, or a so-called foldon. Interestingly, these proteins also contain the collagen-like triple helix, which is one of the few 3D-assembly motifs known to trigger obligatory trimerization [16,17]. Therefore, for the C1q molecule, it remains to be established if it is the C-terminal domain or the collagenous repeats domain which triggers the assembly of the entire C1 molecule. We addressed this question by using the heterotrimeric C-terminal domain of C1q (gC1q) which had been separated from the collagenous repeats by a collagenase treatment. The X-ray structure of human gC1q shows that it is stable as a trimer [18], indicating that whatever roles the collagenous repeats might have in the assembly process, maintaining the gC1q trimeric association is not one of them. In the present work, a protocol has been established to disassemble in vitro the gC1q trimer so that its subsequent ability to reassemble, upon neutralization, could be investigated. After various acidic treatments and subsequent neutralization, the structural states of gC1q have been analyzed using different spectroscopic methods and its stoichiometry has been determined by chemical cross-linking experiments coupled with SDS-PAGE (Sodium dodecyl-sulfate polyacrylamide gel electrophoresis) analysis. The data show that the gC1q modules have the ability to self-assemble into functional trimers, only if some residual tertiary structure is maintained at the end of the dissociation phase.
2. Materials and methods
2.1. Reagents and buffers
Type III collagenase from Clostridium histolyticum and human IgG purified from serum were obtained from SIGMA as well as all other chemicals. The buffers used were McIlVaine buffer (0.2 M di-sodium hydrogen phosphate, 0.1 M citric acid) at pH 3–7 and 0.1 M HCl/KCl at pH 1.0 or 1.7. All buffers were filtered through a sterile 0.22 µM filter before use.
2.2. Purification of the C1q globular domain
The globular domain of human C1q was generated essentially as described by Tacnet et al. [6]. Briefly, C1q was treated with collagenase (C1q:collagenase ratio=15:1, w/w) for 16 h at 37 °C and purification was achieved by high-pressure gel permeation on a TSK-G2000 SW column (LKB).
2.3. Production of recombinant gC1qR
The strategy for the construction of a plasmid containing the full-length gC1qR cDNA was described in detail previously [19,20]. Briefly, the cDNA was subcloned downstream of the glutathione-S-transferase (GST) gene in the expression plasmid pGex-2T (Pharmacia Biotech Inc.), transformed into Escherichia coli BL-21 (DE3) and protein expression was induced by isopropyl β-d-1 thiogalacto pyranoside. The protein was expressed as a GST fusion product with GST at the N-terminus of gC1qR. The fusion product was purified on a glutathione-Sepharose 4B column, then cleaved by thrombin and the GST-free gC1qR was purified by FPLC using a Mono-Q ion exchange column. The single peak containing gC1qR was pooled, concentrated to about 1 mg/ml, and stored at −80 °C.
2.4. N-terminal sequence analysis
N-terminal sequence analysis of the purified C1q globular domain was performed after SDS-PAGE and electrotransfer using an Applied Biosystems model 477 A protein sequencer as described previously [21].
2.5. SDS-PAGE analysis
SDS-PAGE was performed on 15% gels using a Bio-Rad mini-Protean 3 cell system according to Laemmli [22]. Samples containing about 1 µg protein were loaded in each well and gels were silver stained.
2.6. Fluorescence spectroscopy
Fluorescence measurements were performed using a Cary eclipse Varian spectrofluorimeter. Both Tyr and Trp residues were excited at wavelength 280 nm whereas selective excitation of Trp residue was performed at wavelength 295 nm. Slit widths were 5 nm for both excitation and emission. When emission spectra were recorded, the Raman contribution for water was removed by subtraction of a buffer blank. When time-dependent changes in the Trp-fluorescence of gC1q were monitored, the fluorescence intensity was recorded at an emission wavelength of 344 nm. The gC1q trimer contains three tryptophans, two in the A protomer and one in the C protomer. It also contains 20 tyrosine residues, five in the A protomer, seven in the B protomer and eight in the C protomer. Protein samples were directly added to the cuvette containing appropriate buffer at a final protein concentration of 75 µg/ml, otherwise stated. The measurement was started at once and taken for 30 min. The dead time of the instrument is a few seconds (<10 s). The experiments were carried out at 23 °C. Samples were collected at different times of the kinetics of denaturation and of renaturation for subsequent analysis of their trimeric state by chemical cross-linking and SDS-PAGE.
2.7. Circular dichroism (CD)
Circular dichroism spectra were recorded on a Jasco J810 spectropolarimeter. A cell with a path length of 0.1 cm, a spectral band width of 4 nm, and a time constant of 1 s were used, and each spectrum was recorded as an average of 4 scans. Circular dichroism measurements were made on samples diluted from a protein stock solution in McIlVaine buffer at pH 7.0 or at pH 5.0 and in 0.1 M HCl KCl buffer at pH 1.7 to a final protein concentration of 0.2 mg/ml. Samples were mixed directly in the cuvette and scans were taken immediately. The Far-UV spectra of the samples were scanned again, after further 15 min incubation at each pH. The dead time of the instrument is a few seconds (<5 s). The experiments were performed at 23 °C.
2.8. Denaturation of the C1q globular domain
Native gC1q trimer (0.9 mg/ml) was diluted to a final protein concentration of 75 µg/ml in 0.1 M HCl/KCl at pH 1.7 or at pH 1.0 or in McIlVaine buffers at pH ranging from 3.0 to 7.0 and incubated for 15 or 30 min at 23 °C. The pH of the samples was measured with a microelectrode.
2.9. Renaturation of the C1q globular domain
Renaturation was performed after acidification either at pH 1.7 or at pH 5.0, followed by neutralization to provide renaturation conditions. For the treatment at pH 1.7, the gC1q native trimer (0.9 mg/ml) was diluted two-fold in 0.1 M HCl/KCl at pH 1.0 for 10 s, 2 min and 15 min followed by a 6-fold dilution in the McIlVaine buffer at pH 7.0. For the treatment at pH 5.0, the gC1q native trimer (0.9 mg/ml) was diluted two-fold in McIlVaine buffer at pH 5.0 for 10 s, 30 s or 15 min, followed by a 6-fold dilution in the McIlVaine buffer at pH 7.0. To test the role of the buffer in the denaturation and renaturation processes, the gC1q native trimer was diluted two-fold in PBS containing 0.1 M HCl/KCl at pH 1.0 so that after the dilution, the final pH was 5.0. To do so a 10-times dilution of 0.1 M HCl/KCl at pH 1.0 was necessary. This guaranteed that the denaturation occurred at the same protein concentration for both the buffer systems. The sample was incubated for 30 s at pH 5.0 then neutralized by a 6-fold dilution in the McIlVaine buffer at pH 7.0. Otherwise stated, the final protein concentration was 75 µg/ml and all experiments were carried out at 23 °C for 30 min. The return to pH 7.0 was checked by measuring the pH with a microelectrode. To achieve the final pH of 1.7, the two-fold dilution in acid was performed with 0.1 M HCl/KCl at pH 1.0. A two-fold dilution was sufficient to bring the pH to 5.0 when the McIlVaine buffer at pH 5.0 was used. When Trp- and Tyr-fluorescence experiments were performed, the protein was, treated in acid, and then added directly to the cuvette containing McIlVaine buffer at pH 7.0 and the measurement was started immediately. Trp- and Tyr-scans were taken at the end of the 30 min incubation.
2.10. Chemical cross-linking and SDS-PAGE analysis of the oligomeric state of the gC1q trimer
The gC1q trimer is not resistant to SDS and hence each gC1q subunit migrates as a monomer on SDS-PAGE. To assess the oligomeric state of gC1q, chemical cross-linking was performed to covalently attach the three protomers together prior to SDS-PAGE analysis. Incubation of the gC1q trimer in the presence of 0.4% (v/v) glutaraldehyde for 2 min at 23 °C at pH 7.0, led to almost complete cross-linking of the trimer with only little gC1q monomer left not cross-linked. Increasing the amount of cross-linker did not permit complete cross-linking of the gC1q trimer but increased the amount of the cross-linked gC1q higher oligomeric species observed on top of the gel. These cross-linked gC1q higher oligomeric species were the result of an excess of cross-linker. Thus, treatment of gC1q with 0.4% glutaraldehyde for 2 min was the optimum condition for cross-linking. After the treatment of gC1q at different pH, 0.1 M NaOH was added to the samples, just before addition of the glutaraldehyde. As a result, the final pH during the cross-linking reactions was 10.5. Thus, loss of the cross-linking of the gC1q trimer after denaturation could not be due to an inhibition of the cross-linking reaction by a low pH. The amount of the gC1q trimer cross-linked for the native gC1q was similar at pH 7.0 and at 10.5. Glutaraldehyde was chosen as the fastest cross-linker to avoid interference between the time needed for cross-linking and the time leading to reassembly. The cross-linking reaction was stopped by diluting the sample 2-fold in the sample buffer containing β-mercaptoethanol and SDS and then boiling the sample for 5 min. Samples were then analyzed by SDS-PAGE.
2.11. Surface plasmon resonance spectroscopy and data evaluation
Measurements were performed using a BIAcore 3000 instrument (BIAcore AB). The running buffer for protein immobilization was 145 mM NaCl, 5 mM EDTA, 10 mM HEPES, pH 7.4. The protein ligand gC1qR was diluted to 40 µg/ml in 10 mM formate, pH 3.0 and immobilized on the surface of a CM5 sensor chip (BIAcore AB) using the amine-coupling chemistry. Binding of native gC1q and of gC1q samples incubated either at a final pH of 1.7 for 10 s or at pH 5.0 for 15 min and subsequently neutralized at pH 7.0 for 60 min was measured over 2100 resonance units (RU) of immobilized gC1qR at a flow rate of 20 µl/min in McIlVaine buffer at pH 7.0 containing 0.005% surfactant P20 (BIAcore AB). Equivalent volumes of each protein sample were injected over a surface with immobilized BSA to serve as blank sensorgrams for subtraction of the bulk refractive index background. Regeneration of the surface was achieved by injection of 10 µl of 20 mM NaOH. Data were analyzed by global fitting to a 1:1 Langmuir binding model of both the association and dissociation phases for several concentrations simultaneously, using the BIAevaluation 3.1 software (BIAcore AB). The apparent equilibrium dissociation constants (KD) were calculated from the ratio of the dissociation and association rate constants (koff/kon).
2.12. Binding of gC1q to aggregated IgG
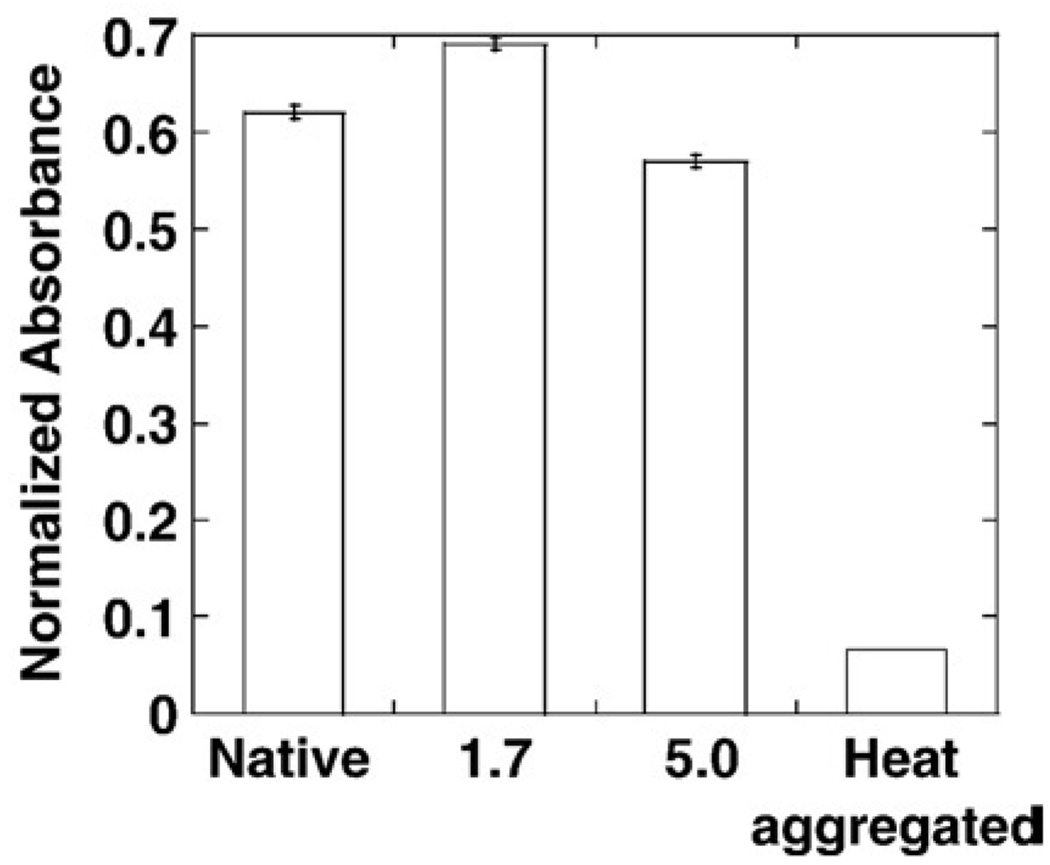
gC1q samples were submitted to treatment at a final pH of 1.7 for 10 s or at pH 5.0 for 15 min and subsequently neutralized for 60 min. These samples were diluted 20-fold in McIlVaine buffer at pH 7.0 and 0.375 µg was coated on a 96-well plate overnight at 4 °C. The same amount of native gC1q trimer was also coated on the plate. In addition, a sample of the native gC1q which had been heated at 120 °C for 10 min was also coated on the plate, under the same condition. This heat-aggregated gC1q was used to test whether aggregated gC1q recognized IgG molecules. Duplicated plates were prepared, one was tested for recognition of aggregated IgG molecule and the other was used to measure the amount of coated gC1q molecule, using anti C1q antibody. After blocking with PBS (phosphate-buffered saline) containing 2% milk for 45 min at 37 °C, and subsequent washings, the wells were incubated for 2 h at 37 °C with either heat-aggregated IgG (10 min at 120 °C) at 10 µg/ml in PBS containing 0.05% tween 20 and 5 mM CaCl2 or with anti gC1q polyclonal antibody (1/4000 dilution). After washing, the wells were incubated for 2 h at 37 °C with goat anti-human IgG conjugated to horse radish peroxidase when the wells had been incubated with IgG molecule or with a secondary antibody conjugated with horse radish peroxidase when the wells had been incubated with anti gC1q antibody. The color was developed using O-phenylene diamine-dihydrochloride (OPD) and the absorbance read at 492 nm after quenching of the reaction with 2 M sulfuric acid. For the sake of comparison between the different gC1q samples, the results were normalized by calculating the ratio of absorbances after reaction with IgG and after reaction with the anti C1q antibody.
3. Results
Our main objective is to investigate whether gC1q, the C-terminal domain of C1q, has alone the ability to fold and assemble into a native heterotrimer, or whether proper assembly is prevented in the absence of the collagenous repeats. To address this question, the gC1q domain generated by collagenase digestion of purified C1q, which is known to remain a heterotrimer after the treatment, was used as a starting material [18]. N-terminal sequence analysis of the purified material indicated that it comprises residues Gly79-Ala223 of the A chain, Gly81-Ala226 of the B chain, and Gly78-Asp217 of the C chain. Each segment therefore only contained three collagenous triplets at its N-terminal end, out of the 26–27 repeats in the full-length A, B, and C chains. Since, formation of a collagen-like triple helix requires multiple collagenous repeats, the three repeats left after collagenase treatment were unlikely to be sufficient for the generation of a triple helix and trimerization of the C-terminal domain [16].
3.1. Disassembly and unfolding of the gC1q trimer
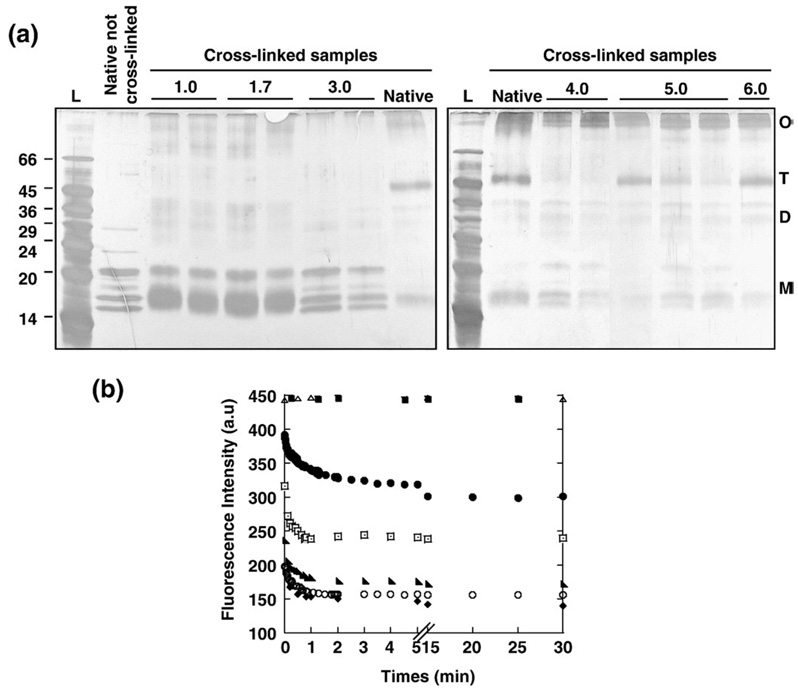
In order to test the capacity of gC1q to reassemble in vitro, we first looked for conditions leading to its disassembly. The native gC1q trimer was treated in different acidic conditions, as follows: samples were incubated for 30 min either in 0.1 M HCl/KCl buffer at pH 1.7 and at pH 1.0 or in McIlVaine buffer at pH from 3.0 to 6.0. To see if the acidic treatments disassembled the gC1q trimer, the oligomeric states of the samples were analyzed by SDS-PAGE, 15 min after incubation at each pH. However, direct analysis of the oligomeric state of gC1q on the gel was not possible because the trimer does not withstand the denaturating activity of SDS, and as a consequence migrates as three dissociated protomers, A, B and C (Fig. 1a, left gel). To detect the trimer by SDS-PAGE, glutaraldehyde was used to covalently cross-link the three chains to one another, prior to addition of the SDS sample buffer. Treatment of the native gC1q trimer for 2 min with 0.4% (v/v) glutaraldehyde followed by SDS-PAGE analysis resulted in a cross-linked gC1q species migrating with an apparent molecular weight of about 48 kDa, in good agreement with the molecular weight of the gC1q trimer calculated from the sequence of the three chains (A, 16 kDa, B, 16.4 kDa, C, 15.5 kDa) (Fig. 1a). After this treatment, only a small amount of the gC1q protomers remained seen on the gel, indicating that most of the gC1q trimer had been successfully cross-linked. Increasing the amount of glutaraldehyde did not lead to a 100% cross-link of the gC1q trimer (not shown). However, it significantly increased the amount of cross-linked gC1q higher oligomers, observed on top of the gel, indicating that those species resulted from an excess of cross-linker. Those cross-linked species were observed not only on the native gC1q sample but after all the pH treatments (see below), indicating the gC1q tendency to aggregate in solution, resulting in intermolecular cross-linking of gC1q trimer when in excess of cross-linker. However, reducing the amount of glutaraldehyde did not prevent intermolecular cross-linking but reduced the amount of the gC1q protomers cross-linked to one another. Therefore, 0.4% (v/v) glutaraldehyde and 2 min incubation was used in all the subsequent experiments to cross-link the gC1q samples. However, to prevent any effect of the pH on the cross-linking reaction, after treatment in acid, the pH of the samples was raised to 10.5 by adding 0.1 M of NaOH, prior to the addition of the glutaraldehyde (see Material and Methods). The presence of the NAOH did not change the cross-linking pattern of the native gC1q trimer.
Fig. 1.
pH-mediated denaturation of the native gC1q trimer. (a) Effect of pH on the oligomeric state of gC1q. gC1q native samples were treated in acid at 75 µg/ml, for 0 and 15 min, left and right lanes, respectively, for each indicated pH. On the right gel, only 15 min incubation time is indicated for the pH 6.0 treatment, whereas 0, 2 and 15 min are indicated for the pH 5.0 treatment. All the samples were cross-linked prior addition of sample buffer and analysis by SDS-PAGE. A sample of the native gC1q was also analyzed on the gel after cross-linking. T, gC1q trimer; D, gC1q heterodimers; M, gC1q protomers, O, cross-linked gC1q higher oligomers. L, molecular weight standards indicated in kDa. The gels were silver stained. (b) Kinetics of the pH-mediated denaturation of the native gC1q trimer. The Trp-fluorescence intensity of the gC1q sample was monitored with excitation and emission wavelengths of 295 and 344 nm, respectively. The native gC1q was diluted to a final concentration of 75 µg/ml in a cuvette containing buffer at pH 7.0 (■), pH 6.0 (△), pH 5.0 (●), pH 4.0 (⊡), pH 3.0 (◣), pH 1.7 (○) and pH 1.0 (◆) and measurement was started immediately. The double bar (//) indicates that the incubation time from 5 to 10 min was omitted for the sake of clarity. a.u. stands for arbitrary unit.
When gC1q was treated in acid at pH 1.0, 1.7 or 3.0, it dissociated rapidly and no trimer could be cross-linked even after only a few seconds of incubation (Fig. 1a, left gel). Mostly protomers and little cross-linked heterodimers could be observed after the short exposure to the acidic pH. A longer incubation time of 15 min at pH 1.0, 1.7 or 3.0 did not lead to further dissociation of the remaining gC1q heterodimers into protomers (Fig. 1a, left gel). After a few seconds at pH 4.0, a small amount of the gC1q trimer could still be cross-linked, suggesting that increasing the pH might slow the dissociation of the gC1q trimer (Fig. 1a, right gel). This was confirmed when gC1q was treated at pH 5.0 for a few seconds, conditions for which little gC1q trimer dissociated (Fig. 1a, right gel). Longer incubation at pH 5.0 decreased the amount of the gC1q trimer that was cross-linked and after 15 min, the gC1q trimer was almost entirely dissociated (Fig. 1a, right gel). The stoichiometry of the dissociated gC1q could however not be established since neither cross-linked heterodimers nor cross-linked protomers accumulated significantly on the SDS-PAGE (Fig. 1a). The easiest explanation was that gC1q had dissociated into three heterodimers (AB, AC and BC) and into three protomers (A, B and C) and as a result the amount of each individual species failed below the detection level of the silver stain. No dissociation happened if gC1q was treated for 15 min at pH 6.0 (Fig. 1a, right gel). At all pH, if gC1q was incubated for 30 min instead of 15 min and subsequently cross-linked, the same pattern was obtained indicating that after 15 min incubation, the maximum dissociation had taken place (data not shown). The cross-linked of higher gC1q oligomers due to an excess of glutaraldehyde was observed after all the treatments and prevented any quantification of the gC1q dissociation based on the cross-linking pattern. However, it is important to note that in order to observe small amount of the cross-linked gC1q trimer, the gels were over stained. As a result, the amount of the cross-linked gC1q higher oligomers detected on the gels is misleading and over-estimated.
Spectroscopic parameters were observed to change in parallel with the dissociation process. At 23 °C and at pH 6.0 no time-dependent changes were seen in the intensity of the fluorescence maxima (excitation 295 nm, emission 344 nm) of gC1q over 30 min (Fig. 1b). Thus, at this pH, the gC1q molecule was neither dissociated nor denaturated. In contrast, at 23 °C and pH values of 5.0 or lower, a time-dependent decrease in the intensity of the fluorescence maxima was observed (Fig. 1b). The fluorescence intensity of free Trp remained unchanged from pH 3.0 to 5.0, showing that the decrease of the fluorescence of the gC1q samples resulted from a change of the protein tertiary structure (not shown and [23]). Similarly, at pH 1.7 and 1.0, the drop of the intensity of the fluorescence maxima also indicated a conformational change of gC1q since addition of 0.1 M Tris at pH 8.0 which quickly raised the pH back to 7.0, did not lead to a fast return of the fluorescence signal to the level of the native fluorescence intensity (not shown). Each pH mediated changes in the tertiary structure of the gC1q to different degree as seen by the different levels of the intensity of the fluorescence maxima reached after the 30 min incubation. At pH 4.0 and lower, the decrease of the fluorescence intensity happened within one minute, when the gC1q was already dissociated (Fig. 1a). It therefore corresponded to a change in the tertiary structure of the gC1q protomers. The dead time of the instrument was of a few seconds, so the changes in the fluorescence intensity due to the dissociation of the gC1q trimer were too fast to be detected under these pH conditions. The same lowest fluorescence intensity was reached at pH 1.7 and 1.0, indicating that the maximum change in the tertiary structure of the gC1q protomers was already induced at pH 1.7. At pH 5.0, the fluorescence intensity decreased slowly within the first 15 min, and only a little further decrease was observed if gC1q was left for another 15 min. The fluorescence level of gC1q remained higher after pH 5.0 than after lower pH suggesting that the gC1q molecules remained more structured. The dissociation of gC1q observed during the 15 min incubation at pH 5.0 was slow (Fig. 1a) and it was therefore likely that the drop of the fluorescence intensity corresponded to changes in the tertiary structure of the gC1q trimer associated with its dissociation. The fluorescence signal of gC1q was stable after 15 min at pH 5.0 suggesting that gC1q did not undergo significant changes in its tertiary structure subsequent to its dissociation. The structural states of gC1q after treatments at pH 5.0 and at pH 1.7 were further investigated.
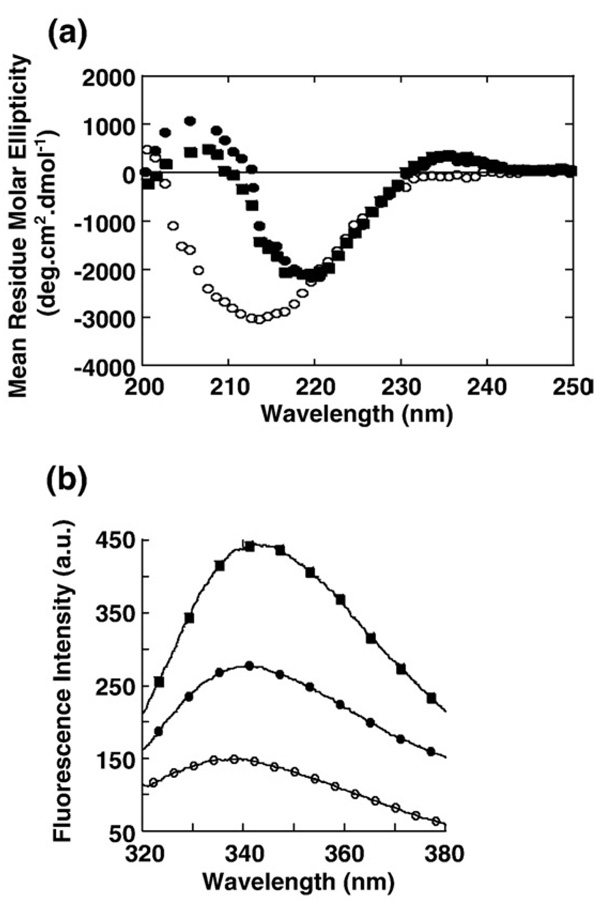
Far-UV circular dichroism was used to analyze the secondary structure of gC1q after both the acidic treatments (Fig. 2a). The native gC1q far-UV CD was typical of a fully β-protein consistent with the X-ray structure of the protein [18,24]. But it had the rare peculiarity of having a positive peak around 235 nm, suggesting the contributions of the side chains of aromatic residues, Trp or Tyr, to the far-UV CD [25]. This seems to occur, in particular in proteins with low α-helical contents [24,25]. No significant modification of the spectrum was observed when gC1q was incubated for 15 min at pH 5.0, indicating that the native secondary structure was maintained even after gC1q was dissociated. In contrast, when gC1q was incubated for 15 min at pH 1.7, it showed a large change in the secondary structure of the protein, consistent with a significant reduction in the β sheet. The change in the gC1q secondary structure was very rapid and the far-UV spectrum recorded immediately after addition of the protein to the buffer at pH 1.7 was identical to the one recorded 15 min later (not shown). Thus, there was no more change in the secondary structure of the gC1q protomers when changes in their tertiary structure still took place (Fig. 1b). But even after 15 min at pH 1.7, the gC1q protomers did not have a CD-spectrum typical of a fully denaturated protein and retained some residual secondary structure [26,27]. The presence of a disulfide bridge in each chain of gC1q (chain A: C150–C168, chain B: C154–C171, chain C: C151–C165) might be responsible for the residual secondary structure in the dissociated molecule. But when the disulfide bridges were reduced using β-mercaptoethanol and the protein treated at pH 1.7, it precipitated. It was therefore not possible to determine if it was the disulfide bridges that maintained some secondary structure in the gC1q protomers. However, it showed that the disulfide bridges were essential to maintain a residual structure in the gC1q protomers that prevented its precipitation in acid. The peak at 235 nm was still present after treatment at pH 5.0 but was absent after treatment at pH 1.7 confirming that the gC1q dissociated at pH 5.0 maintained some residual tertiary structure that was lost after treatment at pH 1.7, as already inferred from the lower fluorescence intensity of the gC1q sample after treatment at pH 1.7.
Fig. 2.
(a) Effect of pH on the secondary structure of gC1q. Far-UV CD spectra of gC1q were recorded after incubation for 15 min at room temperature, at pH 7.0 (■), 5.0 (●) and 1.7 (○). (b) Effect of pH on the tertiary structure of gC1q. Changes in the Trp-fluorescence were monitored at pH 7.0 (■), 5.0 (●) and 1.7 (○). a.u. stands for arbitrary unit.
The Trp-fluorescence scans measured after treatment for 15 min at pH 5.0 and at pH 1.7 showed a shift of the native maximum emission wavelength from 344 nm for the native gC1q to 341 and 339 nm, respectively (Fig. 2b). This again highlighted the two different tertiary structures adopted by gC1q after both pH treatments and showed that if the proteins were not completely unfolded as indicated by the Far-UV spectra, their tertiary structures was however significantly disturbed.
3.2. Reassembly of the gC1q trimer
To summarize, treatment at pH 1.7 led to the fast dissociation of the gC1 trimer into protomers and rapid loss of its native secondary structure followed by subsequent changes in the tertiary structure of the gC1q protomers. In contrast, the treatment at pH 5.0, led to the slow dissociation of the gC1q trimer, into probably a mixture of dimers and protomers, associated with some changes in the gC1q tertiary structure. The native secondary structure was maintained even at the end of the dissociation and no further changes in the tertiary structure of gC1q was detected after the dissociation. We were now interested to test if gC1q could reassemble after these pH treatments and subsequent neutralization and to determine if the different secondary and tertiary structures adopted by the gC1q at the end of the dissociation phase would somehow play a role in the gC1q capacity to reassemble.
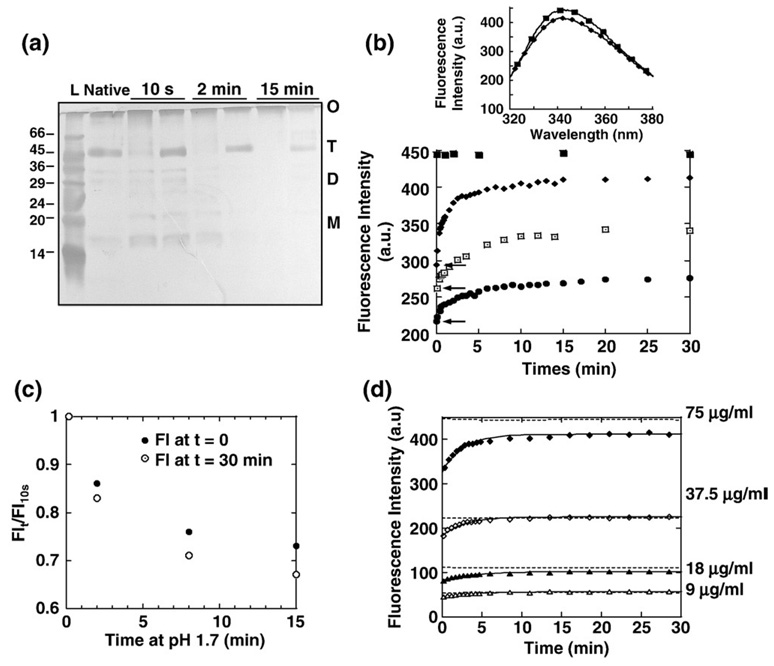
The gC1q native trimer was treated at pH 1.7 for 15 min and subsequently neutralized for 30 min and the oligomeric states of the gC1q samples were assayed by cross-linking and SDS-PAGE analysis (Fig. 3a). Little trimer was cross-linked even 30 min after neutralization indicating that most of the gC1q protomers could not reassemble. Since the tertiary structure of the gC1q protomers changes over the time the molecules spent at pH 1.7, we looked if that had an effect on the capacity of gC1q to reassemble, after neutralization. After 10 s at pH 1.7 and the return to neutral conditions, gC1q protomers could reassemble into trimer and the majority of gC1q trimerized 30 min after neutralization (Fig. 3a). After a 2 min incubation at pH 1.7 and subsequent neutralization, a significant amount of gC1q was still able to trimerize (Fig. 3a). So, it appeared that gC1q lost its capacity to reassemble when left at pH 1.7. The reassembly of gC1q into trimer was then monitored by Trp-fluorescence. After the short treatment of 10 s at pH 1.7 and neutralization, within 30 min, gC1q recovered both the maximum emission wavelength of the native gC1q trimer and an intensity of the fluorescence maxima close to that of the native gC1q (Fig. 3b). This showed that the time-dependent increase of the intensity of the fluorescence maxima was associated with the reassembly of gC1q into trimer. Consistently, the time-dependent increase of the intensity of the fluorescence maxima was inhibited by the increase of the time gC1q spent at pH 1.7, before neutralization (Fig. 3b) as observed in the cross-linking experiment (Fig. 3a).
Fig. 3.
Reassembly of the gC1q, after treatments at pH 1.7 and subsequent neutralization (a). Effect of the time spent at pH 1.7 on the capacity of gC1q to reassemble into trimer. The oligomeric state of gC1q was determined by cross-linking and SDS-PAGE analysis. The native gC1q was treated at pH 1.7 for the indicated times and subsequently neutralized for 0 and 30 min, left and right lanes, respectively. A sample of the native gC1q was also analyzed on the gel after cross-linking. T, gC1q trimer; D, gC1q heterodimers; M, gC1q protomers, O, cross-linked gC1q higher oligomers. L, molecular weight standards indicated in kDa. The gels were silver stained. (b). Effect of the time gC1q native trimer spent at pH 1.7, on the Trp-fluorescence intensity of gC1q after neutralization. The wavelengths parameters are as in Fig. 1b. The native gC1q samples were treated at pH 1.7 for 10 s (◆), 2 min (⊡) and 15 min (●) and subsequently added to a cuvette containing McIlVaine buffer at pH 7.0 and measurements were started immediately. The Trp-fluorescence of the native gC1q was measured in McIlVaine buffer at pH 7.0 (■). Inset. Trp-emission scans. The Trp-scans of the native (■) and the reassembled gC1q trimer (◆) after treatment at pH 1.7 for 10 s and subsequent neutralization for 30 min. a.u. stands for arbitrary unit. (c) Effect of the time gC1q spent at pH 1.7 on the fluorescence of gC1q just after and 30 min after neutralization. For comparison purpose, the fluorescence intensities just after neutralization (0 min, ●) and 30 min later (○) have been normalized as follows: the fluorescence intensity just after neutralization (FIt), was divided by the fluorescence intensity of gC1q after it had spent 10 s at pH 1.7 (FI10 s) and just after neutralization. The same normalization was performed for the fluorescence intensity 30 min after the neutralization. (d) Effect of the protein concentration on the fluorescence increase observed after neutralization. gC1q was acidified for 10 s at pH 1.7 and subsequently neutralized at different concentrations (9 µg/ml (△), 18 µg/ml (▲), 37.5 µg/ml (◇) and 75 µg/ml (◆)) in a cuvette containing McIlVaine buffer at pH 7.0. The wavelengths parameters are as in Fig. 1b. The fluorescence intensity of the native gC1q at different concentrations is indicated by dashed lines. The time-dependent increase of the fluorescence intensity best fits to first order processes (R2 = 0.97, 0.98, 0.99 and 0.98 for 75, 37.5, 18 and 9 µg/ml, respectively). The reassembly happened at a final protein concentration of 75 µg/ml (otherwise mentioned).
The neutralization led to rapid changes in the tertiary structure of the gC1q protomers as illustrated by the increase of the fluorescence intensity maxima from the acidic state (FI ~ 150 a.u.) to the neutral states (Fig. 3b, arrows). The fluorescence intensity maxima detected immediately after the neutralization (Fig. 3b, arrows) were higher than that observed at the end of the acidic phase, indicating that the initial structural changes within the gC1q protomers were too fast to be detected and the data were lost in the dead time of the instrument. Thus, the gC1q protomers had already undergone some conformational changes due to the neutralization when the measurements started. Interestingly, the data showed that those fast changes within the tertiary structure of the gC1q protomers were inhibited by the time gC1q spent at pH 1.7 (Fig. 3b, arrows). When the increase of the fluorescence intensity, just after neutralization (fast changes) and 30 min later (trimerization) were plotted against the time gC1q spent at pH 1.7, the two curves correlated (Fig. 3c). This suggested that the capacity of the gC1q protomers to reassemble was inherent to their capacity to return to an assembly-competent neutral conformation.
The best condition to study the reassembly of gC1q itself was after its treatment for 10 s at pH 1.7 and its subsequent neutralization for 30 min. Under these conditions, analysis of the concentration-dependence of the gC1q reassembly showed that over the range 9–75 µg/ml of protein, the protein always reached approximately the fluorescence level of the native gC1q trimer, indicating that the reassembly of gC1q into trimer occurred at all concentrations (Fig. 3d). The time-dependent increase of the intensity of the fluorescence maxima followed first order kinetics (Fig. 3d). The reassembly appeared only slightly concentration-dependent since the rate constant increased less than twice and the half time decreased only by about three times, over a 8-fold protein concentration increase (Table 1 and Table 2, respectively).
Table 1.
Rate constants for gC1q reassemblya
| gC1q Concentration (µg/ml) |
||||
|---|---|---|---|---|
| 75 | 37.5 | 18 | 9 | |
| k (min−1) | 0.39±0.02 | 0.38±0.01 | 0.26±0.007 | 0.24±0.008 |
Native gC1q was acidified at pH 1.7 for 10 s and subsequently neutralized at pH 7.0 for 30 min.
Table 2.
Half-times of the gC1q reassembly reactionsa
| Acidic treatments |
gC1q Concentration (µg/ml) |
|||
|---|---|---|---|---|
| 75 | 37.5 | 18 | 9 | |
| pH 5.0 | 3±0. 5′ | n.d. | n.d. | n.d. |
| pH 1.7 | 0.8±0. 2′ | 1.8±0. 5′ | 2.1±0. 5′ | 2.7±0. 3′ |
Native gC1q was acidified at pH 5.0 for 15 min or at pH 1.7 for 10 s and subsequently neutralized at pH 7.0 for 30 min. n.d. not determined.
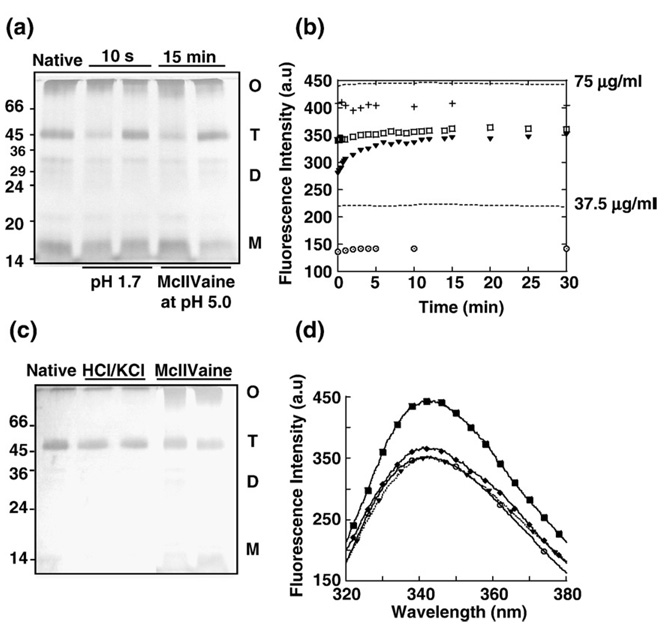
We then studied if after treatment at pH 5.0 and subsequent neutralization, the dissociated gC1q could also reassemble. The reassembly of gC1q was analyzed by cross-linking combined to SDS-PAGE and by Trp-fluorescence (Fig. 4). After 15 min at pH 5.0 and neutralization, gC1q reassembled to the same extent than after treatment at pH 1.7 for 10 s and neutralization, indicating that the capacity of gC1q to reassemble into trimer was independent of the structural state adopted at the end of the dissociation phase (Fig. 4a). It was therefore surprising to observe a lower intensity of the fluorescence maxima when gC1q had been acidified to pH 5.0 and neutralized for 30 min (Fig. 4b: FI ~ 350 a.u.), than when it had been acidified to pH 1.7 and neutralized for 30 min (Fig. 3b: FI ~ 420 a.u.). Since the reassembly yield was similar whatever the pH treatments had been, prior neutralization, it looked as if the reassembly of gC1q into trimer was not involved in quenching the Trp-fluorescence. This was confirmed by a treatment of the gC1q native trimer, for 30 s at pH 5.0, a time, not long enough to dissociate the gC1q trimer (Fig. 4c), but enough to lower the intensity of the fluorescence maxima to 350 a.u. (Fig. 4b and d). Now, because the gC1q trimer treated at the lower pH of 1.7 and neutralized, recovered the native fluorescence, it was important to check if it was the buffer system itself that led to the lower fluorescence intensity observed. To test this possibility, a native gC1q sample was treated at pH 5.0 for 30 s, using PBS containing 0.1 M HCl/KCl at pH 1.0 to lower the pH to 5.0, and gC1q was subsequently neutralized for 30 min. Both the oligomeric state of the gC1q trimer and the Trp-fluorescence scan of the sample were compared to those obtained for the gC1q sample which had been treated at pH 5.0 using the McIlVaine buffer instead (Fig. 4c and d, respectively). For both the buffer systems, gC1q remained trimeric, yet the intensity of the fluorescence maxima was lower than the native gC1q trimer, excluding the role of the buffer system in quenching the fluorescence. Thus, the lower fluorescence intensity was either the result of some artifacts or indicated that the treatment at pH 5.0 led to a change, irreversible even after neutralization, in the gC1q trimeric conformation.
Fig. 4.
Reassembly of the gC1q after treatments at pH 5.0 and subsequent neutralization. (a). Oligomeric state of the gC1q samples after acidic treatment and subsequent neutralization. Native gC1q samples were incubated at the pH, and for the times indicated on the figure, and subsequently neutralized for 0 and 30 min, left and right lanes, respectively. A sample of the native gC1q was also analyzed on the gel after cross-linking. All the samples were cross-linked prior addition of sample buffer and analysis by SDS-PAGE. T, gC1q trimer; D, gC1q heterodimers; M, gC1q protomers, O, cross-linked gC1q higher oligomers. The molecular weight standards were indicated in kDa. The gels were silver stained. (b) Measurements of Trp-fluorescence after the acid pre-treatment at pH 5.0 and subsequent neutralization. The wavelengths parameters are as in Fig. 1b. gC1q was acidified at pH 5.0 for a few seconds (+), for 30 s (□) and for 15 min (▼), and subsequently neutralized at pH 7.0. gC1q was acidified at pH 5.0 for 15 min and subsequently neutralized at pH 7.0 at final protein concentration of 37.5 µg/ml ( ). The fluorescence intensity of the native gC1q at different concentrations is indicated by dashed lines. (c). Effect of the buffer system on the oligomeric state of the gC1q samples. gC1q native samples were treated for 30 s in McIlVaine buffer at pH 5.0 or in PBS containing 10 mM HCl/KCl buffer at a final pH 5.0 and were subsequently neutralized for 0 and 30 min, left and right lanes, respectively. A sample of native gC1q was also analyzed on the gel after cross-linking. All the samples were cross-linked prior addition of sample buffer and analysis by SDS-PAGE. T, gC1q trimer; D, gC1q heterodimers; M, gC1q protomers, O, cross-linked gC1q higher oligomers. The molecular weight standards were indicated in kDa. The gels were silver stained. (d) Trp-emission scans. The Trp-scans of the native gC1q (■), of the gC1q trimer after treatment for 30 s in McIlVaine buffer at pH 5.0 (◆) or in PBS-10 mM HCl/KCl buffer at pH 5.0 (○) and subsequent neutralization for 30 min and of the reassembled gC1q trimer (▼) after treatment at pH 5.0 for 15 min and subsequent neutralization for 30 min. a.u. stands for arbitrary unit. The neutralization step is at a gC1q concentration of 75 µg/ml, otherwise stated.
). The fluorescence intensity of the native gC1q at different concentrations is indicated by dashed lines. (c). Effect of the buffer system on the oligomeric state of the gC1q samples. gC1q native samples were treated for 30 s in McIlVaine buffer at pH 5.0 or in PBS containing 10 mM HCl/KCl buffer at a final pH 5.0 and were subsequently neutralized for 0 and 30 min, left and right lanes, respectively. A sample of native gC1q was also analyzed on the gel after cross-linking. All the samples were cross-linked prior addition of sample buffer and analysis by SDS-PAGE. T, gC1q trimer; D, gC1q heterodimers; M, gC1q protomers, O, cross-linked gC1q higher oligomers. The molecular weight standards were indicated in kDa. The gels were silver stained. (d) Trp-emission scans. The Trp-scans of the native gC1q (■), of the gC1q trimer after treatment for 30 s in McIlVaine buffer at pH 5.0 (◆) or in PBS-10 mM HCl/KCl buffer at pH 5.0 (○) and subsequent neutralization for 30 min and of the reassembled gC1q trimer (▼) after treatment at pH 5.0 for 15 min and subsequent neutralization for 30 min. a.u. stands for arbitrary unit. The neutralization step is at a gC1q concentration of 75 µg/ml, otherwise stated.
The involvement of Trp-quenching processes, which occur in nano- to milliseconds, was ruled out because a short-time exposure of the native gC1q to pH 5.0, followed by neutralization only lowered the signal to ~410 a.u. (Fig. 4b) [28–31]. Next, the possibility of some gC1q aggregation happening as a consequence of the pH 5.0 treatment was investigated. The functional state of the gC1q trimer after reassembly was studied, as it was unlikely that a protein aggregate would still be functional. The capacity of gC1q to recognize aggregated IgG molecules was tested by ELISA (Enzyme-Linked Immunosorbent Assay) following a protocol adapted from Kishore et al. (Fig. 5) [32]. Both the reassembled trimers had similar capacity to recognize aggregated IgG molecules than the native gC1q trimers. But when gC1q was aggregated by a heat-treatment (120 °C for 10 min), as expected, the protein lost its capacity to recognize aggregated IgG (Fig. 5). This argued against the presence of protein aggregates in the sample of the reassembled gC1q trimer, after treatment at pH 5.0 and neutralization. To confirm the functional state of the reassembled gC1q trimers, interaction with the gC1q receptor (gC1qR) was assayed using surface plasmon resonance spectroscopy and the dissociation constants were determined (Table 3). Similar dissociation constants were obtained for native gC1q and gC1q reassembled after treatment at pH 5.0 and subsequent neutralization for 60 min, confirming that the reassembled trimer was functional. Surprisingly, a lower dissociation constant was obtained for gC1q reassembled after treatment at pH 1.7 and subsequent neutralization for 60 min, indicating a higher affinity for the receptor than the native gC1q. The results again supported the idea that the reassembled gC1q trimer failed to recover its native tertiary structure after treatment at pH 5.0 and neutralization, rather than aggregated. Near-UV CD could not be used to show that the native and the reassembled trimer had different tertiary structures because for detection, a protein concentration about 6 times higher was required and a two-fold increase in the gC1q concentration at which reassembly happened led to the precipitation of the protein [24,33]. The protein precipitated whether the gC1q trimer had been treated at pH 1.7 or at pH 5.0 and subsequently neutralized. The effect of protein concentration on the reassembly reaction was then tested to minimize potential aggregation of the gC1q at the higher concentration. The reassembly reaction was studied at a protein concentration (37.5 µg/ml) 2-fold lower than previously and the time-dependent increase of the intensity of the fluorescence maxima was measured (Fig. 4b). The rational was that at the lower protein concentration, protein aggregation would be less and therefore, we should expect an increase of the fluorescence signal close to that of the native gC1q trimer at the same concentration (FI ~ 225 a.u.). Result observed at that concentration, when gC1q had been treated at pH 1.7 and neutralized (Fig. 3c). But on the contrary, the time-dependent increase in the intensity of the fluorescence maxima was entirely abolished by the decrease of the gC1q concentration. This rather indicated that protein assembly was totally abolished by decreasing the protein concentration and argued that protein aggregation was unlikely to happen at the higher protein concentration. The effect of the protein concentration also indicated that the reassembly of gC1q was strongly concentration-dependent after treatment at pH 5.0 and neutralization, in contrast to what was observed for the reassembly of gC1q after treatment at pH 1.7 and neutralization. This showed that the reassembly of gC1q into trimer followed two different pathways depending on the pH used prior neutralization, and that was consistent with the gC1q trimer reassembling into two different conformations. Interestingly, the trimer reassembled after treatment at pH 5.0 also recovered the native maximum emission wavelength after neutralization for 30 min (Fig. 4d), indicating that it shared some structural characteristic with the native trimeric state.
Fig. 5.
Recognition of heat-aggregated IgG. Samples pre-incubated at pH 1.7 for 10 s and at pH 5.0 for 15 min, subsequently neutralized for 60 min were coated on a 96-well plate and allowed to interact with aggregated IgG. Aggregated IgG bound to gC1q samples was probed using goat anti-human IgG conjugated to horse radish peroxidase. The amount of gC1q samples coated on the plate was measured on a duplicate plate using an anti C1q antibody. The absorbance for each sample has been normalized accordingly. Native gC1q trimer and heat-aggregated gC1q were also coated on the plate under the same conditions. The error bar corresponded to triplicate experiments.
Table 3.
Analysis of the interaction between gC1q and its receptor gC1qR by Surface Plasmon Resonancea
| Samples | kon (M−1s−1) | koff (s−1) | KD (nM) |
|---|---|---|---|
| Native | 3.5±0.3 × 103 | 1.3±0.2 × 10−3 | 370±70 |
| Reassembled (pH 5.0/7.0) | 3.0±0.4 × 103 | 1.2±0.01 × 10−3 | 400±50 |
| Reassembled (pH 1.7/7.0) | 7.3 ±0.3 × 103 | 4.5±0.2 × 10−4 | 62±5 |
Native gC1q was treated as described in Table 1 but left for 60 min at pH 7.0. The capacity of the native gC1q and of the reassembled gC1q trimer to bind to gC1q R was tested by measuring both the dissociation (koff) and association (kon) rate constants. The apparent equilibrium dissociation constants (KD) were calculated from the koff/kon ratio.
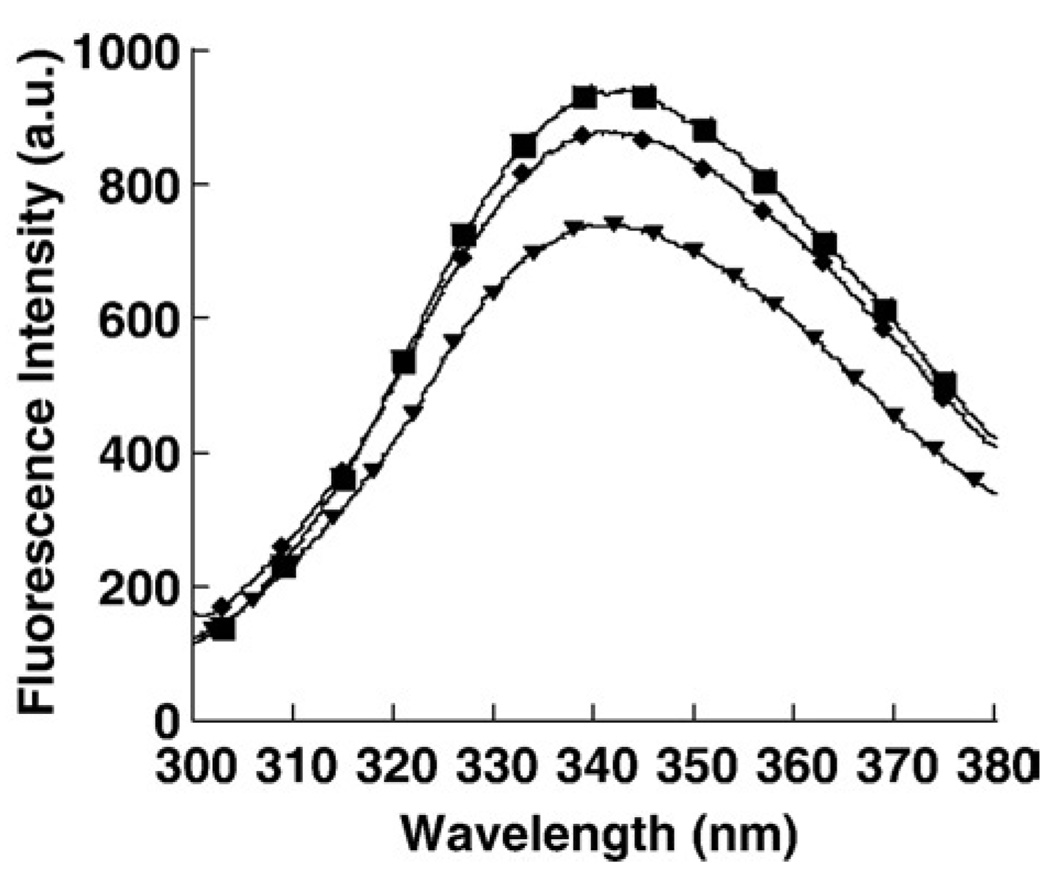
The difference of conformation between the two reassembled trimers was again confirmed using Tyr-spectroscopy. Tyr-scans were performed for the native gC1q trimer and for both reassembled trimers, and similarly to what had been observed on the Trp-scans (Fig. 3b and Fig. 4d), the gC1q trimer recovered native tertiary structure only after treatment at pH 1.7 and 30 min neutralization (Fig. 6). Because gC1q contains 20 Tyr residues located throughout the entire molecule, it showed that the difference in the conformations of the two reassembled trimers was not limited to the vicinity of the Trps but was spread within the overall protein.
Fig. 6.
Tyr-emission scansofreassembled gC1q. Tyr-emission scans were recorded 30 min after neutralization for samples pre-incubated at pH 1.7 for 10 s (◆) or at pH 5.0 for 15 min (▼) and for the native gC1q trimer (■).a.u. stands for arbitrary unit.
4. Discussion
The primary aim of this study is to investigate whether the hetero-trimeric gC1q globular domain located at the C-terminal end of the C1q collagenous triple helices is able to reassemble in vitro in the absence of the N-terminal collagenous domain. The gC1q trimer obtained by collagenase treatment of intact C1q was used as a starting material. To address this question, gC1q was first submitted to different pH treatments to induce its disassembly and subsequently neutralized to provide renaturating conditions. A model describing the disassembly and the reassembly of gC1q is depicted in Fig. 7. The treatment at pH 6.0 induces neither dissociation nor unfolding of the gC1q trimer. At pH 5.0, the gC1q native trimer rapidly lost its native tertiary structure and slowly disassembled, into probably both dimers and protomers (Fig. 7a). Additional changes in the gC1q tertiary structure occurred during the dissociation without changes in the secondary structure which remained native throughout the treatment. After the dissociation had taken place, there was no more structural change that could be detected. Below pH 5.0, the gC1q dissociated more readily into protomers which underwent further structural changes. But it was only after treatment at pH 1.7, that the gC1q protomers adopted the most unfolded structure which, however, still kept a residual secondary structure (Fig. 7b). Interestingly, the Trp-scans showed that the maximum emission wavelength of the Trp blue-shifted upon acidification (pH 5.0 and pH 1.7), whereas one might have expected a red shift due to exposure of the Trp to the water in the acidified gC1q states. According to the gC1q X-ray structure, none of the three Trps are buried in the core of the protein, which might explain the partially-exposed maximum emission wavelength of the native gC1q [18]. Trp 194 and Trp 190 have their benzene ring partially exposed to water whereas that of the Trp-147 is almost fully exposed. Yet, upon the acidic treatments, the Trps could become entirely exposed to water and induce a red shift. Recent work has shown that Trp exposure to water molecules had less influence on the maximum emission wavelength than the residues lying close to the Trp [34,35]. The studies show that the presence of positive charges (Lys, Arg, N-terminal) create a red shift when on the benzene ring end and a blue shift when on the pyrrole ring end, and the reverse for the negative charges (Glu, Asp and C-terminal) [35]. Thus, if the Trps had a particular environment in the native gC1q, the blue shift could arise from its loss upon the acidic treatments. In the native gC1q, there are two opposite charges, Glu 158 and Lys 129 in close proximity to the benzene ring of Trp 190, making it difficult to anticipate what would be the resulting shift of the maximum emission wavelength. The only charged residue in close proximity to the pyrrole ring of Trp 147, is a histidine, His 203, and is therefore not expected to contribute to the shift of the maximum emission wavelength in neutral condition (pKa ~ 6.0). Glu 196 is located near the pyrrole ring of the Trp 194 and two arginine residues, Arg 163 and Arg 128, are located near its benzene ring. In this case, the environment around Trp 194 would be expected to contribute for a red shift of the maximum emission wavelength. Thus, the loss of the environment around Trp 194 might explain the blue shift of the maximum emission wavelength upon the acidic treatments.
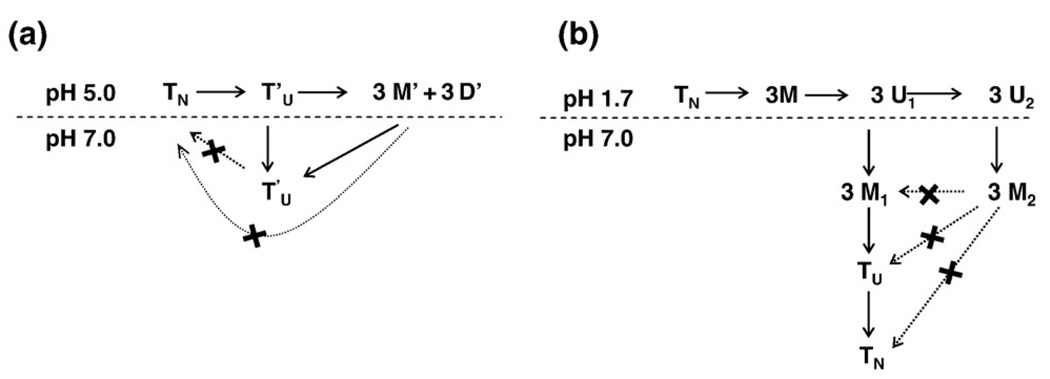
Fig. 7.
Schemes of the disassembly and reassembly of gC1q. (a). After a pre-treatment at pH 5.0 and subsequent neutralization. At pH 5.0, the native gC1q trimer (TN) rapidly loss its native tertiary structure to become (T′U), which subsequently slowly dissociates into probably, both heterodimers and protomers (D′ and M′, respectively). Upon neutralization, D′and M′ re-associates into a non native trimer (T′U). The return to a native trimeric conformation is abolished by the treatment at pH 5.0. (b). After pre-treatment at pH 1.7 and subsequent neutralization. Upon denaturation at pH 1.7, the native gC1q trimer (TN) dissociates rapidly into protomers (M) which further change their tertiary structure to become U1 and U2, depending on the time gC1q spend at pH 1.7. U1 and U2 gC1q protomer conformations can not be distinguished during the acidic phase. Just after subsequent neutralization, U1 and U2 lead to assembly-competent (M1) and assembly-defective (M2) gC1q protomers, respectively. During the neutralization, M1, protomers reassemble into an assembly intermediate, most probably a non native trimer (TU) that subsequently rearranged into a native gC1q trimer (TN). For the sake of clarity and because of the lack of evidences of individual behavior from the three different chains A, B and C, the heterogeneity of the protomer is not indicated. The dimer is indicated, although no experimental conditions have permitted its isolation. Solid arrows indicate permitted transition, dashed arrows non-permitted.
The different structural states reached by gC1q after the pH 5.0 and the pH 1.7 treatments, are probably the result of different protonation states with the likely involvement of glutamic and/or aspartic acid residues (pKa ~ 4.4) as well as of the C-termini (pKa ~ 3.1), respectively. These residues would have to be accessible within the trimer to be responsible for the loss of the native tertiary structure of the gC1q trimer and they would have to be located also nearby interfaces to be responsible for its dissociation. Using the coordinates from the X-ray structure of the gC1q trimer, we looked for residues or domains within the gC1q molecules that could fulfill those requirements [18]. Because there are several accessible aspartic and glutamic acids within the trimer, it was not possible to isolate particular residues or domain of the protein involved in the loss of the native tertiary structure of the gC1q trimer. In contrast, aspartic and glutamic residues at or nearby interfaces and accessible within the trimer are all located in the C-terminal domains of the three protomers (E209A, D211A, E209B, D223B, D206C, D217C). Additionally, there are two residues, D172B and D169A, respectively located at the AB and AC dimeric interfaces which are inaccessible within the trimer. Maybe those are not protonated at pH 5.0 but are at pH 1.7, leading at the end of the acidic phase, to a mixture of dimers and protomers and to fully dissociated protomers, respectively.
After a short time at pH 1.7 and subsequent neutralization, gC1q was able to reassemble into a native trimer as shown by its functional state and the recovery of the fluorescence intensity level of the native trimer (Fig. 7b). The first order kinetics and the weak concentration-dependence for the reassembly are both consistent with the rate-limiting step for reassembly from the acid-denatured state being intramolecular. This may arise from a slow folding state within an assembly intermediate, as occurs in the α-chain of tryptophan synthetase, rather than within a monomeric intermediate, since the gC1q protomers refold rapidly probably within the dead time of the instrument [36]. Cross-linking of the gC1q at early times after the neutralization failed to isolate any heterodimer assembly intermediates suggesting that the rate-limiting step represents an intramolecular rearrangement within a gC1q trimer (not shown). Over the time gC1q spent at pH 1.7, the gC1q protomers lost their capacity to reassemble because they failed to return to an assembly-competent conformation, once neutralized. Thus, at pH 1.7, the gC1q protomers underwent a second-acid mediated step during which the residual structure, which provided the protomers their capacity to reassemble, once neutralized, was lost. As a consequence, the gC1q protomers probably misfolded and became assembly-defective (Fig. 7b). The residual structure required for gC1q assembly did not contain only secondary structural elements since even when the secondary structure remained unchanged (few seconds to 15 min at pH 1.7) the capacity of the gC1q to reassemble decreased. It is therefore a residual tertiary struture that holds the gC1q protomers capacity to reassemble. The loss of this residual structure occurred after treatment at pH 1.7, and might therefore be consecutive to the protonation of glutamic and/or aspartic acid residues, located at the C-terminal of the gC1q protomers and/or to the protonation of the C-termini of the gC1q protomers themselves. This would imply that the C-terminal domain of gC1q provides the molecule its capacity to trimerize.
After a short treatment at pH 5.0, the gC1q trimer changes conformation and lose its capacity to return to the native conformation after subsequent neutralization, as illustrated by a lower fluorescence intensity than the native sample (Fig. 7a). If additionally, the gC1q is dissociated at pH 5.0 and subsequently neutralized, it also loses its capacity to return to the native conformation although it remained able to reassemble as after treatment at pH 1.7 and neutralization (Fig. 7a). Aggregation of the gC1q trimer, consecutive to the treatment at pH 5.0 and neutralization, could explain the lower fluorescence intensity. But it is rather unlikely that a gC1q trimer dissociated and subsequently reassembled, would aggregate similarly to a gC1q trimer which had never been dissociated, so that it gives rise to the same fluorescence intensity level and the same cross-linking pattern. Moreover, the conditions used, limits the possibility of protein aggregation, since the experiments are performed at low protein concentration and far from the pI of chains A (8.87) and B (9.07). The pI of chain C is close to 7 (7.07), but again, it is unlikely that the chain C would aggregate similarly, and hence lead to the same fluorescence level and the same cross-linking pattern, whether it is within the gC1q trimer or dissociated, prior neutralization. So it is more likely that the treatment at pH 5.0 induces a permanent change in the tertiary structure of the gC1q protomers within the trimer and as a consequence prevents the recovery of the native tertiary fold, even after neutralization. After treatment at pH 5.0 and neutralization, the reassembly reaction becomes rate limiting as indicated by the strong concentration-dependence. Once more, the results suggest that the protonations of glutamic and/or aspartic acid residues, located at the C-terminal end of the gC1q protomers are probably involved in controlling the refolding and the reassembly reactions. This would also be consistent with the gC1q trimer being a functional molecule, even in the absence of a native tertiary fold, at least in the recognition of the IgG molecule, since it is residues located on the B protomer, but outside its C-terminal domain that are involved in the gC1q recognition site for IgG [18,32].
In summary, the results show that after the pH 1.7 and 5.0 treatments, gC1q adopts two different structural states and consequently, after neutralization, follows two different refolding and reassembly pathways. Both the pathways lead to the formation of a native trimer and a non native trimer, respectively. In other words, the structural state of gC1q in acid, from which refolding and reassembly start, affects the final structural state reached by gC1q, after neutralization. This is in good agreement with the funnel-shaped energy landscape theory which indicates that multiple folding routes exist and that depending on the pathways and the kinetic of the reactions, different thermodynamically stable functional isomers can be formed [37–39]. The concept of multiple energy minima conformational isomers seems to be particularly appropriate to describe protein with multiple ligand capacities, as are the gC1q and the C1q molecules [2,6,20,32,38]. The structural state of the gC1q protein after the treatment at pH 5.0, keeps some native-like structure, the secondary structure, in particular, suggesting that it has a structure closer to the native state than after the treatment at pH 1.7. Thus, the protein might be in an energy minimum close to that of the native protein and as a consequence lack the driving force to return to the native structural state, after neutralization. Now, the protonation of additional residues at pH 1.7, obviously leads to further unfolding of the gC1q molecule, and hence brings the protein to a different refolding pathway, after neutralization. This pathway leads to the successful reassembly into a native gC1q trimer.
Altogether, the result shows that the gC1q sequence contains the information for the proper assembly of gC1q into trimer so that the fold, the stoichiometry and the chain-selectivity are respected. During the reassembly of gC1q, no other oligomeric species than the gC1q trimers or homotrimers, expected to run at different position than the native gC1q heterotrimer, have been observed on SDS-PAGE (Fig. 3 and Fig. 4).
5. Conclusion
Hence, for gC1q, as for other members of the collagen-containing family such as collagen X, it is the non-collagenous domains which carry the assembly registration [14,15,40]. However, in vitro, the presence of a residual tertiary structure, probably located within the C-terminal domain of gC1q, appears crucial for the protein ability to reassemble and to refold into a native form. This might indicate that the role of the collagenous domain in the assembly of C1q, is to initialize the folding of the gC1q protomers to generate the tertiary structure required for the assembly of the entire molecule.
Acknowledgements
We thank Gisou van der Goot and Anna Mitraki for critical reading of the manuscript. We also thank Martial Rey for numerous challenging discussions and critical reading of the manuscript. Thanks to Vincent Forge for the use of his Circular Dichroism facilities.
References
- 1.Gaboriaud C, Thielens NM, Gregory LA, Rossi V, Fontecilla-Camps JC, Arlaud GJ. Structure and activation of the C1 complex of complement: unraveling the puzzle. Trends Immunol. 2004;25:368–373. doi: 10.1016/j.it.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Blanquet-Grossard F, Thielens NM, Vendrely C, Jamin M, Arlaud GJ. Complement protein C1q recognizes a conformationally modified form of the prion protein. Biochemistry. 2005;44:4349–4356. doi: 10.1021/bi047370a. [DOI] [PubMed] [Google Scholar]
- 3.Bredt W, Wellek B, Brunner H, Loos M. Interactions between Mycoplasma pneumoniae and the first components of complement. Infect Immun. 1977;15:7–12. doi: 10.1128/iai.15.1.7-12.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santoro F, Ouaissi MA, Pestel J, Capron A. Interaction between Schistosoma mansoni and the complement system: binding of C1q to schistosomula. J. Immunol. 1980;124:2886–2891. [PubMed] [Google Scholar]
- 5.Tacnet-Delorme P, Boyer V, Thielens NM, Hernandez JF, Bally I, Sim RB, Desgranges C, Arlaud GJ. In vitro analysis of complement-dependent HIV-1 cell infection using a model system. J. Immunol. 1999;162:4088–4093. [PubMed] [Google Scholar]
- 6.Tacnet-Delorme P, Chevallier S, Arlaud GJ. Beta-amyloid fibrils activate the C1 complex of complement under physiological conditions: evidence for a binding site for A beta on the C1q globular regions. J. Immunol. 2001;167:6374–6381. doi: 10.4049/jimmunol.167.11.6374. [DOI] [PubMed] [Google Scholar]
- 7.Thielens NM, Tacnet-Delorme P, Arlaud GJ. Interaction of C1q and mannan-binding lectin with viruses. Immunobiology. 2002;205:563–574. doi: 10.1078/0171-2985-00155. [DOI] [PubMed] [Google Scholar]
- 8.Arlaud GJ, Colomb MG, Gagnon J. A functional model of the human C1 complex. Immunol. Today. 1987;8:106–111. doi: 10.1016/0167-5699(87)90860-7. [DOI] [PubMed] [Google Scholar]
- 9.Reid KB, Porter RR. Isolation, by partial pepsin digestion, of the three collagen-like regions present in subcomponent C1q of the first component of human complement. Biochem. J. 1976;155:19–23. doi: 10.1042/bj1550005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arlaud GJ, Gaboriaud C, Thielens NM, Rossi V. Structural biology of C1. Biochem. Soc. Trans. 2002;30:1001–1006. doi: 10.1042/bst0301001. [DOI] [PubMed] [Google Scholar]
- 11.Kishore U, Reid KB. C1q: structure, function, and receptors. Immunopharmacology. 2000;49:159–170. doi: 10.1016/s0162-3109(00)80301-x. [DOI] [PubMed] [Google Scholar]
- 12.Kishore U, Gaboriaud C, Waters P, Shrive AK, Greenhough TJ, Reid KB, Sim RB, Arlaud GJ. C1q and tumor necrosis factor superfamily: modularity and versatility. Trends Immunol. 2004;25:551–561. doi: 10.1016/j.it.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Brass A, Kadler KE, Thomas JT, Grant ME, Boot-Handfor RP. The fibrillar collagens, collagen VIII, collagen X and the C1q complement proteins share a similar domain in their C-terminal non collagenous regions. FEBS Lett. 1992;303:126–128. doi: 10.1016/0014-5793(92)80503-9. [DOI] [PubMed] [Google Scholar]
- 14.Mechling DE, Gambee JE, Morris NP, Sakai LY, Keene DR, Mayne R, Bachinger HP. Type IX collagen NC1 domain peptides can trimerize in vitro without forming a triple helix. J. Biol. Chem. 1996;271:13781–13785. doi: 10.1074/jbc.271.23.13781. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Chen Q. The noncollagenous domain 1 of type X collagen. A novel motif for trimer and higher order multimer formation without a triple helix. J. Biol. Chem. 1999;274:22409–22413. doi: 10.1074/jbc.274.32.22409. [DOI] [PubMed] [Google Scholar]
- 16.Kohn WD, Mant CT, Hodges RS. Alpha-helical protein assembly motifs. J. Biol. Chem. 1997;272:2583–2586. doi: 10.1074/jbc.272.5.2583. [DOI] [PubMed] [Google Scholar]
- 17.Engel J, Kammerer RA. What are oligomerization domains good for? Matrix Biol. 2000;19:283–288. doi: 10.1016/s0945-053x(00)00075-5. [DOI] [PubMed] [Google Scholar]
- 18.Gaboriaud C, Juanhuix J, Gruez A, Lacroix M, Darnault C, Pignol D, Verger D, Fontecilla-Camps JC, Arlaud GJ. The crystal structure of the globular head of complement protein C1q provides a basis for its versatile recognition properties. J. Biol. Chem. 2003;278:46974–46982. doi: 10.1074/jbc.M307764200. [DOI] [PubMed] [Google Scholar]
- 19.Ghebrehiwet B, Lim BL, Peerschke EI, Willis AC, Reid KB. Isolation, cDNA cloning, and overexpression of a 33-kD cell surface glycoprotein that binds to the globular “heads” of C1q. J. Exp. Med. 1994;179:1809–1821. doi: 10.1084/jem.179.6.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim BL, Reid KB, Ghebrehiwet B, Peerschke EI, Leigh LA, Preissner KT. The binding protein for globular heads of complement C1q, gC1qR. Functional expression and characterization as a novel vitronectin binding factor. J. Biol. Chem. 1996;271:26739–26744. doi: 10.1074/jbc.271.43.26739. [DOI] [PubMed] [Google Scholar]
- 21.Rossi V, Gaboriaud C, Lacroix M, Ulrich J, Fontecilla-Camps JC, Gagnon J, Arlaud GJ. Structure of the catalytic region of human complement protease C1s: study by chemical cross-linking and three-dimensional homology modeling. Biochemistry. 1995;34:7311–7321. doi: 10.1021/bi00022a004. [DOI] [PubMed] [Google Scholar]
- 22.Laemli U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Eftink MR. Fluorescence techniques for studying protein structure. Methods Biochem. Anal. 1991;35:127–205. doi: 10.1002/9780470110560.ch3. [DOI] [PubMed] [Google Scholar]
- 24.Kelly SM, Price NC. The application of circular dichroismto studies of protein folding and unfolding. Biochim. Biophys. Acta. 1997;1338:161–185. doi: 10.1016/s0167-4838(96)00190-2. [DOI] [PubMed] [Google Scholar]
- 25.Woody RW. Contributions of tryptophan side chains to the far-ultraviolet circular dichroism of proteins. Eur. Biophys. J. 1994;23:253–262. doi: 10.1007/BF00213575. [DOI] [PubMed] [Google Scholar]
- 26.Johnson CW. Protein secondary structure and circular dichroism: a practical guide. Proteins: structure. Functions and Genetics. 1990;7:205–214. doi: 10.1002/prot.340070302. [DOI] [PubMed] [Google Scholar]
- 27.Johnson WC. Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins. 1999;3:307–312. [PubMed] [Google Scholar]
- 28.Chen J, Flaugh SL, Callis PR, King J. Mechanism of the highly efficient quenching of tryptophan fluorescence in human gammaD-crystallin. Biochemistry. 2006;45:11552–11563. doi: 10.1021/bi060988v. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Liu B, Yu HT, Barkley MD. The peptide bond quenches indole fluorescence. J. Am. Chem. Soc. 1996;118:9271–9278. [Google Scholar]
- 30.Chen Y, Barkley MD. Toward understanding tryptophan fluorescence in proteins. Biochemistry. 1998;37:9976–9982. doi: 10.1021/bi980274n. [DOI] [PubMed] [Google Scholar]
- 31.Lakowicz JR. Principles of Fluorescence Spectroscopy. Second Edition. Kluwer Academic/Plenum Publishers; 1999. [Google Scholar]
- 32.Kishore U, Gupta SK, Perdikoulis MV, Kojouharova MS, Urban BC, Reid KB. Modular organization of the carboxyl-terminal, globular head region of human C1q A, B, and C chains. J. Immunol. 2003;171:812–820. doi: 10.4049/jimmunol.171.2.812. [DOI] [PubMed] [Google Scholar]
- 33.Kelly SM, Jess TJ, Price NC. How to study proteins by circular dichroism. Biochim. Biophys. Acta. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 34.Callis PRaB, B K. Tryptophan fluorescence shifts in proteins from hybrid simulations: an electrostatic approach. J. Phys. Chem. B. 1997;101:9429–9432. [Google Scholar]
- 35.Vivian JT, Callis PR. Mechanisms of tryptophan fluorescence shifts in proteins. Biophys. J. 2001;80:2093–2109. doi: 10.1016/S0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blond S, Goldberg ME. Kinetics and importance of the dimerization step in the folding pathway of the beta 2 subunit of Escherichia coli tryptophan synthase. J. Mol. Biol. 1985;182:597–606. doi: 10.1016/0022-2836(85)90245-1. [DOI] [PubMed] [Google Scholar]
- 37.Dill KA. Polymer principles and protein folding. Protein Sci. 1999;8:1166–1180. doi: 10.1110/ps.8.6.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai CJ, Kumar S, Ma B, Nussinov R. Folding funnels, binding funnels, and protein function. Protein Sci. 1999;8:1181–1190. doi: 10.1110/ps.8.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai CJ, Ma B, Nussinov R. Folding and binding cascades: shifts in energy landscapes. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9970–9972. doi: 10.1073/pnas.96.18.9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khoshnoodi J, Cartailler JP, Alvares K, Veis A, Hudson BG. Molecular recognition in the assembly of collagens: terminal noncollagenous domains are key recognition modules in the formation of triple helical protomers. J. Biol. Chem. 2006;281:38117–38121. doi: 10.1074/jbc.R600025200. [DOI] [PubMed] [Google Scholar]