Summary
The pikromycin polyketide synthase (PKS) is unique in its ability to generate both 12-and 14-membered ring macrolactones. As such, dissection of the molecular basis for controlling metabolic diversity in this system remains an important objective for understanding modular PKS function and expanding chemical diversity. Here, we describe a series of experiments designed to probe the importance of the protein-protein interaction that occurs between the final two monomodules, PikAIII (module 5) and PikAIV (module 6), for the production of the 12-membered ring macrolactone 10-deoxymethynolide. The results obtained from these in vitro studies demonstrate that PikAIII and PikAIV generate the 12-membered ring macrocycle most efficiently when engaged in their native protein-protein interaction. Accordingly, the data are consistent with PikAIV adopting an alternative conformation that enables the terminal thioesterase domain to directly off-load the PikAIII-bound hexaketide intermediate for macrocyclization.
Introduction
Polyketides are a diverse class of natural products that are known to possess a wealth of medicinally important biological activities. The biosynthesis of macrolide polyketide compounds occurs via a stepwise mechanism that is catalyzed by type I modular polyketide synthases (PKSs). These modular PKSs are comprised of a number of large, multifunctional enzymes that are responsible for catalyzing the initiation, elongation, and reductive steps that ultimately give rise to the characteristic macrolactone scaffold. For type I PKSs, the extension of a polyketide chain by two carbon atoms requires a minimum of three protein domains, the ketosynthase (KS), acyl transferase (AT), and acyl carrier protein (ACP) that collectively comprise a PKS module. Within the elongation module, the AT domain is responsible for recognizing and loading the appropriate acyl-CoA extender unit onto the ACP domain. Subsequently, the KS domain catalyzes a decarboxylative condensation reaction between the ACP-bound extender unit and the growing polyketide chain, resulting in an ACP-bound elongated polyketide intermediate. In addition to these three core domains, each elongation module may contain up to three additional domains that are responsible for the reductive processing of the β-keto functionality prior to the subsequent extension step. The presence of a ketoreductase (KR) domain gives rise to a β-hydroxyl group, the presence of both a KR and a dehydratase (DH) domain generates an alkene, while the combination of KR, DH, and enoyl reductase (ER) results in complete reduction to the alkane. Finally, termination of polyketide biosynthesis is typically catalyzed by a thioesterase (TE) domain located at the carboxy terminus of the final elongation module.
The pioneering sequence analysis of the biosynthetic gene cluster for the 6-deoxyerythronolide B (DEBS) PKS revealed that both the length and degree of β-keto reduction of the polyketide chain are determined by the sequential arrangement of extension and reductive domains within the PKS system [1, 2]. This co-linear relationship has been exploited to rationally engineer polyketide products via molecular genetic manipulation of PKS systems [3, 4]. However, analysis of several recently identified modular PKS systems has challenged the “one extension per module” paradigm. For example, iterative polyketide extension by a single PKS module has been implicated in the biosynthesis of several natural products, suggesting that alternative activities of modules can lead to structures that cannot be predicted from sequence analysis alone [5–9].
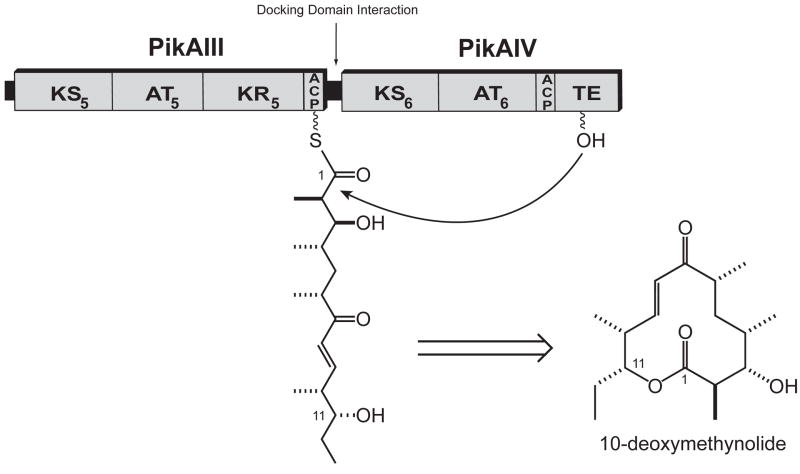
Pikromycin is a naturally occurring ketolide antibiotic that is produced by a type I PKS in Streptomyces venezuelae ATCC® 15439™. Unlike other reported type I PKS systems, the pikromycin PKS demonstrates the unique ability to efficiently generate two macrolactone products of differing ring size. Premature termination of polyketide biosynthesis at the penultimate elongation module, PikAIII, results in generation of the 12-membered ring macrolactone, 10-deoxymethynolide (10-dml), while continued elongation through the final elongation module, PikAIV, yields a heptaketide intermediate that is cyclized to the 14-membered ring macrolactone, narbonolide (Figure 1). Each of these macrolactone products is further elaborated via downstream glycosylation and hydroxylation reactions to yield a series of antibiotics that includes the 12-membered ring macrolides methymycin, neomethymycin, and novamethymycin, as well as the 14-membered ring macrolides pikromycin, neopikromycin, and novapikromycin [10–12].
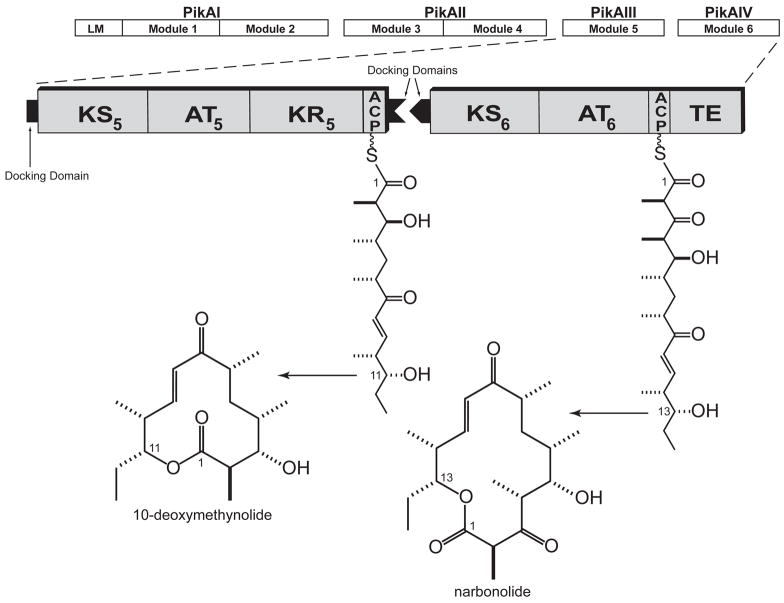
Figure 1. The Pikromycin Polyketide Synthase.
The pikromycin PKS is comprised of a single loading module (LM) and six elongation modules that span four polypeptide chains (PikAI-PikAIV). Interpolypeptide interactions are facilitated by docking domains that reside at the termini of the individual polypeptides. Chain elongation through PikAIII followed by thioesterase catalyzed termination results in the 12-membered macrolactone product, 10-deoxymethynolide, while continued elongation of the polyketide chain produces the 14-membered macrolactone, narbonolide. Both products undergo further processing via a glycosyl transferase and P450 hydroxylase to yield the methymycin and pikromycin suite of antibiotics. Abbreviations: KS, ketosynthase; AT, acyl transferase; ACP, acyl carrier protein; TE, thioesterase.
Insight into the mechanism that directs the pikromycin PKS to produce two distinct macrolactones could facilitate the development of a powerful metabolic engineering tool for assembly of novel biologically active compounds. Despite much effort, the precise catalytic events that give rise to the two different macrolactone products from this system remain unclear. It has previously been established that inactivation of the TE domain of PikAIV eliminates the production of both 10-dml and narbonolide [13]; hence, the PikAIII bound hexaketide intermediate must be presented to the terminal TE domain of PikAIV for 10-dml biosynthesis. Given the monomodular arrangement of PikAIII and PikAIV, several hypothetical paths are possible that guide the hexaketide chain to the TE domain. For example, a direct interaction between the ACP5 domain of PikAIII and the TE domain of PikAIV might facilitate the interdomain transfer of the hexaketide precursor. Such a transfer could be achieved within the context of the native intermodular docking domain mediated protein-protein interaction that naturally occurs between the ACP5 domain and the KS6 domain [14], assuming that the PikAIV monomodule is sufficiently flexible to enable the TE to reach the ACP domain of PikAIII. Alternatively, the monomodular arrangement of PikAIII and PikAIV might enable a direct domain-domain interaction that is independent of the ACP5 and KS6 docking domains. Finally, if PikAIII and PikAIV can only interact through their respective docking domains, the hexaketide chain could reach the TE domain by passing through PikAIV in the absence of a final elongation step (e.g., modular skipping) [15, 16].
Our long standing interest in developing a detailed understanding of the pikromycin PKS biosynthetic system has motivated our continued dissection of the biochemical steps leading to formation of the 12-membered ring macrolactone, 10-dml. Herein, we describe a series of in vitro experiments that probe the importance of the intermodular docking domains between PikAIII and PikAIV for the production of 10-dml. Taken together, the results of this work demonstrate that PikAIII and PikAIV must engage in their docking domain mediated protein-protein interaction in order to facilitate efficient TE-catalyzed lactonization of the PikAIII-bound hexaketide intermediate to generate the 12-membered ring macrolactone product.
Results
Mutation of PikAIV catalytic domains
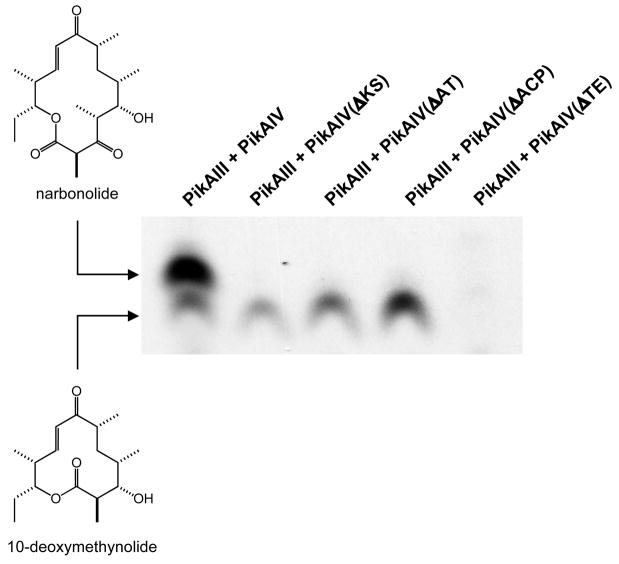
In vitro investigations into multiple macrolactone production by the pikromycin PKS were initiated by first examining whether catalytic competency of PikAIV was a necessary requirement for 10-dml production. Specifically, we sought to systematically probe the importance of each of the four catalytic protein domains of PikAIV in 10-dml biosynthesis. To this end, point mutations, serving to effectively inactivate each individual domain (ΔKS, C207A; ΔAT, S652A; ΔACP, S980A; ΔTE, S1196A) were introduced via PCR mutagenesis into the expression plasmid encoding wild-type pikAIV. Each of the four mutant PikAIV modules was co-expressed in E. coli with the B. subtilis phosphopantetheinyl transferase in order to ensure effective posttranslational modification to the holo-ACP domain [17]. Following purification, apo to holo conversion of the ACP domains was verified by FT-ICR MS analysis (data not shown). Each of the mutants was assessed for its ability to produce both 10-dml and narbonolide when combined with PikAIII and a pentaketide N-acetyl cysteamine (NAC) thioester substrate in vitro [18]. Significantly, none of the four PikAIV mutants was capable of catalyzing synthesis of the 14-membered ring macrolactone narbonolide (Figure 2A). However, each mutant, with the exception of PikAIV(ΔTE), supported production of the smaller macrolactone product 10-dml, suggesting that these domain-level activities are not required for synthesis of the smaller macrocycle (Figure 2A).
Figure 2. Activity of PikAIII + PikAIV Mutants.
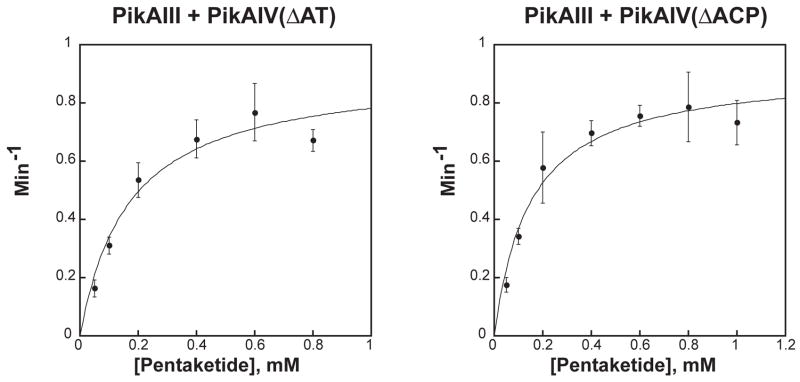
(A) Radio-TLC of reaction products produced by PikAIII + PikAIV mutants. Mutants of PikAIV were each incubated with PikAIII in the presence of the synthetic pentaketide NAC thioester and 2-[14C]-methylmalonyl-CoA as described in the experimental procedures. The reaction products were resolved by TLC (5% MeOH in CHCl3) and subsequently visualized using a Typhoon phosphorimager. (B) Michaelis-Menten plots of PikAIII + PikAIV(ΔAT) and PikAIII + PikAIV(ΔACP). The averages of data points obtained from at least three independent determinations are plotted. The curves represent fits of the data calculated by non-linear regression. Error bars were generated from the standard deviation within each point.
The efficiency of 10-dml synthesis by PikAIV(ΔAT) and PikAIV(ΔACP), in the presence of PikAIII, was further investigated through detailed steady-state kinetic analysis (Figure 2B). For comparison, a parallel analysis was performed with wild-type PikAIII and PikAIV (Supplementary Information, Table 1). Kinetic parameters determined for this control pairing are in close agreement with previous reports (kcat = 3.0 ± 0.03 min−1, KM = 0.31 ± 0.03 mM) [18]. Specifically, both PikAIV(ΔAT) and PikAIV(ΔACP) display similar apparent kcat values for 10-dml production, which are approximately 2-fold lower than the calculated kcat from wild-type PikAIII and PikAIV (Table 1). Likewise, both PikAIV mutants display similar apparent KM values that are also reduced 2-fold when compared to the KM from PikAIII and PikAIV. The equivalent decrease in the apparent kinetic parameters for both mutants results in a specificity constant (kcat/KM) that is comparable to wild-type (Table 1), further emphasizing the non-critical role of these catalytic domains in 10-dml production.
Table 1.
Steady-State Kinetic Parameters for 10-Deoxymethynolide Production by PikAIV Mutants
| Modules | kcat (min−1) | KM (mM) | kcat/KM (min−1·mM−1) | Relative kcat | Relative kcat/KM |
|---|---|---|---|---|---|
| 1,2 PikAIII + PikAIV | 2.0 ± 0.02 | 0.31 ± 0.08 | 6.5 ± 1.7 | 1 | 1 |
| PikAIII + PikAIV(ΔAT) | 0.91 ± 0.09 | 0.17 ± 0.05 | 5.4 ± 1.7 | 0.46 | 0.83 |
| PikAIII + PikAIV(ΔACP) | 0.92 ± 0.05 | 0.15 ± 0.03 | 6.1 ± 1.3 | 0.46 | 0.94 |
Independently determined for this report (see Supplementary Material).
Previously reported values: kcat = 3.0 min−1 and KM = 0.25 mM [18].
PikAIII & PikAIV Docking Domain Truncations
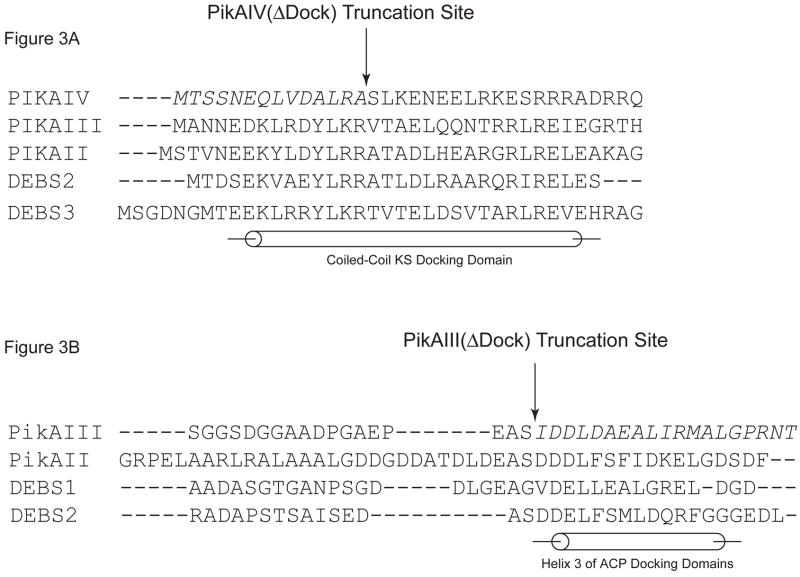
Following analysis of PikAIV catalytic domains in macrocyclic ring formation, we next investigated the role of the intermodular docking domains between PikAIII and PikAIV in 10-dml production. The putative docking domains of PikAIII and PikAIV were predicted from multiple sequence alignment and secondary structural prediction, as well as the NMR solution structure of the docking domain interaction that occurs between DEBS2 and DEBS3 [19]. Guided by this information, partial deletions of both PikAIII C-terminal and PikAIV N-terminal docking domains (hereafter referred to as PikAIII(ΔDock) & PikAIV(ΔDock)) were generated (Figure 3A & 3B).
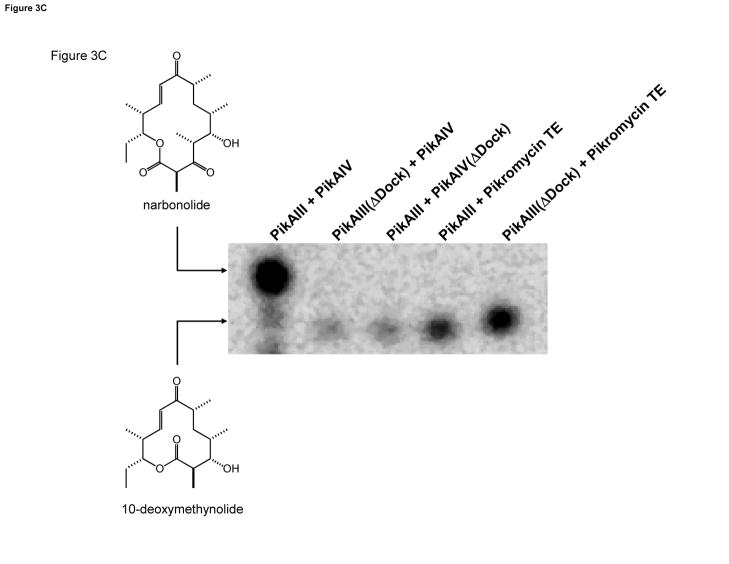
Figure 3. PikAIII(ΔDock) and PikAIV(ΔDock): Design and Activity.
(A). Multiple sequence alignment of the helical N-terminal docking domains from the pikromycin and DEBS PKS. PikAIV(ΔDock) is the result of truncating the first 14 amino acids (in italics) of PikAIV. (B). Multiple sequence alignment of partial C-terminal docking domains from the pikromycin and DEBS PKS. PikAIII(ΔDock) is the result of truncating the final 19 amino acids (in italics) of PikAIII that comprise helix 3. (C). Radio-TLC of reaction products produced by the truncated pikromcyin modules. PikAIII(ΔDock) or PikAIV(ΔDock) was paired with their respective partner wild-type or truncated module in the presence of the synthetic pentaketide NAC thioester and 2-[14C]-methylmalonyl-CoA as described in the experimental procedures. Likewise, PikAIII or PikAIII(ΔDock) was paired with recombinant pikromycin TE under identical experimental condtions. The reaction products were resolved by TLC (5% MeOH in CHCl3) and subsequently visualized using a Typhoon phosphorimager.
PikAIII(ΔDock) and PikAIV(ΔDock) were each co-expressed in E. coli with the phosphopantetheinyl transferase from B. subtilis in order to facilitate efficient phosphopantetheinylation of the ACP domains [17]. Following purification, the extent of apo to holo conversion of the ACP domains in both truncated monomodules was assessed by FT ICR-MS analysis and determined to be comparable to wild-type (>90%, see Supplementary Information). We assessed the ability of both PikAIII(ΔDock) and PikAIV(ΔDock) to produce 10-dml and narbonolide from the synthetic pentaketide NAC thioester substrate when paired with their respective complementary wild-type module. As shown in Figure 3C, exclusive formation of 10-dml, with no detectable 14-membered ring product, was observed in assays that utilized either truncated monomodule. These data reveal that the native protein-protein interaction that typically occurs between PikAIII and PikAIV has been disrupted.
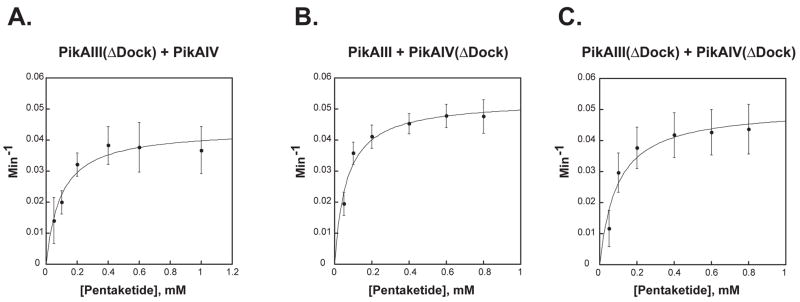
In vitro, both PikAIII(ΔDock) and PikAIV(ΔDock) demonstrate the ability to catalyze the formation of the smaller macrolactone product (Figure 3C, lanes 2 & 3); however, the extended time course required to observe product accumulation (i.e., 75 minutes vs. 15 minutes) suggests that the efficiency of the synthesis of the 12-membered ring macrocycle by the truncated monomodules is compromised signficantly relative to wild-type. Steady-state kinetic parameters were determined for the synthesis of 10-dml by both PikAIII(ΔDock) and PikAIV(ΔDock) when paired with their wild-type partner monomodule (Figure 4A & 4B). As summarized in Table 2, both PikAIII(ΔDock) and PikAIV(ΔDock) display apparent kcat values (0.044 min−1 and 0.053 min−1), which are significantly decreased relative to the kcat observed with wild-type PikAIII and PikAIV. The apparent KM values for PikAIII(ΔDock) and PikAIV(ΔDock) decreased 3–5 fold compared to wild-type, resulting in specificity constants (kcat/KM) that were 14- and 8-fold lower, respectively. Combining PikAIII(ΔDock) and PikAIV(ΔDock) together did not result in an additional rate decrease; the kinetic parameters were comparable to those obtained when either truncated monomodule was paired with its wild-type partner (Figure 4C and Table 2).
Figure 4. Michaelis-Menten Plots of PikAIII(ΔDock) and PikAIV(ΔDock).
The averages of data points obtained from at least three independent determinations are plotted. Error bars were generated from the standard deviation within each point. (A): PikAIII(ΔDock) + PikAIV; (B) PikAIII + PikAIV(ΔDock); (C) PikAIII(ΔDock) + PikAIV(ΔDock).
Table 2.
Steady-State Kinetic Parameters for 10-Deoxymethynolide Production by PikAIII(ΔDock) and PikAIV(ΔDock)
| Modules | kcat (min−1) | KM (mM) | kcat/KM (min−1·mM−1) | Relative kcat | Relative kcat/KM |
|---|---|---|---|---|---|
| PikAIII + PikAIV | 2.0 ± 0.02 | 0.31 ± 0.08 | 6.5 ± 1.7 | 1 | 1 |
| PikAIII(ΔDock) + PikAIV | 0.044 ± 0.003 | 0.095 ± 0.02 | 0.46 ± 0.10 | 0.02 | 0.07 |
| PikAIII + PikAIV(ΔDock) | 0.053 ± 0.002 | 0.064 ± 0.01 | 0.83 ± 0.13 | 0.03 | 0.13 |
| PikAIII(ΔDock)+ PikAIV(ΔDock) | 0.051 ± 0.004 | 0.093 ± 0.03 | 0.55 ± 0.18 | 0.03 | 0.08 |
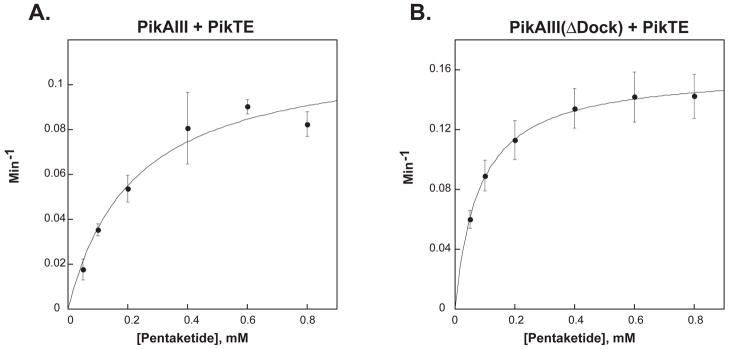
| PikAIII + Pik Thioesterase | 0.12 ± 0.01 | 0.22 ± 0.06 | 0.55± 0.21 | 0.06 | 0.08 |
| PikAIII(ΔDock) + Pik Thioesterase | 0.16 ± 0.002 | 0.08 ± 0.003 | 2 ± 0.8 | 0.08 | 0.3 |
Thioesterase Mediated 10-Deoxymethynolide Production
The observation that 10-dml could be produced, albeit inefficiently, in a reaction mixture containing either truncated monomodule prompted us to explore the possible occurrence of a direct interaction between the penultimate elongation module, PikAIII, and the terminal TE domain of PikAIV. Specifically, we examined the ability of the excised recombinant pikromycin TE domain [20, 21] to catalyze macrolactonization of the PikAIII bound hexaketide intermediate into 10-dml. When assayed in combination with either PikAIII or PikAIII(ΔDock), pikromycin TE catalyzed formation of 10-dml (Figure 3C, lanes 4 & 5). As with the truncated monomodules, steady-state kinetic analysis confirmed the poor efficiency of product formation (Figure 5A & Figure 5B, Table 2) relative to wild-type PikAIV. The apparent kcat for the combination of PikAIII and was 0.12 min−1, while the combination of PikAIII(ΔDock) and pikromcyin TE gave a kcat of 0.16 min−1. Although these values are slightly greater than those observed in assays containing the truncated modules, they represent a rate that is approximately 50-fold slower than the wild-type PikAIII and PikAIV interaction that leads to 10-dml production.
Figure 5. Michaelis-Menten Plots of PikAIII + Pik TE and PikAIII(ΔDock) + Pik TE.
The averages of data points obtained from at least three independent determinations are plotted. Error bars were generated from the standard deviation within each point. (A): PikAIII(ΔDock) + Pik TE; (B) PikAIII(ΔDock) + Pik TE.
Titration of PikAIII Docking Domain: Effect on Macrolactone Production
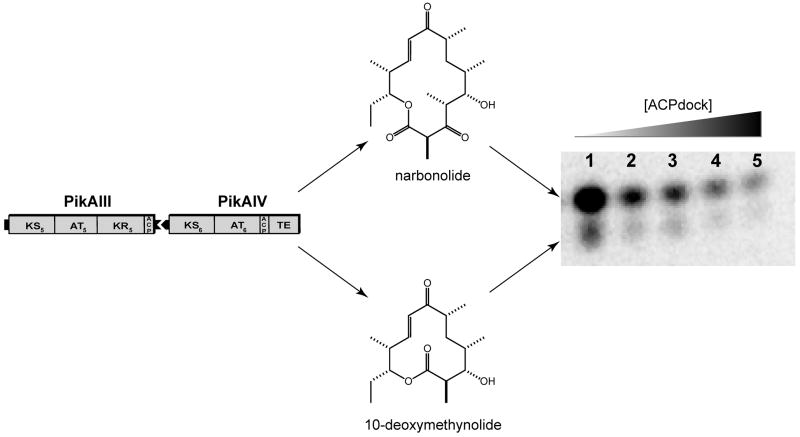
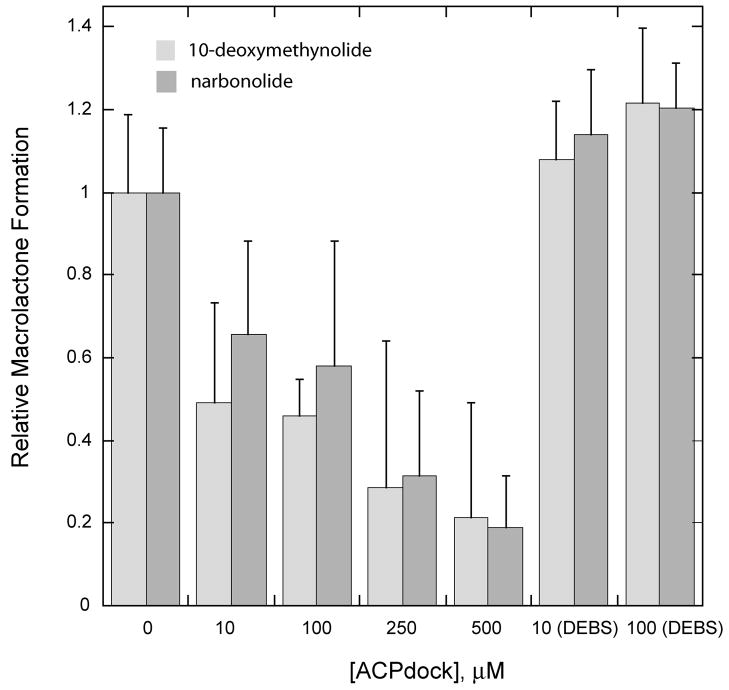
To probe further the significance of the PikAIII and PikAIV interaction in macrolactone production by the pikromycin PKS, competition experiments were performed with the objective of disrupting the native intermodular protein-protein interaction. Previously, Tsuji et al. demonstrated that the synthetic N-terminal docking domain of DEBS2 could effectively compete with DEBS module 3 for interaction with DEBS module 2 [22]. Based on this, it was anticipated that disruption of the PikAIII and PikAIV interaction could be achieved by supplementing reaction mixtures with increasing concentrations of the discrete PikAIII C-terminal docking domain. The C-terminal docking domain of PikAIII (ACPdock) was successfully generated in E. coli as a discrete, soluble protein. Following purification, ACPdock was titrated into reaction mixtures containing PikAIII and PikAIV, and a profound impact on production of both macrolactone products was observed (Figure 6A). The addition of ACPdock in 10- to 50-fold excess relative to PikAIII and PikAIV resulted in an approximate 2- to 4-fold reduction in both narbonolide and 10-dml synthesis (Figure 6B). The observed decrease in macrolactone synthesis was due to specific competition between ACPdock and PikAIII for PikAIV, as the addition of the C-terminal docking domain from DEBS2 (DEBS2 ACPdock) did not affect levels of macrolactone formation (Figure 6B).
Figure 6. ACPdock Competition with PikAIII for Interaction with PikAIV.
Reaction mixtures containing wild type PikAIII and PikAIV were titrated with increasing amounts of the C-terminal docking domain of PikAIII (ACPdock) as described in the experimental procedures. The reaction products were resolved by TLC (5% MeOH in CHCl3) and subsequently visualized using a Typhoon phosphorimager. (A) Radio-TLC of macrolactone production versus increased ACPdock concentration; (B) Plot of relative macrolactone production versus ACPdock concentration.
Discussion
The ability to produce recombinant PikAIII and PikAIV in E. coli [23, 24], coupled with recent advances in the synthesis of late stage pikromycin chain elongation intermediates [18, 20], presents the opportunity to perform detailed in vitro characterization of reactions catalyzed by the final two monomodules of pikromycin biosynthesis. It has recently been demonstrated that the in vitro combination of PikAIII and PikAIV can extend a synthetic pentaketide N-acetyl cysteamine thioester substrate to generate both 10-dml and narbonolide [18]. Development of this in vitro system has enabled detailed biochemical studies of the molecular mechanisms involved in production of both 10-dml and narbonolide by the pikromycin PKS system.
Since elucidation of the pikromycin biosynthetic gene cluster [25], events guiding the fate of the pikromycin hexaketide chain elongation intermediate have been the subject of much investigation. Initially, an in vivo N-terminally truncated form of PikAIV was proposed to catalyze the synthesis of 10-dml due to the inability of the KS domain to catalyze normal condensation activity [13]; however, the observation that full-length recombinant PikAIV can support 10-dml synthesis in vitro does not fully support this model [18]. More recently, a series of in vivo experiments has suggested that all four domains of PikAIV are required for 10-dml production by the pikromycin PKS [15]. This observation led to the hypothesis that the hexaketide intermediate must pass through PikAIV, without undergoing extension, in order to reach the terminal TE domain.
To investigate this proposed domain-skipping hypothesis, each of the four domains that are encompassed within PikAIV was inactivated via site-directed mutagenesis and subsequently assessed for its ability to produce 10-dml when paired with PikAIII in vitro. None of the PikAIV mutants supported narbonolide production; however, with the exception of PikAIV(ΔTE), 10-dml synthesis was observed with all mutant PikAIV modules. These observations imply that the core catalytic domains that comprise PikAIV are not required for generation of the 12-membered ring macrolactone product, a result that contradicts the previously proposed in vivo PikAIV skipping model [15]. Furthermore, detailed kinetic analysis of 10-dml production by PikAIV(ΔAT) and PikAIV(ΔACP) clearly demonstrated that the production efficiency of the smaller macrolactone product, relative to wild-type, is unaffected when the AT and ACP domain are inactive. Such a result would not be expected if these protein domains were, as previously proposed, required for the synthesis of the 12-membered ring macrolactone.
Based on the in vitro activity of the PikAIV mutants, we hypothesized that the generation of 10-dml by the pikromycin PKS system results from the direct transfer and lactonization of the PikAIII bound hexaketide intermediate by the terminal TE domain of PikAIV. If so, the monomodular organization of PikAIII and PikAIV may permit the terminal TE domain to non-specifically interact with the ACP domain of PikAIII. Alternatively, the interaction of PikAIII and PikAIV via their respective docking domains may be required to enable the terminal TE domain to efficiently access the ACP-bound hexaketide intermediate.
The interaction between modules leading to premature lactonization has previously been investigated using the erythromycin PKS system (6-deoxyerythronolide B synthase, DEBS) [26]. In these studies, the combination of DEBS1 and DEBS module 3 containing an engineered C-terminal TE domain was observed to produce a triketide lactone as well as the expected tetraketide lactone product. Moreover, the rate of triketide lactone synthesis was reportedly 10-fold higher when DEBS1 was paired with the DEBS module3-TE fusion protein compared with the excised recombinant DEBS TE alone. This suggests that despite the normal protein-protein interaction between DEBS1 and DEBS module 3 that allows intermodular transfer of the polyketide intermediate, the covalently linked TE domain can compete for the triketide chain and produce the triketide lactone. Extrapolation of this observation to the pikromycin PKS system would suggest that the TE domain of the PikAIV protein must compete for the hexaketide chain as it is being transferred to the KS6 active site.
It has previously been established that within PKS systems, docking domains serve an essential role in interpolypeptide communication [14, 27, 28]. NMR studies of docking domain interactions from the DEBS PKS system have demonstrated that the helical amino-terminal docking domain participates in a coiled-coiled structure in the dimeric module [28]. This structural feature provides a docking site for the helical carboxy-terminal docking domain of the preceding module [28]. Thus, we reasoned that disruption of the overall helical structures of either docking domain through short amino acid truncations would be sufficient to prevent the normal protein-protein interaction with their partner module. When paired with a wild-type partner module, or each other, PikAIII(ΔDock) and PikAIV(ΔDock) failed to generate the expected 14-membered ring macrolactone product, suggesting that the short docking domain truncations disrupted the normal interaction that occurs between the two monomodules. Interestingly, 10-dml production was observed in all cases; however, steady-state kinetic analysis demonstrated that the synthesis of this 12-membered ring macrolactone by either PikAIII(ΔDock) or PikAIV(ΔDock) is much less efficient (ca. 30 to 50-fold based on kcat) than that observed with the combination of wild-type PikAIII and PikAIV (Table 2). One possible explanation for this observation is that the ACP domains are not sufficiently phosphopantetheinylated during protein expression, resulting in an overestimation of the amount of active enzyme in the reaction mixtures. However, rigorous analysis of the apo to holo conversion of the ACP domains of both PikAIII(ΔDock) and PikAIV(ΔDock) by FT MS confirmed that the ACP domains of the truncated modules were comparably modified relative to those of wild-type modules (Supplementary Information). As such, it is reasonable to conclude from this data that production of 10-dml by the pikromycin PKS system requires PikAIII and PikAIV to be engaged in the docking domain protein-protein interaction.
Further evidence for the importance of the PikAIII-PikAIV interaction in mediating macrolactone formation was gained from competition experiments that utilized the discrete C-terminal docking domain of PikAIII (ACPdock) to disrupt the normal PikAIII-PikAIV protein interaction. Given the intrinsic specificity of C-terminal docking domains for their N-terminal docking domain partners [22, 27, 28], we reasoned that the recombinant ACPdock should compete with PikAIII for binding to PikAIV, thereby resulting in an observed decrease in macrolactone production. As expected, titration of ACPdock into reaction mixtures containing wild-type PikAIII and PikAIV gave a dose dependent decrease in the generation of 10-dml and narbonolide (Figure 6), suggesting that ACPdock is indeed capable of precluding PikAIII and PikAIV from participating in the normal protein-protein interaction. Importantly, a similar effect was not observed upon titration of the reaction mixtures with a control peptide, the C-terminal docking domain of DEBS2 (DEBS2 ACPdock), under similar conditions. This indicates that the observed competition is due to a specific interaction between ACPdock and PikAIV. Consistent with the experiments involving PikAIII(ΔDock) and PikAIV(ΔDock), these results further emphasize the key role for docking domains in the maintenance of the PikAIII-PikAIV protein-protein interaction for macrolactone formation by the pikromycin PKS.
Both PikAIII(ΔDock) and PikAIV(ΔDock) were capable of supporting 10-dml synthesis, although at a much reduced efficiency, when paired with each other or their wild-type partner monomodule. Given this, it was compelling to speculate that a direct interaction between the ACP5 domain of PikAIII and the terminal TE domain of PikAIV could occur in a non-specific manner to give low, non-physiological levels of 12-membered ring product formation. The excised pikromycin TE domain demonstrated the ability to cleave and cyclize the hexaketide intermediate from either PikAIII or PikAIII(ΔDock). Furthermore, the calculated kcat values are comparable to those determined from the PikAIII-PikAIV(ΔDock) or PikAIII(ΔDock)-PikAIV pairings. For comparison, 10-dml production by the combination of PikAIII and the excised recombinant TE domain appears to be less efficient compared to an engineered PikAIII-TE fusion protein [18].
Taken together, the data reported here are most consistent with a model for 10-dml biosynthesis that involves TE mediated lactonization of the PikAIII bound hexaketide intermediate within the context of the PikAIII-PikAIV protein-protein interaction (Figure 7). This in vitro model assumes that, upon interaction with PikAIII, PikAIV can adopt a conformation that places the terminal TE domain proximal to the ACP5 domain of PikAIII. Although the dynamics of PKS modules remain poorly understood, recent structural information from both modular PKS systems [29–31] and the related fatty acid synthase [32] suggest that individual modules require inherent flexibility to enable ACP interaction with all necessary catalytic domains. Accordingly, we propose that when docked to PikAIII, PikAIV can adopt a conformation that enables the terminal TE to interact with the ACP of the preceding module.
Figure 7. Proposed Model of 10-Deoxymethynolide Formation by the Pikromyin Polyketide Synthase.
Based on the current in vitro data, the efficient production of 10-deoxymethynolide by the pikromycin PKS system requires that PikAIII and PikAIV be engaged in their docking domain mediated protein-protein interaction. A presumed conformational flexibility of PikAIV enables the thioesterase domain to directly off-load the hexaketide intermediate from PikAIII to generate the 12-membered macrolactone product.
Assuming a PikAIV conformational change is responsible for 10-dml biosynthesis, it is interesting to consider what factors might influence this reorientation. The data presented here highlight the importance of the docking domain interaction between PikAIII and PikAIV for macrolactone production. However, it is likely that this interaction alone is not sufficient to catalyze the PikAIV reorientation, as it is also critical for the intermodular substrate channeling that is necessary for narbonolide production. One possible explanation is that an unloaded ACP6 domain might signal a PikAIV conformational change upon docking with PikAIII. That is, if the activity of the AT domain within PikAIV is not sufficient to maintain a steady supply of the acylated ACP6 domain, PikAIV may convert to the proposed alternative conformation that facilitates 10-dml production. Consistent with this possibility, Wu et al. observed that the kcat value for PikAIV catalyzed macrolactonization of the pikromycin synthetic hexaketide intermediate was approximately 2-fold higher in the absence of methylmalonyl-CoA [33].
Although further work is required to fully establish the mechanistic details that are responsible for 10-dml production by the pikromycin PKS, the in vitro studies reported within provide strong evidence that the docking domain mediated protein-protein interaction between PikAIII and PikAIV is required for efficient generation of the 12-membered ring macrolactone. However, we do recognize that these data remain inconsistent with results obtained from previous in vivo experiments. For example, current and past in vitro data [18] do not support the initial proposal that a truncated PikAIV is necessary for catalyzing 10-dml formation [13]. It is possible that in vivo, the PikAIV protein product suffers N-terminal truncations via several likely mechanisms (e.g., alternative start site or proteolysis) that may be dependent on growth medium. As such, it cannot be ruled out that an N-terminally truncated PikAIV is able to assume an effective alternative interaction with PikAIII that can result in production of 10-dml in vivo. Likewise, previous in vivo experiments suggested the importance of the KS, AT, and ACP domain of PikAIV for 12-membered ring macrolactone production [15], although the in vitro studies reported herein oppose this proposal.
While the results obtained in the current in vitro studies are consistent with a role of docking domain molecular recognition in product formation, the complexity of the in vivo system likely contributed to the previous observations. For example, the role of additional orthogonal factors (regulators, proteases, translational start sites) may promote differential accumulation of macrolactone products in vivo. Consideration of these earlier investigations requires a cautious interpretation of any refined in vitro model as a potential oversimplification of events that operate to specify in vivo production of 10-dml. Nonetheless, the results presented here strongly support a working in vitro model for the mechanism of 10-dml synthesis by the pikromycin PKS system whereby PikAIV serves as a structural scaffold that presents the terminal TE domain to the ACP5 domain of PikAIII (Figure 7). In this respect, both in vivo [13] and in vitro models are coincident; however, a complete understanding of the intricate mechanistic details require significant additional information, such as insights into the macromolecular structure of both PikAIII and PikAIV, as well as their inherent protein dynamics.
Significance
Polyketide natural products continue to be a rich source of important biological activities, and efforts that focus on understanding and manipulating the enzymes responsible for polyketide biosynthesis continue to hold tremendous promise for the discovery and development of new macrolide antibiotics. The unprecedented ability of the pikromycin PKS to generate both 12-and 14-membered ring macrolactones makes this system an important model system for understanding important mechanistic details that can be leveraged for the creation of novel polyketide compounds. While it is well established that the interpolypeptide transfer of polyketide intermediates within PKS systems is facilitated by specific terminal docking domain-mediated interactions, the fate of the hexaketide intermediate produced by PikAIII remained unclear. The results presented in this report clearly demonstrate that the docking domain interaction between PikAIII and PikAIV that facilitates extension to the heptaketide intermediate for narbonolide production is also required for efficient macrolactonization of the hexaketide into 10-dml. To date, this in vitro analysis represents the most comprehensive, molecular study on the ability of the pikromycin PKS to efficiently synthesize multiple macrolactone ring products.
Experimental Procedures
Materials
Unless otherwise specified, all chemical reagents were purchased from Sigma. DNA oligonucleotides were ordered from Integrated DNA Technologies (Coralville, IA). Molecular cloning methods used Pfu Turbo DNA polymerase from Invitrogen (Carlsbad, CA), New England Biolabs (Ipswich, MA) restriction enzymes and T4 DNA polymerase from Novagen (Madison, WI). LB Broth was from EM Sciences (Gibbstown, NJ). Ni-NTA resin was purchased from Qiagen (Valencia, CA) and PD-10 desalting and His-trap columns were from GE Healthcare (Piscataway, NJ). The synthesis of the pentaketide NAC thioester has previously been described [18]. 2-[14C]-methylmalonyl-CoA (55 mCi/mmol) was purchased from American Radiolabeled Chemicals Inc (St. Louis, MO) and Tris [2-carboxyethyl phosphine] (TCEP) was from Hampton Research (Aliso Viejo, CA). Silica TLC plates were from EM Merck.
Construction of Mutant Pikromycin Monomodules
Site-directed mutants of PikAIV were generated using QuikChange® site-directed mutagenesis (Stratagene) following vendor’s protocols. PikAIII(ΔDock) was generated via PCR amplification from the following primers: 5′-gaggaggagcatatggcgaacaacgaagac-3′ and 5′-gagaagctttcacgacgcctccggctc-3′. Following purification, the resulting 4.7 kb amplicon was digested with NdeI and HindIII and subsequently ligated into a similarly digested wild-type pikAIII encoding plasmid (pMPA405) to afford pPikAIII(ΔDock). Similarly, PikAIV(ΔDock) was generated via PCR amplification using the following primer pair, 5′-gaggaggagcatatgtctctcaaggagaacgaa-3′ and 5′-gaggaggaggaattcgggtgagctgtc-3′, to give an approximate 1.5 kb amplicon encoding the desired N-terminal truncation. Following double digestion with NdeI and EcoRI, the PCR product was ligated into similarly digested wild-type pikAIV encoding plasmid (pBB188) to afford pPikAIV(ΔDock).
Protein Overexpression and Purification
The expression of PikAIII, PikAIV, and the pikromycin thioesterase domain has previously been reported [20, 21, 23, 24]. The expression of each mutant pikromycin module (point mutants of individual domains & docking domain truncations) was accomplished in BL21(DE3) E. coli that contain a plasmid for the expression of the Bacillus subtilis phosphopantetheinyl transferase (Sfp) [17]. Co-expression of the phosphopantetheinyl transferase was performed to ensure the efficient post-translational modification of ACP domains. In all cases, the previously reported expression conditions were followed [23].
Each polyhistidine-tagged module was subsequently purified by nickel metal affinity chromatography. Cells from overexpression cultures (typically 1 L) were harvested by centrifugation (5000g, 12 min, 4 °C) and resuspended in approximately 10–15 mL lysis buffer (50 mM NaH2PO4, pH 8.0, 1 mM DTT, 300 mM NaCl, 10 mM imidazole, 10% glycerol). After disruption by sonication (5 × 10 s with 50 s intervening between pulses), the cellular debris was pelleted by centrifugation (40,000g, 30 minutes, 4 °C). The cell free lysate was sterile filtered with a 0.45 μm syringe filter after which Ni-NTA resin (1 mL) was directly added to the filtered lysate and allowed to incubate for at least 30 minutes at 4 °C with shaking. Following transfer to a column, the Ni-NTA resin was washed with additional lysis buffer (5 column volumes), followed by 5 column volumes of wash buffer (50 mM NaH2PO4, pH 8.0, 1 mM DTT, 300 mM NaCl, 30 mM imidazole, 10% glycerol). The polyhistidine-tagged modules were eluted from the Ni-NTA resin with 5 column volumes of elution buffer (50 mM NaH2PO4, pH 8.0, 1 mM DTT, 300 mM NaCl, 300 mM imidazole, 10% glycerol) and fractionated into 0.5 mL fractions. Protein containing fractions were pooled and exchanged into storage buffer (100 mM NaH2PO4, pH 7.2, 1 mM DTT, 1 mM EDTA, 20% glycerol) using PD-10 desalting columns. Protein purity was confirmed to be greater than 95% by SDS-PAGE. Purified proteins were flash frozen in liquid nitrogen and stored at −80 °C. Final protein concentrations were determined by the Bradford method using BSA standards.
In vitro Activity Assay
The activity of the mutant pikromycin monomodules was assessed by monitoring the incorporation of 2-[14C]-methylmalonyl-CoA into the resulting 10-deoxymethynolide and narbonolide macrolactone products using the previously described radio-TLC assay [18]. Reactions were performed in 400 mM NaH2PO4 buffer (pH 7.2) containing 5 mM NaCl, 1 mM TCEP, 1 mM NADPH, 20% glycerol, and 5% DMSO in a final volume of 50 μL. In all cases, the enzyme concentrations were held constant at 1 μM each. For steady-state kinetic determinations, the concentration of the pentaketide NAC thioester substrate ranged from 0.05 – 1 mM. In all cases, enzymes were equilibrated in buffer with the pentaketide NAC thioester substrate for 5 min at 30 °C, after which the reaction was initiated with the addition of 2-[14C]-methylmalonyl-CoA to a final, saturating concentration of 0.8 mM (diluted for a specific activity ca. 1.0 mCi/mmol). Reactions were maintained at 30 °C for 75 minutes, after which they were quenched by vortexing with ethyl acetate (350 μL). The products were extracted once more with ethyl acetate (250 μL). Both ethyl acetate fractions were pooled and subsequently evaporated with a nitrogen stream. The resultant oil was resuspended in dichloromethane (30 μL) and the entire suspension mixture was spotted onto silica gel TLC plates (20 × 20 cm with 2.5 cm concentration zone). Macrolactone products were resolved using a 5% methanol:chloroform solvent system. The TLC plate was exposed to a phosphorimager screen for at least 24 hours and the radiolabeled products were subsequently visualized using a Typhoon phosphoimager (Molecular Dynamics). For kinetic assays, macrolactone product formation was quantified using the ImageQuant software package (Molecular Dynamics) from a standard curve that was generated with 2-[14C]-methylmalonyl-CoA. Apparent kinetic parameters were calculated from a non-linear fit of the initial velocity data to the Michaelis-Menten equation using the KaleidaGraph data analysis software package (Synergy software). All kinetic parameters are calculated from at least three replicate determinations of initial velocity data. The standard deviation from each point was calculated and is represented graphically by the error bars in the data plots. The errors for the fitted parameters are standard errors of the fits.
Docking Domain Competition Experiments
The C-terminal docking domain (ACPdock) that extends from ACP5 of PikAIII was amplified from pMPA405 and cloned into pMCSG7 using ligation independent cloning to generate pDHS9560 [34, 35]. The PikAIII ACPdock insert was generated via PCR amplification from the following primers: 5′-tacttccaatccaatgccctccacgaggcgtacctcgcac-3′ and 5′-ttatccacttccaatgctaggtgttacgggggccgagagc-3′. Similarly, the C-terminal docking domain adjacent to ACP4 of DEBS2 from the erythromycin PKS was cloned into pMCSG7 to produce pDHS9695 using the following primers: 5′-tacttccaatccaatgcctccgcggccagtccggcggtg and 5′-ttatccacttccaatgctactacaggtcctctccccc-3′. Docking domains were overexpressed in BL21(DE3) E. coli and purified by nickel metal affinity chromatography, followed by preparative gel filtration chromatography. Briefly, cells from 1 L overexpression cultures were harvested by centrifugation (3500g, 15 min, 4° C) and resuspended in ~40 mL lysis buffer (20 mM HEPES, pH 7.5, 300 mM NaCl, 20 mM imidazole, 1 mM MgCl2) containing 1 g CelLytic Express. After disruption by sonication (5 × 30 s with 30 s between pulses), clarified cell lysate was generated via centrifugation (20,000g, 30 min, 4 °C). Following filtration of the supernatant with a 0.45 μm filter, the lysate was loaded onto a 5 mL His-trap HP column. After washing the column with 2.5 column volumes of lysis buffer, the polyhistidine-tagged docking domains were eluted using an imidazole gradient (20 mM – 400 mM). Proteins were concentrated using Amicon Ultra devices (5,000 dalton MWCO). Docking domains were further purified by gel fitration on a Pharmacia Superdex S75 column in Hepes buffered saline (HBS: 20 mM HEPES, pH 7.5, 150 mM NaCl). Following concentration, docking domains were stored at −80 °C in HBS. Protein concentration was determined by comparison to a BSA standard using the BCA protein assay method. For competition experiments, ACPdock (0–500 μM) was titrated into reaction mixtures (50 μL) containing 1 μM each PikAIII and PikAIV, 0.4 mM pentaketide NAC thioester, and 0.8 mM 2-[14C]-methylmalonyl-CoA (diluted for a specific activity of 1.0 mCi/mMol) in 400 mM NaH2PO4 buffer (pH 7.2) supplemented with 5 mM NaCl, 1 mM TCEP, 1 mM NADPH, 20% glycerol, and 5% DMSO. Following 20 min incubation at 30 °C, the reactions were quenched and assessed for macrolactone production as described above.
Supplementary Material
Acknowledgments
We thank Mr. Chris Rath and Professor Kristina Håkansson for performing FT-ICR MS analysis of the apo to holo conversion of ACP domains within the recombinant pikromycin monomodules. This research was supported by the National Institutes of Health (R01 GM076477 to DHS), the University of Michigan, College of Pharmacy, Vahlteich Research Award, and the J.G. Searle Professorship in Medicinal Chemistry (University of Michigan, College of Pharmacy). J.D.K. is supported by a NRSA postdoctoral fellowship from the NIH (GM075641). W.S. is supported by the Swiss National Science Foundation (PBBS2-11008).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Donadio S, Staver MJ, McAlpine JB, Swanson SJ, Katz L. Modular organization of genes required for complex polyketide biosynthesis. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- 2.Cortes J, Haydock SF, Roberts GA, Bevitt DJ, Leadlay PF. An unusually large multifunctional polypeptide in the erythromycin-producing polyketide synthase of Saccharopolyspora erythraea. Nature. 1990;348:176–178. doi: 10.1038/348176a0. [DOI] [PubMed] [Google Scholar]
- 3.Weissman KJ, Leadlay PF. Combinatorial biosynthesis of reduced polyketides. Nat Rev Microbio. 2005;3:925–936. doi: 10.1038/nrmicro1287. [DOI] [PubMed] [Google Scholar]
- 4.Kittendorf JD, Sherman DH. Developing tools for engineering hybrid polyketide synthetic pathways. Curr Opin Biotechnol. 2006;17:597–605. doi: 10.1016/j.copbio.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Arakawa K, Sugino F, Kodama K, Ishii T, Kinashi H. Cyclization mechanism for the synthesis of macrocyclic antibiotic lankacidin in Streptomyces rochei. Chem Biol. 2005;12:249–256. doi: 10.1016/j.chembiol.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Olano C, Wilkinson B, Sanchez C, Moss SJ, Sheridan R, Math V, Weston AJ, Brana AF, Martin CJ, Oliynyk M, et al. Biosynthesis of the angiogenesis inhibitor borrelidin by Streptomyces parvulus Tu4055: Cluster analysis and assignment of functions. Chem Biol. 2004;11:87–97. doi: 10.1016/j.chembiol.2003.12.018. [DOI] [PubMed] [Google Scholar]
- 7.He J, Hertweck C. Iteration as programmed event during polyketide assembly; molecular analysis of the aureothin biosynthesis gene cluster. Chem Biol. 2003;10:1225–1232. doi: 10.1016/j.chembiol.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 8.Olano C, Wilkinson B, Moss SJ, Brana AF, Mendez C, Leadlay PF, Salas JA. Evidence from engineered gene fusions for the repeated use of a module in a modular polyketide synthase. Chem Comm. 2003:2780–2782. doi: 10.1039/b310648a. [DOI] [PubMed] [Google Scholar]
- 9.Gaitatzis N, Silakowski B, Kunze B, Nordsiek G, Blocker H, Hofle G, Muller R. The biosynthesis of the aromatic myxobacterial electron transport inhibitor stigmatellin is directed by a novel type of modular polyketide synthase. J Biol Chem. 2002;277:13082–13090. doi: 10.1074/jbc.M111738200. [DOI] [PubMed] [Google Scholar]
- 10.Lee SK, Park JW, Kim JW, Jung WS, Park SR, Choi CY, Kim ES, Kim BS, Ahn JS, Sherman DH, et al. Neopikromycin and novapikromycin from the pikromycin biosynthetic pathway of Streptomyces venezuelae. J Nat Prod. 2006;69:847–849. doi: 10.1021/np060026p. [DOI] [PubMed] [Google Scholar]
- 11.Xue YQ, Wilson D, Sherman DH. Genetic architecture of the polyketide synthases for methymycin and pikromycin series macrolides. Gene. 2000;245:203–211. doi: 10.1016/s0378-1119(00)00003-2. [DOI] [PubMed] [Google Scholar]
- 12.Zhang QB, Sherman DH. Isolation and structure determination of novamethymycin, a new bioactive metabolite of the methymycin biosynthetic pathway in Streptomyces venezuelae. J Nat Prod. 2001;64:1447–1450. doi: 10.1021/np010146r. [DOI] [PubMed] [Google Scholar]
- 13.Xue YQ, Sherman DH. Alternative modular polyketide synthase expression controls macrolactone structure. Nature. 2000;403:571–575. doi: 10.1038/35000624. [DOI] [PubMed] [Google Scholar]
- 14.Gokhale RS, Khosla C. Role of linkers in communication between protein modules. Curr Opin Chem Biol. 2000;4:22–27. doi: 10.1016/s1367-5931(99)00046-0. [DOI] [PubMed] [Google Scholar]
- 15.Beck BJ, Yoon YJ, Reynolds KA, Sherman DH. The hidden steps of domain skipping: Macrolactone ring size determination in the pikromycin modular polyketide synthase. Chem Biol. 2002;9:575–583. doi: 10.1016/s1074-5521(02)00146-1. [DOI] [PubMed] [Google Scholar]
- 16.Rowe CJ, Bohm IU, Thomas IP, Wilkinson B, Rudd BA, Foster G, Blackaby AP, Sidebottom PJ, Roddis Y, Buss AD, et al. Engineering a polyketide with a longer chain by insertion of an extra module into the erythromycin-producing polyketide synthase. Chem Biol. 2001;8:475–485. doi: 10.1016/s1074-5521(01)00024-2. [DOI] [PubMed] [Google Scholar]
- 17.Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT. A new enzyme superfamily - the phosphopantetheinyl transferases. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 18.Aldrich CC, Beck BJ, Fecik RA, Sherman DH. Biochemical investigation of pikromycin biosynthesis employing native penta- and hexaketide chain elongation intermediates. J Am Chem Soc. 2005;127:8441–8452. doi: 10.1021/ja042592h. [DOI] [PubMed] [Google Scholar]
- 19.Broadhurst RW, Nietlispach D, Wheatcroft MP, Leadlay PF, Weissman KJ. The structure of docking domains in modular polyketide synthases. Chem Biol. 2003;10:723–731. doi: 10.1016/s1074-5521(03)00156-x. [DOI] [PubMed] [Google Scholar]
- 20.Aldrich CC, Venkatraman L, Sherman DH, Fecik RA. Chemoenzymatic synthesis of the polyketide macrolactone 10-deoxymethynolide. J Am Chem Soc. 2005;127:8910–8911. doi: 10.1021/ja0504340. [DOI] [PubMed] [Google Scholar]
- 21.Lu HX, Tsai SC, Khosla C, Cane DE. Expression, site-directed mutagenesis, and steady state kinetic analysis of the terminal thioesterase domain of the methymycin/picromycin polyketide synthase. Biochemistry. 2002;41:12590–12597. doi: 10.1021/bi026006d. [DOI] [PubMed] [Google Scholar]
- 22.Tsuji SY, Cane DE, Khosla C. Selective protein-protein interactions direct channeling of intermediates between polyketide synthase modules. Biochemistry. 2001;40:2326–2331. doi: 10.1021/bi002463n. [DOI] [PubMed] [Google Scholar]
- 23.Beck BJ, Aldrich CC, Fecik RA, Reynolds KA, Sherman DH. Substrate recognition and channeling of monomodules from the pikromycin polyketide synthase. J Am Chem Soc. 2003;125:12551–12557. doi: 10.1021/ja034841s. [DOI] [PubMed] [Google Scholar]
- 24.Beck BJ, Aldrich CC, Fecik RA, Reynolds KA, Sherman DH. Iterative chain elongation by a pikromycin monomodular polyketide synthase. J Am Chem Soc. 2003;125:4682–4683. doi: 10.1021/ja029974c. [DOI] [PubMed] [Google Scholar]
- 25.Xue YQ, Zhao LS, Liu HW, Sherman DH. A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: Architecture of metabolic diversity. Proc Natl Acad Sci U S A. 1998;95:12111–12116. doi: 10.1073/pnas.95.21.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gokhale RS, Hunziker D, Cane DE, Khosla C. Mechanism and specificity of the terminal thioesterase domain from the erythromycin polyketide synthase. Chem Biol. 1999;6:117–125. doi: 10.1016/S1074-5521(99)80008-8. [DOI] [PubMed] [Google Scholar]
- 27.Wu N, Tsuji SY, Cane DE, Khosla C. Assessing the balance between protein-protein interactions and enzyme-substrate interactions in the channeling of intermediates between polyketide synthase modules. J Am Chem Soc. 2001;123:6465–6474. doi: 10.1021/ja010219t. [DOI] [PubMed] [Google Scholar]
- 28.Weissman KJ. The structural basis for docking in modular polyketide biosynthesis. ChemBioChem. 2006;7:485–494. doi: 10.1002/cbic.200500435. [DOI] [PubMed] [Google Scholar]
- 29.Keatinge-Clay AT, Stroud RM. The structure of a ketoreductase determines the organization of the beta-carbon processing enzymes of modular polyketide synthases. Structure. 2006;14:737–748. doi: 10.1016/j.str.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 30.Tang Y, Kim CY, Mathews II, Cane DE, Khosla C. The 2.7-Angstrom crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc Natl Acad Sci U S A. 2006;103:11124–11129. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherman DH, Smith JL. Clearing the skies over modular polyketide synthases. ACS Chem Biol. 2006;1:505–509. doi: 10.1021/cb600376r. [DOI] [PubMed] [Google Scholar]
- 32.Maier T, Jenni S, Ban N. Architecture of mammalian fatty acid synthase at 4.5 Å resolution. Science. 2006;311:1258–1262. doi: 10.1126/science.1123248. [DOI] [PubMed] [Google Scholar]
- 33.Wu J, He W, Khosla C, Cane DE. Chain elongation, macrolactonization, and hydrolysis of natural and reduced hexaketide substrates by the picromycin/methymycin polyketide synthase. Angew Chem, Int Ed Engl. 2005;44:7557–7560. doi: 10.1002/anie.200502246. [DOI] [PubMed] [Google Scholar]
- 34.Dieckman L, Gu MY, Stols L, Donnelly MI, Collart FR. High throughput methods for gene cloning and expression. Protein Expr Purif. 2002;25:1–7. doi: 10.1006/prep.2001.1602. [DOI] [PubMed] [Google Scholar]
- 35.Stols L, Gu MY, Dieckman L, Raffen R, Collart FR, Donnelly MI. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.