Abstract
How dopamine (DA) neuronal subtypes are specified remains unknown. In this study we show a robust generation of functional DA neurons from human embryonic stem cells (hESCs) through a specific sequence of application of fibroblast growth factor 8 (FGF8) and sonic hedgehog (SHH). Treatment of hESC-derived Sox1+ neuroepithelial cells with FGF8 and SHH resulted in production of tyrosine hydroxylase (TH)–positive neurons that were mostly bipolar cells, coexpression with γ-aminobutyric acid, and lack of midbrain marker engrailed 1 (En1) expression. However, FGF8 treatment of precursor cells before Sox1 expression led to the generation of a similar proportion of TH+ neurons characteristic of midbrain projection DA neurons with large cell bodies and complex processes and coexpression of En1. This suggests that one mechanism of generating neuronal subtypes is temporal availability of morphogens to a specific group of precursors. The in vitro–generated DA neurons were electrophysiologically active and released DA in an activity-dependent manner. They may thus provide a renewable source of functional human DA neurons for drug screening and development of sustainable therapeutics for disorders affecting the DA system.
Keywords: Neuroepithelial cells, Neural differentiation, Cell replacement, Parkinson’s disease
INTRODUCTION
Dopamine (DA) neurons reside in several brain areas, including the midbrain, hypothalamus, retina, and olfactory bulbs. The most prominent groups of DA neurons are in the substantia nigra (A9 cell group) and the ventral tegmental area (A10) of the midbrain, both of which project to the forebrain [1]. DA neurons in the substantia nigra project to the striatum and form the extrapyramidal motor system that controls postural reflexes and initiation of movement. Degeneration of these DA neurons underlies Parkinson’s disease. The DA neurons in the tegmental area connect to neurons in the limbic system, and defects in the functioning of these cells are thus implicated in psychiatric behaviors. DA neurons in other parts of the brain are mostly interneurons involved in local circuitries. Hence, DA neurons in the midbrain differ significantly from those in the forebrain in morphology, coexpression of neurotransmitters, and function [2, 3].
The ontogeny of DA neurons is not clear and remains a focus of intensive research. Studies to date have focused on the genesis of mesencephalic DA neurons. Information available indicates the involvement of several transcription factors, including Lmx1b, engrailed-1 (En1), Nurr1, and Ptx3, through independent pathways, most of which are shown to be required for survival and/or maturation but not specification of DA neurons [4–12]. How the DA neurons are initially specified from the naive neuroectodermal cells remains largely unknown. Using neuroepithelial explant cultures, Ye et al. [13] have shown that fibroblast growth factor 8 (FGF8), a morphogen involved in the patterning of isthmus, and sonic hedgehog (SHH), a ventralizing molecule, can specify a dopaminergic fate from neuroepithelial cells. The same principle seems to apply to in vitro–generated neuroepithelial cells. Mouse embryonic stem cells (ESCs), derived from the inner cell mass of a preimplantation blastocyst embryo [14, 15], are first differentiated into neuroepithelial cells, expanded in the presence FGF2, and further differentiated into DA neurons in response to FGF8 and SHH [16]. Given that the differentiation of DA neurons in both forebrain and midbrain depends on the signaling of FGF8 and SHH [17, 18], it remains unknown how subtypes of DA neurons are differentially specified.
Like their mouse counterparts, human ESCs (hESCs) [19] offer a system for modeling the early phases of human development, including neural lineage specification. They also provide a renewable source of specialized cells such as DA neurons for systematic studies of the genesis of the DA system, pathogenic process affecting the survival and function of DA neurons, and development of sustainable therapeutics for Parkinson’s disease and other disorders affecting the DA system [20]. We have established a chemically defined system to direct hESCs to neuroepithelial cells that mirrors in vivo human neuroectoderm development in timing and neural tube–like structure formation [21]. Using this model system, we have discovered that FGF8 and SHH efficiently promote the generation of DA neurons from hESCs. However, to generate DA neurons with midbrain projection neuronal phenotypes, early exposure to FGF8 before precursor cells become Sox1-expressing neuroepithelial cells is necessary. Treatment of the Sox1-expressing neuroepithelial cells with FGF8 and SHH still results in an efficient production of DA neurons but with mostly forebrain phenotypes.
MATERIALS AND METHODS
hESC Cultures
hESC lines H9 (p21–56) and H1 (p35–40) were propagated weekly on irradiated mouse embryonic fibroblasts (MEFs) with a daily change of an ESC growth medium that consisted of Dulbecco’s modified Eagle’s medium (DMEM)/F12 (Gibco, Rockville, MD, http://www.invitrogen.com), 20% serum replacer (Gibco), 1 mM glutamine (Sigma, St. Louis, http://sigmaaldrich.com), 0.1 mM nonessential amino acids (Gibco), 2 µg/ml heparin (Sigma), 0.1 mM β-mercaptoethanol (Sigma), and 4 ng/ml FGF2 (R&D Systems, Minneapolis, http://www.rndsystems.com), as previously described [19]. Differentiated colonies were physically removed using a curved Pasteur pipette, and the undifferentiated state of ESCs was confirmed by typical morphology and positive immunostaining for Oct4 and stage-specific embryonic antigen 4 (SSEA4). Cells from both lines exhibited a similar morphology and growth rate under the above culture conditions.
Differentiation and Enrichment of Neuroepithelial Cells
HESC colonies were detached from the MEF layer by treatment of the culture with 0.2 mg/ml of dispase (Roche Diagnostics, Indianapolis, http://www.roche-diagnostics.com) and grown as floating cell aggregates (embryoid bodies) for 4 days with a daily change of ESC medium. They were then grown in an adherent substrate in a neural medium consisting of DMEM/F12 (2:1), supplemented with N2 (Gibco), 0.1 mM nonessential amino acids, and 2 µg/ml heparin, with a medium change every other day. The ESC aggregates attached and formed individual colonies at approximately day 6. Neuroepithelial cells, exhibited by columnar cells organizing into neural tube–like rosettes, were developed at approximately day 14–16 [21]. The neural rosettes were isolated through differential enzymatic response [21]. Growth factors were added during the course of differentiation to influence regionalization (see Results).
DA Neuron Differentiation
The enriched neuroepithelial cells were dissociated by 0.025% trypsin and 0.27 mM EDTA in phosphate-buffered saline (PBS) at 37°C for 10–15 minutes and plated onto 12-mm coverslips (precoated with 100 µg/ml polyornithine and 10 µg/ml laminin) at a density of 40,000 to 50,000 cells per coverslip. The neuronal differentiation medium consisted of neurobasal medium (Gibco) supplemented with N2 (Gibco), 0.1 mM nonessential amino acids, 0.5 mM glutamine, 1 µg/ml laminin (Sigma), 1 µM cAMP (Sigma), 200 µM ascorbic acid (Sigma), 10 ng/ml brain-derived neurotrophic factor (R&D Systems), and 10 ng/ml glial cell line–derived neurotrophic factor (GDNF) (R&D Systems). The cells were cultured for 3–4 weeks, with medium changes every other day.
Immunocytochemistry and Cell Quantification
Coverslip cultures were fixed in 4% paraformaldehyde in PBS for 10–20 minutes or methanol (−20°C) for 5 minutes and processed for immunostaining [21]. The following primary antibodies were used: mouse anti-SSEA4 (1:40), mouse anti-En1 (1:50), and mouse anti-Pax6 (1:5,000, all from Developmental Studies Hybridoma Bank, Iowa City, IA, http://www.uiowa.edu/~dshbwww); rabbit anti-Sox1 (1:500), rabbit anti-human nestin (1:200), rabbit anti-aromatic amino acid decarboxylase (AADC) (1:1,000), rabbit anti-phenylethanolamine N-methyltransferase (PNMT) (1:200), sheep anti-dopamine β hydroxylase (DβH) (1:400), mouse anti-synaptophysin (1:500), and rabbit anti-cholecystokinin octapeptide (CCK8) 1:2,000 (all from Chemicon, Temecula, CA, http://chemicon.com); mouse anti-tyrosine hydroxylase (TH) (1:1,000), mouse anti–βIII-tubulin (1:500), rabbit anti–γ-aminobutyric acid (GABA) (1:5,000), and mouse anti-calbindin (1:400, all from Sigma); rabbit anti-TH (1:500) and rabbit anti-vesicular monoamine transporter 2 (VMAT2) (1:500, all from Pel-Freez, Rogers, AK, http://www.pel-freez.com); goat anti-Ret (1:400) and mouse anti-Oct4 (1:1,000, both from Santa Cruz Biotechnology, Santa Cruz, CA, http://www.scbt.com); rabbit anti-brain factor 1 (Bf1) (1:5,000; gift from Lorenz Studer, Sloan-Kettering Institute) and Otx2 (1:5,000, gift from F. Vaccarino, Yale University). Antibody-antigen reaction was revealed by Alexa Flour 488- or 594-conjugated secondary antibody (1:1,000, Molecular Probes, Eugene, OR, http://probes.invitrogen.com). Cell nuclei were stained with Hoechst 33342 (Sigma). Staining was visualized with a Nikon fluorescence microscope. Brain sections from adult rats and E38 embryonic monkeys were used as positive controls for many of the antibodies against neuronal types and neurotransmitters. Negative controls were also set by omitting the primary or secondary antibodies in the immunostaining procedures. Cell counting was achieved blindly by using a reticule on an eyepiece and a ×40 objective. The cells in 10 visual fields were randomly selected and counted from each coverslip. Data were presented as mean ± SD. Values of p< .05 were considered significant by the unpaired Student’s t-test.
Reverse Transcription–Polymerase Chain Reaction
Total RNA was extracted from cultured cells using RNA Stat-60 (Tel-Test, Friendswood, TX, http://www.isotexdiagnostics.com), followed by the treatment with DNase I (DNA-free, Ambion, Austin, TX, http://www.ambion.com). Synthesis of cDNA was carried out with the Superscript First-Strand Synthesis System for reverse transcription–polymerase chain reaction (RT-PCR) (Invitrogen Corporation, Carlsbad, CA, http://www.invitrogen.com) according to the manufacturer’s directions. PCR amplification was performed using a standard procedure with Taq Polymerase (Promega, Madison, WI, http://www.promega.com). The number of cycles varied from 25 to 35 cycles depending on the particular mRNA abundance with denaturation at 94°C for 15 seconds, annealing temperatures at 55°C or 60°C for 30 seconds according to the primers, and elongation at 72°C for 45 seconds. Negative control was achieved by omitting transcriptase during RT or cDNA sample during PCR. The primers and product lengths were as follows:
GAPDH (5'-ACCACAGTCCATGCCATCAC-3', 5'-TCCACCACCCTGTTGCTGTA-3', 450 bp)
Nurr1 (5'-CGATGCCTTGTGTTCAGGCGCAG-3', 5'-AGCCTTTGCAGCCCTCACAGGTG-3', 858 bp)
Ptx3 (5'-GTGGGTGGAGAGGAGAACAA-3', 5'-TTCCTCCCTCAGGAAACAATG-3', 175 bp)
Lmx1b (5'-GGGATCGGAAACTGTTACTGC-3', 5'-GTAGTCACCCTTGCACAGCA-3', 218 bp)
En1 (5'-CCCTGGTTTCTCTGGGACTT-3', 5'-GCAGTCTGTGGGGTCGTATT-3', 162 bp)
Bf1 (5'-ACTCAAAACTCGCTGGGCAAC-3', 5'-CGTGGGGGAAAAAGTAACTGG-3', 226 bp)
Gbx2 (5'-CACCACGTCTACGGGCAAGAAC-3', 5'-AGCTGCTGATGCTGACTTCTGA-3', 308 bp)
SHH (5'-CCAATTACAACCCCGACATC-3', 5'-CCGAGTTCTCTGC TTTCACC-3', 339 bp)
Nkx6.1 (5'-ACACGAGACCCACTTTTTCCG-3', 5'-TGCTGGACTTGTGCTTCTTCAAC-3', 335 bp)
Wnt1 (5'-CGGGCAACAACCAAAGTCG-3', 5'-GACAGTTCCAGCGGCGATTC-3', 321 bp)
Pax2 (5'-ATGTTCGCCTGGGAGATTCG,-3' 5'-GCAAGTGCTTCCGCAAACTG-3', 361 bp).
DA Measurement
After 21 days of DA neuronal differentiation, media conditioned for 48 hours were collected. Activity-dependent DA release from the cultured cells was measured by first conditioning cultured cells in Hanks’ balanced salt solution (HBSS) for 15 minutes and then replacing it with HBSS containing 56 mM KCl for 15 minutes at 37°C. DA in the culture media or in HBSS was stabilized by adding 20 µl stabilization buffer (900 mg EGTA and 700 mg gluthatione in 10 ml of 0.1 M NaOH), and samples were stored at −80°C. A high-performance liquid chromatography ( HPLC) kit (Chromsystems, München, Germany, http://www.chromsystems.de) was used to extract monoamines. The levels of monoamines were determined by HPLC (Model 508 autosampler and model 118 pump, Beckman Coulter, Fullerton, CA, http://www.beckmancoulter.com) coupled to an electrochemical detector (Coulochem II, ESA Inc., Chelmsford, MA, http://www.esainc.com) by using MD-TM mobile phase (ESA Inc.). The cultures in each group were triplicated, and data were collected from three separate experiments.
Electrophysiological Recording
Electrophysiological properties of the hESC-generated DA neurons were investigated using whole-cell patch-clamp recording techniques [22]. Pipettes were filled with intracellular solutions containing (in mM) KCl 140 or K-gluconate 140, Na+-HEPES 10, BAPTA 10, and Mg2+-ATP 4 (pH 7.2, 290 mOsm, 2.3–5.0 MΩ). Biocytin (0.5%, Sigma) was added to the recording solution, and subsequent labeling with streptavidin-Alex Fluor 488 (1:1,000, Molecular Probes) and an antibody against TH was used to identify DA neurons. The bath solution contained (in mM) NaCl 127, KH2PO4 1.2, KCl 1.9, NaHCO3 26, CaCl2 2.2, MgSO4 1.4, and glucose 10 and 95% O2/5% CO2 (pH 7.3; 300 mOsm). For some experiments, tetrodotoxin (TTX) (1 µM; Sigma) was applied in the bath solution to block voltage-gated sodium currents.
Current-clamp and voltage-clamp recordings were performed using a MultiClamp 700A amplifier (Axon Instruments, Inverurie, Scotland, U.K., http://www.moleculardevices.com). Signals were filtered at 4 kHz, sampled at 10 kHz using a Digidata 1322A analogue-digital converter (Axon Instruments), and acquired and stored on a computer hard disk using commercially available software (pClamp9, Axon Instruments). Access resistance was typically 8–18 MΩ and was compensated by 50%–80% using amplifier circuitry. Voltages were corrected for liquid junction potential of +13 mV [23]. Vrest and action potentials (APs) were examined in current-clamp mode. Spontaneous excitatory (inward) and inhibitory (outward) synaptic currents were characterized in voltage-clamp mode using K-gluconate–based pipette solution and Vhold = −40 mV. Synaptic events were detected using a template detection algorithm (Mini Analysis Program 5.6.28, Synaptosoft, Decatur, GA, http://www.synaptosoft.com), and deactivation phase was fitted to a biexponential function using the Levenberg-Marquardt algorithm. Data were presented as mean ± SE.
RESULTS
hESC-Derived Neuroepithelial Cells Display a Forebrain Character
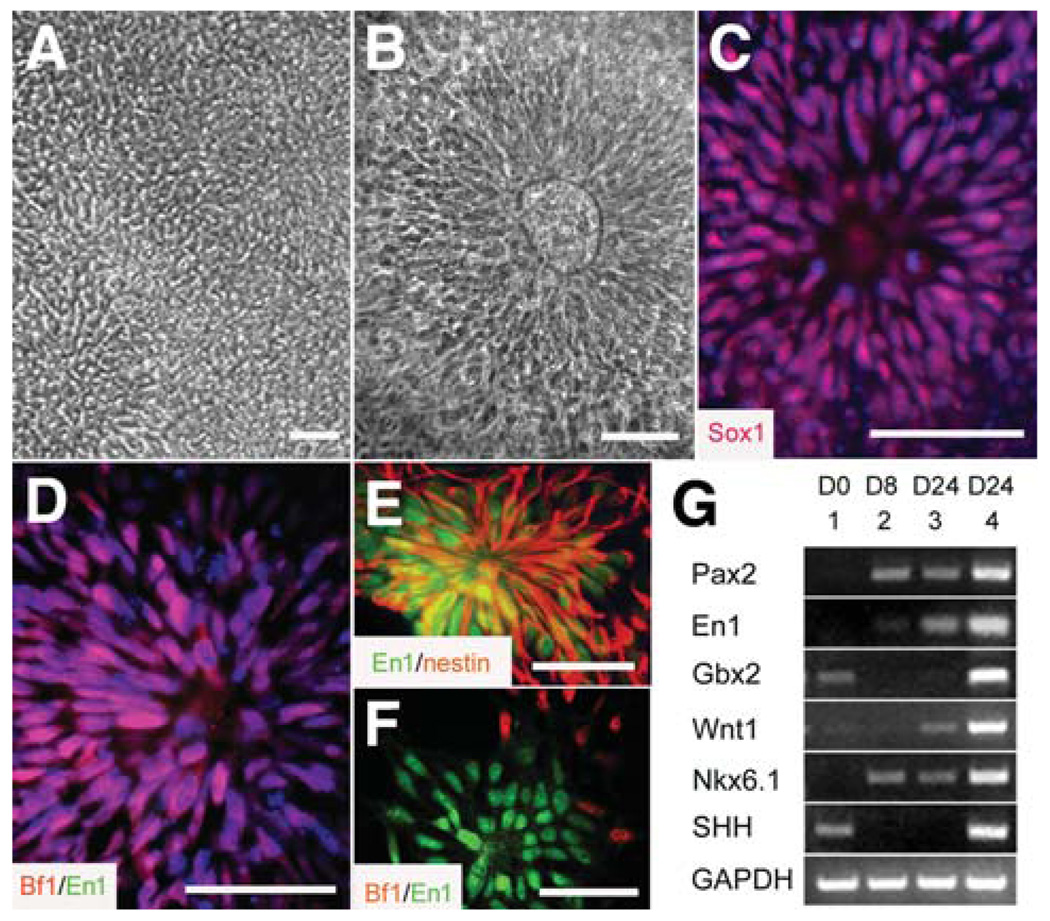
ESC colonies, detached from a feeder layer, were cultured in suspension as aggregates for 4 days in the ESC growth medium and then grown in an adhesive culture dish in a chemically defined neural medium containing FGF2 (20 ng/ml). Cells in the colony center developed a columnar morphology and lined up in a rosette formation around days 8–10 (Fig. 1A). These columnar cells were positive for Pax6 but negative for the pan-neural transcription factor Sox1 [24], indicative of early neuroepithelial cells. Over another 4–6 days (days 14–16), the columnar cells expanded and organized into neural tube–like rosettes (Fig. 1B) and expressed Sox1 (Fig. 1C), a transcription factor expressed by definitive neuroepithelial cells during neural tube closure [25]. They were positive for Otx2 [24] and Bf1, transcription factors expressed by forebrain cells [26], but negative for En1 (Fig. 1D), a transcription factor expressed by midbrain cells [27, 28], suggesting a forebrain identity of the in vitro–generated neuroepithelial cells.
Figure 1.
Specification of midbrain neural progenitors. Columnar cells (A) appeared in the differentiating human embryonic stem cell colony at days 8–10, (B) formed neural tube–like rosettes at day 14, and (C) expressed Sox1. (D): The neuroepithelial cells in FGF2-treated cultures expressed Bf1 (red) but not En1 (green). (E): En1 (green) expression was observed in the nestin+ (red) neural progenitors after 6 days of treatment with FGF8 (100 ng/ml) at day 10, expansion in FGF8 for 2 days, and subsequent treatment with FGF8 and SHH (200 ng/ml) for another 6 days on laminin substrate. (F): These En1+ cells (green) were negative for Bf1 (red). The cell nuclei were stained with Hoechst (bluein C, D). Bar = 50 µm. (G): Reverse transcription–polymerase chain reaction showed a higher expression level of Pax2, En1, Wnt1, and Gbx2, as well as Nkx6.1 and SHH mRNAs, when the embryonic stem cell (lane 1)–derived early neuroepithelial cells (lane 2) were treated with FGF8 followed by FGF8 and SHH (lane 4) compared with the cells that were treated with FGF2 followed by FGF8 and SHH for the same period (lane 3). Abbreviations: FGF, fibroblast growth factor; SHH, sonic hedgehog.
Induction of Midbrain Progenitors Requires Early Action of FGF8
For specification of a ventral midbrain fate, neuroepithelial cells in the neural tube–like rosettes were enriched through differential enzymatic and adhesion treatment [21], expanded for 2 days as aggregates in suspension with FGF2, and then plated onto a laminin substrate and treated with SHH (50–200 ng/ml) and FGF8 (20–100 ng/ml) for 6 days (Fig. 2A). Immunocytochemical analyses revealed that most of the neural progenitors remained positive for Bf1 and Otx2 but not for En1 (not shown). In long-term differentiation cultures, En1+ cells were observed (see below).
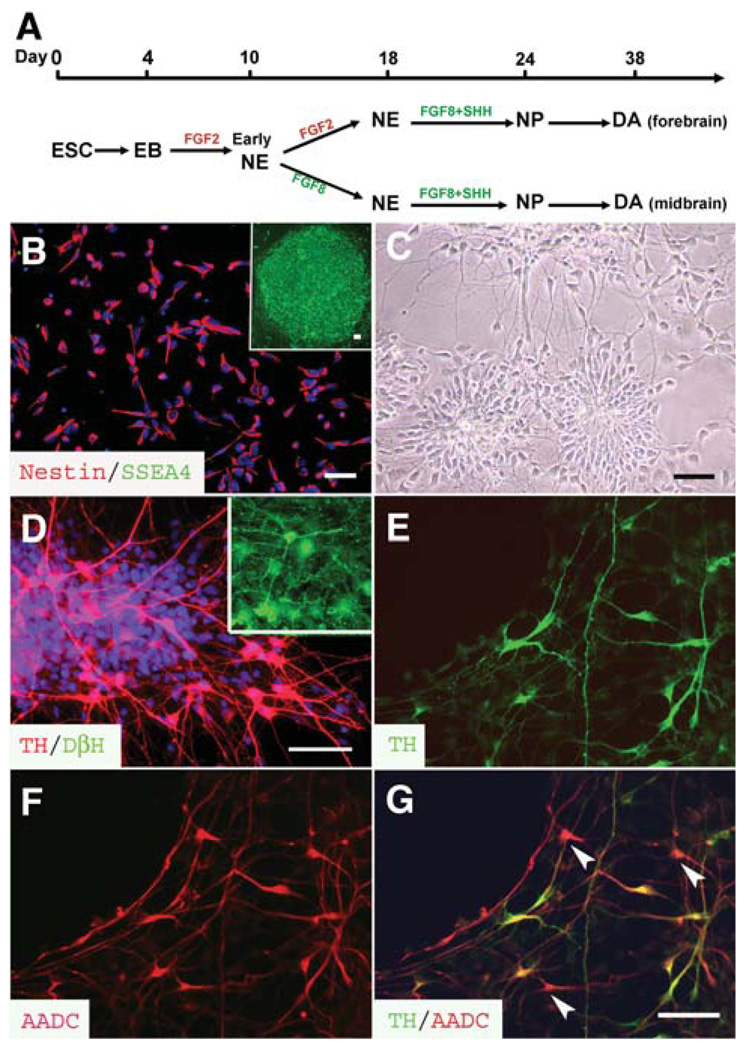
Figure 2.
Differentiation of DA neurons. (A): Schematic procedures of DA neuron differentiation. hESCs were differentiated to early neuroepithelial cells in the form of early rosettes at 8–10 days, followed by formation of neural tube–like rosettes for another week in the presence of either FGF2 (late FGF8 treatment) or FGF8 (early FGF8 treatment). They were then differentiated to NPs in the presence of FGF8 and SHH for 6 days before proceeding to DA neurons in the next 2–3 weeks after withdrawal of morphogens. (B): The neural progenitors derived form hESCs were positive for nestin (red) and negative for SSEA4 (green), which was expressed in the undifferentiated hESCs (inset). (C): The dissociated NPs reformed rosettes and began to extend neurorites 5 days after plating. (D): Approximately one third of the differentiated cells were TH+ (red) but DβH− (green) in the early FGF8 treatment group at 3 weeks of differentiation. The inset indicates that DβH positively stained cells in the section of adult rat brainstem. (E–G): All TH+ cells (E, green) were positively stained with AADC (F and G, red), but some AADC+ cells were negative for TH (G, arrowheads). The cell nuclei were stained with Hoechst (B and D, blue). Bar = 50 µm. Abbreviations: A ADC, amino acid decarboxylase; DA, dopamine; EB, embryoid body; ESC, embryonic stem cell; FGF, fibroblast growth factor; hESC, human embryonic stem cell; NE, neuroepithelia; NP, neural progenitor; SHH, sonic hedgehog; SSEA, stage-specific embryonic antigen; TH, tyrosine hydroxylase.
The failure of FGF8 to induce Sox1+ neuroepithelial cells to express En1 suggests that the Sox1-expressing neuroepithelial cells may be refractory to patterning signals. Because the Sox1-expressing cells are generated 2 weeks after differentiation of hESCs (equivalent to a 6-day-old embryo) [19] and form neural tube–like structures, they may correspond to the neuroepithelial cells at the neural tube closure, during which neuroepithelial cells express Sox1 and are regionally specified [29]. This led us to hypothesize that FGF8 may promote midbrain specification before neuroepithelial cells express Sox1. We thus applied FGF8 (100 ng/ml) at the time when the cells in the colony center became columnar at day 10 (Fig. 2A). After 6 days, cells in the colony center developed neural tube–like formations, as seen in the presence of FGF2. These neuroepithelial cells were similarly enriched and treated with FGF8 and SHH (200 ng/ml) for 6 days on the laminin substrate (Fig. 2A). Under this culture condition, En1 expression was observed in the nestin-expressing neural progenitors, often in the form of rosettes (Fig. 1E), although there were still cells that expressed Bf1 (Fig. 1F). RT-PCR analyses revealed a more effective induction of midbrain-related transcription factors En1, Pax2, and Wnt1 by early FGF8 exposure (Fig. 1G). Early exposure to FGF8 also induced the expression of SHH mRNA and a more robust mRNA expression of the ventral gene Nkx6.1 (Fig. 1G). Thus, neuroepithelial cells are more efficiently regionalized to a ventral midbrain fate by exposure to FGF8 before precursor cells become Sox1+.
Neural Progenitors Differentiate to DA Neurons in the Presence of FGF8 and SHH
The neural progenitors, treated with FGF8 before (at day 10) or after (day 18) Sox1 expression, were dissociated and differentiated in a neuronal differentiation medium (Fig. 2A). The enriched neural progenitors were essentially nestin+ (>95%) and did not express SSEA4 (Fig. 2B), a glycoprotein highly expressed by undifferentiated hESCs. The disaggregated neural progenitors, initially distributed evenly, reformed rosettes 3–5 days after plating. They then extended processes and exhibited polar morphology (Fig. 2C). At 3 weeks after differentiation, approximately one third of the total differentiated cells were positive for TH (Fig. 2D; 31.8% ± 3.1% in the early FGF8 treatment group, 17,965 cells counted from four experiments; 29.8% ± 1.6% in the late FGF8 treatment group, 7,954 cells counted from three experiments). All of the TH+ cells were stained positively for a neuronal marker βIII-tubulin (Figs. 3A, 3B), among which 50%–60% were TH+. Morphologically, the TH-expressing neurons in the early FGF8 treatment group were 10–20 µm in diameter. They exhibited multipolar morphology, with differentiable axons and dendrites (Fig. 3A). In the late FGF8 treatment group, the TH+ neurons were relatively uniform, with a size of approximately 8–13 µm. Most of them displayed a bipolar morphology (Fig. 2B). A similar proportion of TH+ neurons was also obtained from the H1 hESC line.
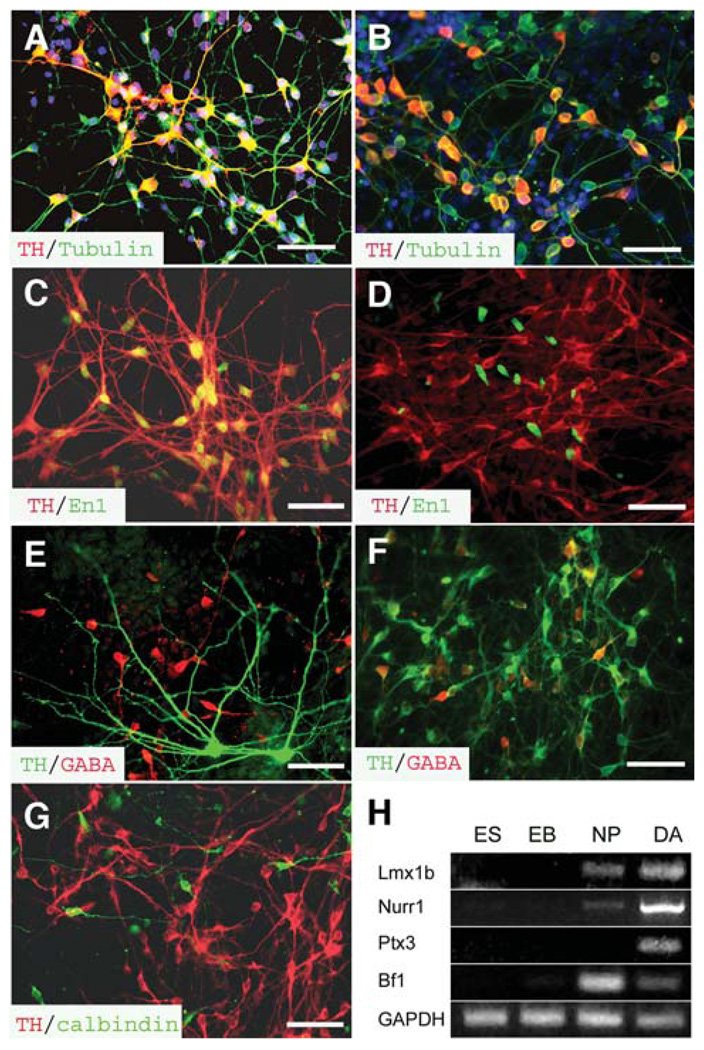
Figure 3.
Characterization of human ES cell–derived DA neurons. (A, B): All TH+ cells (red) in the (A) early and (B) late FGF8-treated cultures were positively stained for βIII-tubulin (green). The TH+ cells exhibited (A) multipolar morphology in the early FGF8 treatment culture and (B) bipolar morphology in the late FGF8 treatment culture. (C, D): Coexpression of TH+ (red) and En1 (green) was observed in (C) the early FGF8 treatment cultures but not in (D) the late FGF8 treatment culture. (E, F): Few TH+ neurons (green) coexpressed GABA (red) in the (E) early FGF8 treatment cultures compared with the (F) late FGF8 treatment group. (G): The TH+ cells (red) in the early FGF8 treatment culture were negative for calbindin (green). The cell nuclei were stained with Hoechst (A and B, blue). Bar = 50 µm. (H): Reverse transcription–polymerase chain reaction analyses indicated the expression of other dopaminergic-related genes when the ES cells were differentiated into DA neurons through EB and NP stages. Abbreviations: DA, dopamine; EB, embryoid body; ES, embryonic stem; FGF, fibroblast growth factor; GABA, γ-aminobutyric acid; NP, neural progenitor; TH, tyrosine hydroxylase.
In the biosynthesis of monoamines, TH hydroxylates tyrosine to levodopa, which is subsequently decarboxylated by AADC to become DA. Two other enzymes, DβH and PNMT, transform DA to norepinephrine and catalyze norepinephrine to epinephrine, respectively. Immunostaining showed that all TH+ neurons were AADC+, although some AADC+ cells were negative for TH (Figs. 2E–2G). These TH+ cells were negative for DβH (Fig. 2D) and PNMT (not shown), although DβH strongly stained noradrenergic neurons in the adult rat and embryonic monkey brainstem (inset in Fig. 2D). These suggest that the TH-expressing neurons, whether they were generated with early or late FGF8 treatment, possess both enzymes that are necessary for DA synthesis and that these neurons are DA neurons rather than noradrenergic or adrenergic neurons.
DA Neurons Produced with FGF8 Treatment Before or After Sox1 Expression Display Different Regional Identities
In addition to morphological difference of the TH+ neurons, immunocytochemical analyses indicated that most TH+ cells with multiple processes in the early FGF8 treatment group coexpressed the midbrain marker En1 in the nuclei (Fig. 3C). In contrast, few TH+ neurons coexpressed En1, although En1 was expressed by some cells in the late FGF8 treatment group (Fig. 3D). Thus, DA neurons generated with the early FGF8 treatment possess a midbrain positional identity.
DA neurons in the forebrain, especially those in the olfactory bulb, often coexpress GABA [2, 3]. Double immunostaining of TH and GABA indicated that most of the DA neurons in the early FGF8 treatment group were negative for GABA, although GABA+ neurons were found in the culture (Fig. 3E). Among all TH+ cells, 8% (8.7% ± 3.9%, 6,520 TH+ cells counted from four experiments) of TH+ cells coexpressed GABA. Most of these double-positive cells were small bipolar cells. However, 22.5% ± 8.9% of the TH+ neurons in the late FGF8 treatment group (5,032 TH+ cells counted from four experiments, p< .05 compared with the early FGF8 treatment group) coexpressed GABA (Fig. 2F). Thus, DA neurons generated in the late FGF8 treatment group are likely forebrain DA neurons.
Some midbrain DA neurons, especially those in the ventral tegmental area, contain CCK8 or calbindin [30, 31]. Immunohistochemical analyses indicated that the TH+ neurons generated after early FGF8 treatment were not labeled with calbindin, although calbindin-expressing neurons were observed (Fig. 3G). These calbindin neurons were mostly small cells. No CCK8-positive cells were detected in the cultures. RT-PCR analyses indicated that the transcription factors, such as Lmx1b, Nurr1, and Ptx3, which are involved in midbrain DA neuron development [4–7, 9, 10, 12], were expressed in the DA neuron cultures that were induced with early FGF8 treatment (Fig. 3H). The forebrain transcription factor Bf1 was detectable in the DA culture (Fig. 3H)
hESC-Generated Midbrain DA Neurons Are Biologically Functional
The above studies indicate that the DA neurons generated after the early neuroepithelial cells are exposed to FGF8 and SHH are likely midbrain DA neurons. Further analyses showed that all of these TH+ neurons expressed c-Ret, a component of the receptor for GDNF (Figs. 4A–4C). Most of the TH+ cells, especially those with branched neurites, expressed VMAT2 (Figs. 4D–4F), which is responsible for packaging DA into subcellular compartments in monoamine neurons [32]. In addition, TH+ neurons expressed synaptophysin (Figs. 4G–4I), a membrane glycoprotein essential to synapse formation.
Figure 4.
Expression of receptors and transporters in the human embryonic stem cell–derived midbrain dopamine neurons. (A–C): All TH+ cells (A, green) expressed c-Ret (B and C, red). (D–F): TH+ cells (D, green) coexpressed VMAT2 (E and F; red). (G–I): TH+ neurons (G, green) expressed synaptophysin (H and I, red). Bar = 25 µm. Abbreviations: TH, tyrosine hydroxylase; VMAT, vesicular monoamine transporter.
DA release is a functional hallmark of DA neurons. HPLC analyses revealed the presence of DA in the medium of DA neuron differentiation cultures (230.8 ± 44.0 pg/ml). In the control cultures without the presence of SHH and FGF8, few TH+ neurons (3%–5%) were generated [21]. DA was detectable in the control cultures but at a lower level (46.3 ± 9.2 pg/ml) (Fig. 5A). When cultured cells were washed and incubated in HBSS for 15 minutes, the DA level was similar between the two cultures (Fig. 5A). However, depolarization of the cultured neurons by 56 mM KCl in HBSS significantly increased the amount of DA (35.8 ± 9.2 and 111.0 ± 15.0 pg/ml in the control and DA differentiation cultures, respectively; Fig. 5A). These observations suggest that the in vitro–generated DA neurons can secrete DA and that the release of DA is activity-dependent.
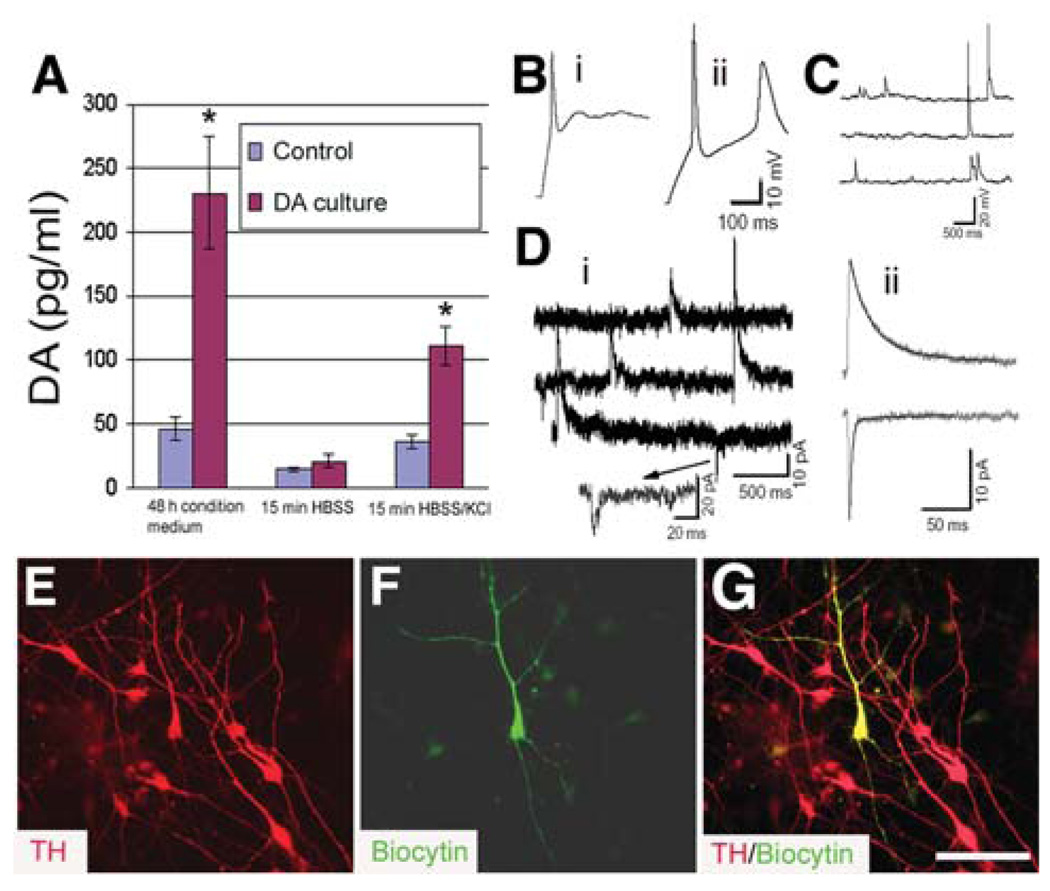
Figure 5.
Functional characteristics of the in vitro–generated midbrain DA neurons. (A): Spontaneous and depolarization (56 mM KCl in HBSS)–induced DA release in the non-DA cultures and the early fibroblast growth factor 8–treated cultures at 3 weeks of differentiation. Data were presented as mean ± SD from three experiments. *p < .05 versus control by the unpaired Student’s t-test. (B): Action potentials evoked by depolarizing current steps (0.2 nA) in two neurons differentiated for 30 days. Passive membrane properties: (i) Vrest −49 mV, Cm 15.5 pF, Rm 5.0 GΩ; (ii) Vrest −72 mV, Cm 45 pF, Rm 885 MΩ. (C): Spontaneous postsynaptic potentials in a neuron differentiated for 36 days. (D): Spontaneous postsynaptic currents in a neuron differentiated for 30 days in culture. The neuron was voltage clamped at −40 mV using a K gluconate–based pipette solution. The outward currents reflect inhibitory events, and the inward currents reflect excitatory events in this low-chloride recording solution. (ii): Averaged events from the cell illustrated in panel (i). The weighted decay time constants are 61.4 ms and 9.9 ms for inhibitory (n = 17 events) and excitatory (n = 14 events) currents, respectively. (E–G): Immunostaining showed that the recorded neuron (F, green) was positive for TH (E and G, red). Bar = 50 µm. Abbreviations: DA, dopamine; HBSS, Hanks’ balanced salt solution; TH, tyrosine hydroxylase.
Electrophysiological recordings were used to determine whether ESC-generated DA neurons were physiologically active. In cells differentiated in culture for 30–38 days (n = 14), the resting membrane potential (Vrest) ranged from −32–−72 mV (−54 ± 2.9 mV), cell capacitance (Cm) ranged from 11–45 pF (21 ± 2.7 pF), and input resistance (Rm) ranged from 480–3,500 MΩ (1,506 ± 282 MΩ). Depolarizing current steps (0.2 nA × 200–500 ms) usually elicited single APs, but in several cases, decrementing trains of APs were observed (Figs. 5Bi, 5Bii). AP threshold ranged from −26–−5.2 mV (−17.4 ± 2.1 mV) and peaked at −9.6–30 mV. APs up to 50.2 mV were observed (32 ± 2.8 mV). AP duration ranged from 3–20.6 ms (7.2 ± 1.3 ms). Spontaneous firing was observed in two cells (Fig. 5C).
In voltage-clamp mode, both inward and outward currents were observed in all cells (not shown), but their relative magnitudes varied considerably. Inward currents were activated rapidly (<1 ms) and peaked within 1–3 ms. Activation threshold was −30 ± 1 mV, maximal peak current amplitude was obtained at a mean voltage of −13 ± 1.9 mV, and currents were completely blocked by TTX (n = 3). These properties are consistent with the presence of voltage-gated sodium channels that underlie AP generation. In three cells, we observed spontaneous transient currents that had the characteristics of synaptic currents, including a rapid rise and slower decay phase. One of these recordings was made with a K-gluconate–based pipette solution, and holding this cell at −40 mV allowed us to observe both outward (inhibitory) and inward (excitatory) currents (Figs. 5Di, 5Dii). Although all 14 cells were injected with biocytin, only five cells were recovered after the completion of the subsequent immunostaining procedures. However, all of the 5 biocytin-filled cells were TH+ (Figs. 5E–5G).
DISCUSSION
Neuronal subtype specification depends on interplay of the intrinsic program of precursor cells and extrinsic morphogens that are present in the surroundings [33]. Although the intrinsic control of dopaminergic fate specification remains to be clarified, FGF8 and SHH play an instructive role [17, 18]. FGF8 and SHH efficiently promote differentiation of DA neurons from primary [13] and ESC-derived neuroepithelia [16, 34, 35]. We have demonstrated in the present study that DA neurons with either midbrain or forebrain characters can be efficiently generated from hESCs in response to a simple administration of FGF8 and SHH. By characterizing the process of neuroepithelial differentiation, we have discovered that it is the time window of FGF8 and SHH application or the intrinsic program of the precursor cells that determines the specification of midbrain versus forebrain DA neurons. Early exposure to FGF8 during the process of neuroepithelial specification directs the precursors preferentially to a midbrain fate and subsequent differentiation to midbrain DA neurons. Exposure of FGF2-expanded neuroepithelial cells to FGF8 and SHH promotes differentiation of DA neurons but mostly to cells with a forebrain phenotype.
Mouse ESCs have been differentiated to neuroepithelial cells, expanded with FGF2, regionalized or specified with FGF8 and SHH, and subsequently differentiated into DA neurons [16]. We hypothesized that the same principle should apply to human primates. Indeed, we are able to generate a large number of DA neurons by differentiating hESCs into neuroepithelial cells that express Sox1 and organize into neural tube–like rosettes in the presence of FGF2 [21], treating the neuroepithelial cells with FGF8 and SHH to induce a ventral midbrain fate and finally differentiating the progenitors to DA neurons. However, most of the DA neurons generated in this way lack some of the key characteristics of midbrain projection DA neurons such as large size with complex morphology and expression of midbrain transcription factors at the protein level. Given that Sox1 is usually expressed by neuroepithelial cells at the neural tube closure stage in animals [25] and that it is expressed by cells in the neural tube–like rosettes that correspond to the neural tube formation stage in human development [20], we reasoned that the Sox1-expressing neuroepithelial cells are regionally specified and are thus less responsive to morphogens for generating DA neurons with midbrain phenotypes.
Our hypothesis that FGF8 may instruct the early precursors to adopt a midbrain identity is confirmed by the efficient induction of its downstream transcription factors, such as En1, Wnt1, Pax2, and Gbx2, all of which are crucial in patterning the mid-hindbrain junction [36, 37], and by the generation of DA neurons that possess characteristics of projection neurons such as large cell bodies with complex processes and expression of midbrain markers. Developmentally, FGF8 is first expressed by epiblasts or ectodermal cells during gastrulation with a proximal boundary around the primitive streak (E6.5 in mouse). After the neural plate/tube forms (E8.5–9), FGF8 is present in the telencephalic commissural plate, the ventral midline of the hypothalamus, and the optic stalks in addition to the mid-hindbrain boundary (isthmus) [38]. Another major event during gastrulation is the formation of the notochord, which produces the ventralizing morphogen SHH. Hence, from a pure mechanical view, this would mean that both FGF8 and SHH are made available first to the ectodermal or perhaps primitive neuroectodermal cells in the mid-hindbrain region. Consequently, these progenitors differentiate into DA neurons with early born, large-projection neuron phenotypes. This is in line with our observation that early application of FGF8 and SHH induces predominantly midbrain DA neurons. Theoretically, SHH needs to be present at the same time. However, we found that early addition of SHH did not increase the production of DA neurons, which may be due to the induction of endogenous SHH by FGF8 at this particular window. Alternatively, but not mutually exclusive, the intrinsic program of the neuroepithelial cells, such as expression of Pax6 but not Sox1, may endow the precursors with the machinery to respond to FGF8 and SHH for production of large-projection DA neurons. This notion parallels our recent finding that only the Pax6+ but not the Sox1+ early neuroepithelial cells can be efficiently instructed to differentiate into spinal motor neurons by retinoic acid and SHH [24]. Therefore, specification of early born projection neurons such as midbrain DA neurons and spinal motor neurons requires morphogen instruction while the neuroectodermal fate is still being determined.
It is presently not clear why FGF2-expanded mouse ESC-derived, but not human ESC-derived, neuroepithelial cells can be efficiently specified to a ventral midbrain fate. There is recent evidence that the dorsal or ventral identity of neural progenitors isolated from mouse spinal cord may be deregulated upon culture, especially in the presence of FGF2 [39], which may partially account for the capability of expanded mouse ESC-derived neuroepithelial cells to be respecified. We have observed that the hESC-derived neuroepithelial cells, after expansion in FGF2, can still generate cells that express En1 but rarely coexpress TH after further differentiation in the presence of SHH and FGF8. This suggests that human neuroepithelial cells may not be as plastic as their mouse counterparts to be regionally respecified. Human neural stem cells retain their regional phenotypes after long-term expansion in the presence of mitogens, including FGF2 [40], whereas those from mouse brain seem to be respecified, at least from the gene expression standpoint [41]. Technically, most differentiation protocols including our present one generate mixed progenies that include undifferentiated or partially differentiated precursors [42]. These precursors may respond to morphogens at a later stage. Besides, FGF2 can also caudalize neuroepithelial cells. Due to a much shorter cell-cycle time in mouse cells, these newly specified progenitors might readily take over the differentiated population, thus masking the true mechanism. In our present study, undifferentiated stem cells are removed during the neuroepithelial enrichment process, which minimizes the chance of the less-differentiated precursors being regionally specified at a later stage.
The differentiation of DA neurons from hESCs has recently been reported by using a coculture system with stromal cells [35, 43, 44] or in an aggregated suspension culture system [45]. The signals associated with stromal cells are strong inducers of neuroectodermal and dopaminergic differentiation [46]. However, the nature of the signals remains unknown. In contrast, the entire differentiation process in our present study is achieved under a chemically defined system. This allows delineating environmental factors that influence cell-fate decision at each individual step along the pathway. It is because of this chemically defined system and the well-characterized neuroectodermal induction process that we are able to differentiate the requirements for production of midbrain versus forebrain DA neurons. From the application standpoint, coculture with cell lines that are often tumorigenic introduces risk components as well as unknown factors, whereas our chemically defined system will allow direct translation to clinical application should new hESC lines be created without contact with animal products.
The early requirement of morphogen action on neuroepithelial cells for production of projection neurons like midbrain DA neurons may explain why stem cells or progenitors expanded from embryonic and adult mammalian brains do not produce projection neurons [47–49]. Our study also indicates a slow maturation process of the human DA neurons in vitro, as revealed by the lack of typical electrophysiological properties of midbrain DA neurons [50] and the poor DA uptake activity (unpublished observation). This may partly reflect the nature of human cells as well as culture environment that lacks targets and further signals important for full maturation. Nonetheless, the electrophysiological properties and the activity-dependent DA release do indicate that the in vitro–generated DA neurons are functional. The ability to efficiently generate functional human DA neurons from a renewable source offers a system for toxicological and pharmaceutical screening for chemicals and drugs that may affect human DA neurons as well as potential cell therapy in the future.
CONCLUSION
By examining the neuroectoderm differentiation process, we have shown that DA neurons with the forebrain or midbrain phenotypes can be differentially specified under a chemical-defined culture system. Although DA phenotype can be specified by exposure to FGF8 and SHH, it is the time window in which precursors are exposed to the morphogens that determines the regional identity. Exposure of the hESC-derived Sox1+ neuroepithelial cells to FGF8 and SHH results in the generation of DA neurons with a forebrain identity such as bipolar morphology, coexpression with GABA, and absence of midbrain marker En1 expression. Early application of FGF8 and SHH during the process of neuroectoderm specification leads to the differentiation of midbrain neuroepithelial cells and subsequent production of midbrain DA neurons, which have large cell bodies and complex processes and express En1. The in vitro–generated DA neurons are biologically functional and may provide a renewable source for developing sustainable therapies for disorders that affect the DA system, such as Parkinson’s disease.
Acknowledgments
This project was supported by The Michael J. Fox Foundation, NIH/NINDS U01 (NS 046587), and partly by a core grant to the Waisman Center from the National Institute of Child Health and Human Development (P30 HD03352). We thank Lorenz Studer for suggestions in HPLC analysis and the Bf1 antibody. We are grateful to Elizabeth Lyons and Patricia Moreno for the help with hESC cultures.
REFERENCES
- 1.Bjorklund A, Lindvall O. Dopamine-containing systems in the CNS. In: Bjorklund A, Hokfelt T, editors. Handbook of Chemical Neuroanatomy. Vol. 2. Amsterdam: Elsevier Science publishers; 1984. pp. 55–111. Classical Transmitters in the CNS. [Google Scholar]
- 2.Gall CM, Hendry SH, Seroogy KB, et al. Evidence for coexistence of GABA and dopamine in neurons of the rat olfactory bulb. J Comp Neurol. 1987;266:307–318. doi: 10.1002/cne.902660302. [DOI] [PubMed] [Google Scholar]
- 3.Kosaka T, Kosaka K, Hataguchi Y, et al. Catecholaminergic neurons containing GABA-like and/or glutamic acid decarboxylase-like immunore-activities in various brain regions of the rat. Exp Brain Res. 1987;66:191–210. doi: 10.1007/BF00236215. [DOI] [PubMed] [Google Scholar]
- 4.Zetterstrom RH, Solomin L, Jansson L, et al. Dopamine neuron agenesis in Nurr1-deficient mice. Science. 1997;276:248–250. doi: 10.1126/science.276.5310.248. [DOI] [PubMed] [Google Scholar]
- 5.Smidt MP, van Schaick HS, Lanctot C, et al. A homeodomain gene Ptx3 has highly restricted brain expression in mesencephalic dopaminergic neurons. Proc Natl Acad Sci U S A. 1997;94:13305–13310. doi: 10.1073/pnas.94.24.13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saucedo-Cardenas O, Quintana-Hau JD, Le WD, et al. Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc Natl Acad Sci U S A. 1998;95:4013–4018. doi: 10.1073/pnas.95.7.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smidt MP, Asbreuk CH, Cox JJ, et al. A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat Neurosci. 2000;3:337–341. doi: 10.1038/73902. [DOI] [PubMed] [Google Scholar]
- 8.Simon HH, Saueressig H, Wurst W, et al. Fate of midbrain dopaminergic neurons controlled by the engrailed genes. J Neurosci. 2001;21:3126–3134. doi: 10.1523/JNEUROSCI.21-09-03126.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nunes I, Tovmasian LT, Silva RM, et al. Pitx3 is required for development of substantia nigra dopaminergic neurons. Proc Natl Acad Sci U S A. 2003;100:4245–4250. doi: 10.1073/pnas.0230529100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van den Munckhof P, Luk KC, Ste-Marie L, et al. Pitx3 is required for motor activity and for survival of a subset of midbrain dopaminergic neurons. Development. 2003;130:2535–2542. doi: 10.1242/dev.00464. [DOI] [PubMed] [Google Scholar]
- 11.Alberi L, Sgado P, Simon HH. Engrailed genes are cell-autonomously required to prevent apoptosis in mesencephalic dopaminergic neurons. Development. 2004;131:3229–3236. doi: 10.1242/dev.01128. [DOI] [PubMed] [Google Scholar]
- 12.Smidt MP, Smits SM, Bouwmeester H, et al. Early developmental failure of substantia nigra dopamine neurons in mice lacking the homeodomain gene Pitx3. Development. 2004;131:1145–1155. doi: 10.1242/dev.01022. [DOI] [PubMed] [Google Scholar]
- 13.Ye W, Shimamura K, Rubenstein JL, et al. FGF and Shh signals control dopaminergic and serotonergic cell fate in the anterior neural plate. Cell. 1998;93:755–766. doi: 10.1016/s0092-8674(00)81437-3. [DOI] [PubMed] [Google Scholar]
- 14.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 15.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SH, Lumelsky N, Studer L, et al. Efficient generation of midbrain and hindbrain neurons from mouse embryonic stem cells. Nat Biotechnol. 2000;18:675–679. doi: 10.1038/76536. [DOI] [PubMed] [Google Scholar]
- 17.Hynes M, Rosenthal A. Specification of dopaminergic and serotonergic neurons in the vertebrate CNS. Curr Opin Neurobiol. 1999;9:26–36. doi: 10.1016/s0959-4388(99)80004-x. [DOI] [PubMed] [Google Scholar]
- 18.Goridis C, Rohrer H. Specification of catecholaminergic and serotonergic neurons. Nat Rev Neurosci. 2002;3:531–541. doi: 10.1038/nrn871. [DOI] [PubMed] [Google Scholar]
- 19.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 20.Zhang SC. Embryonic stem cells for neural replacement therapy: prospects and challenges. J Hematother Stem Cell Res. 2003;12:625–634. doi: 10.1089/15258160360732650. [DOI] [PubMed] [Google Scholar]
- 21.Zhang SC, Wernig M, Duncan ID, et al. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 22.Hamill OP, Marty A, Neher E, et al. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Arch. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- 23.Neher E. Correction for liquid junction potentials in patch clamp experiments. Methods Enzymol. 1992;207:123–131. doi: 10.1016/0076-6879(92)07008-c. [DOI] [PubMed] [Google Scholar]
- 24.Li XJ, Du ZW, Zarnowska E, et al. Specification of motoneurons from human embryonic stem cells. Nat Biotechnol. 2005;23:215–221. doi: 10.1038/nbt1063. [DOI] [PubMed] [Google Scholar]
- 25.Pevny LH, Sockanathan S, Placzek M, et al. A role for SOX1 in neural determination. Development. 1998;125:1967–1978. doi: 10.1242/dev.125.10.1967. [DOI] [PubMed] [Google Scholar]
- 26.Tao W, Lai E. Telencephalon-restricted expression of BF-1, a new member of the HNF-3/fork head gene family, in the developing rat brain. Neuron. 1992;8:957–966. doi: 10.1016/0896-6273(92)90210-5. [DOI] [PubMed] [Google Scholar]
- 27.Davidson D, Graham E, Sime C, et al. A gene with sequence similarity to Drosophila engrailed is expressed during the development of the neural tube and vertebrae in the mouse. Development. 1988;104:305–316. doi: 10.1242/dev.104.2.305. [DOI] [PubMed] [Google Scholar]
- 28.Wurst W, Bally-Cuif L. Neural plate patterning: upstream and downstream of the isthmic organizer. Nat Rev Neurosci. 2001;2:99–108. doi: 10.1038/35053516. [DOI] [PubMed] [Google Scholar]
- 29.Lumsden A, Krumlauf R. Patterning the vertebrate neuraxis. Science. 1996;274:1109–1115. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- 30.Hokfelt T, Skirboll L, Rehfeld JF, et al. A subpopulation of mesencephalic dopamine neurons projecting to limbic areas contains a cholecystokinin-like peptide: evidence from immunohistochemistry combined with retrograde tracing. Neuroscience. 1980;5:2093–2124. doi: 10.1016/0306-4522(80)90127-x. [DOI] [PubMed] [Google Scholar]
- 31.McRitchie DA, Hardman CD, Halliday GM. Cytoarchitectural distribution of calcium binding proteins in midbrain dopaminergic regions of rats and humans. J Comp Neurol. 1996;364:121–150. doi: 10.1002/(SICI)1096-9861(19960101)364:1<121::AID-CNE11>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 32.Nirenberg MJ, Chan J, Liu Y, et al. Ultrastructural localization of the vesicular monoamine transporter-2 in midbrain dopaminergic neurons: potential sites for somatodendritic storage and release of dopamine. J Neurosci. 1996;16:4135–4145. doi: 10.1523/JNEUROSCI.16-13-04135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 34.Barberi T, Klivenyi P, Calingasan NY, et al. Neural subtype specification of fertilization and nuclear transfer embryonic stem cells and application in parkinsonian mice. Nat Biotechnol. 2003;21:1200–1207. doi: 10.1038/nbt870. [DOI] [PubMed] [Google Scholar]
- 35.Perrier AL, Tabar V, Barberi T, et al. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci U S A. 2004;101:12543–12548. doi: 10.1073/pnas.0404700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu A, Joyner AL. Early anterior/posterior patterning of the midbrain and cerebellum. Annu Rev Neurosci. 2001;24:869–896. doi: 10.1146/annurev.neuro.24.1.869. [DOI] [PubMed] [Google Scholar]
- 37.Liu A, Joyner AL. EN and GBX2 play essential roles downstream of FGF8 in patterning the mouse mid/hindbrain region. Development. 2001;128:181–191. doi: 10.1242/dev.128.2.181. [DOI] [PubMed] [Google Scholar]
- 38.Crossley PH, Martin GR. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- 39.Gabay L, Lowell S, Rubin LL, et al. Deregulation of dorsoventral patterning by FGF confers trilineage differentiation capacity on CNS stem cells in vitro. Neuron. 2003;40:485–499. doi: 10.1016/s0896-6273(03)00637-8. [DOI] [PubMed] [Google Scholar]
- 40.Ostenfeld T, Joly E, Tai YT, et al. Regional specification of rodent and human neurospheres. Brain Res Dev Brain Res. 2002;134:43–55. doi: 10.1016/s0165-3806(01)00291-7. [DOI] [PubMed] [Google Scholar]
- 41.Hitoshi S, Tropepe V, Ekker M, et al. Neural stem cell lineages are regionally specified, but not committed, within distinct compartments of the developing brain. Development. 2002;129:233–244. doi: 10.1242/dev.129.1.233. [DOI] [PubMed] [Google Scholar]
- 42.Du ZW, Zhang SC. Neural differentiation from embryonic stem cells: which way? Stem Cells Dev. 2004;13:372–381. doi: 10.1089/scd.2004.13.372. [DOI] [PubMed] [Google Scholar]
- 43.Buytaert-Hoefen KA, Alvarez E, Freed CR. Generation of tyrosine hydroxylase positive neurons from human embryonic stem cells after coculture with cellular substrates and exposure to GDNF. Stem Cells. 2004;22:669–674. doi: 10.1634/stemcells.22-5-669. [DOI] [PubMed] [Google Scholar]
- 44.Zeng XM, Cai JL, Chen J, et al. Dopaminergic differentiation of human embryonic stem cells. Stem Cells. 2004;22:925–940. doi: 10.1634/stemcells.22-6-925. [DOI] [PubMed] [Google Scholar]
- 45.Schulz TC, Noggle SA, Palmarini GM, et al. Differentiation of human embryonic stem cells to dopaminergic neurons in serum-free suspension culture. Stem Cells. 2004;22:1218–1238. doi: 10.1634/stemcells.2004-0114. [DOI] [PubMed] [Google Scholar]
- 46.Morizane A, Takahashi J, Takagi Y, et al. Optimal conditions for in vivo induction of dopaminergic neurons from embryonic stem cells through stromal cell-derived inducing activity. J Neurosci Res. 2002;69:934–939. doi: 10.1002/jnr.10363. [DOI] [PubMed] [Google Scholar]
- 47.Svendsen CN, Caldwell MA, Shen J, et al. Long-term survival of human central nervous system progenitor cells transplanted into a rat model of Parkinson’s disease. Exp Neurol. 1997;148:135–146. doi: 10.1006/exnr.1997.6634. [DOI] [PubMed] [Google Scholar]
- 48.Daadi MM, Weiss S. Generation of tyrosine hydroxylase-producing neurons from precursors of the embryonic and adult forebrain. J Neurosci. 1999;19:4484–4497. doi: 10.1523/JNEUROSCI.19-11-04484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Storch A, Paul G, Csete M, et al. Long-term proliferation and dopaminergic differentiation of human mesencephalic neural precursor cells. Exp Neurol. 2001;170:317–325. doi: 10.1006/exnr.2001.7706. [DOI] [PubMed] [Google Scholar]
- 50.Grace AA, Onn SP. Morphology and electrophysiological properties of immunocytochemically identified rat dopamine neurons recorded in vitro. J Neurosci. 1989;9:3463–3481. doi: 10.1523/JNEUROSCI.09-10-03463.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]