Abstract
Analysis of variants in three genes encoding oxysterol-binding protein (OSBP) homologues (OSBPL2, OSBPL9, OSBPL10) in Finnish families with familial low high-density lipoprotein (HDL) levels (N = 426) or familial combined hyperlipidemia (N = 684) revealed suggestive linkage of OSBPL10 single-nucleotide polymorphisms (SNPs) with extreme end high triglyceride (TG; >90th percentile) trait. Prompted by this initial finding, we carried out association analysis in a metabolic syndrome subcohort (Genmets) of Health2000 examination survey (N = 2,138), revealing association of multiple OSBPL10 SNPs with high serum TG levels (>95th percentile). To investigate whether OSBPL10 could be the gene underlying the observed linkage and association, we carried out functional experiments in the human hepatoma cell line Huh7. Silencing of OSBPL10 increased the incorporation of [3H]acetate into cholesterol and both [3H]acetate and [3H]oleate into triglycerides and enhanced the accumulation of secreted apolipoprotein B100 in growth medium, suggesting that the encoded protein ORP10 suppresses hepatic lipogenesis and very-low-density lipoprotein production. ORP10 was shown to associate dynamically with microtubules, consistent with its involvement in intracellular transport or organelle positioning. The data introduces OSBPL10 as a gene whose variation may contribute to high triglyceride levels in dyslipidemic Finnish subjects and provides evidence for ORP10 as a regulator of cellular lipid metabolism.
Electronic supplementary material
The online version of this article (doi:10.1007/s00109-009-0490-z) contains supplementary material, which is available to authorized users.
Keywords: Cholesterol, High-density lipoprotein, Microtubule, Oxysterol-binding protein, Single-nucleotide polymorphism, Triglyceride
Introduction
Families of proteins with homology to the carboxy-terminal ligand binding domain of oxysterol binding protein (OSBP) are suggested to function in various cellular processes such as lipid metabolism, intracellular lipid transport, membrane trafficking, and cell signaling [1, 2]. In humans, the gene family consists of 12 members, which are denoted OSBP-like genes (OSBPL), while the encoded proteins are called either OSBP-like (OSBPL) or OSBP-related proteins (ORP) [3, 4].
OSBP is the most extensively studied member of the mammalian ORP protein family. OSBP acts as a sterol sensor that integrates the cellular sterol status with sphingomyelin metabolism [5] and controls the dephosphorylation and hence the activity of extracellular signal-regulated kinases, ERK [6]. We recently showed that OSBP overexpression in mouse liver impacts serum very-low-density lipoprotein triglyceride (TG) levels and hepatic TG synthesis through up-regulation of the expression and activity of sterol regulatory element binding protein 1c (SREBP-1c) [7], a major insulin-responsive regulator of hepatic and adipose tissue lipogenesis. Over-expression of human ORP1L in mouse macrophages resulted in impaired cholesterol efflux and increased size of atherosclerotic lesions in low-density lipoprotein (LDL)-receptor deficient mice [8]. Furthermore, our latest data demonstrates a role of the OSBP homologue ORP8 as factor that suppresses, most likely via an indirect mechanism, the expression of ATP-binding cassette transporter A1 (ABCA1) [9], a key mediator of high-density lipoprotein (HDL) biogenesis. Moreover, ORP2, ORP4S, and ORP9 have been implicated in cellular sterol or neutral lipid metabolism [10–12], lending further support to the view that ORP family members play important roles in cellular lipid metabolism.
In the present study, we assessed whether allelic diversity of the OSBPL2, OSBPL9, or OSBPL10 genes contributes to extreme serum lipid levels in Finnish families ascertained for low HDL-cholesterol or familial combined hyperlipidemia (FCHL, common dyslipidemias associated with increased risk of premature coronary heart disease [13–16]. Variants of OSBPL10 displayed suggestive linkage with high triglyceride levels in the families, and association analysis in a metabolic syndrome study sample (N = 2,138) strengthened the notion that variation in OSBPL10 may influence serum TG levels in Finnish dyslipidemic subjects. RNA interference experiments in cultured human hepatoma cells suggested that the encoded protein, ORP10, acts as a regulator of cellular lipid biosyntheses and apolipoprotein B-100 (apoB100) secretion, lending functional support to the genetic findings.
Materials and methods
Dyslipidemic cohorts and genotyping strategy
The FCHL families (53 families, 684 subjects) were recruited through the Helsinki, Turku, and Kuopio University Central Hospitals and the inclusion/exclusion criteria for the probands have been described [17]. The low-HDL families (39 families, 426 subjects) were collected in the Helsinki and Turku University Hospitals, and probands were required to have HDL-cholesterol (HDL-C) levels <10th age- and sex-specific percentile of the Finnish population and angiographically verified coronary heart disease and are more fully described in [18]. Phenotypic information on the study subjects and the distributions of their serum HDL-C and TG concentrations are displayed in Table 1 and in Electronic Supplementary Materials Fig. 1, respectively. DNA extraction, single-nucleotide polymorphism (SNP) genotyping, and data analysis were carried out as described in Electronic Supplementary Materials method descriptions. Detailed information on the SNPs analyzed is displayed in Electronic Supplementary Materials Table 1 and in Fig. 1.
Table 1.
Phenotypic characteristics of the low-HDL and FCHL study samples
| Trait | Males | Females | Total | |
|---|---|---|---|---|
| Low-HDL | N | 327 | 338 | 665 |
| Agea (years) | 45.30 ± 22.08 | 45.61 ± 20.5 | 45.47 ± 21.2 | |
| BMI (kg/m2) | 25.47 ± 4.25 | 24.76 ± 4.66 | 25.09 ± 4.5 | |
| TC (mmol/l) | 5,40 ± 1.16 | 5,47 ± 1.07 | 5,44 ± 1.11 | |
| HDL (mmol/l) | 1.02 ± 0.3 | 1.35 ± 0.38 | 1.20 ± 0.38 | |
| TG (mmol/l) | 1.59 ± 0.8 | 1.45 ± 0.79 | 1.51 ± 0.8 | |
| FCHL | N | 687 | 632 | 1,319 |
| Age (years) | 37.60 ± 20.44 | 37.91 ± 20.35 | 37.7 ± 20.4 | |
| BMI (kg/m2) | 25.56 ± 4.46 | 24.90 ± 5.29 | 25.22 ± 5.02 | |
| TC (mmol/l) | 5.84 ± 1.43 | 5.85 ± 1.32 | 5.84 ± 1.37 | |
| HDL (mmol/l) | 1.15 ± 0.36 | 1.39 ± 0.41 | 1.27 ± 0.41 | |
| TG (mmol/l) | 2.11 ± 1.88 | 1.59 ± 1.27 | 1.85 ± 1.6 |
The data represent mean ± SD
BMI body mass index, TC total cholesterol, HDL high-density lipoprotein cholesterol, TG triglycerides
aSubjects under the age of 20 years were excluded
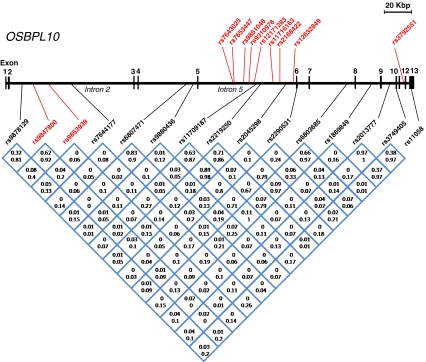
Fig. 1.
Location and linkage disequilibrium (LD) relationships of the investigated SNPs in OSBPL10. Exons 1–13 as well as introns 2 and 5 are indicated. The SNPs analyzed in the low-HDL and FCHL family materials are shown below the gene structure; those with the highest LOD scores (>2.0) for linkage with high-TG trait are indicated in red. The LD relationships between the SNPs are depicted at the bottom; Numbers represent r2, the top one for FCHL and the bottom one for low-HDL families. The SNPs showing strongest association (p < 0.01) with high TG in the Genmets data set are displayed (in red) above the gene structure
The Genmets sample is subset of 2,212 individuals of the Health2000 study (http://www.terveys2000.fi/doc/methodologyrep.pdf) collected as metabolic syndrome cases and their matched controls. A total of 2,173 of these individuals have been genotyped on Illumina 610K arrays. In this study, 102 SNPs located in OSBPL10 or its flanking regions were analyzed. For the association analysis, individuals with the TG levels above the 95th population, sex, and age-specific percentile (>3.4 mmol/l) were coded as cases and the rest as controls. The same coding was done for the individuals with the high-density lipoprotein cholesterol values below the 5th percentile (<0.8 mmol/l). For these two dichotomous phenotypes, logistic regression was applied with age and sex as covariates (for details, see Electronic Supplementary Materials method descriptions).
The study materials were collected according to the Helsinki declaration, and the ethics committees of the participating centers approved the study design.
Generation of a rabbit antibody against ORP10
A cDNA fragment encoding amino acid residues 1–80 of the full-length ORP10 protein (NM_017784) was inserted in pGEX1λT (GE Healthcare, Uppsala, Sweden) for production of a glutathione-S-transferase fusion protein in Escherichia coli BL21. The protein was purified on Glutathione-Sepharose 4B (GE Healthcare) and used for subcutaneous immunization of New Zealand white rabbits by a standard procedure.
Cell culture
The human hepatoma cell line Huh7 was cultured in Eagle’s minimal essential medium with Earle’s salts (EMEM, Sigma-Aldrich, St. Louis, MO, USA), 20 mM Hepes, pH 7.4, 10% fetal bovine serum (FBS; Gibco/Invitrogen, Grand Island, NY, USA), 100 U/ml penicillin, and 100 μg/ml streptomycin. The pancreatic adenocarcinoma cell line PANC-1 was cultured in RPMI 1640 (BioWhittaker, Walkersville, MD, USA), 10% FBS, and the above antibiotics. Caco-2 cells were cultured in EMEM supplemented with 10% FBS, non-essential amino acids, and the above antibiotics.
RNA interference
Huh7 cells were transfected with ORP10-specific (siORP10.1, sense strand CCACAGCCUCAAUCUUGUAdTdT; siORP10.2, GAGAAUUUCCUGUGGAU-UAdTdT) or nontargeting control siRNA (siNT, sense strand UAGCGACUAAACACAUCAAdTdT) using INTERFERin (Polyplus, Illkirch, France) or HiPerFect (Qiagen, Valencia, CA, USA) according to the manufacturers’ instructions. After 48 h transfection, the cells were either subjected to total RNA isolation by using the RNeasy Mini kit (Qiagen) or used for analysis of cholesterol and triglyceride biosynthesis.
Analysis of cholesterol and triglyceride biosynthesis
Huh7 cells treated with ORP10-specific or nontargeting control siRNAs were labeled with [3H]acetic acid or [3H]oleic acid, and incorporation of the radioactive tracers into cholesterol or TG was carried out, as specified in Electronic Supplementary Materials method descriptions.
Assays for apoA-I and apoB100 secretion by Huh7 cells
Huh7 cells treated for 48 h with nontargeting or ORP10-specific siRNAs as specified above were washed with phosphate-buffered saline and transferred into serum-free culture medium. The medium and the cells were harvested at 6, 12, and 24 h. The apoA-I [19] and apoB100 (Mabtech, Nacka Strand, Sweden) concentrations in the medium were determined with specific sandwich enzyme-linked immunosorbent assay (ELISA) assays and normalized for total cell protein.
mRNA quantification
Total RNA was isolated from Huh7 cells treated with ORP10 or control siRNAs, and quantitative real-time polymerase chain reaction (PCR) analysis of specific mRNAs was carried out using SYBR-green (Applied Biosystems) as described in Electronic Supplementary Materials method descriptions. The primers used are specified in Electronic Supplementary Materials Table 2.
Assay for 3-hydroxy-3-methylglutaryl coenzyme A reductase activity
Huh7 cells treated with ORP10-specific or control siRNAs as specified above were harvested, and 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase activity in total membranes of the cells was assayed as detailed in Electronic Supplementary Materials method descriptions.
Analysis of the subcellular localization of ORP10
The human ORP10 open reading frame (NM_017784) was subcloned into the pEGFP-C (BD Biosciences Clontech, Palo Alto, CA, USA) or pcDNA4HisMax (Invitrogen) vectors for mammalian cell expression. Huh7, PANC-1, or Caco-2 cells cultured in normal serum-containing growth medium were transfected for 24 h using Lipofectamine 2000 (Invitrogen), fixed with paraformaldehyde and processed for immunofluorescence microscopy essentially as described previously [20]. The immunofluorescence double stainings carried out, and pretreatments of the cells are described in Electronic Supplementary Materials method descriptions. Fluorescence recovery after photobleaching (FRAP) in Huh7 cells transfected with EGFP-ORP10 was carried out as detailed in Electronic Supplementary Materials method descriptions.
Results
Single-nucleotide polymorphisms in OSBPL10 show suggestive linkage with serum triglyceride levels
We used carefully phenotyped Finnish families ascertained for low HDL or FCHL to investigate the contribution of variation in OSBPL genes to extreme ends of lipid traits. Phenotypic information on the study subjects is summarized in Table 1, and the distributions of HDL and TG measurements used as traits in the genetic analyses are depicted in Electronic Supplementary Materials Fig. 1. Genotyping of tag SNPs of OSBPL2, -9, and -10 in 39 families (N = 426 genotyped individuals) with low-HDL and 53 Finnish FCHL families (N = 684 genotyped individuals) revealed indications of linkage between multiple SNPs in OSBPL10 and the status for high TG, which was treated as an affection status and defined as exceeding the population sex- and age-specific 90th percentile (Table 2). The indications of linkage emerged mostly in the FCHL families. No evidence for linkage or association could be observed for SNPs of OSBPL2 and -9, so they were excluded from further studies (data not shown). Combining the study materials (92 pedigrees, N = 1,110 individuals) increased the linkage logarithm of the odds (LOD) scores for a number of SNP variants with the high-TG trait (Table 2). The strongest evidence for linkage with the high TG trait was observed for SNP variants rs9853939 (LOD 2.40) and rs9847800 (LOD 2.07), located in intron 2 of OSBPL10 (Fig. 1).
Table 2.
Linkage and association results for the dichotomized HDL-cholesterol and TG traits in the low-HDL and FCHL families
| Low-HDL | FCHL | Combined | ||||
|---|---|---|---|---|---|---|
| SNP (OSBPL10) | <10th %a HDL | >90th % TG | <10th % HDL | >90th % TG | <10th % HDL | >90th % TG |
| rs9878129 | 0.02 (1)b | 0.00 (0.07) | 0.25 (1) | 0.78c(0.53) | 0.26 (0.24) | 0.71 (0.77) |
| rs9847800 | 0.43 (0.22) | 0.15 (0.53) | 0.06 (0.54) | 1.36 (0.23) | 0.80 (0.21) | 2.07 (0.50) |
| rs9853939 | 0.66 (0.19) | 0.00 (0.89) | 0.09 (0.49) | 1.79 (0.82) | 0.98 (0.21) | 2.40 (1) |
| rs7644177 | 0.01 (0.42) | 0.18 (0.0005) | 0.01 (0.74) | 0.12 (0.64) | 0.23 (0.84) | 0.16 (0.03) |
| rs6807471 | 0.40 (0.92) | 0.00 (0.69) | 0.54 (0.26) | 0.01 (0.14) | 0.34 (0.44) | 0.00 (0.39) |
| rs9880436 | 0.00 (0.67) | 0.36 (0.60) | 0.59 (0.07) | 0.50 (0.03) | 0.46 (0.33) | 0.80 (0.14) |
| rs11709187 | 0.16 (0.17) | 0.00 (0.56) | 0.08 (0.84) | 0.28 (0.03) | 0.23 (0.32) | 0.22 (0.08) |
| rs2219250 | 0.55 (1) | 0.08 (0.75) | 0.74 (0.79) | 0.20 (0.88) | 0.94 (0.84) | 0.41 (1) |
| rs2045298 | 0.04 (0.14) | 0.00 (0.55) | 0.04 (0.79) | 0.51 (0.03) | 0.09 (0.47) | 0.38 (0.07) |
| rs2290531 | 0.13 (0.6) | 0.02 (0.63) | 0.00 (0.69) | 0.65 (0.84) | 0.00 (0.75) | 1.22 (0.92) |
| rs6803685 | 0.49 (0.32) | 0.77 (0.04) | 0.03 (0.34) | 0.01 (0.72) | 0.52 (0.20) | 0.10 (0.25) |
| rs1869849 | 0.72 (1) | 0.35 (0.29) | 0.04 (0.76) | 0.10 (0.11) | 0.52 (0.77) | 0.12 (0.75) |
| rs2013777 | 0.65 (0.53) | 0.00 (0.92) | 0.01 (0.09) | 0.01 (0.48) | 0.03 (0.21) | 0.05 (0.37) |
| rs3749405 | 0.91 (0.54) | 0.00 (0.92) | 0.01 (0.14) | 0.01 (0.37) | 0.00 (0.19) | 0.004 (0.36) |
| rs11058 | 0.00 (0.14) | 0.00 (0.58) | 0.00 (0.78) | 0.45 (0.15) | 0.00 (0.57) | 0.12 (0.13) |
| NAffected | 167 | 46 | 255 | 205 | 422 | 251 |
aThe percentiles used to diagnose represent age- and sex-specific population cut-off values
bThe first value represents the LOD score, the number in parentheses represents p value for association, given linkage
cLOD scores >0.7 and p values <0.05 are indicated in bold
We next studied whether the SNP alleles with the best evidence for linkage with the extreme end of TG concentration show correlation with the OSBPL10 mRNA expression level in fat biopsies withdrawn from 47 individuals belonging to the low-HDL and FCHL families, using transcriptome data acquired with Affymetrix U133 Plus 2.0 microarrays. No significant correlation was detected (data not shown), suggesting that rather than affecting the OSBPL10 mRNA levels, the SNPs may be in linkage disequilibrium (LD) with a yet unidentified functional variant.
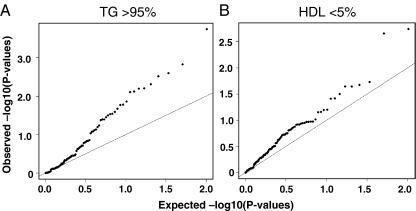
Prompted by the suggestive linkage observation, we moved on to analyze the extreme ends of the HDL and TG distribution in a subcohort (Genmets) of the Health2000 population cohort, consisting of subjects with metabolic syndrome and matched healthy controls. Similar to the analysis done in the family sample, in this data set (N = 2,138) individuals with the TG above the 95th percentile were coded as cases and the rest as controls. A corresponding dichotomous division was made using HDL cholesterol <5th percentile as the cut-off. The analysis revealed association of multiple SNPs (21 SNPs with p < 0.05, nine SNPs with p < 0.01) in OSBPL10 or its flanking regions with high TG levels (Fig. 2a, Table 3). Also, a tendency of association with low HDL was observed, but this failed to reach statistical significance after multiple testing correction (Fig. 2b). The SNPs showing strongest association with high TG in the Genmets data set were, with the exception of one, located in OSBPL10 intron 5 and contained within a single LD block (Fig. 1).
Fig. 2.
Association of OSBPL10 SNPs with high TG (>95th percentile) of low HDL (<5th percentile) in the Genmets study material. Expected and observed p values are depicted. The axes represent −log10 of the p values, the straight line indicating the expected p values assuming no association
Table 3.
Genmets study material: OSBPL10 SNPs associated with high TG (>95th percentile), p < 0.01
| SNP | Position, bp | Effect allele | Effect all. frequency | Odds ratio | P-value |
|---|---|---|---|---|---|
| rs11716163 | 31782580 | T | 0.2633 | 1.771 | 0.000179 |
| rs7653447 | 31814139 | A | 0.3383 | 0.5882 | 0.001484 |
| rs2168422 | 31779846 | T | 0.2921 | 0.5827 | 0.002511 |
| rs9851048 | 31806124 | T | 0.3324 | 0.6091 | 0.003025 |
| rs7643025 | 31814246 | A | 0.2527 | 0.5911 | 0.004931 |
| rs12171283 | 31797341 | G | 0.2659 | 0.6077 | 0.00621 |
| rs9310978 | 31801771 | T | 0.2656 | 0.6085 | 0.006334 |
| rs3792551 | 31681904 | T | 0.09962 | 0.4152 | 0.007563 |
| rs12632849 | 31767922 | T | 0.3582 | 1.464 | 0.00786 |
ORP10 modulates hepatocellular lipogenesis
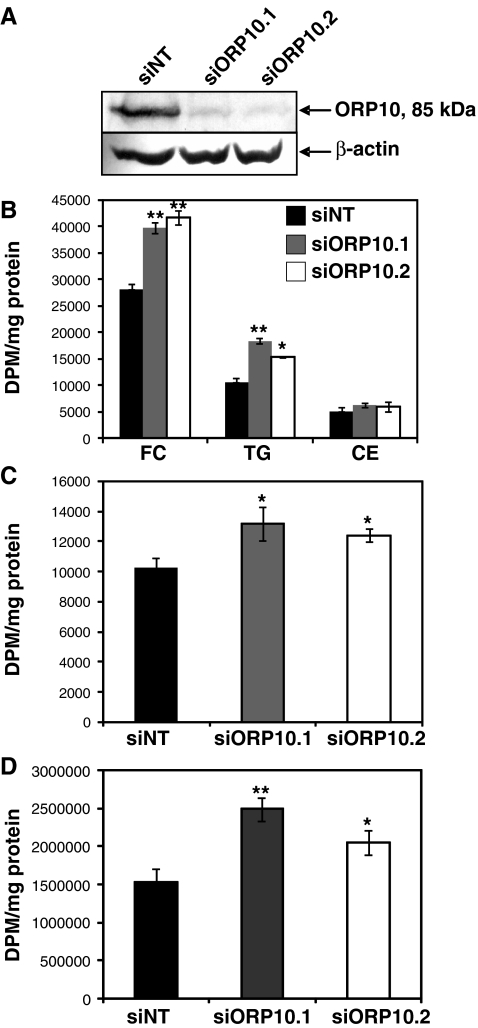
We employed siRNA-mediated silencing of ORP10 expression in a human hepatoma cell line Huh7 to investigate the role of ORP10 in hepatocellular lipid biosyntheses. Transfection of the two independent siRNAs used, siORP10.1 and siORP10.2, resulted in approximately 90% reduction of the endogenous Huh7 cell ORP10 protein (Fig. 3a). To assess the role of ORP10 in hepatocellular cholesterol biosynthesis, we determined the incorporation of [3H]acetic acid into cholesterol in cells transfected with scrambled control or ORP10-specific siRNAs. Silencing of ORP10 resulted in a significant, 40–50% increase of [3H]free cholesterol and a mild elevation of [3H]cholesterol esters, suggesting enhancement of cholesterol biosynthetic activity (Fig. 3b). We also quantified from the same experiments the [3H]radioactivity incorporated into TG, revealing a significant elevation of [3H]TG in cells treated with the ORP10 siRNAs (Fig. 3b).
Fig. 3.
Silencing of ORP10 enhances cholesterol and triglyceride biosynthesis in Huh7 cells. a Western blot analysis of ORP10 protein level in cells transfected for 48 h with nontargeting control (siNT) or ORP10-specific siRNAs (siORP10.1, siORP10.2) using INTERFERin. Anti-β-actin was used as a control for equal protein loading. The apparent molecular mass of the protein detected by the antibody (85 kDa) matches the ORP10 mass deduced from the cDNA sequence (83.97 kDa). b Incorporation of [3H]acetate into free cholesterol (FC), cholesteryl esters (CE), and triglycerides (TG) in cells treated with control or ORP10-specific siRNAs (N = 4). c HMG-CoA reductase activity (expressed as [14C]HMG-CoA incorporation into mevalonic acid, DMP per milligrams membrane protein) in cells treated with control or ORP10-specific siRNAs (N = 4). d Incorporation of [3H]oleic acid into triglycerides (TG) in cells treated with control or ORP10-specific siRNAs (N = 4). The results represent mean ± s.e.m. Statistical significance of the differences between values obtained for siNT and the ORP10-specific siRNAs was evaluated using Student’s t test; *p < 0.05, **p < 0.01
To investigate whether the elevation of [3H]cholesterol biosynthesis in Huh7 cells subjected to ORP10 silencing reflects a change in the activity of the rate-limiting enzyme 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase, we determined in vitro the activity of this enzyme in membranes isolated from cells treated with ORP10-specific or control siRNAs. Incorporation of [14C]HMG-CoA into mevalonate was significantly (siORP10.1, 29%; siORP10.2, 21%) enhanced in cells subjected to ORP10 silencing as compared to those treated with the nontargeting control siRNA (Fig. 3c). Furthermore, to test if the effect of OSBPL10 silencing on TG biosynthesis occurs at the level of fatty acid biosynthesis or at the level of their activation or incorporation into TG, we assessed TG synthesis using [3H]oleic acid as the radioactive precursor. The results revealed enhancement in its incorporation into TG in cells subjected to ORP10 silencing, similar in its extent to the effect observed upon [3H]acetic acid labeling (Fig. 3d).
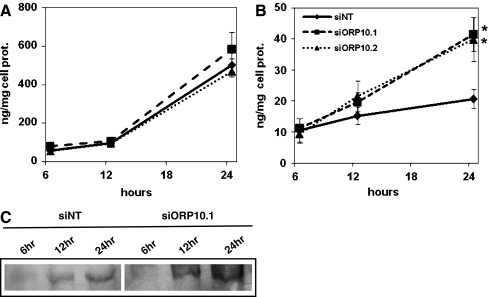
To investigate whether ORP10 might impact lipoprotein secretion by hepatic cells, we transferred Huh7 cells subjected to ORP10 silencing into serum-free medium and quantified, using specific ELISA assays, the secretion of apoA-I and apoB100. During a time course of up to 24 h, reduction of the cellular ORP10 induced a significant increase in the amount of apoB100 that accumulated in the growth medium, while no effect was detected on the accumulation of apoA-I (Fig. 4a, b). At 12 h, the increase in apoB100 was 22% (siORP10.1) or 40% (siORP10.2) as compared to siNT-transfected cells; At 24 h, the corresponding numbers were 59% and 85%. Western analysis of the growth media was consistent with this finding (Fig. 4c; Electronic Supplementary Materials Fig. 2).
Fig. 4.
Silencing of ORP10 increases the accumulation of apoB100 in the growth medium. Accumulation of apoA-I (a) and apoB100 (b) into the growth medium of Huh7 cells treated with nontargeting (siNT) or ORP10-specific (siORP10.1, siORP10.2) siRNAs during a 24-h time course was quantified with specific sandwich ELISA assays. The data represents a mean ± s.e.m. (aN = 3, bN = 5; *p < 0.05, **p < 0.01, t test). c Analysis of apoB100 accumulation into growth medium at the indicated time points by Western blotting. An experiment representative of three independent analyses with similar results is shown
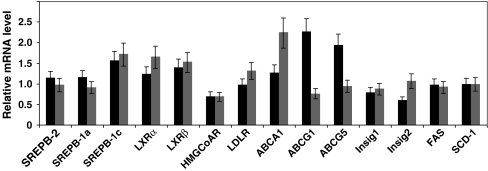
To obtain clues of the mechanism by which ORP10 impacts cellular lipid metabolism, we analyzed by quantitative real-time RT-PCR the mRNAs for LXRα, LXRβ, ABCA1, ABCG1, ABCG5, SREBP-1a, SREBP-1c, SREBP-2, HMG-CoA reductase, LDL-receptor, fatty acyl synthetase, stearoyl-CoA desaturase 1, Insig-1, and Insig-2. No significant change consistent with both ORP10-specific siRNAs was observed in these mRNAs upon silencing of OSBPL10 (Fig. 5).
Fig. 5.
OSBPL10 silencing has no significant effect on the mRNAs for key factors involved in lipid metabolism. Huh7 cells were transfected for 48 h with control (siNT) or ORP10-specific siRNAs (siORP10.1, black bars; siORP10.2, gray bars), followed by total RNA isolation and quantitative RT-PCR analysis of mRNA levels. SREBP sterol regulatory element binding protein, LXR liver X receptor, HMGCoR 3-hydroxy-3-methylglutaryl coenzyme A reductase, LDLR low-density lipoprotein receptor, ABC ATP-binding cassette transporter, Insig insulin-induced gene, FAS fatty acyl synthase, SCD stearoyl coenzyme A desaturase-1. The data depict mRNA levels in ORP10 siRNA-treated cells relative to those in cells transfected with the scrambled control siRNA (siNT) and represents a mean ± s.e.m. (N = 4–5)
ORP10 localizes on microtubules
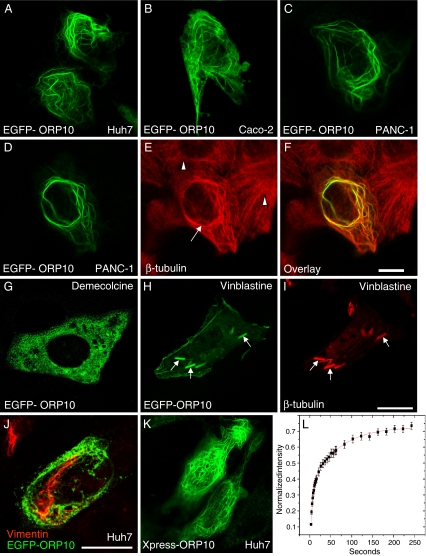
To obtain clues of ORP10 function, we determined its intracellular localization. We transfected the human cell lines Huh7 (hepatoma), Caco-2 (colon carcinoma), and PANC-1 (pancreatic carcinoma) with EGFP- or Xpress epitope-tagged cDNA constructs and visualized the expressed proteins by confocal microscopy. The expressed ORP10 distributed between a cytosolic-appearing pool and fibril-like structures (Huh7, Fig. 6a; Caco-2, Fig. 6b; PANC-1, Fig. 6c). Especially in cells expressing the protein at high levels, the fibrils often showed a cable-like, undulating morphology. The ORP10 fluorescence co-localized extensively with β-tubulin, identifying the filaments as microtubules. Localization of ORP10 on cable-like microtubule elements and the morphologic change induced by excess ORP10 were especially prominent in the human pancreatic carcinoma cell line PANC-1 (Fig. 6d–f; note the normal microtubule morphology in untransfected cells). However, ORP10 silencing using siRNA did not significantly affect the overall morphology of the microtubule cytoskeleton (data not shown). The identity of the ORP10 fibrils was further confirmed by treatment of transfected cells with the microtubule depolymerizing agent demecolcine, resulting in a dispersed, cytosolic distribution of ORP10 (Fig. 6g). To investigate further the microtubule association of ORP10, PANC-1 cells expressing ORP10 were treated with vinblastine, which binds to α- and β-tubulin, prevents their polymerization, and allows formation of paracrystals consisting of both tubulins. ORP10 was found to decorate tubulin crystals in the vinblastine-treated cells, suggesting an intimate association with tubulin (Fig. 6h, i). No colocalization of ORP10 with vimentin (Fig. 6j) or cytokeratin (data not shown) intermediate filaments was observed. Identical results were obtained with the EGFP- and Xpress-epitope-tagged (Fig. 6k) constructs. To investigate whether manipulation of the cellular lipid status affects the localization of ORP10, we incubated Huh7 cells expressing EGFP-ORP10 for 48 h in lipoprotein-deficient medium. For some specimens, 50 μM lovastatin was added for 24 h to potentiate the lipid/sterol depletion. For a comparison, cells were also incubated for 48 h with 100 μg/ml of human LDL to increase the cellular lipid/sterol content or treated for 4 h with 5 μM 25-hydroxycholesterol. Fluorescence microscopy analysis of EGFP-ORP10 revealed no significant effect of the lipid manipulations on the steady-state subcellular distribution of the protein (data not shown).
Fig. 6.
ORP10 localizes on microtubules. Huh7, Caco-2, and PANC-1 cells transfected with EGFP-ORP10 were visualized by confocal fluorescence microscopy. a Transfected Huh7 cells. b A transfected Caco-2 cell. c A transfected PANC-1 cell. d–f A transfected PANC-1 cell (arrow) surrounded by untransfected ones (arrowheads), costained with an antibody against β-tubulin. Note the aberrant bundled morphology of the EGFP-ORP10-positive microtubules in the transfected cell as compared to the normal microtubule cytoskeleton in the untransfected cells. g A transfected PANC-1 cell treated for 1 h with demecolcine (10 μg/ml) to disrupt microtubules. h, i A transfected PANC-1 cell after 2 h treatment with vinblastine (10 μg/ml), an agent that binds to α- and β-tubulin, prevents their polymerization and allows formation of tubulin paracrystals (indicated with arrows). j Channel merge image of a Huh7 cell expressing EGFP-ORP10 (green), stained for vimentin (red). k Huh7 cells transfected with ORP10/pcDNA4HisMaxC encoding ORP10 devoid of the EGFP fusion partner (staining with Xpress™ antibody). Bars 10 μm. l. Huh7 cells transfected with EGFP-ORP10 were subjected to fluorescence recovery after photobleaching analysis using a live cell confocal microscopy setup. The recovery curve representing microtubule associated fluorescence in a bleached 6 × 6 μm area as a function of time is displayed. The data represents a mean (N = 8 cells) ± s.e.m.; R2 of the fit = 0.9993. Recovery half times for the two kinetic components are 4.5 ± 0.94 s and 64.3 ± 11.6 s
To investigate the dynamic parameters of EGFP-ORP10 microtubule association, transfected Huh7 cells were subjected to fluorescence recovery after photobleaching analysis using a confocal microscope set up for live cell analysis. After photobleaching the microtubule-associated EGFP-ORP10, fluorescence recovered to approximately 70% of the starting value during the 6-min analysis period (Fig. 6l; Electronic Supplementary Materials: Time lapse video). The recovery followed a kinetics which curve-fitting suggested to arise from two components. The t1/2 for the fast component was 4.5 ± 0.94 s, and that for the slow one was 64.3 ± 11.6 s.
Discussion
In the present study, we investigated a putative function of OSBP-related proteins in the regulation of serum lipids and lipoproteins in humans. Genotyping of OSBPL gene SNPs in a dyslipidemic family material was initially used to evaluate the contribution of OSBPL genes to extreme lipid levels predisposing to cardiovascular disease, high triglycerides, and low HDL. Although recent genome-wide association studies [21–23] have initially identified common variants affecting lipid concentrations at the population level, data are also emerging for the impact of multiple rare variants influencing the extreme lipid levels [24]. Here, we wanted to study such potential loci by testing the involvement of select OSBPL genes in carefully phenotyped Finnish families ascertained for dyslipidemias, low HDL, or FCHL. The FCHL study set revealed suggestive linkage of several SNPs in OSBPL10 to the high TG trait, and analysis of the combined material strengthened the evidence. Importantly, association analysis using an independent study sample consisting of subjects with metabolic syndrome and matched healthy controls provided substantial proof for association of OSBPL10 SNPs with the extreme end (>95th percentile) high-TG trait.
The genetic analyses suggest that variation within the chromosome 3p23 region containing OSBPL10 may contribute to the high TG levels in Finnish dyslipidemic subjects. The present data do not allow us to firmly conclude that the underlying gene is OSBPL10, but they prompted us to investigate in more detail the function of the encoded protein, ORP10, in cultured cell models. The metabolic studies in cultured cells subjected to ORP10 silencing identify the protein as a novel regulator of hepatocellular lipid metabolism and apoB100 secretion, consistent with the notion that OSBPL10 may be the gene underlying the genetic findings. The likelihood of this is further increased by the lack of other genes plausibly connected with lipid metabolism within the chromosome region (10 Mb interval).
Hepatoma cells subjected to ORP10 silencing displayed a significant increase in the incorporation of [3H]acetate into cholesterol and of [3H]acetate and [3H]oleic acid into TG, suggesting that the intrinsic function of ORP10 involves suppression of hepatocellular lipid biosyntheses. Consistent with this observation, the cells showed increased HMG-CoA reductase activity, demonstrating an effect already at an early stage of the pathway, on conversion of HMG-CoA to mevalonic acid. An independent, systematic esiRNA silencing screen of all ORP family members carried out in HeLa cells supported a role of ORP10 in the regulation of cholesterol biosynthesis (data not shown). Labeling with [3H]oleic acid suggested that the impact of ORP10 on TG biosynthesis occurs at the levels of fatty acid activation or acyl transfer to glycerol. Importantly, we also found that ORP10 silencing enhances the accumulation of apoB100 in the hepatoma cell growth medium. One can envision that ORP10 could affect the synthesis of TG and cholesterol as well as the assembly of TG-rich lipoproteins in the intestine and the liver, both tissues in which OSBPL10 mRNA is expressed abundantly [4].
ORP10 was found to distribute between a cytosolic localization and microtubules, a major apparatus determining the spatial distribution of organelles and forming tracks for intracellular transport events [25]. FRAP analysis of live Huh7 cells with EGFP-ORP10 on microtubules revealed a recovery with two kinetic components. We find it likely that the fast component (t1/2 = 4.5 s) reflects the diffusion of EGFP-ORP10 in the cytosolic compartment, while the slow component (t1/2 = 64.3 s) could represent the actual microtubule association of the protein. The latter half-time is in the same range as the life-times determined for transport carriers moving along microtubules from the endoplasmic reticulum (ER) to the Golgi complex [26] and from the trans-Golgi network to the plasma membrane [27, 28]. The data are thus consistent with the notion that ORP10 could be involved in microtubule-dependent membrane trafficking or organelle motility.
Microtubule-dependent membrane trafficking is intimately connected with the control of lipid metabolism. Importantly, the membrane compartments that play key roles in the cellular metabolism of cholesterol and triglycerides, the ER and cytoplasmic lipid droplets, are connected to microtubules, which determine the spatial organization of the ER [29] and control the distribution and motility of lipid droplets [30, 31]. Pertinent to serum lipoproteins, also the Golgi complex playing a central role in lipoprotein secretion is organized by microtubules [32]. One can therefore envision that a protein associated with microtubules, such as ORP10, is in the position to modulate cellular lipid metabolism and lipoprotein secretion.
Electronic Supplementary Materials
Below is the link to the electronic supplementary material.
(AVI 3486 kb)
(PDF 27 kb)
(PDF 735 kb)
(PDF 44 kb)
Acknowledgments
We are grateful to Pipsa Kaipainen, Sari Nuutinen, Marja-Leena Piironen, Seija Puomilahti, and Pirjo Ranta for expert technical assistance. Dr. Markku Lehto is thanked for kindly providing the full-length ORP10 cDNA, Pekka Ellonen for help with the genotyping work, and Dr. Matti Jauhiainen for valuable comments on the manuscript. This study was supported by The Academy of Finland (grants 113013, 118720, and 121457 to V.M.O.), the European Union FP7 (LipidomicNet, agreement no. 202272), the Sigrid Juselius Foundation, the Finnish Foundation for Cardiovascular Research (M.-R.T., V.M.O.), the Magnus Ehrnrooth Foundation (V.M.O.), the Finnish Cultural Foundation (V.M.O.), and Helsinki University Central Hospital: A special governmental subsidy for health sciences research (M.-R.T., I.V.). Further support was provided by The Center of Excellence in Complex Disease Genetics (L.P., K.M., J.N.), The Research Program of Molecular Medicine, University of Helsinki, Biomedicum Foundation, and Biocentrum Helsinki Foundation (L.P., M.-R.T., K.M., J.N.).
Disclosure statement Concerning this work, the authors have no conflicts of interest to declare.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Julia Perttilä and Krista Merikanto contributed equally to this work.
Electronic supplementary material
The online version of this article (doi:10.1007/s00109-009-0490-z) contains supplementary material, which is available to authorized users.
References
- 1.Fairn GD, McMaster CR (2007) Emerging roles of the oxysterol-binding protein family in metabolism, transport, and signaling. Cell Mol Life Sci 65:228–236 [DOI] [PMC free article] [PubMed]
- 2.Yan D, Olkkonen VM (2008) Characteristics of oxysterol binding proteins. Int Rev Cytol 265:253–285 [DOI] [PubMed]
- 3.Jaworski CJ, Moreira E, Li A, Lee R, Rodriguez IR (2001) A family of 12 human genes containing oxysterol-binding domains. Genomics 78:185–196 [DOI] [PubMed]
- 4.Lehto M, Laitinen S, Chinetti G, Johansson M, Ehnholm C, Staels B, Ikonen E, Olkkonen VM (2001) The OSBP-related protein family in humans. J Lipid Res 42:1203–1213 [PubMed]
- 5.Perry RJ, Ridgway ND (2006) Oxysterol-binding protein and vesicle-associated membrane protein-associated protein are required for sterol-dependent activation of the ceramide transport protein. Mol Biol Cell 17:2604–2616 [DOI] [PMC free article] [PubMed]
- 6.Wang PY, Weng J, Anderson RG (2005) OSBP is a cholesterol-regulated scaffolding protein in control of ERK 1/2 activation. Science 307:1472–1476 [DOI] [PubMed]
- 7.Yan D, Lehto M, Rasilainen L, Metso J, Ehnholm C, Ylä-Herttuala S, Jauhiainen M, Olkkonen VM (2007) Oxysterol binding protein induces upregulation of SREBP-1c and enhances hepatic lipogenesis. Arterioscler Thromb Vasc Biol 27:1108–1114 [DOI] [PubMed]
- 8.Yan D, Jauhiainen M, Hildebrand RB, Willems van Dijk K, Van Berkel TJ, Ehnholm C, Van Eck M, Olkkonen VM (2007) Expression of human OSBP-related protein 1L in macrophages enhances atherosclerotic lesion development in LDL receptor-deficient mice. Arterioscler Thromb Vasc Biol 27:1618–1624 [DOI] [PubMed]
- 9.Yan D, Mäyränpää MI, Wong J, Perttilä J, Lehto M, Jauhiainen M, Kovanen PT, Ehnholm C, Brown AJ, Olkkonen VM (2007) OSBP-related protein 8 (ORP8) suppresses ABCA1 expression and cholesterol efflux from macrophages. J Biol Chem 283:332–340 [DOI] [PubMed]
- 10.Hynynen R, Suchanek M, Spandl J, Back N, Thiele C, Olkkonen VM (2009) OSBP-related protein 2 (ORP2) is a sterol receptor on lipid droplets that regulates the metabolism of neutral lipids. J Lipid Res 50:1305–1315 [DOI] [PMC free article] [PubMed]
- 11.Ngo M, Ridgway ND (2009) Oxysterol binding protein-related Protein 9 (ORP9) is a cholesterol transfer protein that regulates Golgi structure and function. Mol Biol Cell 20:1388–1399 [DOI] [PMC free article] [PubMed]
- 12.Wang C, JeBailey L, Ridgway ND (2002) Oxysterol-binding-protein (OSBP)-related protein 4 binds 25-hydroxycholesterol and interacts with vimentin intermediate filaments. Biochem J 361:461–472 [DOI] [PMC free article] [PubMed]
- 13.Genest JJ Jr, Martin-Munley SS, McNamara JR, Ordovas JM, Jenner J, Myers RH, Silberman SR, Wilson PW, Salem DN, Schaefer EJ (1992) Familial lipoprotein disorders in patients with premature coronary artery disease. Circulation 85:2025–2033 [DOI] [PubMed]
- 14.Goldstein JL, Schrott HG, Hazzard WR, Bierman EL, Motulsky AG (1973) Hyperlipidemia in coronary heart disease. II. Genetic analysis of lipid levels in 176 families and delineation of a new inherited disorder, combined hyperlipidemia. J Clin Invest 52:1544–1568 [DOI] [PMC free article] [PubMed]
- 15.Nikkilä EA, Aro A (1973) Family study of serum lipids and lipoproteins in coronary heart-disease. Lancet 1:954–959 [DOI] [PubMed]
- 16.Stein O, Stein Y (1999) Atheroprotective mechanisms of HDL. Atherosclerosis 144:285–301 [DOI] [PubMed]
- 17.Pajukanta P, Nuotio I, Terwilliger JD, Porkka KV, Ylitalo K, Pihlajamäki J, Suomalainen AJ, Syvanen AC, Lehtimäki T, Viikari JS et al (1998) Linkage of familial combined hyperlipidaemia to chromosome 1q21–q23. Nat Genet 18:369–373 [DOI] [PubMed]
- 18.Soro A, Pajukanta P, Lilja HE, Ylitalo K, Hiekkalinna T, Perola M, Cantor RM, Viikari JS, Taskinen MR, Peltonen L (2002) Genome scans provide evidence for low-HDL-C loci on chromosomes 8q23, 16q24.1–24.2, and 20q13.11 in Finnish families. Am J Hum Genet 70:1333–1340 [DOI] [PMC free article] [PubMed]
- 19.van Haperen R, van Tol A, Vermeulen P, Jauhiainen M, van Gent T, van den Berg P, Ehnholm S, Grosveld F, van der Kamp A, de Crom R (2000) Human plasma phospholipid transfer protein increases the antiatherogenic potential of high density lipoproteins in transgenic mice. Arterioscler Thromb Vasc Biol 20:1082–1088 [DOI] [PubMed]
- 20.Johansson M, Olkkonen VM (2005) Assays for interaction between Rab7 and oxysterol binding protein related protein 1L (ORP1L). Methods Enzymol 403:743–758 [DOI] [PubMed]
- 21.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS et al (2008) Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet 40:189–197 [DOI] [PMC free article] [PubMed]
- 22.Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T et al (2009) Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet 41:56–65 [DOI] [PMC free article] [PubMed]
- 23.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM et al (2008) Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet 40:161–169 [DOI] [PMC free article] [PubMed]
- 24.Cohen JC, Pertsemlidis A, Fahmi S, Esmail S, Vega GL, Grundy SM, Hobbs HH (2006) Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc Natl Acad Sci U S A 103:1810–1815 [DOI] [PMC free article] [PubMed]
- 25.Welte MA (2004) Bidirectional transport along microtubules. Curr Biol 14:R525–R537 [DOI] [PubMed]
- 26.Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJ, Lippincott-Schwartz J (1997) ER-to-Golgi transport visualized in living cells. Nature 389:81–85. doi:10.1038/38001 [DOI] [PubMed]
- 27.Hirschberg K, Miller CM, Ellenberg J, Presley JF, Siggia ED, Phair RD, Lippincott-Schwartz J (1998) Kinetic analysis of secretory protein traffic and characterization of golgi to plasma membrane transport intermediates in living cells. J Cell Biol 143:1485–1503 [DOI] [PMC free article] [PubMed]
- 28.Toomre D, Keller P, White J, Olivo JC, Simons K (1999) Dual-color visualization of trans-Golgi network to plasma membrane traffic along microtubules in living cells. J Cell Sci 112(Pt 1):21–33 [DOI] [PubMed]
- 29.Vedrenne C, Hauri HP (2006) Morphogenesis of the endoplasmic reticulum: beyond active membrane expansion. Traffic 7:639–646 [DOI] [PubMed]
- 30.Cohen RS (2005) Microtubule motors: LSD2 trips the toggle. Curr Biol 15:R651–R653 [DOI] [PubMed]
- 31.Welte MA, Cermelli S, Griner J, Viera A, Guo Y, Kim DH, Gindhart JG, Gross SP (2005) Regulation of lipid-droplet transport by the perilipin homolog LSD2. Curr Biol 15:1266–1275 [DOI] [PubMed]
- 32.Rios RM, Bornens M (2003) The Golgi apparatus at the cell centre. Curr Opin Cell Biol 15:60–66 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
(AVI 3486 kb)
(PDF 27 kb)
(PDF 735 kb)
(PDF 44 kb)