Abstract
To investigate the role of phosphatidylglycerol (PG) in photosynthesis, we constructed a mutant defective in the CDP-diacylglycerol synthase gene from a cyanobacterium, Synechocystis sp. PCC6803. The mutant, designated as SNC1, required PG supplementation for growth. Growth was repressed in PG-free medium concomitantly with the decrease in cellular content of PG. These results indicate that PG is essential, and that SNC1 is defective in PG synthesis. Decrease in PG content was accompanied by a reduction in the cellular content of chlorophyll, but with little effect on the contents of phycobilisome pigments, which showed that levels of chlorophyll–protein complexes decreased without alteration of those of phycobilisomes. Regardless of the decrease in the PG content, CO2-dependent photosynthesis by SNC1 was similar to that by the wild type on a chlorophyll basis, but consequently became lower on a cell basis. Simultaneously, the ratio of oxygen evolution of photosystem II (PSII) measured with p-benzoquinone to that of CO2-dependent photosynthesis, which ranged between 1.3 and 1.7 in the wild type. However, it was decreased in SNC1 from 1.3 to 0.4 during the early growth phase where chlorophyll content and CO2-dependent photosynthesis were little affected, and then finally to 0.1, suggesting that PSII first lost its ability to reduce p-benzoquinone and then decreased in its level and actual activity. These results indicate that PG contributes to the accumulation of chlorophyll–protein complexes in thylakoid membranes, and also to normal functioning of PSII.
Biological membranes with special functions show specific protein and lipid compositions. The membrane lipids form a barrier between the inside and the outside of the membranes, supply the membrane proteins with sites for their action, and regulate the functions of the membrane proteins. So far, research on the functioning of membrane systems has focused mainly on the proteins with actual catalytic activities themselves and to a much smaller extent on the membrane lipids that surround or are integrated into the membrane proteins. Studies on the latter kind include (i) examination of the effects of the removal or addition of specific lipids on some purified membrane protein complexes, (ii) analysis of lipids bound to the complexes, and (iii) characterization of the membrane functions of mutants defective in some particular lipid. Although the former two strategies have led to the accumulation of a substantial amount of information on the possible roles of lipids, a final conclusion as to their roles in vivo cannot be drawn owing to potential artifacts produced by, for example, the use of detergents with in vitro experimental systems. In contrast, the last strategy has been successfully used to obtain in vivo evidence regarding the roles of lipids.
Thylakoid membranes, sites for photosynthetic electron transport and photophosphorylation, contain monogalactosyl diacylglycerol (MGDG), digalactosyl diacylglycerol (DGDG), sulfoquinovosyl diacylglycerol (SQDG), and phosphatidylglycerol (PG) as the major lipid constituents (e.g., 1). Mutants devoid of SQDG and DGDG have been isolated and characterized for some photosynthetic organisms. A mutant devoid of SQDG of a green alga, Chlamydomonas reinhardtii, showed an ≈40% decrease in photosystem II (PSII) activity with little damage to photosystem I (PSI) activity, despite little effect on the content of the PSII complex or its subunit composition (2, 3). Taken together with the observation of specific association of SQDG with the PSII complex, but not with the PSI complex, in wild-type C. reinhardtii, the results can be interpreted as demonstrating contribution of SQDG for full expression of PSII activity through maintenance of the normal conformation of the PSII complex through specific binding (3). On the other hand, a mutant of a higher plant, Arabidopsis thaliana, with a 90% reduction of DGDG content exhibited complex effects on photosynthesis (i.e., an increase in the antenna size of PSII and a decrease in the PSII/PSI ratio), suggesting the involvement of DGDG in structural organization of the photosynthetic apparatus (4, 5).
For clarification of the function of PG, the sole phospholipid in thylakoid membranes, it is necessary to produce mutants defective in PG. However, deduction of the physiological role of PG through mutant analyses has been successful so far only for nonphotosynthetic organisms (6–14). The null mutation of phosphatidylglycerol phosphate (PGP) synthase, for example, brought about a loss of PG and cardiolipin (CL) syntheses, and eventually lethality in Escherichia coli and Saccharomyces cerevisiae under nonfermentation growth conditions (9, 14). These observations can be explained by the occupation of a major fraction of the anionic lipids by PG and/or CL in the membrane systems of E. coli and mitochondria of eukaryotic cells (14, 15). It was recently reported that the null mutant of E. coli became viable with a background of deficiency in the major lipoprotein of the outer membranes (16). The anionic lipid constituents of photosynthetic membrane systems differ from those of the nonphotosynthetic membrane ones of E. coli and S. cerevisiae, being composed of PG and a nonphospholipid (SQDG). It is thus of particular interest to establish whether PG is dispensable for photosynthetic organisms. CDP-diacylglycerol synthase (CDS), located in the biosynthetic pathway for PG, has been characterized for several organisms through the cloning of genes and/or cDNAs. A similarity search of the DNA sequence of the genome of a cyanobacterium, Synechocystis sp. PCC6803, revealed one ORF encoding a CDS homolog (17). We have now produced a mutant defective in PG synthesis from Synechocystis sp. PCC6803 through disruption of this ORF by homologous recombination, and have characterized the effects of this mutation on the photosynthetic apparatus.
Materials and Methods
Cell Culture.
Wild-type and mutant cells of Synechocystis sp. PCC6803 were grown photoautotrophically in BG-11 medium (18) under constant fluorescent lamp illumination (10 W⋅m−2) at 30°C, in oblong glass vessels aerated with air containing 2% CO2, or in flasks with shaking. Dioleoyl-PG (Sigma) was sonicated in BG-11 medium for liposome production (6 mM) to be filtered for sterilization. The culture medium was supplemented with the PG liposomes at given concentrations where indicated.
Isolation of DNA.
Cells of Synechocystis sp. PCC6803 were grown as described above and harvested by centrifugation (8,000 × g, 10 min, 4°C). DNA was isolated from these cells as described in ref. 19.
Amplification of a Putative Gene for CDS of Synechocystis sp. PCC6803.
For amplification of the coding region of a putative CDS gene found on the genome of Synechocystis sp. PCC6803, PCR was performed with primer 1 (5′-GGGTCATATGCCCACCCAGCGAATTATTAG-3′) and primer 2 (5′-GGGTGGATCCTCACAAATTATTCAACACAG-3′). The underlined codons in primers 1 and 2 are the start and stop codons, respectively. PCR was performed with a DNA Thermal Cycler (Perkin-Elmer) with the following thermocycle program: 2 min at 95°C, followed by 25 cycles of 30 s at 95°C, 60 s at 55°C, and 90 s at 72°C. A major product of 1.1 kbp was purified from the agarose gel after electrophoresis.
Screening of Genomic DNA Library of Synechocystis sp. PCC6803.
A genomic DNA library of Synechocystis sp. PCC6803 was constructed in bacteriophage λDASHII (Stratagene) through integration of the genomic DNA partially digested with Sau3AI into the phage vector. The genomic DNA was screened with the putative CDS gene as a probe through plaque hybridization, as previously described (20).
Construction of a Mutant with Disruption of the Putative CDS Gene.
The plasmid containing the disrupted putative gene for CDS was used to transform wild-type Synechocystis sp. PCC6803 by homologous recombination, as described in ref. 21. The disruption of the putative gene was confirmed by PCR with primers 1 and 2 described above.
Lipid Analysis.
The total lipids were extracted from the cells and then separated into individual lipid classes by two-dimensional TLC, as previously described (2). The spots of lipids were visualized by spraying with primulin. Fatty acid methyl esters were then prepared from the total lipids and each lipid class with 5% anhydrous methanolic HCl for analysis by capillary GLC, as described previously (2). The lipid contents were estimated with arachidonic acid as an internal standard.
Determination of Pigment Contents and Photosynthesis.
The content of chlorophyll was determined after its extraction from the cells with 100% methanol (22), whereas the contents of phycocyanin, allophycocyanin, and phycoerythrin were determined as described in ref. 23. Photosynthetic O2 evolution coupled with CO2 fixation was measured in BG-11 medium containing cells equivalent to 2.67 μg of Chl⋅ml−1 and 10 mM NaHCO3 with a Clark-type electrode (Rank Brothers, Cambridge, U.K.). The electrode chamber was filled with 5 ml of reaction mixture, kept at 30°C, and illuminated with a tungsten projector lamp (180 W⋅m−2).
Results and Discussion
Disruption of the Putative Gene for CDS in Synechocystis sp. PCC6803.
Through a similarity search of the DNA sequence of the genome of a cyanobacterium, Synechocystis sp. PCC6803, one ORF, slr1369, was found to encode a putative CDS composed of 293 amino acids with a molecular mass of 31.9 kDa (16). The amino acid sequence deduced from this ORF exhibited 38% identity with that of E. coli CDS, and 27–30% identity with the sequences of CDS of S. cerevisiae, Drosophila, rat, and A. thaliana (refs. 24–28 and data not shown). The putative CDS of Synechocystis sp. PCC6803, like the CDSs of E. coli and the eukaryotic organisms, possessed a sequence corresponding to a CDS motif, SMMKRDAGVKDSGQLIPGHGGILDRTD, at its C terminus (PROSITE database, http://www.expasy.ch/sprot/prosite.html), and several potential membrane-spanning regions (data not shown) as found on hydrophobicity analysis according to the method of Kyte and Doolittle (29). On the other hand, a hydrophilic N terminus commonly present in the eukaryotic CDSs was not found in the cyanobacterial sequence or in that of E. coli CDS (data not shown). These features suggest that this ORF actually encodes the CDS of Synechocystis sp. PCC6803.
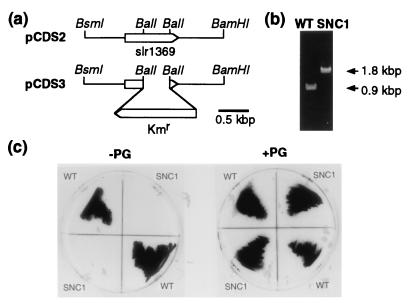
We then constructed a null mutant of the putative CDS gene of Synechocystis sp. PCC6803 to determine the role of this putative gene as to lipid and photosynthetic properties. A 0.9-kbp DNA fragment corresponding to slr1369 was amplified by PCR with the genomic DNA of Synechocystis sp. PCC6803 as a template, and then ligated into pUC119 to yield a plasmid, pCDSP. The insert of pCDSP showed restriction sites expected from the genome sequence of Synechocystis sp. PCC6803 (data not shown), and was used as a probe for screening of the genomic DNA library of Synechocystis sp. PCC6803 constructed in λDASH II. As a result, we obtained a λ phage clone containing a 14-kbp insert. A 4.4-kbp BamHI/HindIII fragment in the insert was expected to contain the putative CDS gene from the genome sequence of Synechocystis sp. PCC6803. We then excised the 4.4-kbp fragment for ligation into pUC119 to yield a plasmid, pCDS1. The insert of pCDS1 was confirmed to contain the putative gene with its 5′- and 3′-flanking regions by restriction mapping (data not shown). pCDS1 was then cut with BsmI/BamHI, the resultant 2.4-kbp fragment being blunt-ended and ligated again into pUC119 to yield a plasmid, pCDS2 (Fig. 1a). Plasmid pCDS2 was digested with BalI to remove a 0.5-kbp fragment within the putative CDS gene, which was replaced by a 1.4-kbp fragment carrying the kanamycin-resistance (Kmr) cassette of pHSG298 for the production of a plasmid, pCDS3 (Fig. 1a). Disruption of the putative gene of the wild-type Synechocystis sp. PCC6803 was conducted with pCDS3 through homologous recombination in the presence of PG liposomes in case of PG requirement by the mutant for its growth, which was checked by PCR with primers for the 5′- and 3′-terminal regions of the ORF (Fig. 1b). A 0.9-kbp fragment was amplified as expected for the wild type, whereas only a 1.8-kbp fragment corresponding to the ORF∷Kmr gene was observed with the mutant, which proved the complete replacement of the ORF with the ORF∷Kmr gene. We designated the null mutant of the putative CDS gene as SNC1.
Figure 1.
Construction of a disruptant of slr1369 from Synechocystis sp. PCC6803. (a) Plasmid construction for disruption of slr1369 in Synechocystis sp. PCC6803. (b) Confirmation by PCR of the disruption of slr1369 by insertion of the kanamycin-resistance (Kmr) gene in SNC1. Sense and antisense primers, corresponding to the 5′- and 3′-terminal regions of slr1369, respectively, were used for the PCR with genomic DNA of the wild type or SNC1 as a template. (c) PG-requiring phenotype of SNC1. Wild-type and SNC1 cells were cultured on agar plates of BG-11 with or without PG supplementation.
Growth and Lipid Phenotypes of the Mutant.
SNC1 was lethal when cultured photoautotrophically on an agar plate of BG-11, but could grow with PG supplementation (Fig. 1c), indicating that PG is essential for the cyanobacterium, but is not produced by SNC1, at least not to a level that supports its growth. The quantitative requirement of PG by SNC1 was then examined by culturing SNC1 in a liquid medium containing 0, 1.5, 6, 30, or 150 μM PG. With respect to growth rate and cell density at the stationary phase, similar growth curves were observed with 6, 30, and 150 μM PG, whereas lowered curves were observed with 0 or 1.5 μM PG (data not shown). We thereafter added 60 μM PG to the medium for the culturing of SNC1 cells.
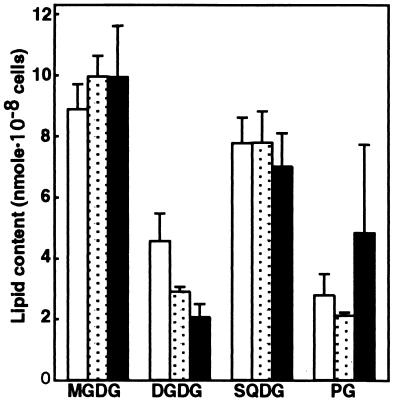
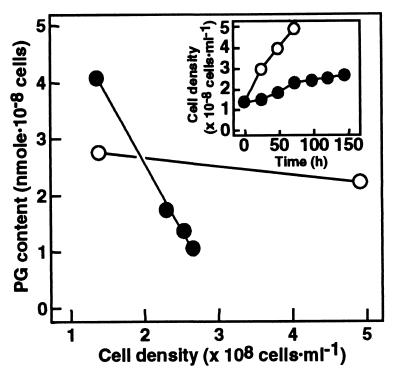
The contents of individual lipid classes were then determined for the wild type and SNC1 grown in the presence or absence of PG liposomes (Fig. 2). Little effect of PG supplementation was observed on the individual lipid contents of the wild type—i.e., MGDG was the most abundant lipid, followed by SQDG, DGDG, and PG in that order (Fig. 2). The respective contents of MGDG, DGDG, and SQDG of SNC1 grown with PG liposomes were similar to those of the wild type, indicating that SNC1 is little affected as to glycolipid metabolism, at least when PG is exogenously supplied, whereas the PG content of SNC1, ranging from 0.1 to 5 times of the wild type level (data not shown), was, on average, about twice that of the wild type (Fig. 2). The wide PG content range of SNC1 could depend on the various levels of incorporated PG with the background of a defect in PG synthesis. To investigate the possible lack of PG synthesis by SNC1 in more detail, we shifted SNC1 cells, grown in advance with a supply of PG liposomes, to medium devoid of PG, and then monitored the changes in the cellular content of PG (Fig. 3). SNC1 showed a slower growth rate and a lower cell density at the stationary phase than the wild type (Fig. 3 Inset), which coincided with the PG requirement of SNC1 for growth. The PG content, which remained at an almost constant level in the wild type throughout growth, decreased in SNC1 with growth in the absence of PG. These results indicate that SNC1 is defective in PG synthesis because of the deletion of the CDS gene. Contents of the other three glycolipids were little changed in SNC1 (data not shown), indicating that glycolipid metabolism is little affected by the defect in PG metabolism in Synechocystis sp. PCC6803.
Figure 2.
Individual lipid contents of the wild type and SNC1 cells. Individual lipid contents were determined on a cell basis for the wild type grown with or without PG supplementation, and SNC1 grown with PG supplementation. The data are presented as the means ± SE for three and five experiments with the wild type and SNC1, respectively. □, wild type without PG;  , wild type with PG; ■, SNC1 with PG.
, wild type with PG; ■, SNC1 with PG.
Figure 3.
Defect in PG synthesis of SNC1. Wild-type cells were grown in the absence of PG, while SNC1, pregrown with PG supplementation, was shifted to PG-free medium, for analyses of the effects on the cellular content of PG. The data are presented as the means for three measurements in one experiment representative of three independent experiments. (Inset) Growth curve. ○, Wild type; ●, SNC1.
Several photosynthetic organisms have been shown under various circumstances to exhibit inverse changes in the levels of PG and SQDG, which resulted in little alteration of the level of the sum of these anionic lipids: (i) Exposure to phosphorus-limiting conditions during growth induced a decrease in the level of PG with a concomitant increase in the level of SQDG in Rhodobacter sphaeroides, Synechococcus sp. PCC7942, and A. thaliana (30–32). (ii) Mutations which impair SQDG synthesis increased the PG levels in R. sphaeroides, Synechococcus sp. PCC7942, and C. reinhardtii (2, 30, 31). It has thus been thought that photosynthetic organisms require a certain summed level of SQDG and PG, and that each of these anionic lipids has common roles, to some extent, in the photosynthetic apparatus, which can be taken over, if necessary, by the other. In contrast to the above findings, the SQDG level in SNC1 remained almost constant, despite various levels of incorporation of exogenously supplemented PG (Fig. 3) or the decrease in the PG content after the shift to PG-free medium (data not shown). One possible explanation for this discrepancy might be that the incorporated PG in SNC1 was located at sites that, although sufficient for some recovery of the repressed growth, were improper for regulation of the levels of anionic lipids. Indispensability of PG for the growth of Synechocystis sp. PCC6803, and the fact that PG did not disappear but remained at a lower level under phosphorus limitation (30–32), indicate some essential role of PG which cannot be replaced by SQDG.
Effect of the CDS Mutation on the Photosynthetic Pigments.
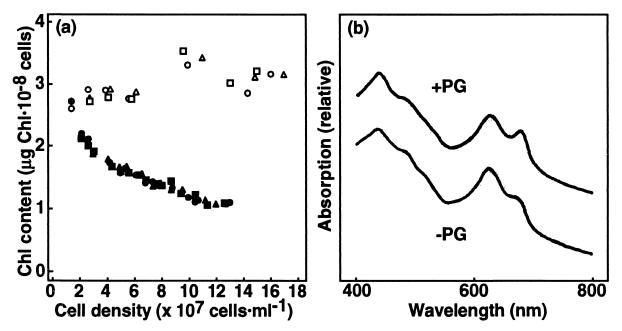
To determine the function of PG in the photosynthetic apparatus, SNC1 grown with PG supplementation was transferred to PG-free medium, and the effects of the subsequent decrease in PG content on the photosynthetic pigments and activities were investigated. The cellular content of chlorophyll (Chl) a, the sole Chl species of PSI and PSII complexes, remained almost constant during growth in the wild type, whereas it decreased in SNC1 to 40% of the level in the wild type (Fig. 4a). These results indicate that the levels of Chl-protein complexes decreased in SNC1 cells with decreasing PG content.
Figure 4.
Changes in Chl content and absorption spectra of SNC1 after the shift to PG-free medium. (a) The wild-type cells were grown in the absence of PG, while SNC1, pregrown with PG supplementation, was shifted to PG-free medium. The precultured cells were divided into three cultures for independent determination of Chl contents (circle, triangle, and square). Open symbols, wild type; filled symbols, SNC1. (b) SNC1 cells were grown with PG supplementation, and then were shifted to PG-free medium to grow until Chl content on a per-cell basis was reduced by 45%. The spectra were normalized at 730 nm.
On the other hand, contents of phycobilisome pigments such as phycocyanin and allophycocyanin were similar for SNC1 grown with PG liposomes and the wild type, and remained at almost the same levels in both SNC1 and the wild type throughout growth in the PG-free medium [e.g., phycocyanin amounted finally to 103–106% and 122–125% in SNC1 and the wild type, respectively, as compared with the contents at the start of the culture in PG-free medium (data not shown)]. These results indicate that the levels of phycobilisomes remained at certain levels in SNC1 and wild type cells. The phycobilisomes of SNC1 would be bound on the thylakoid membranes rather than released into the cytoplasm, in view of the report that dissociation of phycobilisomes from thylakoid membranes occurred with destabilization of the subunit assembly to cause decreases in the subunit levels (33).
Chl absorption of the SNC1 cells, consistent with the above results, decreased in the ratio of peak height of Chl absorption at 678 nm to that of phycocyanin absorption at 628 nm after the shift to PG-free medium (Fig. 4b). Peripheral membrane protein complexes like phycobilisomes, the subunits of which are not integrated into the membranes, might not require PG of the thylakoid membranes for the construction of the complexes, in contrast to the integral membrane protein complexes like Chl-protein complexes.
Effect of the CDS Mutation on the PSII Activity.
CO2-dependent photosynthesis rate on a Chl basis decreased during growth in the wild type and SNC1 (Fig. 5a). Reduction in the cellular content of Chl a in SNC1 resulted in a much more drastic decrease on a per cell basis—i.e., as much as a 70% decrease for SNC1, in comparison to a 30% decrease for the wild type (Fig. 5b). These results indicate that decrease in PG content brought about reductions in cellular content of Chl a and, consequently, in the photosynthetic activity of a cell. The CO2-dependent photosynthetic activity of SNC1 cells at the stationary phase, which remained as high as 30% of its maximal level, does not appear to be a direct cause of the growth arrest of SNC1 after transfer to PG-free medium.
Figure 5.
Changes in the photosynthetic activities of SNC1 after a shift to PG-free medium. Wild-type cells were grown in the absence of PG, while SNC1 cells, pregrown with PG supplementation, were shifted to PG-free medium. The precultured cells were divided into three cultures for independent measurements of the photosynthetic activity with NaHCO3 as an electron acceptor (circle, triangle, and square) on a Chl basis (a), or on a per-cell basis (b). Photosynthetic activity per cell (b) was normalized to the respective maximal values. Open symbols, wild type; filled symbols, SNC1.
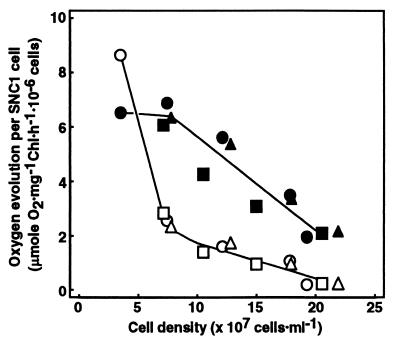
To determine the effect of the decrease in the PG content on the photosynthetic apparatus in more detail, we examined the oxygen evolution rate of PSII on a per cell basis with an artificial electron acceptor, p-benzoquinone (Fig. 6). The oxygen evolution rate of PSII, compared with that of CO2-dependent photosynthesis, was 1.3- to 1.7-fold higher in the wild type throughout growth (data not shown). At the early growth phase of SNC1, where the cell density increased to ≈2 times the initial level, the oxygen evolution rate of PSII was decreased to as low as 27–33% of the original level, although the rate of CO2-dependent photosynthesis was little affected and cellular content of Chl was reduced only to 82–87% (data not shown). The PSII activity using p-benzoquinone was consequently surpassed by the rate of CO2-dependent photosynthesis. Because 2-fold increase in the density of SNC1 cells corresponded to a reduction in the cellular content of PG to 25% of the original level (Fig. 3), these results indicate that the decrease in PG content to that extent is enough to largely deprive PSII of its ability to use p-benzoquinone as an electron acceptor, with only a slight decrease, if any, in the level and actual activity of PSII complex. The inability of PSII to use p-benzoquinone efficiently indicates a conformational change of the QB-binding site on the D1 protein at which p-benzoquinone should originally act. Further cell growth (i.e., a much greater decrease in the cellular content of PG) was accompanied by decrease in the rate of oxygen evolution of PSII as well as that of Chl content and the CO2-dependent photosynthesis rate, indicating that the more drastic reduction in PG content decreased the level and actual PSII activity owing probably to the instability of the abnormally conformed PSII complex (Fig. 6). Initial Chl fluorescence, which was almost steady in the wild type, was elevated finally to 4.5 times of the initial level in SNC1 (data not shown), indicating functional damage to the photosynthetic apparatus. A disruptant of the pgsA gene for PGP synthase of Synechocystis sp. PCC6803 was recently isolated and shown, as in SNC1, to require PG supplementation for growth and to lower PG content after removal of PG from the medium (34). The mutant, similar to SNC1, could not exhibit PSII activity with artificial electron acceptors such as p-benzoquinone. In addition, the mutant was damaged in its electron transport system from H2O to methylviologen, but not in that from reduced diaminodurene to methylviologen (i.e., PSI activity). This revealed a decrease in PSII activity.
Figure 6.
Rates of p-benzoquinone-dependent O2 evolution and CO2-dependent photosynthesis of SNC1 after a shift to PG-free medium. SNC1, pregrown with PG supplementation, was shifted to PG-free medium. The precultured cells were divided into three cultures for independent measurements of oxygen evolution (circle, triangle, and square). Open symbols, PSII; filled symbols, CO2-dependent photosynthesis.
PG was previously shown to be tightly associated with the D1 subunit of PSII in a cyanobacterium, Oscillatoria chalybea, and the binding site was assigned to the hydrophobic amino acid section between Arg 27 and Arg 225 on the basis of the immunoreaction of peptide fragments of the trypsin-digested D1 subunit with antibodies against PG (35). In addition, the activity of PSII particles from the cyanobacterium was inhibited on treatment with phospholipase A2, which modified PG, but was restored upon PG supplementation (35). Alteration in the conformation of the PSII complex including that of the QB-binding site may be induced in SNC1 and the pgsA mutant through depletion of PG from PSII subunits such as the D1 protein, bringing about damage to the function of the PSII reaction center.
In nonphotosynthetic organisms, anionic phospholipids such as PG appear to contribute to the process of localization of some proteins in appropriate membrane systems. The reduced levels of both PG and cardiolipin (CL) in an E. coli mutant with an altered pgsA gene (PGP synthase) showed the in vivo inhibition of translocation of an outer membrane protein, PhoE, across the inner membranes, and in vitro inhibition through the use of its inner membrane vesicles (6, 36). Because a deficiency of only CL (i.e., not of PG) in a null mutant of CL synthase of E. coli brought about few remarkable phenotypes other than abnormal lipid compositions (12), the conclusion was drawn that PG is responsible for the translocation of proteins such as PhoE across the inner membranes in E. coli. On the other hand, the decreased contents of PG and CL in the pgsA mutant were shown to affect the orientation of derivatives of Lep, an inner membrane protein, which led to the conclusion that the topology of the membrane protein depends on the contents of these anionic lipids (7). Furthermore, involvement of PG in the maintenance of a certain level of membrane protein complexes was proposed for a protein complex of the inner membranes of mitochondria such as cytochrome c oxidase, because the mutant of S. cerevisiae lacking both PG and CL showed a reduced level of the protein complex (9, 10), whereas a null mutant of the CL synthase gene of this organism demonstrated no remarkable deleterious phenotypes other than a lack of CL (13). Because a lack of some individual subunits of PSI and PSII complexes is known to induce pleiotropic effects on other subunits, leading to disassembly of these complexes (37), it is probable that, in Synechocystis sp. PCC6803, PG is required for the normal integration of some subunits of PSI and/or PSII complexes into the thylakoid membranes, and that the normal construction of the Chl-protein complexes is hampered by the lack of PG. Further work is necessary to clarify how PG contributes to the maintenance of Chl content in thylakoid membranes and the conformation of the PSII complex.
Acknowledgments
This work was supported by grants from the Ministry of Education, Science, Sports and Culture, Japan (10554049) and from the Promotion and Mutual Aid Corporation for Private Schools of Japan.
Abbreviations
- CDS
CDP-diacylglycerol synthase
- Chl
chlorophyll
- CL
cardiolipin
- DGDG
digalactosyl diacylglycerol
- MGDG
monogalactosyl diacylglycerol
- PG
phosphatidylglycerol
- PGP
phosphatidylglycerol phosphate
- PSI
photosystem I
- PSII
photosystem II
- SQDG
sulfoquinovosyl diacylglycerol
References
- 1.Douce R, Holz R B, Benson A A. J Biol Chem. 1973;248:7215–7222. [PubMed] [Google Scholar]
- 2.Sato N, Tsuzuki M, Matsuda Y, Ehara T, Osafune T, Kawaguchi A. Eur J Biochem. 1995;230:987–993. doi: 10.1111/j.1432-1033.1995.tb20646.x. [DOI] [PubMed] [Google Scholar]
- 3.Sato N, Sonoike K, Tsuzuki M, Kawaguchi A. Eur J Biochem. 1995;234:16–23. doi: 10.1111/j.1432-1033.1995.016_c.x. [DOI] [PubMed] [Google Scholar]
- 4.Dörmann P, Hoffmann-Benning S, Balbo I, Benning C. Plant Cell. 1995;7:1801–1810. doi: 10.1105/tpc.7.11.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Härtel H, Lokstein H, Dörmann P, Grimm B, Benning C. Plant Physiol. 1997;115:1175–1184. doi: 10.1104/pp.115.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Vrije T, de Swart R L, Dowhan W, Tommassen J, de Kruijff B. Nature (London) 1988;334:173–175. doi: 10.1038/334173a0. [DOI] [PubMed] [Google Scholar]
- 7.van Klompenburg W, Nilsson I, von Heijne G, de Kruijff B. EMBO J. 1997;16:4261–4266. doi: 10.1093/emboj/16.14.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia W, Dowhan W. Proc Natl Acad Sci USA. 1995;92:783–787. doi: 10.1073/pnas.92.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang S-C, Heacock P N, Clancey C J, Voelker D R, Dowhan W. J Biol Chem. 1998;273:9829–9836. doi: 10.1074/jbc.273.16.9829. [DOI] [PubMed] [Google Scholar]
- 10.Subik J. FEBS Lett. 1974;42:309–313. doi: 10.1016/0014-5793(74)80753-2. [DOI] [PubMed] [Google Scholar]
- 11.Ohtsuka T, Nishijima M, Suzuki K, Akamatsu Y. J Biol Chem. 1993;268:22914–22919. [PubMed] [Google Scholar]
- 12.Hiraoka S, Matsuzaki H, Shibuya I. FEBS Lett. 1993;336:221–224. doi: 10.1016/0014-5793(93)80807-7. [DOI] [PubMed] [Google Scholar]
- 13.Chang S-C, Heacock P N, Mileykovskaya E, Voelker D R, Dowhan W. J Biol Chem. 1998;273:14933–14941. doi: 10.1074/jbc.273.24.14933. [DOI] [PubMed] [Google Scholar]
- 14.Heacock P N, Dowhan W. J Biol Chem. 1987;262:13044–13049. [PubMed] [Google Scholar]
- 15.Daum G. Biochim Biophys Acta. 1985;822:1–42. doi: 10.1016/0304-4157(85)90002-4. [DOI] [PubMed] [Google Scholar]
- 16.Kikuchi S, Shibuya I, Matsumoto K. J Bacteriol. 2000;182:371–376. doi: 10.1128/jb.182.2.371-376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 18.Stanier R Y, Kunisawa R, Mandel M, Cohen-Bazire G. Bacteriol Rev. 1971;35:171–205. doi: 10.1128/br.35.2.171-205.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch F F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab.; 1989. [Google Scholar]
- 20.Sato N, Fujiwara S, Kawaguchi A, Tsuzuki M. J Biochem (Tokyo) 1997;122:1224–1232. doi: 10.1093/oxfordjournals.jbchem.a021885. [DOI] [PubMed] [Google Scholar]
- 21.Golden S S, Brusslan J, Haselkorn R. Methods Enzymol. 1987;153:215–231. doi: 10.1016/0076-6879(87)53055-5. [DOI] [PubMed] [Google Scholar]
- 22.Arnon D I, McSwain B D, Tsujimoto H Y, Wada K. Biochim Biophys Acta. 1974;357:231–245. doi: 10.1016/0005-2728(74)90063-2. [DOI] [PubMed] [Google Scholar]
- 23.Tandeau de Marsac N, Houmard J. Methods Enzymol. 1988;167:318–328. doi: 10.1016/0076-6879(88)67092-3. [DOI] [PubMed] [Google Scholar]
- 24.Icho T, Sparrow C P, Raetz C R H. J Biol Chem. 1985;260:12078–12083. [PubMed] [Google Scholar]
- 25.Shen H, Heacock P N, Clancey C J, Dowhan W. J Biol Chem. 1996;271:789–795. doi: 10.1074/jbc.271.2.789. [DOI] [PubMed] [Google Scholar]
- 26.Wu L, Niemeyer B, Colley N, Socolich M, Zuker C S. Nature (London) 1995;373:216–222. doi: 10.1038/373216a0. [DOI] [PubMed] [Google Scholar]
- 27.Saito S, Goto K, Tonosaki A, Kondo H. J Biol Chem. 1997;272:9503–9509. doi: 10.1074/jbc.272.14.9503. [DOI] [PubMed] [Google Scholar]
- 28.Kopka J, Ludewig M, Müler-Röber B. Plant Physiol. 1997;113:997–1002. doi: 10.1104/pp.113.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 30.Benning C, Beatty J T, Prince R C, Somerville C R. Proc Natl Acad Sci USA. 1993;90:1561–1565. doi: 10.1073/pnas.90.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Güler S, Seeliger A, Härtel H, Renger G, Benning C. J Biol Chem. 1996;271:7501–7507. doi: 10.1074/jbc.271.13.7501. [DOI] [PubMed] [Google Scholar]
- 32.Essigmann B, Güler S, Narang R A, Linke D, Benning C. Proc Natl Acad Sci USA. 1998;95:1950–1955. doi: 10.1073/pnas.95.4.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen G, Boussiba S, Vermaas W F J. Plant Cell. 1993;5:1853–1863. doi: 10.1105/tpc.5.12.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hagio, M., Gombos, M., Varkonyi, Z., Masamoto, K., Sato, N., Tsuzuki, M. & Wada, H. (2000) Plant Physiol., in press. [DOI] [PMC free article] [PubMed]
- 35.Kruse O, Schmidt G H. Z Naturforsch. 1995;49c:115–124. [Google Scholar]
- 36.Miyazaki C, Kuroda M, Ohta A, Shibuya I. Proc Natl Acad Sci USA. 1985;82:7530–7534. doi: 10.1073/pnas.82.22.7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hippler M, Redding K, Rochaix J-D. Biochim Biophys Acta. 1998;1367:1–62. doi: 10.1016/s0005-2728(98)00136-4. [DOI] [PubMed] [Google Scholar]