Abstract
Fatty acid synthesis in bacteria is catalyzed by a set of individual enzymes known as the type II fatty acid synthase. Acyl carrier protein (ACP) shuttles the acyl intermediates between individual pathway enzymes. In this study, we determined the solution structures of three different forms of ACP, apo-ACP, ACP, and butyryl-ACP under identical experimental conditions. The structural studies revealed that attachment of butyryl acyl intermediate to ACP alters the conformation of ACP. This finding supports the more general notion that the attachment of different acyl intermediates alters the ACP structure to facilitate their recognition and turnover by the appropriate target enzymes.
Keywords: acyl carrier protein, type II fatty acid synthase, NMR, protein structure
Introduction
Fatty acid biosynthesis in bacteria and plants is carried out by a series of enzymes that are collectively known as type II fatty acid synthase (FAS II), and the system is most extensively studied in the Escherichia coli system.1 Acyl carrier protein (ACP) is a small acidic protein with a molecular mass of ∼9 kDa that plays an essential role as a carrier of the pathway acyl intermediates for FAS II enzymes with different biochemical functions and structures.2 ACP also delivers acyl intermediates to enzymes of other metabolic pathways such as the biosynthesis of lipid A,3 phospholipids,4 and activation of toxins.5 The acpP gene product is the inactive apo-protein that is converted to the active holo form (termed as ACP) by the attachment of a 4′-phosphopantetheine moiety from CoA to Ser36 of apo-ACP catalyzed by holo-(acyl carrier protein) synthase (AcpS).6,7 The various acyl intermediates are attached to the sulfhydryl of ACP's 4′-phosphopantetheine prosthetic group (4′-PP group) via a thioester linkage.
ACPs are highly conserved in nature, particularly at the attachment site for 4′-PP group and the following helix.8 NMR solution structures of E. coli ACP,9,10 frenolicin ACP,11 Mycobacterium tuberculosis ACP (AcpM),12 Bacillus subtilis apo-ACP, and ACP,13 as well as the crystal structures of B. subtilis ACP in the ACP•AcpS complex7 and butyryl-ACP (C4:0-ACP) of E. coli14 have been reported. In addition, a comparison of the E. coli butyryl-ACP structure to the crystal structure of B. subtilis holo-ACP structure determined in the complex with AcpS14 and a comparison of the AcpM structure to the B. subtilis holo-ACP structure12 have been presented. Futhermore, structures of other ACP family proteins, actinorhodin apo-ACP from the polyketide synthase of Streptomyces coelicolor15 and Bacillus brevis nonribosomal peptidyl carrier protein (PCP),16 are also known. The overall structures of these proteins are very similar with three long helices and one short helix arranged in a right-twisted four helical bundle. The conserved serine residue, to which the 4′-PP group is attached, locates at the amino-terminal end of the conserved helix II.
Despite of the extensive effort focused on solving ACP structures, the molecular detail of the interaction between ACP and its target proteins is poorly understood; especially, how a particular acyl-ACP is able to recognize its specific partner enzyme for catalysis is not clear. One of the possible models is that the attachment of each acyl intermediate alters the structure of ACP in such a way that the acyl-ACP intermediate is selectively recognized by its target protein.12 Availability of many ACP structures of different forms makes it feasible to test such hypothesis by pinpointing how the acyl chain modification influences the protein structure by comparing these reported structures. But, a potential drawback in doing the structural comparison is the uncertainty of the observed structure differences which may reflect either the real conformational changes or artifacts arising from different structure determination conditions or approaches. Therefore, although the NMR and crystal structures of E. coli apo-ACP, ACP, and C4:0-ACP have been reported,14,17 we redetermined their solution structures by using NMR methodology under the same condition for data collection and structure calculation to eliminate any experimental differences in elucidating structures. Our structural studies demonstrated that ACP experienced significant conformational change in C4:0-ACP and revealed the potential relationship between the structural modification of ACP and the function.
Results and Discussion
NMR spectroscopy studies and structure determination of different forms of ACP from E. coli
The solution structures of ACP, apo-ACP, and C4:0-ACP were studied under identical conditions. In our studies, we chose to use the butyryl ester of ACP as opposed to other chain lengths, because the conversion efficiency of apo-ACP to acyl-ACP is the best with the four-carbon acyl-CoA substrates using the ACP synthase. Highly purified samples were used in the structural studies. Backbone resonances of 15N/13C-labeled proteins were determined on the basis of HNCO, CBCANH, and HN(CO)CBCA experiments, and side-chain proton resonances were assigned mainly by 15N-edited HSQC-TOCSY and 13C-edited HCCH-TOCSY experiments.18–20 The assignments of side-chain amide protons in some Asp, Glu, and Arg residues were obtained using 15N-edited and 13C-edited HSQC-NOESY experiments. The NOE constraints were obtained from 15N-edited and 13C-edited HSQC-NOESY experiments. All NOE assignments were confirmed by 4D 15N/13C-edited and 4D 13C/13C-edited HMQC-NOESY-HSQC experiments. A total of 1017, 1129, and 1062 NOE distance restraints were identified for apo-ACP, ACP, and C4:0-ACP, respectively. The patterns of short- and medium-range NOEs observed in the 15N-edited and 13C-edited HSQC-NOESY spectra of apo-ACP, ACP, and C4:0-ACP clearly indicate that all three forms of ACP possess three relatively long helices and one short helix.
In the final structural calculations, a total of 300 structures for ACP, apo-ACP, and C4:0-ACP, respectively, were calculated using the program CYANA,21 and 20 structures with the lowest target functions were selected and superimposed as ensembles (see Fig. 1). All experimental NMR constraints were satisfied, and there were no NOE violations above 0.3 Å. Although no dihedral angle data were applied for structural calculations, the backbone conformations of the three structures fit within the Ramachandran plots. The structural statistics for the structures of ACP, apo-ACP, and C4:0-ACP was summarized in Table I. The average root-mean-square deviation (RMSD) values for the backbone atoms (C′, Cα, and N) of residues Glu4–Gly74 of ACP, apo-ACP, and C4:0-ACP are 0.566, 0.546, and 0.503 Å, respectively; the average RMSD values for the heavy atoms of the same residues of ACP, apo-ACP, and C4:0-ACP are 1.07, 1.021, and 0.932 Å, respectively. The structures with the lowest target function were selected from the structural ensembles as the representatives of ACP, apo-ACP, and C4:0-ACP for further analyses.
Figure 1.
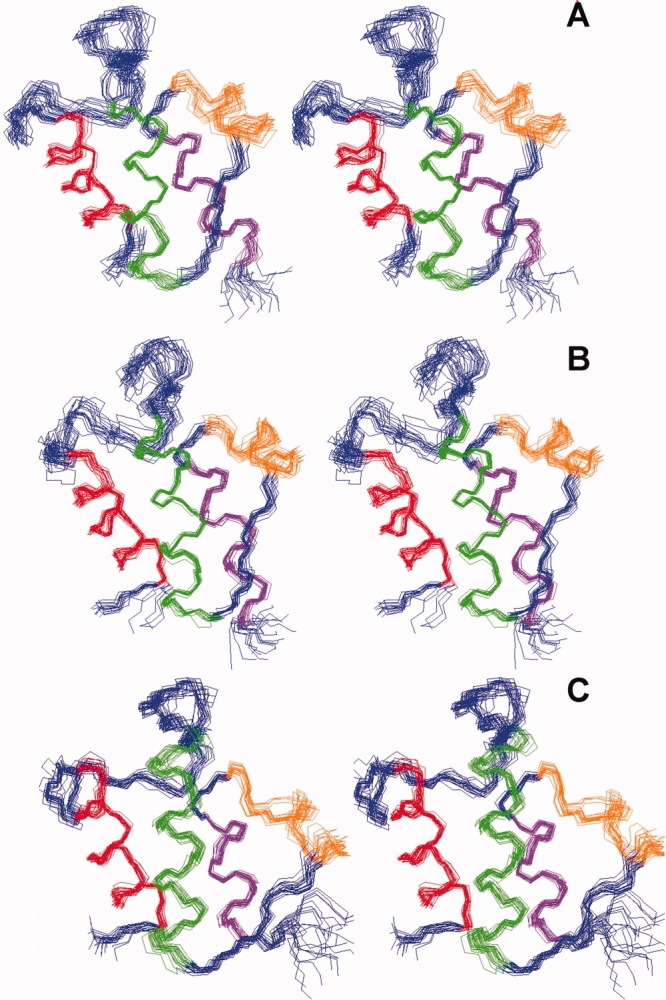
Solution strucures of ACPs. Stereo view of the ensemble of the 20 best structures of apo-ACP (A), ACP (B), and C4:0-ACP (C). Helices I–IV are shown in red, green, orange, and purple, respectively.
Table I.
Statistics for Structures of Apo-ACP, ACP, and C4:0-ACP
| Parameter | apo-ACP | ACP | C4:0-ACP |
|---|---|---|---|
| No. of NOE distance restraints | |||
| Intraresidues | 299 | 343 | 322 |
| Interresidues | |||
| Sequential | 264 | 297 | 277 |
| Medium range ([I, I+2] to [I, I+5]) | 252 | 275 | 281 |
| Long range | 202 | 214 | 177 |
| Total | 1017 | 1129 | 1062 |
| RMSDs from the meansa (Å) | |||
| Overall structure, backbone (4–74) | 0.546 | 0.566 | 0.503 |
| Overall structure, heavy atoms (4–74) | 1.021 | 1.07 | 0.932 |
| Helix region, backbone (helix I, II, IV) | 0.323 | 0.321 | 0.358 |
| Helix region, heavy stoms (helix I, II, IV) | 0.912 | 0.842 | 0.777 |
| No. of residues in Ramachandran plotb (%) | |||
| Most favorable regions | 82.3 | 85.4 | 78 |
| Additionally allowed regions | 14.6 | 14.6 | 22 |
| Generously allowed regions | 2.4 | 0 | 0 |
| Disallowed regions | 0 | 0 | 0 |
The average root-mean-square deviation (RMSD) between the 20 structures of the lowest target functions and the mean coordinates of the protein.
Including 4-helices (4–15, 36–50, 56–62, 66–74) and calculated using Ramachandran macro in DYANA corresponding to the background in the program Procheck.22
E. coli ACP structure has been reported before.17 Superimposing the backbone atoms of 71 residues (residues Glu4–Gly74) of our structure and the previous structure yielded an RMSD value of 4.191 Å. Superposition with only the backbone atoms of the four well-defined helices gave an RMSD value of 3.514 Å. These values are quite large for the same protein. However, it is not surprising to observe this discrepancy because the quality of the previous ACP structure was limited by the available NMR methodology in 1990. In fact, this discrepancy emphasizes the importance of determining NMR structures of ACP under identical conditions for comparison analysis, which allowed us to find subtle conformational variations of ACPs. Recently, the crystal structure of E. coli C4:0-ACP was also determined,14 and we compared the solution structure of C4:0-ACP with the crystal structure of E. coli C4:0-ACP. Superposition of the backbone atoms of residues Glu4–Gly74 of the two structures revealed that the NMR and crystal structures of C4:0-ACP are very similar with an RMSD value of 1.01 Å [Fig. 2 (A)]. Even the long loop I adopts a similar conformation in both structures. The high degree of consistency between the NMR and the crystal C4:0-ACP structures reflects the high quality of our NMR structures.
Figure 2.
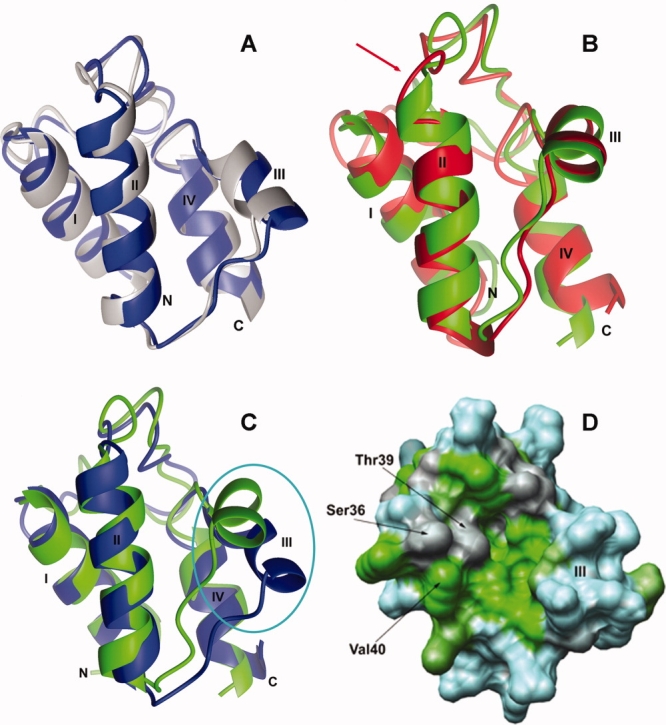
Comparisons of the structures of different forms of ACP. (A) Superposition of the solution (blue) and crystal (gray) structures of C4-0-ACP. (B) The representative structures of apo-ACP (red) and ACP (green) are superimposed by fitting the backbone atoms of the three long helices (residues 4–15, 36–50, and 66–74). The four helices are labeled by Roman numerals and the location of Ser36 is indicated by a red arrow. (C) The representative structures of C4:0-ACP (blue) and ACP (green) were superimposed. The movement of loop II/helix III was circled in cyan. (D) Hydrophobicity of each residue is calculated based on Kyte–Doolittle-hydrophobicity value23 and then visualized on the surface map of solution structure of C4:0-ACP using Chimera program.24 The hydrophobic residues are colored in green, the neutral residues are colored in gray, and the hydrophilic residues are colored in cyan. Three residues in helix II are pointed by black arrow, and helix III is labeled on the surface.
Effects of 4′-phosphopantetheine group on ACP
Active ACP requires a 4′-PP group linked to a conserved serine residue, and the sulfhydryl of the 4′-PP group enables ACP to carry fatty acyl intermediates of different types and lengths. We first examined the effects of 4′-PP group on ACP by analyzing the structures of ACP and apo-ACP. The topologies of ACP and apo-ACP are similar to the other known ACPs,12 consisting of a four-helix bundle with helix I antiparallel to helices II and IV and a short helix III perpendicular to the other three helices. Comparison of the tertiary structures of ACP and apo-ACP was carried out by superimposing the backbone atoms of the three long helices [Fig. 2(B)]. The superposition gave a low RMSD value of 1.133 Å, indicating that ACP and apo-ACP had similar tertiary structures, with the four helices adopting the same packing angle. The significant conformational differences are located in the residues surrounding the 4′-PP group attachment site [Fig. 2(B)]. Analysis of the helical unique NOE patterns indicated Ser36 is a component of the helix II in ACP, whilst this residue is excluded from the helix II in apo-ACP.
In our studies, because of weak interactions as have been previously documented,12,13 we were unable to fully detect NOEs between ACP and the 4′-PP group in the NOESY spectra. However, by comparing the 3D and 4D NOESY spectra, two possible NOEs between the amine group in the unlabeled 4′-PP group and the 13C/15N-labeled residues in helix II (Thr39 and Leu42) were observed. These observations suggest that the 4′-PP prosthetic group is positioned adjacent to the helix II in ACP. Such conclusion is consistent to the reported interactions between 4′-PP group and the protein moiety of holo-ACP from Plasmodium falciparum.25
Effects of acylation on protein conformation of acyl-ACP
ACP carries fatty acyl chains of various types and lengths at the end of 4′-PP group during de novo fatty acid biosynthesis. How an acylation of ACP affects the protein conformation is critical to understand a specific recognition mechanism between acyl-ACPs and their target proteins. To address this question, C4:0-ACP was used in our studies, and a comparison between C4:0-ACP and ACP was performed. Superimposing only the three-long-helix scaffolds in C4:0-ACP onto ACP gave an RMSD value of 1.2 Å [Fig. 2(C)]. These values are similar to the value of 1.1 Å from the superposition of apo-ACP and ACP, demonstrating that the conformations of the three long helices are essentially identical among ACP and C4:0-ACP. However, comparing with ACP, there is a distinct conformational difference in the segment of helix III of C4:0-ACP, which moves away from the helix II [Fig. 2(C)]. Using the distance between the Cα atoms of Thr39 (on helix II) and Asp56 (on helix III) as a “ruler,” the distance increases from 6.86 Å in the ACP structure to 13.53 Å in the C4:0-ACP structure. The observed structural differences between C4:0-ACP and ACP were further verified by analyzing the NOE assignments. Indeed, several long-range NOEs between the helix II and the helix III segment were observed in ACP and apo-ACP, but not in C4:0-ACP. These unique NOE connections indicated that, in ACP and apo-ACP, the helix III is packed close to helix II, resulting in a compact center in the proteins. However, in C4:0-ACP, the helix III moved away from helix II and the “core” of the protein. An important consequence of the movement of helix III in C4:0-ACP structure is the appearance of a flat and broad hydrophobic cleft extending across the surface of C4:0-ACP structure [Fig. 2(D)]. The bottom of the cleft is composed of hydrophobic residues Thr39, Val43, Ile62, and Ile54. The walls of the hydrophobic cleft are lined by, on one side, Ala34-Ser36 around the prosthetic group and Asp56-Thr63 in helix III on the other side. In addition, a hydrophobic cavity is visible in C4:0-ACP structure buried in the helix bundle and is lined by the hydrophobic residues Val7 and Ile11 in helix I, Phe28 and Val29 in loop I and Val65, Gln68, and Ile69 in helix IV. These unique structural features of C4:0-ACP are also noted in the crystal structure.14 Such movement is likely caused by the butyrylation of the 4′-PP group. On the basis of these results, we speculate an accommodation model for the butyrylated prosthetic group on the protein; first, hydrophobic effect spontaneously drives the insertion of hydrophobic butyryl group into the inner pocket of the protein core; second, the insertion leads to the movement of the helix III which in turn forms a hydrophobic cleft to accommodate the exposed part of the butyrylated prosthetic group.
Mechanism of recognition and specificity
ACP has two opposing functions in fatty acid synthesis. As an acyl carrier, it not only must interact with the growing acyl chain and sequester it from the aqueous environment but also has to specifically deliver the appropriate intermediates to enzymes with different biochemical functions. Our study provides new structural information that contributes to the understanding of how ACP performs these two roles. A consequence of the movement of the helix III in the C4:0-ACP structure is the appearance of a flat and broad hydrophobic cleft extending across the surface of C4:0-ACP structure, which otherwise is blocked by the helix III in the other two forms of ACP structures. The walls of the hydrophobic cleft are lined by Ala34-Ser36 of helix II on one side and the Asp56-Thr63 of helix III on the other side, and the bottom of this cleft is composed of hydrophobic residues Thr39, Val43, Ile62, and Ile54. Clearly, such hydrophobic cleft provides protection to the butyrylated prosthetic group.
The movement of the helix III also leads to an alteration in the electrostatic surface properties of ACP; the movement of the helix III in the butyryl-ACP structure leads to the appearance of a negative protruding point formed by Glu57, Glu58, and Glu60. We speculate that such alteration is likely to match the surface features of β-ketoacyl-ACP synthease I or II (FabB or FabF), the enzymes that utilize butyryl-ACP,2 and to facilitate the interaction between C4:0-ACP and FabB or FabF. Typical ACP binding surface on the enzymes consists of positively charged residues imbedded in a hydrophobic patch26,27; therefore, this selectivity mechanism may exist extensively in all acyl-ACP derivatives. Indeed, in a recent report, the importance of conformational diversity between apo and holo forms of PCP, a protein similar to ACP, in enzyme recognition was discussed.28 Our studies extend this “conformational diversity” model and further suggest that each acyl-ACP derivative causes subtle changes in protein conformation that are important in protein recognition. Indeed, these conformational differences may be crucial for ACP to transfer the acyl chains by recognizing proper pathway enzymes. To complete the understanding of this selectivity mechanism, more structures of acyl-ACP derivatives need to be determined and analyzed in the future.
Methods
Expression and purification of apo-ACP
The acpP gene was cloned into the pET11a vector and the uniformly 15N/13C-labeled apo-ACP was expressed in E. coli strain BL21 (DE3) growing in the MOPS minimal medium containing 0.1% 15NH4Cl plus 0.2% [U-13C]-d-glucose. The apo-ACP in the cell-free extract was purified by anion-exchange DE-52 cellulose column, with a step gradient of LiCl followed by gel filtration using 40 mM potassium phosphate buffer pH 6.5. In general, the extent of heavy isotope incorporation averages greater than 99.5% based on mass spectra and the samples yield excellent NMR spectra.
Conversion of apo-ACP to ACP and C4:0-ACP
The purified E. coli AcpS with a His-tag was used to convert apo-ACP to ACP and C4:0-ACP. The reaction mixture for the conversion to ACP contained 2 mM apo-ACP, 10 mM CoA, 2.5 mM DTT, 10 mM MgCl2, and 4 μM AcpS in 50 mM Tris, pH 8.5. After incubation at 37°C for 30 min, the His-tagged AcpS was removed by Ni-NTA agarose (Qiagen) and ACP was purified by gel filtration chromatography. The same method was used to convert apo-ACP to C4:0-ACP, except that 10 mM C4:0-CoA was used instead of CoA and DTT was omitted from the reaction mixture.
NMR spectroscopy
All NMR data were acquired with Varian Inova 600-MHz spectrometers at 27°C. Data were processed and displayed by the program package NMRPipe and NMRDraw29 on an SGI Octane work station. The programs XEASY30 and Sparky31 were used for assignment and data analysis. Similar to previously described ones,12,20 backbone resonances were assigned on the basis of three-dimensional HNCACB and CBCA(CO)NH spectra, and side-chain resonance was assigned on the basis of three-dimensional 15N-edited HSQC-TOCSY and 13C-edited HCCH-TOCSY spectra.
Structural calculation
The NOE connections were assigned based on three-dimensional 15N-edited and 13C-edited NOESY with the help of four-dimensional 15N/13C NOESY and 13C/13C NOESYHMQC-NOESY-HSQC. A total of 1017, 1129, and 1062 meaningful distance constraints for apo-ACP, ACP, and C4:0-ACP, respectively, were derived from NMR data. Integrated NOE peaks were calibrated and converted to distance constraints with the program CALIBA.21 In the final structural determination, the program DYANA32 was used.
Accession codes
The coordinates for apo-ACP, ACP, and C4:0-ACP have been deposited in the Protein Data Bank with accession codes 2K92, 2K93, and 2K94 respectively.
Acknowledgments
We thank Dr. Weixing Zhang for giving support with NMR experiments, and Dr. Charles Ross for giving computer support.
References
- 1.Rock CO, Jackowski S. Forty years of bacterial fatty acid synthesis. Biochem Biophys Res Commun. 2002;292:1155–1166. doi: 10.1006/bbrc.2001.2022. [DOI] [PubMed] [Google Scholar]
- 2.White SW, Zheng J, Zhang YM, Rock CO. The structural biology of type II fatty acid biosynthesis. Annu Rev Biochem. 2005;74:791–831. doi: 10.1146/annurev.biochem.74.082803.133524. [DOI] [PubMed] [Google Scholar]
- 3.Anderson MS, Raetz CR. Biosynthesis of lipid A precursors in Escherichia coli. J Biol Chem. 1987;262:5159–5169. A cytoplasmic acyltransferase that converts UDP- N-acetylglucosamine to UDP-3- O-(R-3-hydroxymyristoyl)- N-acetylglucosamine. [PubMed] [Google Scholar]
- 4.Rock CO, Jackowski S. Regulation of phospholipid synthesis in Escherichia coli. J Biol Chem. 1982;257:10759–10765. Composition of the acyl–acyl carrier protein pool in vivo. [PubMed] [Google Scholar]
- 5.Issartel JP, Koronakis V, Hughes C. Activation of Escherichia coli prohaemolysin to the mature toxin by acyl carrier protein-dependent fatty acylation. Nature. 1991;351:759–761. doi: 10.1038/351759a0. [DOI] [PubMed] [Google Scholar]
- 6.Flugel RS, Hwangbo Y, Lambalot RH, Cronan JE, Jr, Walsh CT. Holo-(acyl carrier protein) synthase and phosphopantetheinyl transfer in Escherichia coli. J Biol Chem. 2000;275:959–968. doi: 10.1074/jbc.275.2.959. [DOI] [PubMed] [Google Scholar]
- 7.Parris KD, Lin L, Tam A, Mathew R, Hixon J, Stahl M, Fritz CC, Seehra J, Somers WS. Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites. Structure Fold Des. 2000;8:883–895. doi: 10.1016/s0969-2126(00)00178-7. [DOI] [PubMed] [Google Scholar]
- 8.Zhang YM, Wu B, Zheng J, Rock CO. Key residues responsible for acyl carrier protein and beta-ketoacyl-acyl carrier protein reductase (FabG) interaction. J Biol Chem. 2003;278:52935–52943. doi: 10.1074/jbc.M309874200. [DOI] [PubMed] [Google Scholar]
- 9.Holak TA, Kearsley SK, Kim Y, Prestegard JH. Three-dimensional structure of acyl carrier protein determined by NMR pseudoenergy and distance geometry calculations. Biochemistry. 1988;27:6135–6142. doi: 10.1021/bi00416a046. [DOI] [PubMed] [Google Scholar]
- 10.Kim Y, Prestegard JH. A dynamic model for the structure of acyl carrier protein in solution. Biochemistry. 1989;28:8792–8797. doi: 10.1021/bi00448a017. [DOI] [PubMed] [Google Scholar]
- 11.Li Q, Khosla C, Puglisi JD, Liu CW. Solution structure and backbone dynamics of the holo form of the frenolicin acyl carrier protein. Biochemistry. 2003;42:4648–4657. doi: 10.1021/bi0274120. [DOI] [PubMed] [Google Scholar]
- 12.Wong HC, Liu G, Zhang YM, Rock CO, Zheng J. The solution structure of acyl carrier protein from Mycobacterium tuberculosis. J Biol Chem. 2002;277:15874–15880. doi: 10.1074/jbc.M112300200. [DOI] [PubMed] [Google Scholar]
- 13.Xu GY, Tam A, Lin L, Hixon J, Fritz CC, Powers R. Solution structure of B. subtilis acyl carrier protein. Structure. 2001;9:277–287. doi: 10.1016/s0969-2126(01)00586-x. [DOI] [PubMed] [Google Scholar]
- 14.Roujeinikova A, Baldock C, Simon WJ, Gilroy J, Baker PJ, Stuitje AR, Rice DW, Slabas AR, Rafferty JB. X-ray crystallographic studies on butyryl-ACP reveal flexibility of the structure around a putative acyl chain binding site. Structure. 2002;10:825–835. doi: 10.1016/s0969-2126(02)00775-x. [DOI] [PubMed] [Google Scholar]
- 15.Crump MP, Crosby J, Dempsey CE, Parkinson JA, Murray M, Hopwood DA, Simpson TJ. Solution structure of the actinorhodin polyketide synthase acyl carrier protein from Streptomyces coelicolor A3(2) Biochemistry. 1997;36:6000–6008. doi: 10.1021/bi970006+. [DOI] [PubMed] [Google Scholar]
- 16.Weber T, Baumgartner R, Renner C, Marahiel MA, Holak TA. Solution structure of PCP, a prototype for the peptidyl carrier domains of modular peptide synthetases. Structure Fold Des. 2000;8:407–418. doi: 10.1016/s0969-2126(00)00120-9. [DOI] [PubMed] [Google Scholar]
- 17.Kim Y, Prestegard JH. Refinement of the NMR structures for acyl carrier protein with scalar coupling data. Proteins. 1990;8:377–385. doi: 10.1002/prot.340080411. [DOI] [PubMed] [Google Scholar]
- 18.Grzesiek S, Bax A. Amino acid type determination in the sequential assignment procedure of uniformly 13C/15N-enriched proteins. J Biomol NMR. 1993;3:185–204. doi: 10.1007/BF00178261. [DOI] [PubMed] [Google Scholar]
- 19.Gronenborn AM, Clore GM. Structures of protein complexes by multidimensional heteronuclear magnetic resonance spectroscopy. Crit Rev Biochem Mol Biol. 1995;30:351–385. doi: 10.3109/10409239509083489. [DOI] [PubMed] [Google Scholar]
- 20.Wong HC, Mao J, Nguyen JT, Srinivas S, Zhang W, Liu B, Li L, Wu D, Zheng J. Structural basis of the recognition of the dishevelled DEP domain in the Wnt signaling pathway. Nat Struct Biol. 2000;7:1178–1184. doi: 10.1038/82047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guntert P, Braun W, Wuthrich K. Efficient computation of three-dimensional protein structures in solution from nuclear magnetic resonance data using the program DIANA and the supporting programs CALIBA, HABAS and GLOMSA. J Mol Biol. 1991;217:517–530. doi: 10.1016/0022-2836(91)90754-t. [DOI] [PubMed] [Google Scholar]
- 22.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 23.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 24.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 25.Sharma AK, Sharma SK, Surolia A, Surolia N, Sarma SP. Solution structures of conformationally equilibrium forms of holo-acyl carrier protein (PfACP) from Plasmodium falciparum provides insight into the mechanism of activation of ACPs. Biochemistry. 2006;45:6904–6916. doi: 10.1021/bi060368u. [DOI] [PubMed] [Google Scholar]
- 26.Zhang YM, Rao MS, Heath RJ, Price AC, Olson AJ, Rock CO, White SW. Identification and analysis of the acyl carrier protein (ACP) docking site on beta-ketoacyl-ACP synthase III. J Biol Chem. 2001;276:8231–8238. doi: 10.1074/jbc.M008042200. [DOI] [PubMed] [Google Scholar]
- 27.Zhang YM, Marrakchi H, White SW, Rock CO. The application of computational methods to explore the diversity and structure of bacterial fatty acid synthase. J Lipid Res. 2003;44:1–10. doi: 10.1194/jlr.r200016-jlr200. [DOI] [PubMed] [Google Scholar]
- 28.Koglin A, Mofid MR, Lohr F, Schafer B, Rogov VV, Blum MM, Mittag T, Marahiel MA, Bernhard F, Dotsch V. Conformational switches modulate protein interactions in peptide antibiotic synthetases. Science. 2006;312:273–276. doi: 10.1126/science.1122928. [DOI] [PubMed] [Google Scholar]
- 29.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 30.Xia T-H, Bartels C, Wuthrich K. XEASY ETH automated spectroscopy for X window system user manual. Zurich: ETH-Honggerberg; 1993. [Google Scholar]
- 31.Goddard TD, Kneller DG. SPARKY, 3.0. San Francisco: University of California; 1998. [Google Scholar]
- 32.Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]


