Abstract
Osteogenesis imperfecta (OI) is a genetic disease characterized by fragile bones, skeletal deformities and, in severe cases, prenatal death that affects more than 1 in 10,000 individuals. Here we show by full atomistic simulation in explicit solvent that OI mutations have a significant influence on the mechanical properties of single tropocollagen molecules, and that the severity of different forms of OI is directly correlated with the reduction of the mechanical stiffness of individual tropocollagen molecules. The reduction of molecular stiffness provides insight into the molecular-scale mechanisms of the disease. The analysis of the molecular mechanisms reveals that physical parameters of side-chain volume and hydropathy index of the mutated residue control the loss of mechanical stiffness of individual tropocollagen molecules. We propose a model that enables us to predict the loss of stiffness based on these physical characteristics of mutations. This finding provides an atomistic-level mechanistic understanding of the role of OI mutations in defining the properties of the basic protein constituents, which could eventually lead to new strategies for diagnosis and treatment the disease. The focus on material properties and their role in genetic diseases is an important, yet so far only little explored, aspect in studying the mechanisms that lead to pathological conditions. The consideration of how material properties change in diseases could lead to a new paradigm that may expand beyond the focus on biochemical readings alone and include a characterization of material properties in diagnosis and treatment, an effort referred to as materiomics.
Keywords: steered molecular dynamics, osteogenesis imperfecta, Young's modulus, collagen, nanomechanics, genetic disease, protein material, materiomics
Introduction
Patients affected by osteogenesis imperfecta (OI) exhibit an array of associated symptoms, including short stature, loose joints, blue sclearae, dentinogenesis imperfecta, hearing loss, and neurological and pulmonary complications.1,2 The classification of OI is commonly based on clinical features that led to four different groups, from mild (OI type I) to severe (OI type III and IV) to perinatal lethal (OI type II).3 The genetic basis for about 90% of the form of this disease lies in mutations of type I collagen genes,4 as tabulated in the database of human collagen mutations (URL: http://www.le.ac.uk/genetics/collagen).5 Missense mutations that alter a glycine (Gly) codon in the genes encoding the characteristic collagen triple helix are the most common causes of OI.4 The replacement of either guanine (G) residue in the glycine codon (GGC) can theoretically result in the replacement of eight different amino acids: serine (Ser), cysteine (Cys), alanine (Ala), valine (Val), aspartic acid (Asp), glutamic acid (Glu), arginine (Arg), and tryptophan (Trp). All possibilities have been described in conjunction with OI, although the frequency with which the different mutations occur varies considerably, with tryptophan replacements being exceedingly rare.6 Although a general correspondence between the specific mutations and the severity of OI has been reported, the molecular mechanisms of how a single point mutation can alter the susceptibility of an entire bone to brittle fracture are still unknown.
Many studies have attempted to correlate glycine mutation types and locations with phenotypic severity. Some trends are apparent, such as OI severity increasing with an amino to carboxyl terminal orientation and with substitution by large and charged amino acids.7–9 At present, however, genotype–phenotype correlations are too weak to accurately predict the phenotypic effect of a particular glycine mutation. Indeed, the answer to the question how a single point mutation in the tropocollagen molecule can cause the failure of the entire skeletal system still remains a mystery. In particular, it remains unclear at what level in the tissue structure the mutations influence the behavior. To address these points, investigations at all hierarchical levels must be carried out, beginning at the molecular scale. This should begin with an investigation of the effects at the level of single molecules, followed by studies of the effects at the microfibrillar level (interaction of different peptides, effects on mineral crystal growth and distribution), eventually incorporating fibrillar and larger-scale levels (investigation of composite materials featuring protein and mineral components).
Here we report a series of systematic molecular scale experiments, carried out using a molecular dynamics simulation approach that provides us with the ability to probe molecular mechanics at physiologically relevant ultraslow loading rates (details about the computational experiments see Materials and Methods, as well as Ref. 10). We consider seven collagen-like peptides with a glycine mutation as shown in Figure 1(a). The goal of the study reported here is to investigate the effect of these OI mutations on the mechanical properties of a single tropocollagen molecule under tensile stretch [Fig. 1(b)], a physiologically relevant mechanical loading condition. Individual tropocollagen molecules are subjected to tensile stretch when the load is applied at larger hierarchical length scales in tissues, such as in collagen fibrils or fibers.11–13 This illustrates the significance of considering the tensile elastic properties of tropocollagen molecules to provide a fundamental description of the effect of OI on the material properties. The mechanical stiffness of individual molecules is measured by the Young's modulus, which is defined as the proportionality between stress and strain.
Figure 1.
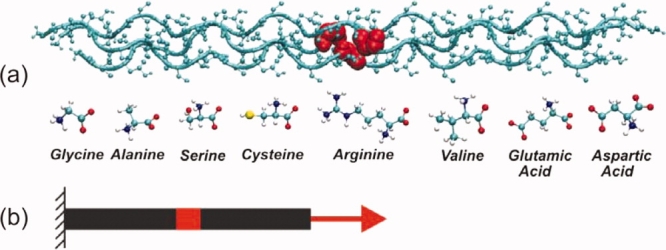
Tropocollagen molecular model and loading conditions. Panel (a): Molecular geometry of the tropocollagen molecule, indicating the residues that are replaced in the mutation. All tropocollagen-like peptides considered in this study share the same structure, consisting of three identical chains made of gly-pro-hyp triplets: [(GPO)5-(XPO)-(GPO)4]3. The X position of each chain (highlighted in red in the upper part) is one of seven replacing residues related to OI (lower part). Panel (b) depicts the loading condition, subjecting the tropocollagen molecule to tensile deformation. The N-terminus of the molecule is kept fix with a strong position restrain, while the C-terminus is linked to a moving spring. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Results and Discussion
The Young's modulus of the reference tropocollagen molecule is determined to be 3.96 ± 0.21 GPa (see Table I), in agreement with corresponding experimental results.14,15 However, molecules with glycine mutation display softer mechanical properties than the reference peptide, with values ranging from 3.37 ± 0.32 GPa to 3.78 ± 0.29 GPa. A decrease in Young's modulus up to 15% is observed, depending on the specific type of replacing residue [see Table I and Fig. 2(a)]. Since the average values are close and standard deviations large, we performed an ANOVA analysis to assess the statistical significance of the results. Considering a significance α = 0.05, the critical F value is equal, in this case, to 2.947. The test gives for our data an F value of 3.500, which is higher than the critical F value, and thus we can reject the null hypothesis (i.e., that there is no effect of mutations on Young's modulus). On the other hand, the null hypothesis would have a probability P = 0.011.
Table I.
Type of Glycine Mutation, Tropocollagen Young's Modulus, and Severity of OI
| Replacing residue | Young's modulus (GPa) | Severity (%) |
|---|---|---|
| Glycine10 | 3.96 ± 0.21 | N/A |
| Alanine | 3.78 ± 0.29 | 41.6 (5/12)a |
| Arginine | 3.69 ± 0.23 | 45.8 (22/48) |
| Cysteine | 3.74 ± 0.11 | 47.8 (22/46) |
| Serine | 3.51 ± 0.10 | 65.0 (41/63) |
| Valine | 3.91 ± 0.24 | 65.2 (15/23) |
| Glutamic acid | 3.38 ± 0.24 | 66.6 (6/9) |
| Aspartic acid | 3.37 ± 0.32 | 87.5 (35/40) |
All peptides featuring a glycine replacement show slight but statistically significant lower Young's modulus, which are correlated with the severity of the OI forms because of the different mutations. The severity is here defined as the ratio between the severe occurrences (leading to OI type II and type III) and the total occurrences of each considered glycine replacement. A total of 241 mutations are considered in this analysis.5
Values in parentheses indicate ratio of severe/total occurrences.5
Figure 2.
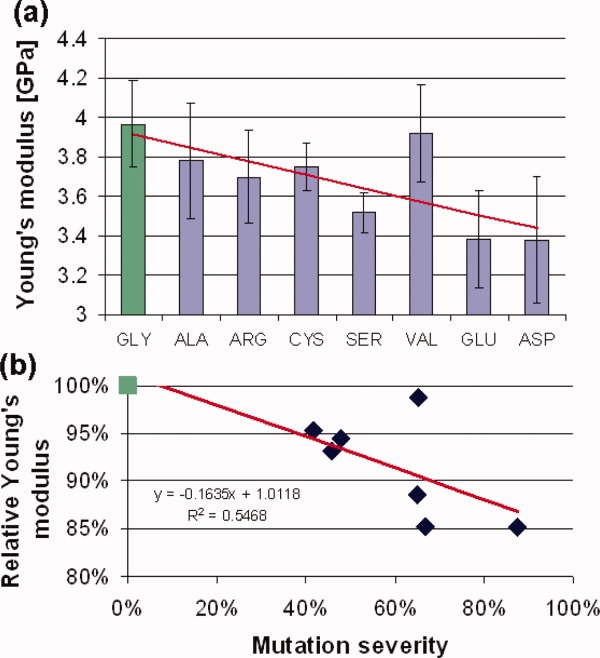
Young's modulus of the different peptides as a function of the glycine replacement [panel (a)] and as a function of OI severity [panel (b)]. In panel (a), the mechanical properties of each different tropocollagen molecule are depicted as a function of the replacing amino acid residues, which are ordered based on the resulting disease severity, that is, from the physiological glycine (left) to the most severe OI mutation, aspartic acid (right). Panel (b) shows the Young's modulus relative to that of the reference (glycine) as a function of the mutation severity. The severity parameter, introduced for the first time in this work, is meant to quantify the degree of the OI severity due to specific mutations. It is generally accepted that some mutations leads to more severe phenotype.8 The severity parameter quantify this trend, using all available data on OI mutations, that is more than 200 occurrences catalogued in the Database of Human Collagen Mutations (http://www.le.ac.uk/genetics/collagen).5 [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
The observed mechanical properties are local, because of the truncated model considered in our study, and therefore the mechanical properties of the overall molecule are probably unaffected. However, local changes as observed in this work could have a tremendous impact (in ways yet to be established) on the onset of the disease, which is characterized by catastrophic failure of bone. Catastrophic failure of materials typically begins with a very small defect, which grows until catastrophic failure occurs. This is a hallmark of brittle fracture, as it is well established in the materials science field (see, e.g. Refs. 16–18). In such models, stress concentrations develop at small crack-like flaws, which represent regions of structurally soft and weak material. Thereby, the crack tip provides a mathematical singularity for stresses even at finite applied load, 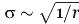 , where r is the distance from a flaw and σ is the resulting stress. In this sense, one can understand the significance of local effects in inducing catastrophic failure of a material due to a local defect, as they magnify a small stress applied at the overall tissue level to a much greater value at a local scale, where it can lead to a permanent damage of tissue and grow into macroscopic failure.16–18 Furthermore, these peptides feature only Gly-Pro-Hyp triplets around the mutations, and these triplets are considered the most stabilizing. These triplets are also the most common, but still represents only ≈10% of the total amount (in human collagen type I). Thus, mutations surrounded by different (and in particular, less stabilizing) triplets could lead to much stronger effects, and therefore, the results presented here may be a conservative lower-bound estimate of the effects.
, where r is the distance from a flaw and σ is the resulting stress. In this sense, one can understand the significance of local effects in inducing catastrophic failure of a material due to a local defect, as they magnify a small stress applied at the overall tissue level to a much greater value at a local scale, where it can lead to a permanent damage of tissue and grow into macroscopic failure.16–18 Furthermore, these peptides feature only Gly-Pro-Hyp triplets around the mutations, and these triplets are considered the most stabilizing. These triplets are also the most common, but still represents only ≈10% of the total amount (in human collagen type I). Thus, mutations surrounded by different (and in particular, less stabilizing) triplets could lead to much stronger effects, and therefore, the results presented here may be a conservative lower-bound estimate of the effects.
We find that mutations related to more severe phenotypes are associated with softer tropocollagen mechanical properties, as illustrated in Figure 2(b). This is confirmed by a linear curve fit to the Young's modulus results over the severity of the disease for the particular mutations. The severity parameter is calculated as the ratio between the severe occurrences (leading to OI type II and type III) and the total occurrences of each considered glycine replacement.5 Through this analysis, we obtained a numerical parameter based on a well-established classification. We note that a correlation does not necessarily imply causality. Nonetheless, it is known that OI mutations affect the mechanical properties of collagenous tissues, and thus it provides the basis for the hypothesis that an effect might also be observed at the molecular level. The ANOVA analysis, which indeed suggests an effect of mutations on the Young's modulus, confirm that the observed correlation is likely due to causality.
We observe that the mechanical properties of collagen peptides are affected, to a varying degree, as a consequence of glycine mutations. Thus, we hypothesize that some properties of the replacing amino acids must affect the peptide mechanical properties, in this case the Young's modulus. The amino acids can in principle be characterized through a variety of physical properties. However, here we focus on the hydropathy index (H) (a measure of how hydrophobic or hydrophilic an amino acid is19) and the residue volume (V). These two parameters have been chosen, since there exists evidence in the literature that these physical properties are related to the severity degree of the OI disease, as pointed out in earlier work.8,9 We find that the molecule's stiffness can be expressed as a function of these two factors. This is achieved in an empirical relation with five parameters, in a simple model:
| (1) |
We fit the parameters a, b, c, d, and e in Eq. (1) based on the results for six peptides (Gly, Ala, Ser, Cys, Arg, Val), for a total of 24 independent data points. This then enables us to predict the Young's modulus for the remaining two peptides (Glu and Asp) that were not included in the initial fitting. A good agreement between the results of the simulations and the value predicted using Eq. (1) is found (see Fig. 3). This suggests that the physical parameters of side-chain volume and hydropathy index of the replacing residue control the loss of mechanical stiffness, and thus our results corroborate the hypothesis that these two parameters are important in determining the severity degree of the disease (albeit future studies could focus on other physical parameters).
Figure 3.
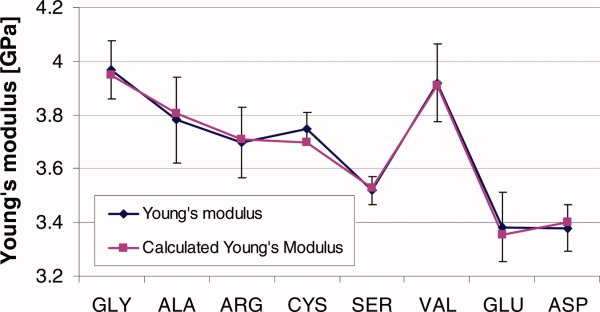
Measured and predicted value of Young's modulus. The plot shows Young's modulus as measured from atomistic simulation for each tropocollagen molecule (blue) is compared with the mechanical properties calculated using Eq. (1) (purple). The results from the first six peptides (Gly-Val) are use to fit the parameters of Eq. (1), which then predicted the Young's modulus of the remaining two peptides (Glu and Asp). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Figure 4 represents a contour graph of Young's modulus as predicted from the side-chain volume and the hydropathy index of the replacing residue. This plot illustrates that the stiffness decreases when glycine is replaced by larger residues. This observation is consistent with the fact that each third residue in collagen triple helix turn is tucked into the sterically constricted internal space of the molecule. Glycine, being the smallest amino acid, is the only one that can fit this position without destabilizing the triple helix8 (see Ramachandran analysis shown in Fig. 5). Figure 4 illustrates also that the mechanical properties of a single molecule are reduced with mildly hydrophilic replacing residues. This could be explained taking into account that residues with a high hydropathy index fit well in the hydrophobic protein core of tropocollagen; on the other hand, highly hydrophilic residues could partially compensate the destabilizing effect on the triple helix through a larger number of water-mediated H bonds.20 Indeed, the valine mutation is the only one that does not follow the general trend, being a mutation that usually leads to a severe phenotype while not reducing the mechanical properties of the peptide. The reason is not obvious, but could be related to the high hydropathy index: since the position usually occupied by glycine is tucked into the core of the protein, a substitution by a highly hydrophobic residue such as valine would lead to lower disruption of the triple helix. This is indeed observed in the molecular structure of the peptides at the end of the equilibration stage. While the other mutations affect the coiling of the molecule, the valine mutation does not (see Fig. 5).
Figure 4.
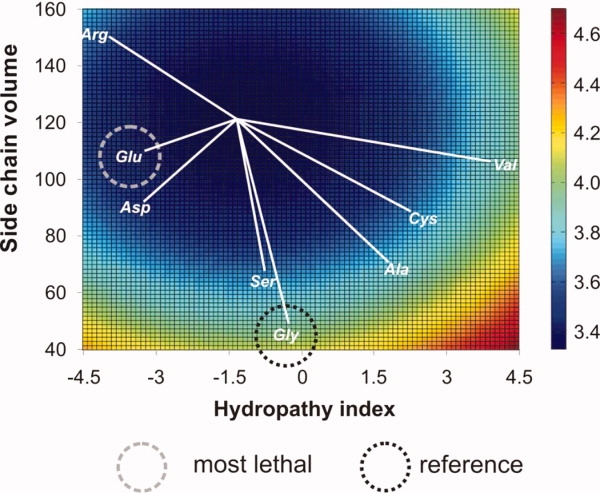
Influence of the hydropathy index and the side-chain volume of replacing residues on tropocollagen Young's modulus. Most severe cases of OI fall into the dark blue domain, with a hydropathy index of −3.75 and a side-chain volume of ≈100 Å3. The most severe mutations feature the smallest distance from the center of the dark blue domain (distances of individual cases to the center of the dark blue domain are indicated by white lines). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Figure 5.
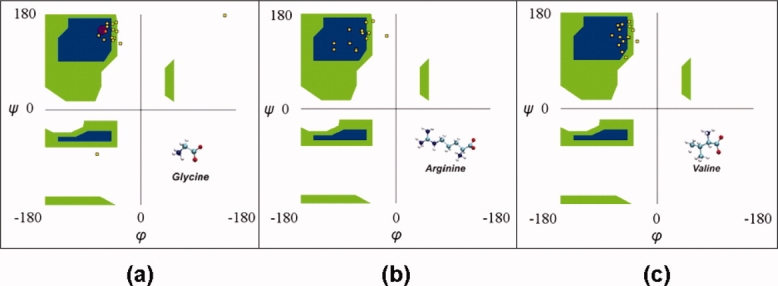
Ramachandran plot relative to the mutation positions and to the two adjacent amino acids in each direction. In the reference case [panel (a)] the configuration is close to that of the polyproline II chain [Psi = 150°; Phi = −75°; red circle in panel (a)]. The presence of the mutations alters the coiling of the molecule, shown by the scattering in the Phi and Psi values [panel (b)]. In the case of valine [panel (c)] this scattering is reduced, leading to a coiling similar to the reference case. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
The results of our investigation show that the mechanical properties of collagen peptides are affected by glycine mutations, and that the degree of this reduction is related to the type of mutation. The underlying physical reason seems to be related to the degree of helical disruption, since glycine substitutions lead to local disruption of the triple helical arrangement. This is not observed in the case of valine mutations, and indeed this is reflected in the mechanical properties of the peptide, which are similar to those of the reference peptide. However, despite the structural changes, no effect on the number of interchain H-bonds is observed, which can explain the relative slight reduction of mechanical properties for mutated peptides.
It has been speculated earlier that the type of mutation, together with the position along the triple helix, influences the phenotypic severity of OI. In particular it has been observed that large and charged residues generally lead to more severe OI types.7–9 Here we have shown that the local mechanical properties of single tropocollagen molecules can be predicted based on the physical properties of the replacing residue.
Conclusions
The results reported here may contribute to a better understanding of the molecular mechanisms underlying OI and could eventually enable us to establish a direct link from the scale of genetic mutations to the macroscale phenomenon of brittle bones. The present study shows that the loss of stiffness in tropocollagen molecules and the severity of OI are correlated. Our findings suggest that the changes in the molecular structure due to the OI mutations change properties already at the single molecule level. This is in contrast to recent studies of mutations related to muscle dystrophies21 that have shown that the properties of individual α-helical protein molecules are not affected, suggesting that the disease has its origin at larger hierarchical levels. The mechanism involved, however, is still unclear. The development of understanding of how structural modifications lead to the changes in the mechanical properties, as observed in the present work, is critically important on the path of forming a more holistic picture of OI. Extending this work to consider larger length scales, multiple hierarchical levels or other structural modifications is a complex task that will be the aim of future works. Predicting the folded structure of collagenous protein structures, for example, using approaches developed in Pande's lab,22 could be used in conjunction with mechanical analyses as reported here.
We note that the results reported here alone cannot fully explain the molecular mechanism of the disease, and other effects must be taken into consideration, such as effects at larger hierarchical scales. However, this is a first step, which shows that a single point mutation (i.e., 1 residue out of 30, corresponding to 3.3% of the total length) can affect the molecular mechanical properties by as much as 15%. In order to evaluate the importance of these results, it is important to consider that OI itself is based on apparently negligible effects: a single point mutation in a triple helix of more than 1000 residues (i.e., less than 0.1% of the total residues) may lead to failure of the entire skeletal system. Thus, in order to understand the disease mechanism of OI, we should not neglect local effects simply based on the hypothesis that they cannot immediately explain the entire phenomenon. Rather, we should expand from these findings to continue the investigation to include more structural length scales (fibrils, fibers, mineralized fibrils, etc.). Along similar lines, as discussed earlier, it is well established in the materials science field that local defects and flaws can lead to catastrophic brittle failure, because of large interatomic/intermolecular forces that generate local shear regions because of localized stress magnifications that greatly exceed the stress applied at a global, tissue scale. This concept, together with the findings put forward in this article provides a possible avenue of future investigation.16–18
The present study has been focused solely at molecular scale phenomena. For understanding the more macroscopic and therefore clinically relevant aspects of OI, one must consider the effects of the change of single molecule properties on larger-scale tissue properties. This could be addressed by using a multiscale scheme of collagen modeling as reported in earlier studies,11,13,23,24 which could be based on the results reported here. A careful atomistic-scale molecular dynamics study of the properties of the material's basic constituents as reported here is crucial for the advancement of these models, as they provide the fundamental input parameter of the constitutive behavior of the material's basic building blocks. The effect of locally softened domains on the behavior of collagen fibrils could be explored.
After identifying the entire genetic code of several species, an outstanding grand challenge in the life sciences is the understanding of the multiscale behavior of hierarchical protein assemblies. The advancement of this field is crucial for studies of biological systems, disease diagnosis, and treatment, as well as the design of novel biomaterials. The focus on material properties and their role in diseases is an important, up to date little explored aspect in studying the mechanisms that lead to pathological conditions. Thereby, the consideration of how material properties change in diseases could lead to a new paradigm that may expand beyond the focus on biochemical readings alone, an effort referred to as materiomics. This could eventually be important for both disease diagnosis and treatment.
Methods
Single molecule full atomistic models in explicit water
The mechanical properties of tropocollagen molecules are investigated using steered molecular dynamics (SMD) simulations, submitting the tropocollagen molecule to traction along the principal axis, that is, from the N-terminus to the C-terminus [see inlay in Fig. 1(b)].
We build tropocollagen molecule models with various sequences using the software THeBuScr (Triple-Helical collagen Building Script).25,26 We choose the simplest model of tropocollagen, with only Gly-Pro-Hyp (GPO) triplets on each of the three chains as the reference system (Hyp and O are, respectively, the three-letter code and single-letter code for the amino acid hydroxy proline). The central triplet is used to introduce the Gly replacement. The peptide structure is [(GPO)5-(XPO)-(GPO)4]3, where the X position is occupied by alanine, serine, cysteine, arginine, valine, glutamic acid, or aspartic acid [see Fig. 1(a)]. For all the peptides, the N-terminals are assumed protonated while the C-terminals are assumed deprotonated. The tropocollagen models we use are truncated at 30 amino acids per chain to reduce computational costs. This leads to short-length tropocollagen segments with a length of ∼8 nm. For reasons of comparison, peptides of comparable length were used both in computational and experimental studies.8,27–30
Because of the short length (the molecule considered here are shorter than its persistence length of ∼10–20 nm), entropic elasticity is a minor aspect and its effects are not significant in the present model. This approach enables us to focus on energetic elastic effects15,23 and their change under OI mutations.
Model equilibration
Molecular dynamics simulations are performed using the GROMACS code31,32 and the GROMOS96 43a1 force field, as used in earlier studies,10 which includes also parameters for the hydroxy proline (HYP) residue. The protein molecules are entirely solvated in a 15 nm × 3 nm × 3 nm periodic water box (which ensure a minimum distance of 0.8 nm between the protein and the box edge). Single point charge water molecules is used for the solvent, leading to a total of ≈12,800 atoms for each system. SETTLE (for water) and LINCS algorithms are used to constrain covalent bond lengths involving hydrogen atoms, thus allowing a time step of 2 fs. Nonbonding interactions are computed using a cutoff for neighbor list at 1 nm, with a switching function between 0.8 and 0.9 nm for Van der Waals interactions, while the Particle-Mesh Ewald sums method is applied to describe electrostatic interactions. In the case of charged peptides, counter ions (Cl− or Na+) were added to keep the system neutral. The preliminary system energy minimization is performed by using a steepest descent algorithm until convergence or for a maximum of 10,000 steps. The system is then equilibrated at a temperature of 310 K (37°C) for 1200 ps of molecular dynamics. The entire protein is held fixed for the first 200 ps by restraining the atomic positions, and thereafter only the first and the last Cα-atoms of each chain are restrained for the following 1000 ps. We observe that, even in the presence of large mutant amino acids, the root-mean-square deviation of the proteins reaches a stable value within the 1-ns simulation time. Thus, we assume that the peptides are equilibrated properly.
SMD approach
Four different configurations are extracted during the last 300 ps of equilibration for each peptide, and are then used as independent starting points for subsequent SMD simulations. This leads to a total of 32 cases (i.e., four replicas of eight peptides). In each case, the center of mass of the three N-terminal Cα atoms is kept fixed by means of a strong harmonic restrain with a spring constant of 3 × 105 kJ mol−1 nm−2, while the center of mass of the three C-terminal Cα atoms (the pulled group of atoms) is linked to a spring with an elastic constant kspring = 4000 kJ mol−1 nm−2, which is moved along the direction of the molecular axis with a velocity of 0.1 m s−1. Each SMD simulation is carried out to model a time span of 70 ns for each peptide, leading to a total simulation time of more than 2 μs. The choice of the pulled and reference groups is arbitrary, but since the models are symmetric, we do not expect a different behavior if the N-terminus was pulled instead of the C-terminus. The pulling rate is chosen based on previous results, in which we showed that the mechanical properties of tropocollagen molecules are rate dependent and converge to a finite value for pulling velocities <1 m s−1 (please see Ref. 10 and discussion mentioned later in the section “Rate dependence in molecular simulation studies”).
All molecular dynamics simulations are carried out in an NPT ensemble, with the systems coupled to a heat bath at 310 K (coupling constant of 0.1 ps and Berendsen thermostat) and to an hydrostatic bath at 1 atm (coupling constant of 0.5 ps and Berendsen barostat). The force applied to the tropocollagen molecule by the virtual spring is
| (2) |
where xspring and xpull represent the spring's and the pulled group's positions, respectively.
Definition of elastic properties: Young's modulus, stress, and strain
Young's modulus relates stress and strain and is a commonly used engineering measure to characterize the stiffness of a material. The molecular elastic spring constant of tropocollagen, kTC, is calculated by fitting the force F versus ΔL relationship (the parameter L is the tropocollagen molecular length) and by considering the value of its derivative. Since the stress–strain relationship is nonlinear there are different possible choices of how to calculate kTC and thus Young's modulus. We chose to consistently consider the value at 8% applied strain. For strains smaller than 8%, the tropocollagen molecules are crimped, so they are not under tension (the molecules are slightly bent in the initial configuration). The result thus represents Young's modulus at small strains.
The Young's modulus Y is calculated based on
| (3) |
The strain is given by
| (4) |
where ɛ is the engineering strain in the measured direction. In Eqs. (3) and (4), L and L0 are the current and the initial tropocollagen length, respectively; F is the force inducing the molecule to elongate by ΔL = L − L0. The parameter A denotes the cross-sectional area of the tropocollagen molecule, obtained from the ratio between the molecular volume and L0, by assuming a cylindrical shape. Note that the molecular stress is defined as σ = F / A.
In order to estimate the statistical accuracy of the determined elastic moduli, we perform four simulations with different initial configurations for each case studied (i.e., for each of the eight peptides) and determine the average values and standard deviations.
Empirical relationship between biochemical and mechanical properties
We express the molecule's stiffness as a function of two main biochemical properties of mutant amino acids, that is the residue volume and the hydropathy index. This is achieved in an empirical relation with five parameters [see Eq. (1)]. We fit the parameters a, b, c, d, and e in Eq. (1) based on the results for six peptides (Gly, Ala, Ser, Cys, Arg, Val), for a total of 24 independent data points. This then enables us to predict the Young's modulus for the remaining two peptides (Glu and Asp) that are not included in the initial fitting. We use five parameters, since it is the minimum number in order to capture the quadratic trend relative to both side-chain volume and hydropathy index. Indeed, if fewer parameters are used, it leads to greater errors in the least square analysis and poorer prediction of the Young's moduli of the peptides not included in the fitting.
Rate dependence in molecular simulation studies
Molecular dynamics simulations are typically carried out at very high deformation rates. We have therefore taken special care in the interpretation of the results, and ensured that the measured elastic properties do not depend on the deformation speed. We previously studied the mechanical behavior of tropocollagen in terms of its Young's modulus for under a systematic variation of loading rates.10 It was found that for pulling rates lower than 1 m s−1 the Young's modulus of single tropocollagen molecules is independent on the loading rate. All simulations reported in this paper are thus carried out at a loading rate of 0.1 m s−1 and therefore in the regime in which the Young's modulus has converged.
Acknowledgments
The authors are most grateful for helpful suggestions put forth by the reviewers of this manuscript.
References
- 1.Primorac D, Rowe DW, Mottes M, Barisic I, Anticevic D, Mirandola S, Lira MG, Kalajzic I, Kusec V, Glorieux FH. Osteogenesis imperfecta at the beginning of bone and joint decade. Croat Med J. 2001;42:393–415. [PubMed] [Google Scholar]
- 2.Rauch F, Glorieux FH. Osteogenesis imperfecta. Lancet. 2004;363:1377–1385. doi: 10.1016/S0140-6736(04)16051-0. [DOI] [PubMed] [Google Scholar]
- 3.Sillence DO, Senn A, Danks DM. Genetic heterogeneity in osteogenesis imperfecta. J Med Genet. 1979;16:101–116. doi: 10.1136/jmg.16.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozloff KM, Carden A, Bergwitz C, Forlino A, Uveges TE, Morris MD, Marini JC, Goldstein SA. Brittle IV mouse model for osteogenesis imperfecta IV demonstrates postpubertal adaptations to improve whole bone strength. J Bone Miner Res. 2004;19:614–622. doi: 10.1359/JBMR.040111. [DOI] [PubMed] [Google Scholar]
- 5.Dalgleish R. The human type I collagen mutation database. Nucleic Acid Res. 1997;25:181–187. doi: 10.1093/nar/25.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roughley PJ, Rauch F, Glorieux FH. Osteogenesis imperfecta—Clinical and molecular diversity. Eur Cells Mater. 2003;5:41–47. doi: 10.22203/ecm.v005a04. [DOI] [PubMed] [Google Scholar]
- 7.Yang W, Battineni ML, Brodsky B. Amino acid environment modulates the disruption by osteogenesis imperfecta glycine substitutions in collagen-like peptides. Biochemistry. 1997;36:6930–6935. doi: 10.1021/bi970051h. [DOI] [PubMed] [Google Scholar]
- 8.Beck K, Chan VC, Shenoy N, Kirkpatrick A, Ramshaw JAM, Brodsky B. Destabilization of osteogenesis imperfecta collagen-like model peptides correlates with the identity of the residue replacing glycine. Proc Natl Acad Sci USA. 2000;97:4273–4278. doi: 10.1073/pnas.070050097. doi: 10.1016/j.jmbbm.2008. 03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byers PH. Folding defects in fibrillar collagens. Philos Trans Royal Soc Lond B Biol Sci. 2001;356:151–157. doi: 10.1098/rstb.2000.0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gautieri A, Buehler MJ, Redaelli A. Deformation rate controls elasticity and unfolding pathway of single tropocollagen molecules. J Mech Behav Biomed Mater. doi: 10.1016/j.jmbbm.2008.03.001. (in press) doi: 10.1016/j.jmbbm.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Buehler MJ. Nature designs tough collagen: explaining the nanostructure of collagen fibrils. Proc Natl Acad Sci USA. 2006;103:12285–12290. doi: 10.1073/pnas.0603216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta HS, Seto J, Wagermaier W, Zaslansky P, Boesecke P, Fratzl P. Cooperative deformation of mineral and collagen in bone at the nanoscale. Proc Natl Acad Sci USA. 2006;103:17741–17746. doi: 10.1073/pnas.0604237103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buehler MJ. Molecular architecture of collagen fibrils: a critical length scale for tough fibrils. Curr Appl Phys. 2008;8:440–442. [Google Scholar]
- 14.Sasaki N, Odajima S. Stress–strain curve and Young's modulus of a collagen molecule as determined by the X-ray diffraction technique. J Biomech. 1996;29:655–658. doi: 10.1016/0021-9290(95)00110-7. [DOI] [PubMed] [Google Scholar]
- 15.Sun YL, Luo ZP, Fertala A, An KN. Stretching type II collagen with optical tweezers. J Biomech. 2004;37:1665–1669. doi: 10.1016/j.jbiomech.2004.02.028. [DOI] [PubMed] [Google Scholar]
- 16.Hirth JP, Lothe J. Theory of dislocations. New York: Wiley-Interscience; 1982. [Google Scholar]
- 17.Broberg KB. Cracks and fracture. San Diego: Academic Press; 1999. [Google Scholar]
- 18.Phillips R. Crystal, defects and microstructure—modeling across scales. Cambridge: Cambridge University Press; 2001. [Google Scholar]
- 19.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 20.Melacini G, Bonvin AJJ, Goodman M, Boelens R, Kaptein R. Hydration dynamics of the collagen triple helix. J Mol Biol. 2000;300:1041–1048. doi: 10.1006/jmbi.2000.3919. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, Ackbarow T, Buehler MJ. Muscle dystrophy single point mutation in the 2B segment of lamin A does not affect the mechanical properties at the dimer level. J Biomech. 2008;41:1295–1301. doi: 10.1016/j.jbiomech.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 22.Park S, Klein T, Pande V. Folding and misfolding of the collagen triple helix: Markov analysis of molecular dynamics simulations. Biophys J. 2007;93:4108–4115. doi: 10.1529/biophysj.107.108100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buehler MJ, Wong SY. Entropic elasticity controls nanomechanics of single tropocollagen molecules. Biophys J. 2007;93:37–43. doi: 10.1529/biophysj.106.102616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buehler MJ. Nanomechanics of collagen fibrils under varying cross-link densities: atomistic and continuum studies. J Mech Behav Biomed Mater. 2008;1:59–67. doi: 10.1016/j.jmbbm.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Rainey J, Goh M. A statistically derived parameterization for the collagen triple-helix. Protein Sci. 2004;13:2276. doi: 10.1110/ps.0218502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rainey J, Goh M. An interactive triple-helical collagen builder. Bioinformatics. 2004;20:2458–2459. doi: 10.1093/bioinformatics/bth247. [DOI] [PubMed] [Google Scholar]
- 27.Bella J, Eaton M, Brodsky B, Berman HM. Crystal-structure and molecular-structure of a collagen-like peptide at 1.9-angstrom resolution. Science. 1994;266:75–81. doi: 10.1126/science.7695699. [DOI] [PubMed] [Google Scholar]
- 28.Lorenzo AC, Caffarena ER. Elastic properties, Young's modulus determination and structural stability of the tropocollagen molecule: a computational study by steered molecular dynamics. J Biomech. 2005;38:1527–1533. doi: 10.1016/j.jbiomech.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 29.Vesentini S, Fitie CFC, Montevecchi FM, Redaelli A. Molecular assessment of the elastic properties of collagen-like homotrimer sequences. Biomech Model Mechanobiol. 2005;3:224–234. doi: 10.1007/s10237-004-0064-5. [DOI] [PubMed] [Google Scholar]
- 30.Buehler MJ. Atomistic and continuum modeling of mechanical properties of collagen: elasticity, fracture and self-assembly. J Mater Res. 2006;21:1947–1961. [Google Scholar]
- 31.Berendsen HJC, Vanderspoel D, Vandrunen R. Gromacs—a message-passing parallel molecular-dynamics implementation. Comput Phys Commun. 1995;91:43–56. [Google Scholar]
- 32.Vandrunen R, Vanderspoel D, Berendsen HJC. Gromacs—a software package and a parallel computer for molecular-dynamics. Abstr Pap Am Chem Soc. 1995;209:49. [Google Scholar]


