Abstract
Ions can significantly modulate the solution interactions of proteins. We aim to demonstrate that the salt-dependent reversible heptamerization of a fusion protein called peptibody A or PbA is governed by anion-specific interactions with key arginyl and lysyl residues on its peptide arms. Peptibody A, an E. coli expressed, basic (pI = 8.8), homodimer (65.2 kDa), consisted of an IgG1-Fc with two, C-terminal peptide arms linked via penta-glycine linkers. Each peptide arm was composed of two, tandem, active sequences (SEYQGL PPQGWK) separated by a spacer (GSGSATGGSGGGASSGSGSATG). PbA was monomeric in 10 mM acetate, pH 5.0 but exhibited reversible self-association upon salt addition. The sedimentation coefficient (sw) and hydrodynamic diameter (DH) versus PbA concentration isotherms in the presence of 140 mM NaCl (A5N) displayed sharp increases in sw and DH, reaching plateau values of 9 s and 16 nm by 10 mg/mL PbA. The DH and sedimentation equilibrium data in the plateau region (>12 mg/mL) indicated the oligomeric ensemble to be monodisperse (PdI = 0.05) with a z-average molecular weight (Mz) of 433 kDa (stoichiometry = 7). There was no evidence of reversible self-association for an IgG1-Fc molecule in A5N by itself or in a mixture containing fluorescently labeled IgG1-Fc and PbA, indicative of PbA self-assembly being mediated through its peptide arms. Self-association increased with pH, NaCl concentration, and anion size (I− > Br− > Cl− > F−) but could be inhibited using soluble Trp-, Phe-, and Leu-amide salts (Trp > Phe > Leu). We propose that in the presence of salt (i) anion binding renders PbA self-association competent by neutralizing the peptidyl arginyl and lysyl amines, (ii) self-association occurs via aromatic and hydrophobic interactions between the..xx
PPQGWK) separated by a spacer (GSGSATGGSGGGASSGSGSATG). PbA was monomeric in 10 mM acetate, pH 5.0 but exhibited reversible self-association upon salt addition. The sedimentation coefficient (sw) and hydrodynamic diameter (DH) versus PbA concentration isotherms in the presence of 140 mM NaCl (A5N) displayed sharp increases in sw and DH, reaching plateau values of 9 s and 16 nm by 10 mg/mL PbA. The DH and sedimentation equilibrium data in the plateau region (>12 mg/mL) indicated the oligomeric ensemble to be monodisperse (PdI = 0.05) with a z-average molecular weight (Mz) of 433 kDa (stoichiometry = 7). There was no evidence of reversible self-association for an IgG1-Fc molecule in A5N by itself or in a mixture containing fluorescently labeled IgG1-Fc and PbA, indicative of PbA self-assembly being mediated through its peptide arms. Self-association increased with pH, NaCl concentration, and anion size (I− > Br− > Cl− > F−) but could be inhibited using soluble Trp-, Phe-, and Leu-amide salts (Trp > Phe > Leu). We propose that in the presence of salt (i) anion binding renders PbA self-association competent by neutralizing the peptidyl arginyl and lysyl amines, (ii) self-association occurs via aromatic and hydrophobic interactions between the..xx ..xxx..
..xxx.. xx.. motifs, and (iii) at >10 mg/mL, PbA predominantly exists as heptameric clusters.
xx.. motifs, and (iii) at >10 mg/mL, PbA predominantly exists as heptameric clusters.
Keywords: Hofmeister series, electroselectivity series, self-association, ion-protein interactions, anion binding, dynamic light scattering, analytical ultracentrifugation
Introduction
Although the differential effects of ions on protein precipitation were observed more than a century ago by Hofmeister,1,2 the role of ions in the modulation of the solution behavior and interactions of proteins is increasingly well appreciated. Ions have been observed to impact protein stability,3 fibril formation,4–6 reversible self-association,7–9 and even protein solution rheology.10,11 An ion-specific dependence is often discussed in terms of the Hofmeister effect or the electroselectivity series;3–5,12–14 however, the exact nature of ion-protein interactions remains unresolved. The classical view holds that such interactions are mediated indirectly through the alteration of bulk water structure,15 although there is increasing recognition that ions interact directly with proteins.16 Significantly, Collins has proposed17–21 that ionic interactions in aqueous solution are governed by the relative affinities of ions for the molecular dipole of water. This qualitative description known as the “Law of Matching Water Affinities” provides a good basis for discussing interactions of dissolved salt-ions with charged protein side chains.
Therapeutic proteins often exhibit reversible self-association under different solution conditions of pH, excipients, and ionic strength. This can have a significant impact on drug development, affecting pharmacokinetics, manufacturing processes, and delivery. There are several such examples reported in literature. The hormone insulin can be stabilized as a reversible hexamer when formulated with zinc.22,23 However, the hexameric form causes a delayed onset of glucose reduction following subcutaneous injection, which consequently has led to the development of genetically engineered, monomeric, insulin analogues such as Humalog® and Novolog®.23 The reversible self-association of some monoclonal antibodies at high concentrations has been implicated in dramatically increasing solution viscosity, leaving significant challenges to be resolved during manufacturing and delivery of the drug product.10,24 There also have been concerns about oligomeric forms of a drug presenting an increased immunogenic risk.25,26 For these reasons, reversible self-association interactions have been studied in almost all classes of protein therapeutics, including peptides,27 hormones,28–30 growth factors,31 cytokines,32 antibodies,33,34 and recently even in single-chain Fv molecules.35
This report is focused on characterizing the salt-dependent reversible self-association pathway of a fusion protein called peptibody A (PbA). Peptibodies form a unique class of recombinant proteins consisting of active peptide sequences attached to the Fc domain of an immunoglobulin G (IgG). In particular, PbA (see Fig. 1) is an E. coli expressed, basic (pI = 8.8), covalent homodimer with a sequence molecular weight of 65,223 Da and consists of an Fc domain of an IgG1 with “peptide arms” linked to each of its C-termini. Each peptide arm consists of two active peptide sequences [SEYQGL PPQGWK] separated by a spacer group [GSGSATGGSGGGASSGSGSATG] and is attached to the Fc with a flexible, penta-glycine linker.
PPQGWK] separated by a spacer group [GSGSATGGSGGGASSGSGSATG] and is attached to the Fc with a flexible, penta-glycine linker.
Figure 1.
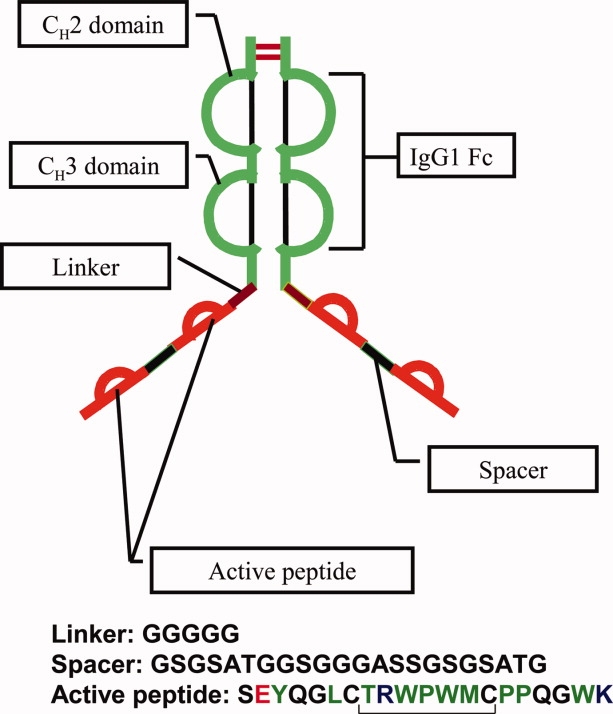
Structural cartoon of Peptibody A. PbA is an E. coli expressed, basic (pI = 8.8), covalent homodimer with a sequence molecular weight of 65,223 Da and consists of an Fc domain of an IgG1 with “peptide arms” linked to each of its C-termini. Each peptide arm consists of two active peptide sequences separated by a spacer group and is attached to the Fc with a flexible, penta-glycine linker.
Peptibody A exhibited peculiar solution behavior. Although it was distinctly monomeric in a low-ionic strength pH 5.0 solution buffered with 10 mM acetate, it appeared to undergo rapid, reversible self-association with the addition of sodium chloride. To understand the underlying ion-protein interactions and gain insights into the assembly process, we employed a set of monovalent ions from the Hofmeister series, varied ionic strength and pH and studied their effects on PbA self-association using sedimentation and dynamic light scattering (DLS) techniques. The self-association pathway of PbA was anticipated to be complex given the symmetrical nature of the molecule (see Fig. 1) allowing for the propagation of the self-assembly process to occur via a combination of peptide–peptide, peptide-Fc, and Fc–Fc interactions (see Fig. 2). Through the systematic elucidation of the self-association pathway of PbA, our studies shed light on the importance of ion-specific protein interactions and their role in modulating the solution behavior of proteins.
Figure 2.
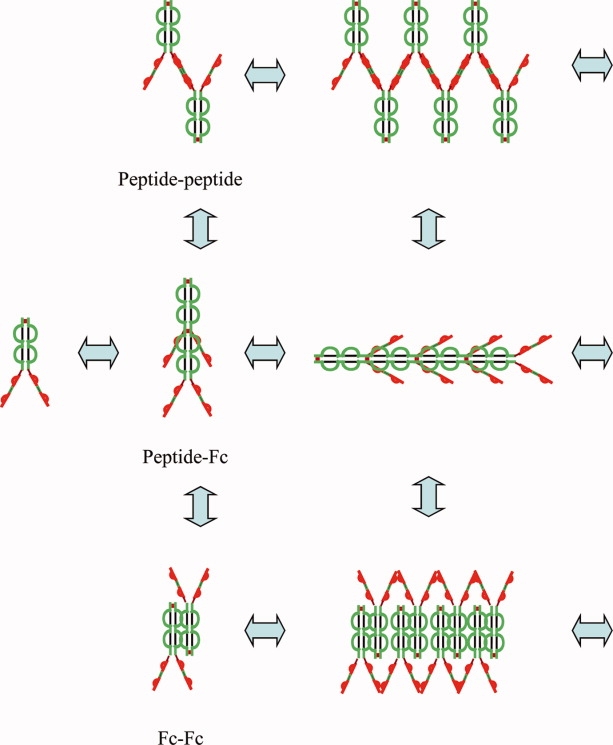
Generalized self-association model for PbA depicting possible association pathways mediated via peptide–peptide, peptide-Fc, or Fc–Fc interactions.
Results
Solution behavior of peptibody A
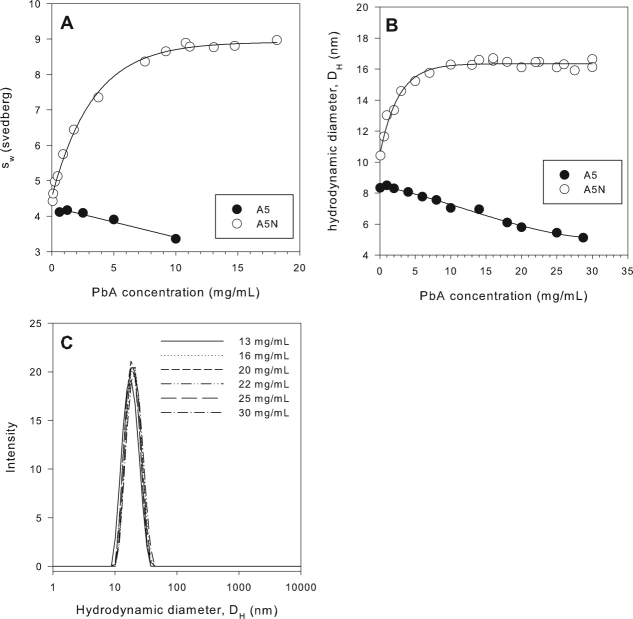
Sedimentation velocity and DLS experiments with PbA in a low ionic strength, pH 5.0 solution buffered with 10 mM acetate (A5 buffer) revealed it to be monomeric.* There was no evidence of any concentration dependent self-association of PbA in the A5 buffer from SV and DLS experiments over a wide PbA concentration range of 0.1–35 mg/mL [Fig. 3(A–C)]. An increase in weight average sedimentation coefficient (sw) or the hydrodynamic diameter (DH) with increasing PbA concentration would be expected if significant self-association interactions were present. In fact, a decrease in the monomer sw and DH values, attributable to nonideality effects, was observed as a function of PbA concentration.
Figure 3.
SV and DLS data indicate that PbA self-associates in the presence of salt. SV experiments (50,000 rpm) show the sw of PbA to increase with PbA concentration in A5N reaching a plateau value of ∼9 s above 10 mg/mL (monitored by absorbance at 230 nm for PbA concentrations <0.4 mg/mL). A decrease in sw is observed in A5 with increasing PbA concentration (Panel A). The hydrodynamic diameter for PbA increases rapidly with protein concentration in A5N reaching a plateau value of 16 nm above 10 mg/mL. A decrease in hydrodynamic diameter is observed in A5 with increasing peptibody concentration (Panel B). The DLS data show that the PbA oligomeric ensemble in A5N from 13–30 mg/mL is monodisperse with an average pDI value of 0.052 ± 0.011 over the entire concentration range (Panel C).
With the addition of an isotonic amount (140 mM) of NaCl to the A5 buffer, however, a concentration dependent increase in sw and DH was observed over the 1000-fold PbA concentration range examined [0.03–18 mg/mL—SV, 0.1–30 mg/mL—DLS, Fig. 3(A,B)]. This strongly indicated that PbA was undergoing reversible self-association. A key feature of the sw and DH versus protein concentration plots was that both isotherms appeared to asymptotically approach plateau sw and DH values of ∼9 s and 16 nm, respectively, by 10 mg/mL PbA. In addition, the very low polydispersity indices (average = 0.05) observed in the plateau region (13–30 mg/mL) of the DLS data [Fig. 3(C)] were indicative of a monodisperse oligomeric ensemble. Generally, size distributions with polydispersity indices of less than 0.1 are considered to be monodisperse.36,37
Effect of ion identity, ionic strength, and pH on the self-association of peptibody A
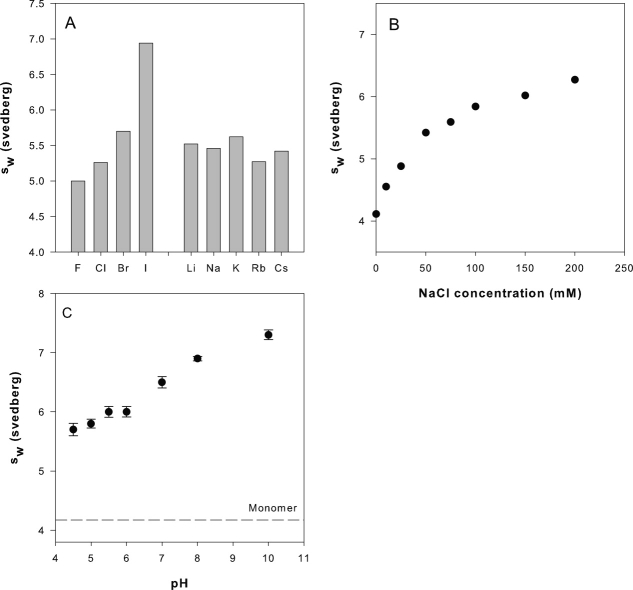
The PbA sw values (and hence self-association) were observed to be very sensitive to ionic strength as well as anion identity. For a fixed PbA concentration (1 mg/mL) and a given NaX (X = halogen ion) concentration, the sw values increased with the atomic size of the monovalent anion [Fig. 4(A)]. The extent of self-association was highest in the presence of 50 mM sodium iodide (6.9 s) and lowest in 50 mM NaF (5.0 s). Monovalent cations (Li → Cs), on the other hand, did not appear to affect the self-association equilibrium appreciably. Although the observed sw values (5.4 ± 0.2 s) were much higher than those expected for the monomer (4.2 s), indicative of self-association in the presence of all the cation-chloride salts, no significant differences in the extent of self-association between the cations could be discerned [Fig. 4(A)]. The sodium chloride concentration had a significant impact on the self-association interaction. For a given PbA concentration (1 mg/mL) and pH (5.0), the PbA sw values increased with increasing NaCl concentration [Fig. 4(B)]. An experiment designed to study the effect of pH on PbA self-association revealed that the extent of association also increased with pH. The sw values for 1 mg/mL PbA solutions prepared in an acetate-histidine-phosphate-Tris (5 mM each) buffer with 75 mM NaCl increased from 5.7 s at pH 4.5 to 7.4 s at pH 10.0 [Fig. 4(C)].
Figure 4.
Factors influencing PbA self-association. Interference SV experiments (50,000 rpm) on 1 mg/mL PbA in 140 mM salt solutions show peptibody self-association to have a strong dependence on the identity of the halogen anion, but not the alkali cation (Panel A). Self-association increased with NaCl concentration (0 and 200 mM) (Panel B) and with pH (Panel C).
Solution behavior of Fc in the presence of salt and peptibody A
Sedimentation velocity experiments with Fc in the presence of 140 mM NaCl at pH 5.0 indicated that it remained monomeric over the entire Fc concentration range (0.3–10 mg/mL) examined. The monomer was observed to sediment at a rate of 3.6 s under dilute conditions (0.3 mg/mL). A linear decrease in sw, attributable to hydrodynamic nonideality effects, was observed with increasing Fc concentration [Fig. 5(A)]. Fluorescent labeling had no effect on the association state of Fc; FITC-Fc was observed to sediment as a monomer in PBS at pH 7.5 as indicated by the sharp peak at 3.7 s in its c(s) distribution [Fig. 5(B)]. In the presence of PbA, Fc displayed no self-associative behavior. The c(s) distribution of FITC-Fc (monitored at 495 nm) in a mixture of FITC-Fc and PbA (0.3 mg/mL each) in PBS at pH 7.5 was super-imposable upon the control of only FITC-Fc in PBS. These experiments indicated that Fc did not participate in the PbA self-association interaction.
Figure 5.
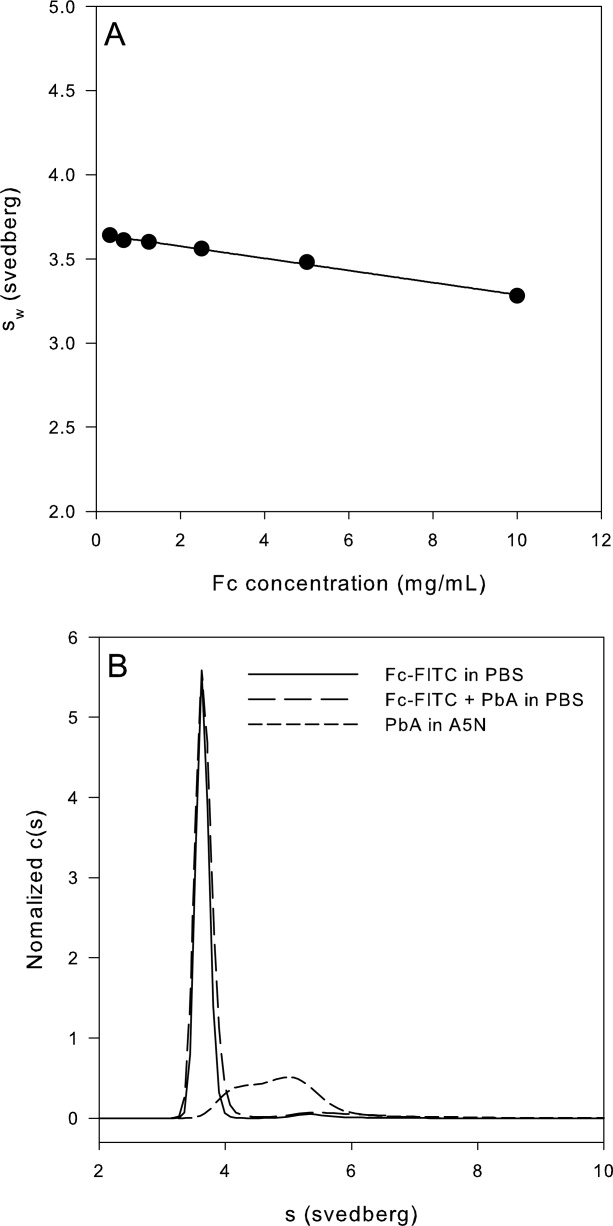
AUC experiments with Fc-FITC indicated PbA does not self-associate through Fc–Fc or Fc-peptide interactions. The sw of 0.3 mg/mL FITC-Fc obtained from SV absorbance (495 nm) experiments at 60,000 rpm matched those of the unlabeled Fc (Panel A); the c(s) distribution of FITC-Fc was unchanged by the addition of PbA to 0.3 mg/mL (Panel B).
Effect of aromatic and hydrophobic amino acids on the self-association of peptibody A
The importance of aromatic and hydrophobic interactions in mediating self-association was probed by attempting to inhibit the interaction with appropriate amino acids. Soluble amide-hydrochloride salts of tryptophan, phenylalanine, and leucine were employed for this purpose with alanine amide hydrochloride being used as a control. Inhibition of self-association was measured in terms of the decrease in sw values of a 1 mg/mL PbA solution formulated in A5N with additional amounts (50–150 mM) of the hydrochloride salt of a given amino acid. It should be noted that in the absence of any inhibition, an increase in the extent of self-association with increasing amino acid-NH2-HCl concentration was expected due to the concomitant increase in chloride concentration. For example, the total chloride concentration resulting from the addition of 150 mM Ala-NH2-HCl to A5N was 300 mM. Consistent with this expectation, a slight increase (to 6.6 s) in sw values of PbA was observed in the case of alanine-amide. However, even with the additional chloride content, significant inhibition of the self-association interaction was observed in the presence of both aromatic compounds. With the addition of 150 mM phenylalanine amide or tryptophan amide, the sw value of 6.0 s for 1 mg/mL PbA in A5N decreased to ∼5.0 or 4.5 s, respectively (see Fig. 6). Inhibition of self-association was also observed in the presence of leucine amide; however, it did not appear to be as effective as the aromatic amino acid derivatives. Tryptophan amide was observed to be the most effective in inhibiting the salt induced self-association of PbA.
Figure 6.
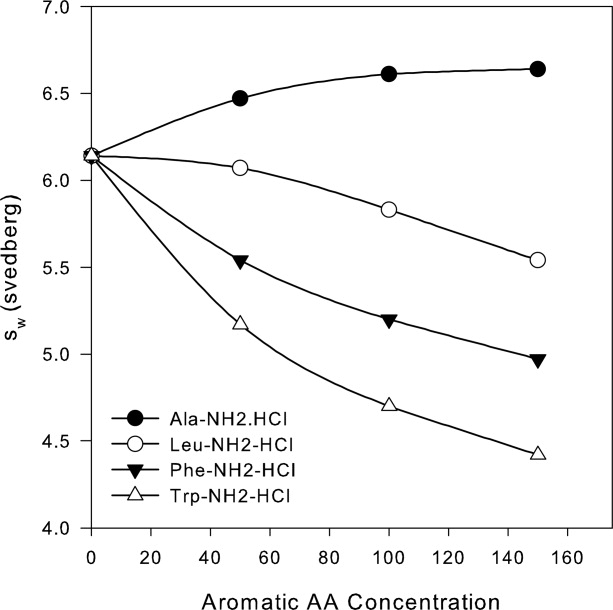
Aromatic and hydrophobic amino acids inhibit self-association of PbA. The addition of chloride salt of tryptophan-, phenylalanine-, or leucine-amide decreases the degree of self-association of 1 mg/mL PbA in A5N, as measured by interference, SV experiments (50,000 rpm). The degree of self-association increases in the presence of alanine amide hydrochloride due to the concomitant increase in chloride concentration.
Sedimentation equilibrium experiments with peptibody A
Sedimentation equilibrium (SE) studies were performed to determine the molecular weight of the PbA oligomeric ensemble in the plateau region of the DLS isotherm [Fig. 3(B)]. Experiments were conducted at 12–20 mg/mL PbA in A5N. The resulting equilibrium concentration distributions could be fit to the single, ideal-species model and yielded a z-average molecular weight (Mz) value of 433 kDa (see Fig. 7) with an RMSD of 0.012.
Figure 7.
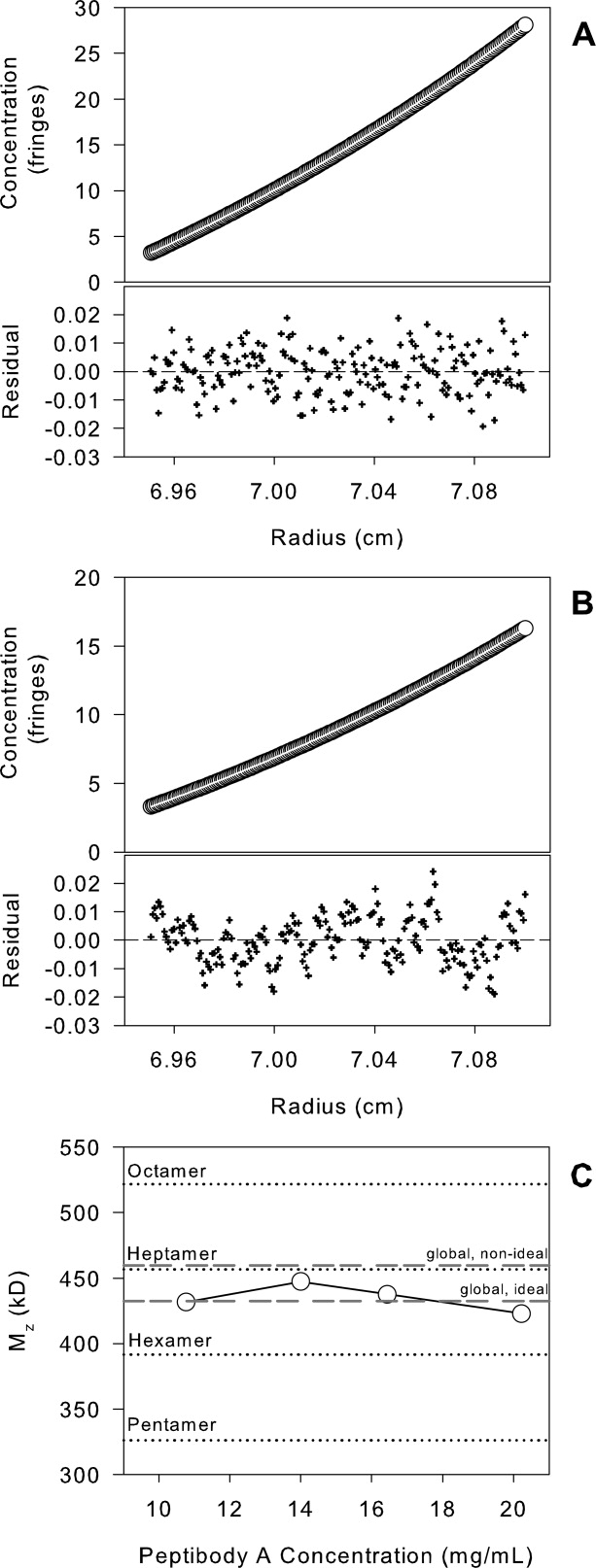
Sedimentation equilibrium studies of PbA (12–20 mg/mL) in A5N. Experiments were performed at 3000 rpm and 20°C using interference detection. The data could be described well by the single ideal species model for the entire PbA concentration examined (20 mg/mL—Panel A, 12 mg/mL—Panel B). The molecular weight from individual fits (single, ideal) and from the global single ideal and nonideal species fits are presented in Panel C.
Discussion
Self-association of peptibody A occurs via its peptide arms
Peptibody A is a basic (pI = 8.8), symmetric molecule composed of two peptide arms attached to the C termini of a human IgG1 Fc. The Fc itself is a symmetric, covalent dimer composed of two CH2 and CH3 domains. The structural symmetry of PbA led to construction of a generalized assembly model in which reversible self-association of the peptibody was proposed to occur via a combination of intermolecular peptide-peptide, peptide-Fc, and/or Fc–Fc interactions (see Fig. 2). The symmetry allowed for the possibility that self-association could propagate indefinitely.
The sedimentation velocity experiments with Fc and FITC-Fc were useful in demonstrating that self-association of PbA was likely to be mediated through its peptide arms. Over the wide Fc concentration range of 0.3–10 mg/mL examined, there was no indication of any rapid, reversible self-association of Fc under the A5N conditions [Fig. 5(A)]. The observed monotonic decrease in sw with increasing Fc concentration was a result of hydrodynamic nonideality, which becomes significant at higher protein concentration.38 Based on this, the possibility of PbA self-association being mediated by Fc–Fc interactions could be eliminated. Because PbA is expressed in E. coli, the Fc CH2 domain is not glycosylated. It was important to ensure that the lack of glycosylation was not causing the reversible self-association observed in PbA.
With no evidence of Fc–Fc mediated self-association, we were able to simplify the PbA reversible self-association model, leaving peptide-Fc and peptide–peptide interactions as the only two possible pathways. The FITC labeling of Fc enabled us to selectively follow its sedimentation in the presence of PbA by setting the detection wavelength to 495 nm. It is important to note here that the SV experiment with Fc + PbA mixture was conducted at pH 7.5 in PBS (which contained 140 mM NaCl), conditions in which the extent of self-association was expected to be even greater than at pH 5.0 [Fig. 4(C)]. The c(s) analysis of the 495-nm signal indicated that the sedimentation coefficient of FITC-Fc was unaffected by the presence of PbA and that it migrated as a monomer (from its corresponding c(M) analysis, data not shown). Further, the c(s) distribution of FITC-Fc from the mixture experiment was super-imposable with that of the control (c(s) distribution of FITC-Fc alone). If peptide-Fc interactions were significant in mediating the reversible self-association of PbA, then the c(s) distribution should be broad, similar to that observed for PbA in A5N [Fig. 5(B)]. Given that Fc did not appear to self-associate in the presence of PbA, the peptibody was most likely to self-associate via its peptide arms.
Anion binding is essential and self-association is mediated via a combination of aromatic and hydrophobic interactions
The salt dependence of the reversible self-association of PbA indicated that charge interactions were important in the mediation of the interaction. Combined with the evidence that self-association of PbA occurred only through its peptide arms, a model which included ion binding to specific residues in the peptide arms as a necessary step for self-association could be constructed. Upon inspection of the amino acid sequence of the peptide arm, the possible roles of anion binding and tryptophan (aromatic and hydrophobic) interactions in mediating PbA self-association became apparent. Each active sequence in a single peptide arm of PbA (see Fig. 1) contains two basic amino acids, lysine and arginine, with the latter near two tryptophan residues in a disulfide-closed loop (- -). We tested our hypothesis that charge neutralization of the arginyl and lysyl side chains rendered the peptibody self-association competent and that self-association was mediated through the peptide loops via hydrophobic and aromatic interactions. In the absence of a significant anion concentration, repulsion between intermolecular, positively charged arginine and lysine residues prevented self-association.
-). We tested our hypothesis that charge neutralization of the arginyl and lysyl side chains rendered the peptibody self-association competent and that self-association was mediated through the peptide loops via hydrophobic and aromatic interactions. In the absence of a significant anion concentration, repulsion between intermolecular, positively charged arginine and lysine residues prevented self-association.
The positively charged guanidino group of arginines and the ɛ-amine of lysines are weakly hydrated and expected to preferentially bind weakly hydrated anions.17–21,39 The responsiveness of PbA self-association to halogen anion identity and its relative unresponsiveness to alkali cation identity [Fig. 4(A)] supported anion binding as being critical for self-association. The binding affinity of halogen ions to the arginyl and lysyl side-chains is expected to increase with increasing anion size.18 Indeed, the greatest extent of self-association was observed in the presence of iodide, and it progressively decreased with decreasing anion size, from I− → Br− → Cl− → F− [Fig. 4(A)].
The observed pH dependence of the PbA sw value (at a fixed NaCl concentration of 75 mM) also supported anion binding being an important step in the reversible self-association process [Fig. 4(C)]. With increasing pH, the effective charge of PbA (pI = 8.8) was expected to decrease progressively, requiring a proportionately lower anion concentration to achieve equivalent charge reduction. For a given ionic strength, this would free-up an increasing concentration of anions (with increasing pH) for binding to the arginyl side chains in the peptide loops (which were expected to be always positively charged, pKa = 12). Consequently, an increase in the extent of self-association would be expected with increasing pH. This expectation was consistent with our data, in which the sw values of PbA steadily increased from 5.7 s at pH 4.5–7.4 s at pH 10.0.
Anion binding has been reported to be important in several biological systems, for the modulation of ion transport across membranes,40 enzymatic activity, and even self-association.7 Anion binding to arginine and lysine residues on the interaction interface of hemoglobin (from Archid Mollusks) has been reported to be critical for its ionic strength dependent reversible polymerization.7 Although there was valid evidence to suggest that anion binding enabled self-association of PbA, the specific residue-level interactions through which self-association was mediated remained to be investigated. It was recognized that the peptide loop - - was fairly hydrophobic (except for R) and that self-association via this hydrophobic patch was likely. Further, because there were two such patches per peptide arm (see Fig. 1), a stable interaction interface involving both peptide loops […xxx
- was fairly hydrophobic (except for R) and that self-association via this hydrophobic patch was likely. Further, because there were two such patches per peptide arm (see Fig. 1), a stable interaction interface involving both peptide loops […xxx xxx…xxx
xxx…xxx xxx] was likely to be favored. However, at least five types of interactions between two intermolecular peptide arms were theoretically possible and considered: (i) R→W, cation-π interactions,41 (ii) K ←→ E ion-pair interactions, (iii) W←→W aromatic interactions, (iv) W-X-W type interactions, coordinated by anions, especially halides,42 and (v) intermolecular hydrophobic interactions.
xxx] was likely to be favored. However, at least five types of interactions between two intermolecular peptide arms were theoretically possible and considered: (i) R→W, cation-π interactions,41 (ii) K ←→ E ion-pair interactions, (iii) W←→W aromatic interactions, (iv) W-X-W type interactions, coordinated by anions, especially halides,42 and (v) intermolecular hydrophobic interactions.
Both R→W and K←→ E are ionic interactions and expected to be screened with increasing ionic strength. Contrary to this expectation, PbA self-association was enhanced with increasing ionic strength suggesting R→W, and K←→ E interactions were unlikely to be responsible for PbA self-association. Evidence in support of W←→W aromatic interactions and/or hydrophobic interactions came from the ability of tryptophan-amide, phenylalanine-amide, and leucine-amide to inhibit the self-association interaction (see Fig. 6). And, because both phenylalanine and leucine could also inhibit the self-association (albeit to a lesser extent), the fourth possibility of W-X-W interactions was considered unlikely. The ability of the weakly charged indole nitrogen of tryptophan to interact with halides has been reported in literature. The haloalkane dehalogenase enzyme is known to employ this type of interaction to coordinate a halide ion between two adjacent tryptophan residues in its active site.42 The tryptophan coordination “strength” of halides is expected to increase with ionic size (I− better than Cl−), which in the case of PbA could potentially have helped explain the observation of increased self-association with increasing halide ion size. However, if W-X-W were the only interaction involved in mediating PbA self-association, then inhibition of the interaction by phenylalanine and leucine (with no expectation to coordinate halides) would not be observed. On the other hand, the inhibition of W ←→ W aromatic interactions by free phenylalanine was possible, as formation of similar W ←→ F aromatic interactions is well known. Given the abundance of hydrophobic residues in the peptide loops (- -), self-association could also simply be mediated by hydrophobic interactions upon neutralization of arginines and lysines. In this event, greater inhibition of self-association by phenylalanine and leucine, being more hydrophobic than tryptophan, would be expected. In contrast, tryptophan was observed to be most effective at inhibiting PbA self-association. Although W ←→W interactions appear to be significant, the self-association of PbA probably occurs through a combination of aromatic and hydrophobic interactions. The active peptide sequence is largely hydrophobic, with 20% of the amino acids having aromatic side chains. From our studies with tryptophan- and phenylalanine-amide, we have strong evidence that tryptophan-aromatic interactions play a significant role. Furthermore, the sensitivity of the self-association interaction to salt concentration is consistent with side-chain charge neutralization of the arginyl and lysyl residues within the active peptides being essential. Neutralization would reduce the energetic cost of bringing the residues into close proximity with like-charges and/or burying them in a hydrophobic/aromatic interaction. Although there is no evidence for the residue-level interactions to be highly specific or structured, our proposition of cluster formation (next section) is more consistent with nonspecific, aromatic/hydrophobic interactions.
-), self-association could also simply be mediated by hydrophobic interactions upon neutralization of arginines and lysines. In this event, greater inhibition of self-association by phenylalanine and leucine, being more hydrophobic than tryptophan, would be expected. In contrast, tryptophan was observed to be most effective at inhibiting PbA self-association. Although W ←→W interactions appear to be significant, the self-association of PbA probably occurs through a combination of aromatic and hydrophobic interactions. The active peptide sequence is largely hydrophobic, with 20% of the amino acids having aromatic side chains. From our studies with tryptophan- and phenylalanine-amide, we have strong evidence that tryptophan-aromatic interactions play a significant role. Furthermore, the sensitivity of the self-association interaction to salt concentration is consistent with side-chain charge neutralization of the arginyl and lysyl residues within the active peptides being essential. Neutralization would reduce the energetic cost of bringing the residues into close proximity with like-charges and/or burying them in a hydrophobic/aromatic interaction. Although there is no evidence for the residue-level interactions to be highly specific or structured, our proposition of cluster formation (next section) is more consistent with nonspecific, aromatic/hydrophobic interactions.
Oligomer geometry, stoichiometry, and a model for peptibody A self-association
The plateau in the DLS and SV isotherms in A5N [Fig. 3(A,B)] at high PbA concentrations (>10 mg/mL) was suggestive of a termination step in the PbA oligomerization pathway. With increasing protein concentration, PbA self-association may propagate via its peptide arms and terminate with the formation of cyclic oligomers. Alternatively, PbA may form heptameric clusters in which peptide arms are buried in a central core. The monodispersity of the PbA oligomers in the plateau region [Fig. 3(C)] is consistent with both models.
The determination of the molecular weight of PbA oligomers (and hence stoichiometry) was complicated by potential thermodynamic nonideality effects, which can be significant at high protein concentrations. Self-association and nonideality have opposite effects on the measured molecular weight by SE in a reversibly interacting system. While self-association leads to an increase in buoyant molecular weight, nonideality effects will lower the value. Still, the sedimentation equilibrium data between 12 and 20 mg/mL PbA in A5N could be described well using the single-ideal species model as supported by the randomness of the residuals to the fits and yielded a z-average molecular weight of 433 kDa (Fig. 7, Panel C global ideal fit), consistent with a molecular stoichiometry of ∼7.
It is indeed tempting to interpret the sharp increases and plateauing of sw and DH in their respective isotherms in terms of a cooperative, monomer ←→ heptamer, self-association interaction. However, the propagation of PbA self-association via a sequential oligomerization pathway terminating at a heptamer is also likely and thus cannot be ruled out. On the basis of the evidence presented, and employing parsimony, we propose the following self-association model for PbA in A5N (see Fig. 8)
Figure 8.
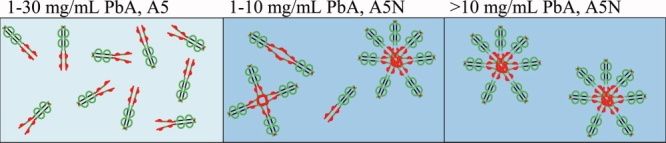
Cartoon depicting the proposed self-association model for PbA in A5N. In 10 mM acetate, PbA remains monomeric. Upon addition of an isotonic amount of NaCl, at low to intermediate concentrations PbA (< 10 mg/mL), it exists as a mixture of dimeric to heptameric oligomers. At higher concentrations, PbA self-associates to form heptameric clusters.
Upon salt addition, anion binding renders PbA self-association competent by neutralizing the peptidyl arginyl and lysyl amines
Self-association propagates via a combination of intermolecular, aromatic, and hydrophobic interactions between …xx
 ..xxx..
..xxx..  xx.. motis on the peptide arms
xx.. motis on the peptide armsAt low to intermediate concentrations (<10 mg/mL), PbA exists as a mixture of dimeric to heptameric oligomers. At higher concentrations, PbA self-associates to form heptameric clusters with a DH of 16 nm.
Materials and Methods
Peptibody A
The bulk drug substance lots of PbA were supplied by Amgen Process Sciences & Protein Sciences groups. The lot from Protein Sciences was formulated at 10 mg/mL in 10 mM acetate, 5% sorbitol at pH 5.0 (A5S), whereas the lot from Process Sciences was received at 30 mg/mL in 10 mM acetate, 9.25% sucrose at pH 4.8. The E. coli expressed human IgG1 Fc (FC) was received at a concentration of 25 mg/mL in A5S from Amgen's Protein Sciences group. The l-tryptophan amide and l-phenylalanine amide hydrochloride salts were purchased from CalBiochem (San Diego, CA). All chemicals were of analytical grade or better.
Preparation of solutions
For all experiments, the PbA bulk was first dialyzed into 10 mM acetate, pH 5.0 (A5) buffer and concentrated to 30 mg/mL. When conducting experiments in the 10 mM acetate, 140 mM NaCl, pH 5.0 (A5N) buffer, appropriate volumes of a 3M sodium chloride stock solution prepared in 10 mM acetate, A5 buffer, and the PbA bulk in A5 were mixed proportionately to achieve the desired protein and NaCl concentrations. All other solutions were prepared similarly by dilution and/or addition of the stocks of the desired chemicals to the bulk in A5 buffer.
Analytical ultracentrifugation: sedimentation velocity and sedimentation equilibrium
Sedimentation experiments were conducted in a Beckman Coulter XL-I analytical ultracentrifuge at 20°C, using either the absorbance or interference optical system. Experiments were performed using cells fitted with sapphire (specifically used in interference experiments) or quartz windows, and 12-mm path-length, charcoal-epon centerpieces. Samples were centrifuged at 3000 rpm for equilibrium experiments and 50,000 rpm or 60,000 rpm for velocity experiments. With the absorbance optical system, sedimentation was monitored using a detection wavelength of 280 nm in all experiments except for those involving the FITC labeled Fc (FITC-Fc), in which case a wavelength of 495 nm was used to selectively detect the sedimentation of FITC-Fc. When employing the absorbance optical system for SV experiments, scans were gathered using a radial step size of 0.003 in continuous mode, with no replicates.
The software program SEDFIT (kindly provided by Dr. Peter Schuck, NIH, Bethesda, USA) were employed to analyze the SV data (using the c(s) and c(M) distribution analysis).43,44 The program HETEROANALYSIS (kindly provided by Prof. James Cole, University of Connecticut, Storrs, USA) was employed to determine the apparent z average molecular weights from the SE data by applying the single ideal and nonideal species models.
Dynamic light scattering experiments
DLS studies were conducted using the Zetasizer Nano ZS® instrument (Malvern Instruments, Worcestershire, UK) at 20°C to determine the hydrodynamic diameter (DH) of PbA. One hundred microliters of a given solution was placed in a disposable DTS1060, folded capillary cuvette (also from Malvern Instruments) and analyzed in the DLS mode. Prior to any DLS measurements, the PbA bulk was desalted and eluted into a given buffer (A5N or A5) using NAP-5® columns (GE Healthcare, Piscataway, NJ) and then concentrated to ∼30 mg/mL using 10,000 MWCO Amicon Ultra-15® centrifugal filters (Millipore Corporation, Billerica, MA). The DH was measured as a function of PbA concentration from 0.1 to 30 mg/mL.
FITC labeling of FC
Fluorescent labeling of the FC with fluroescein isothiocyanate (FITC) was accomplished using the FLUOROTAG™ FITC Conjugation Kit (Sigma Aldrich # FITC-1) and following the protocol (FITC-1) provided with the kit. The reaction conditions were optimized (as prescribed in the protocol) to achieve a FITC/protein (F/P) conjugation ratio of 1. The actual F/P ratio in the case FITC-Fc was 1.1. After the removal of the unreacted FITC, the purified FITC-Fc was transferred into Dulbecco's PBS and used immediately in sedimentation experiments.
Acknowledgments
The authors thank Himanshu Gadgil, Gaya Ratnaswamy, and Songpon Deechongkit for their helpful suggestions and insights.
Glossary
Abbreviations
- A5
10 mM acetate buffer at pH 5.0
- A5N
10 mM acetate buffer at pH 5.0 containing 140 mM sodium chloride
- AUC
analytical ultracentrifugation
- CH2
constant heavy chain domain 2
- CH3
constant heavy chain domain 3
- c(M)
continuous molecular mass distribution function
- DNA-PK
DNA-dependent protein kinase
- c(s)
continuous sedimentation coefficient distribution function
- DH
hydrodynamic diameter; DLS, dynamic light scattering
- Fc
fraction crystallized
- Fc
fragment crystallizable
- FITC
fluroescein isothiocyanate
- Fv
fragment variable
- IAA
iodoacetic acid
- IgG1
immunoglobulin G type 1
- IPA
isopropyl alcohol
- Mz
z-average molecular weight
- NaX
sodium halogen salt
- PbA
peptibody A
- PBS
phosphate buffered saline
- PdI
polydispersity index
- RPM
revolutions per minute
- SE
sedimentation equilibrium
- SV
sedimentation velocity
- sw
weight-average sedimentation coefficient
- X
halogen ion
Footnotes
It should be noted that PbA itself is a covalent dimer consisting of two CH2 and CH3 domains of the human Fc with two peptide arms attached to the C termini of the Fc. The sequence molecular weight of the PbA monomer is 65,223 Da. The word “monomer” in this report refers to the full length PbA molecule with a molecular weight of 65,223 Da.
References
- 1.Hofmeister F. Zur Lehre von der Wirkung der Salze. Naunyn Schmiedebergs Arch Pharmacol. 1888;24:247–260. [Google Scholar]
- 2.Washabaugh MW, Collins KD. The systematic characterization by aqueous column chromatography of solutes which affect protein stability. J Biol Chem. 1986;261:12477–12485. [PubMed] [Google Scholar]
- 3.Ramos CH, Baldwin RL. Sulfate anion stabilization of native ribonuclease A both by anion binding and by the Hofmeister effect. Protein Sci. 2002;11:1771–1778. doi: 10.1110/ps.0205902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munishkina LA, Henriques J, Uversky VN, Fink AL. Role of protein-water interactions and electrostatics in alpha-synuclein fibril formation. Biochemistry. 2004;43:3289–3300. doi: 10.1021/bi034938r. [DOI] [PubMed] [Google Scholar]
- 5.Raman B, Chatani E, Kihara M, Ban T, Sakai M, Hasegawa K, Naiki H, Rao ChM, Goto Y. Critical balance of electrostatic and hydrophobic interactions is required for beta 2-microglobulin amyloid fibril growth and stability. Biochemistry. 2005;44:1288–1299. doi: 10.1021/bi048029t. [DOI] [PubMed] [Google Scholar]
- 6.Pedersen JS, Flink JM, Dikov D, Otzen DE. Sulfates dramatically stabilize a salt-dependent type of glucagon fibrils. Biophys J. 2006;90:4181–4194. doi: 10.1529/biophysj.105.070912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boffi A, Vecchini P, Chiancone E. Anion-linked polymerization of the tetrameric hemoglobin from Scapharca inaequivalvis. Characterization and functional relevance. J Biol Chem. 1990;265:6203–6209. [PubMed] [Google Scholar]
- 8.Wolff J, Knipling L, Sackett DL. Charge-shielding and the “paradoxical” stimulation of tubulin polymerization by guanidine hydrochloride. Biochemistry. 1996;35:5910–5920. doi: 10.1021/bi9527395. [DOI] [PubMed] [Google Scholar]
- 9.Wolff J, Sackett DL, Knipling L. Cation selective promotion of tubulin polymerization by alkali metal chlorides. Protein Sci. 1996;5:2020–2028. doi: 10.1002/pro.5560051008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, Nguyen MD, Andya JD, Shire SJ. Reversible self-association increases the viscosity of a concentrated monoclonal antibody in aqueous solution. J Pharm Sci. 2005;94:1928–1940. doi: 10.1002/jps.20347. [DOI] [PubMed] [Google Scholar]
- 11.Kanai S, Liu J, Patapoff TW, Shire SJ. Reversible self-association of a concentrated monoclonal antibody solution mediated by Fab-Fab interaction that impacts solution viscosity. J Pharm Sci. 2008;97:4219–4227. doi: 10.1002/jps.21322. [DOI] [PubMed] [Google Scholar]
- 12.Gjerde DT, Schmuckler G, Fritz JS. Anion chromatography with low-conductivity eluents. J Chromatogr. 1980;187:35–45. II. [Google Scholar]
- 13.Muzammil S, Kumar Y, Tayyab S. Anion-induced stabilization of human serum albumin prevents the formation of intermediate during urea denaturation. Proteins. 2000;40:29–38. doi: 10.1002/(sici)1097-0134(20000701)40:1<29::aid-prot50>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 14.Muzammil S, Kumar Y, Tayyab S. Anion-induced refolding of human serum albumin under low pH conditions. Biochim Biophys Acta. 2000;1476:139–148. doi: 10.1016/s0167-4838(99)00226-5. [DOI] [PubMed] [Google Scholar]
- 15.Baldwin RL. How Hofmeister ion interactions affect protein stability. Biophys J. 1996;71:2056–2063. doi: 10.1016/S0006-3495(96)79404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Cremer PS. Interactions between macromolecules and ions: the Hofmeister series. Curr Opin Chem Biol. 2006;10:658–663. doi: 10.1016/j.cbpa.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 17.Collins KD. Sticky ions in biological systems. Proc Natl Acad Sci USA. 1995;92:5553–5557. doi: 10.1073/pnas.92.12.5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins KD. Charge density-dependent strength of hydration and biological structure. Biophys J. 1997;72:65–76. doi: 10.1016/S0006-3495(97)78647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins KD. Ions from the Hofmeister series and osmolytes: effects on proteins in solution and in the crystallization process. Methods. 2004;34:300–311. doi: 10.1016/j.ymeth.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 20.Collins KD. Ion hydration: implications for cellular function, polyelectrolytes, and protein crystallization. Biophys Chem. 2006;119:271–281. doi: 10.1016/j.bpc.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 21.Collins KD, Neilson GW, Enderby JE. Ions in water: characterizing the forces that control chemical processes and biological structure. Biophys Chem. 2007;128:95–104. doi: 10.1016/j.bpc.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 22.Brems DN, Alter LA, Beckage MJ, Chance RE, DiMarchi RD, Green LK, Long HB, Pekar AH, Shields JE, Frank BH. Altering the association properties of insulin by amino acid replacement. Protein Eng. 1992;5:527–533. doi: 10.1093/protein/5.6.527. [DOI] [PubMed] [Google Scholar]
- 23.Pang Y, Sakagami M, Byron PR. Insulin self-association: effects on lung disposition kinetics in the airways of the Isolated Perfused Rat Lung (IPRL) Pharm Res. 2007;24:1636–1644. doi: 10.1007/s11095-007-9292-6. [DOI] [PubMed] [Google Scholar]
- 24.Shire SJ, Shahrokh Z, Liu J. Challenges in the development of high protein concentration formulations. J Pharm Sci. 2004;93:1390–1402. doi: 10.1002/jps.20079. [DOI] [PubMed] [Google Scholar]
- 25.Cromwell ME, Hilario E, Jacobson F. Protein aggregation and bioprocessing. AAPS J. 2006;8:E572–E579. doi: 10.1208/aapsj080366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenberg AS. Effects of protein aggregates: an immunologic perspective. AAPS J. 2006;8:E501–E507. doi: 10.1208/aapsj080359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ralston G, Cronin T, Branton D. Self-association of spectrin's repeating segments. Biochemistry. 1996;35:5257–5263. doi: 10.1021/bi952224d. [DOI] [PubMed] [Google Scholar]
- 28.Havel HA, Kauffman EW, Plaisted SM, Brems DN. Reversible self-association of bovine growth hormone during equilibrium unfolding. Biochemistry. 1986;25:6533–6538. doi: 10.1021/bi00369a029. [DOI] [PubMed] [Google Scholar]
- 29.Richards JP, Stickelmeyer MP, Flora DB, Chance RE, Frank BH, DeFelippis MR. Self-association properties of monomeric insulin analogs under formulation conditions. Pharm Res. 1998;15:1434–1441. doi: 10.1023/a:1011961923870. [DOI] [PubMed] [Google Scholar]
- 30.Dannies P. Manipulating the reversible aggregation of protein hormones in secretory granules: potential impact on biopharmaceutical development. BioDrugs. 2003;17:315–324. doi: 10.2165/00063030-200317050-00002. [DOI] [PubMed] [Google Scholar]
- 31.Pellaud J, Schote U, Arvinte T, Seelig J. Conformation and self-association of human recombinant transforming growth factor-beta3 in aqueous solutions. J Biol Chem. 1999;274:7699–7704. doi: 10.1074/jbc.274.12.7699. [DOI] [PubMed] [Google Scholar]
- 32.Banks WA, Kastin AJ. Reversible association of the cytokines MIP-1 alpha and MIP-1 beta with the endothelia of the blood-brain barrier. Neurosci Lett. 1996;205:202–206. doi: 10.1016/0304-3940(96)12410-1. [DOI] [PubMed] [Google Scholar]
- 33.Panka DJ. Glycosylation is influential in murine IgG3 self-association. Mol Immunol. 1997;34:593–598. doi: 10.1016/s0161-5890(97)00080-1. [DOI] [PubMed] [Google Scholar]
- 34.Moore JM, Patapoff TW, Cromwell ME. Kinetics and thermodynamics of dimer formation and dissociation for a recombinant humanized monoclonal antibody to vascular endothelial growth factor. Biochemistry. 1999;38:13960–13967. doi: 10.1021/bi9905516. [DOI] [PubMed] [Google Scholar]
- 35.Lee YC, Boehm MK, Chester KA, Begent RH, Perkins SJ. Reversible dimer formation and stability of the anti-tumour single-chain Fv antibody MFE-23 by neutron scattering, analytical ultracentrifugation, and NMR and FT-IR spectroscopy. J Mol Biol. 2002;320:107–127. doi: 10.1016/S0022-2836(02)00403-5. [DOI] [PubMed] [Google Scholar]
- 36.Jule E, Nagasaki Y, Jiskoot W. Surface plasmon resonance study on the interaction between lactose-installed poly(ethylene glycol)-poly(d,l-lactide) block copolymer micelles and lectins immobilized on a gold surface. Langmuir. 2002;18:10334–10339. [Google Scholar]
- 37.Amidi M, Romeijn SG, Borchard G, Junginger HE, Hennink WE, Jiskoot W. Preparation and characterization of protein-loaded N-trimethyl chitosan nanoparticles as nasal delivery system. J Control Release. 2006;111:107–116. doi: 10.1016/j.jconrel.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 38.Solovyova A, Schuck P, Costenaro L, Ebel C. Non-ideality by sedimentation velocity of halophilic malate dehydrogenase in complex solvents. Biophys J. 2001;81:1868–1880. doi: 10.1016/S0006-3495(01)75838-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collins KD, Washabaugh MW. The Hofmeister effect and the behaviour of water at interfaces. Q Rev Biophys. 1985;18:323–422. doi: 10.1017/s0033583500005369. [DOI] [PubMed] [Google Scholar]
- 40.Wahl-Schott C, Baumann L, Zong X, Biel M. An arginine residue in the pore region is a key determinant of chloride dependence in cardiac pacemaker channels. J Biol Chem. 2005;280:13694–13700. doi: 10.1074/jbc.M413197200. [DOI] [PubMed] [Google Scholar]
- 41.Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proc Natl Acad Sci USA. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verschueren KH, Kingma J, Rozeboom HJ, Kalk KH, Janssen DB, Dijkstra BW. Crystallographic and fluorescence studies of the interaction of haloalkane dehalogenase with halide ions. Studies with halide compounds reveal a halide binding site in the active site. Biochemistry. 1993;32:9031–9037. doi: 10.1021/bi00086a008. [DOI] [PubMed] [Google Scholar]
- 43.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]