Abstract
We tabulated 541 measured pK values reported in the literature for the Asp, Glu, His, Cys, Tyr, and Lys side chains, and the C and N termini of 78 folded proteins. The majority of these values are for the Asp, Glu, and His side chains. The average pK values are Asp 3.5 ± 1.2 (139); Glu 4.2 ± 0.9 (153); His 6.6 ± 1.0 (131); Cys 6.8 ± 2.7 (25); Tyr 10.3 ± 1.2 (20); Lys 10.5 ± 1.1 (35); C-terminus 3.3 ± 0.8 (22) and N-terminus 7.7 ± 0.5 (16). We compare these results with the measured pK values of these groups in alanine pentapeptides, and comment on our overall findings.
Keywords: pK values, protein ionizable groups, pH titration, peptide model compounds, NMR spectroscopy
Introduction
About 25% of the residues in proteins contain ionizable side chains that are of crucial importance to the mechanism of action of enzymes and to the binding of proteins to other small and large molecules. In addition, the ionization state of these residues determines the net charge on a protein, and this is important to the structure, function, stability, and solubility of a protein. Consequently, biochemists have a strong interest in the pK values of the ionizable groups of proteins and in the factors that perturb them.
We previously published pK values for the ionizable groups of proteins determined with uncharged, alanine pentapeptides.1 These values reflect the inductive effects of neighboring peptide bonds, but will not be influenced by charge–charge interactions or burial of the ionizable group. Consequently, we think these pKs will serve as reasonable models for the unperturbed pK values of the ionizable groups in proteins. Most of these pKs do not differ significantly from the intrinsic pK values estimated earlier by Nozaki and Tanford.2 It will be interesting to compare these values to the pKs determined in unfolded proteins.3–10
Most of the pK values for folded proteins have been measured directly using techniques based on nuclear magnetic resonance (NMR).11–13 A smaller number have been measured using indirect techniques.14,15 In several recent papers, summaries of these results have been given: 212 carboxyl groups13; 37 histidine16; 260 values from 41 proteins17; 314 values.18
These previous surveys have provided new insight into the relationship between protein structure and pK values,13,16 as well as helping those who use theory to predict the pK values of the groups in folded proteins.17–21 This is interesting because the pK values of many of the ionizable groups in folded proteins are strongly influenced by the local environment.14,22–29
In this article, we have listed all of the measured pK values that we could find in the literature. The data were taken directly as published from experiments on folded proteins. We summarize our findings and give a few brief comments. We hope this collection of data will be useful to those seeking pK values for proteins, and to those who are interested in how intrinsic pK values can be perturbed in folded proteins.
Results and Discussion
Table I summarizes the measured pK values for 541 ionizable groups from 78 proteins. The individual values, references, and our data analysis are given in the Supporting Information. Data have been collected for 139 Asp, 153 Glu, 131 His, 25 Cys, 20 Tyr, 35 Lys, 22 C-termini, and 16 N-termini. For comparison, the pK values for the same groups measured in uncharged alanine pentapeptides are also given in the table. Here we have corrected the pK for Asp in the pentapeptide from 3.67, as we previously reported 1 to 3.94. Our new value is based on further measurements under the same conditions on a newly synthesized peptide, where a pK = 3.90 ± 0.02 was measured using a potentiometric titration similar to the technique described in the reference mentioned earlier, and a pK = 3.98 ± 0.05 was determined using NMR (these measurements, as well as others using NMR, will be discussed in an upcoming publication). Therefore, a pK = 3.94 for Asp in the alanine pentapeptide seems most reasonable.
Table I.
Summary of 541 pK Values for 78 Proteins Tabulated from the Literaturea
| Group | pK value in alanine pentapeptidesb | Average pK value | Low pK value | High pK value | Number of measurements |
|---|---|---|---|---|---|
| Asp | 3.9c | 3.5 ± 1.2 | 0.5 | 9.2 | 139 |
| Glu | 4.3 | 4.2 ± 0.9 | 2.1 | 8.8 | 153 |
| His | 6.5 | 6.6 ± 1.0 | 2.4 | 9.2 | 131 |
| Cys | 8.6 | 6.8 ± 2.7 | 2.5 | 11.1 | 25 |
| Tyr | 9.8 | 10.3 ± 1.2 | 6.1 | 12.1 | 20 |
| Lys | 10.4 | 10.5 ± 1.1 | 5.7 | 12.1 | 35 |
| C-term | 3.7 | 3.3 ± 0.8 | 2.4 | 5.9 | 22 |
| N-term | 8.0 | 7.7 ± 0.5 | 6.8 | 9.1 | 16 |
The majority of the measurements reported in the literature (78%) are for Asp, Glu, and His (Table I and Fig. 1), so most of our comments will be limited to these groups. The average pKs are Asp = 3.5, Glu = 4.2, and His = 6.6. In the case of Asp, the average is 0.4 below the measured value in the alanine pentapeptide (3.5 vs. 3.9). This is clearly reflected by the distribution in Figure 1(A), where ≈77% of the values are lower than the peptide pK of 3.9. In the case of Glu, the average is close to the value measured in the pentapeptide (4.2 vs. 4.3), and 53% of the values are between pH 4–5 [Fig. 1(B)]. The case for His is similar, where the average is again close to the measured value in the alanine pentapeptide (6.6 vs. 6.5). Figure 1(C) shows that ≈44% of the pK measurements fall between pH 6–7, and ≈90% of the His pKs are in the range of pH 5–8.
Figure 1.
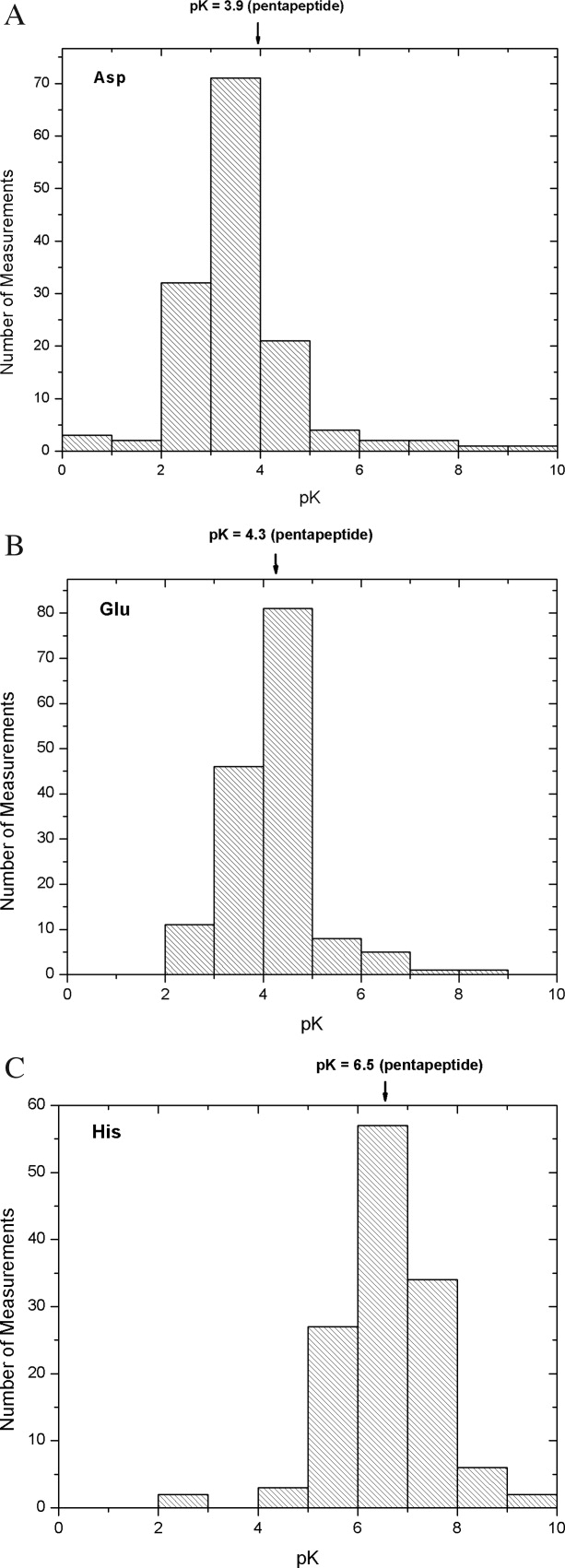
Distribution of the measured pK values for Asp (A), Glu (B), and His (C). The arrow on the top axis of each figure indicates the intrinsic pK value as measured in alanine pentapeptides.
The three main factors that will influence the pKs of the ionizable groups in folded proteins are charge–charge interactions, charge–dipole interactions (hydrogen bonding), and the Born effect (dehydration). The pK shift due to the Born effect will depend mainly on the effective dielectric constant of the environment of the ionizable group in the protein compared with the dielectric constant of water (78 at 25°C). The pKs of carboxyl, hydroxyl, and sulfhydryl groups will be increased and the pKs of amino groups decreased in the presence of the lower dielectric constant typically encountered when the groups are partially or completely buried in folded proteins. The presence of a positively charged environment will lower the pKs of all the ionizable groups in a protein, and the presence of a negatively charged environment will raise the pKs. The effect of hydrogen bonding on the pK values will depend on whether the hydrogen bonding is better to the protonated or the deprotonated form of the ionizable group.
The effect of the local environment on the pKs due to the factors mentioned earlier is clearly supported by the low and high values given in Table I. In the case of Asp, the lowest value (0.5) is for Asp70 of T4 lysozyme and for Asp76 of RNase T1. In T4 lysozyme, Asp70 forms a salt bridge with His31, stabilizing the protein by ≈4 kcal mol−1.30 In RNase T1, Asp76 is buried and forms four hydrogen bonds, which account for more than half of the net stability of the protein at pH 7.14 The highest value (9.2) is Asp 26 in E. coli thioredoxin, where the side chain is completely buried near the active site.31,32 Another Asp with a pK value near 9 is the V66D variant of staph nuclease, where the side chain is buried in the hydrophobic core of the protein.29 In the case of Glu, the lowest value (2.1) is Glu73 in barnase. This residue participates as a general base at the catalytic site and is close to the positively charged groups of Lys27, Arg83, and Arg 87.33 The highest value (8.8) is the V66E substitution in staph nuclease that again buries the Glu in the hydrophobic interior of the protein.34 In the case of His, the lowest value (2.4) is for His18 in horse cytochrome c. His18 is coordinated to the iron in the completely buried heme group, and additionally forms two good hydrogen bonds to Ala15 and Pro3035 and this no doubt contributes to its unusually low pK. The highest value (9.2) is for His72 in bovine protein tyrosine phosphatase. Here the imidazole group is buried, but participates in electrostatic interactions with several nearby negatively charged groups.36
There are relatively few pK values reported for Cys (25), Tyr (20), Lys (35), and the N and C termini (16 and 22, respectively). For Cys, the average pK (6.8) is considerably lower than the value measured in alanine pentapeptides (8.6). The Cys residues with the lowest pKs are generally in the active sites of enzymes. For example, the Cys25-S−/His159-I-imidazole(Im)+ ion pair is found in the active site of cysteine proteinases (see Ref. 18 and Ref. 25 for detailed discussions of these topics). These residues are often studied, and this leads to the low average pK. The lowest value (2.5) is Cys 25 in ficin, a cysteine proteinase, and the highest value (11.1) is the active site Cys in a ubiquitin-conjugating enzyme, Ubc13, where the Cys is surrounded by acidic residues.37,38
The limited number of measurements for Tyr and Lys is because many proteins unfold in the higher pH range where these groups ionize. The average value for Lys (10.5) does not differ significantly from the pK measured in the pentapeptide (10.4). This is not surprising because the amino group of Lys is usually exposed to solvent in folded proteins (avg % exposed = 6639). The lowest value (5.7) results when a Lys is substituted for Val (V66K) and buried in the hydrophobic core of staph nuclease.40 The highest value (12.1) is Lys55 in the Apo-form of calbindin D9k, where the perturbation is due to the large net negative charge on the protein.41 For Tyr, the average pK value is 0.5 units above the measured value in the alanine pentapeptide (10.3 vs. 9.8). Again, this is not surprising because the —OH group of Tyr is usually buried in folded proteins (avg % buried = 6739), which would raise the average value.
For the C-termini, the average pK value (3.3) is 0.4 units lower than that measured in our model peptide (3.7). This is not surprising because at low pH where the carboxyl group ionizes, most proteins will have a large positive charge, consequently lowering the pK. The lowest value (2.4) is found in RNase Sa 5K, a basic variant of RNase Sa, with a pI of 10.2.42 Furthermore, the C-terminus is a Cys that forms a disulfide bond (Cys96-Cys7).43 This and the large positive charge on the protein are expected to lower the pK significantly.27,42,44,45 The highest value (5.9) is for Ala79, the C-terminus of subunit c of F1F0 ATP Synthase. The high pK results because it is partially buried (see the discussion in Ref. 46). For the N-termini, the average pK value (7.7) does not differ significantly from the pK measured in the peptide (8.0), and the range of low and high values is not as great as that observed for the other groups. The low value (6.8) was measured for the β chain of human deoxyhemoglobin.47 The high value (9.1) was measured in RNase Sa, an acidic protein.27 In this case, the large negative charge in the pH range where the N-terminus ionizes undoubtedly contributes to the higher pK.
Methods
The measured pK values for the ionizable groups of Asp, Glu, His, Tyr, Lys, C-termini, and N-termini in 78 proteins were taken from the primary reference given (see Supporting Information for the actual measurements and our data analysis). The technique most used for determining the pK values was NMR, but some measurements were based on less direct techniques. The experiments were performed over a range of temperatures and ionic strength, but in each case, the values were thought to be measured on the folded form of the protein. The significant figures for the measured pK values and the identity of the ionizable groups are given as reported in the original reference. The majority of the data are for wild-type proteins that have been well studied. A few well characterized variants are also included.
Acknowledgments
The authors intend to continue to update Table I as more pK values become available and encourage readers to inform us regarding new pK values, or relevant pK values that we inadvertently omitted from Table I.
References
- 1.Thurlkill RL, Grimsley GR, Scholtz JM, Pace CN. pK values of the ionizable groups of proteins. Protein Sci. 2006;15:1214–1218. doi: 10.1110/ps.051840806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nozaki Y, Tanford C. Examination of titration behavior. Methods Enzymol. 1967;11:715–734. [Google Scholar]
- 3.Tollinger M, Crowhurst KA, Kay LE, Forman-Kay JD. Site-specific contributions to the pH dependence of protein stability. Proc Natl Acad Sci USA. 2003;100:4545–4550. doi: 10.1073/pnas.0736600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho JH, Raleigh DP. Mutational analysis demonstrates that specific electrostatic interactions can play a key role in the denatured state ensemble of proteins. J Mol Biol. 2005;353:174–185. doi: 10.1016/j.jmb.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 5.Marti DN. Apparent pKa shifts of titratable residues at high denaturant concentration and the impact on protein stability. Biophys Chem. 2005;118:88–92. doi: 10.1016/j.bpc.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 6.Pujato M, Bracken C, Mancusso R, Cataldi M, Tasayco ML. pH dependence of amide chemical shifts in natively disordered polypeptides detects medium-range interactions with ionizable residues. Biophys J. 2005;89:3293–3302. doi: 10.1529/biophysj.105.060384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho JH, Raleigh DP. Electrostatic interactions in the denatured state and in the transition state for protein folding: effects of denatured state interactions on the analysis of transition state structure. J Mol Biol. 2006;359:1437–1446. doi: 10.1016/j.jmb.2006.04.038. [DOI] [PubMed] [Google Scholar]
- 8.Pujato M, Navarro A, Versace R, Mancusso R, Ghose R, Tasayco ML. The pH-dependence of amide chemical shift of Asp/Glu reflects its pKa in intrinsically disordered proteins with only local interactions. Biochim Biophys Acta. 2006;1764:1227–1233. doi: 10.1016/j.bbapap.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 9.Matousek WM, Ciani B, Fitch CA, Garcia-Moreno EB, Kammerer RA, Alexandrescu AT. Electrostatic contributions to the stability of the GCN4 leucine zipper structure. J Mol Biol. 2007;374:206–219. doi: 10.1016/j.jmb.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quijada J, Lopez G, Versace R, Ramirez L, Tasayco ML. On the NMR analysis of pKa values in the unfolded state of proteins by extrapolation to zero denaturant. Biophys Chem. 2007;129:242–250. doi: 10.1016/j.bpc.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 11.Markley JL. Observation of histidine residues in proteins by means of nuclear magnetic resonance spectroscopy. Acc Chem Res. 1975;8:70–80. [Google Scholar]
- 12.Matthew JB, Gurd FR, Garcia-Moreno B, Flanagan MA, March KL, Shire SJ. pH-dependent processes in proteins. CRC Crit Rev Biochem. 1985;18:91–197. doi: 10.3109/10409238509085133. [DOI] [PubMed] [Google Scholar]
- 13.Forsyth WR, Antosiewicz JM, Robertson AD. Empirical relationships between protein structure and carboxyl pKa values in proteins. Proteins. 2002;48:388–403. doi: 10.1002/prot.10174. [DOI] [PubMed] [Google Scholar]
- 14.Giletto A, Pace CN. Buried, charged, non-ion-paired aspartic acid 76 contributes favorably to the conformational stability of ribonuclease T1. Biochemistry. 1999;38:13379–13384. doi: 10.1021/bi991422s. [DOI] [PubMed] [Google Scholar]
- 15.Baran KL, Chimenti MS, Schlessman JL, Fitch CA, Herbst KJ, Garcia-Moreno BE. Electrostatic effects in a network of polar and ionizable groups in staphylococcal nuclease. J Mol Biol. 2008;379:1045–1062. doi: 10.1016/j.jmb.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 16.Edgcomb SP, Murphy KP. Variability in the pKa of histidine side-chains correlates with burial within proteins. Proteins. 2002;49:1–6. doi: 10.1002/prot.10177. [DOI] [PubMed] [Google Scholar]
- 17.Wisz MS, Hellinga HW. An empirical model for electrostatic interactions in proteins incorporating multiple geometry-dependent dielectric constants. Proteins. 2003;51:360–377. doi: 10.1002/prot.10332. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 19.Antosiewicz J, McCammon JA, Gilson MK. Prediction of pH-dependent properties of proteins. J Mol Biol. 1994;238:415–436. doi: 10.1006/jmbi.1994.1301. [DOI] [PubMed] [Google Scholar]
- 20.Schutz CN, Warshel A. What are the dielectric “constants” of proteins and how to validate electrostatic models? Proteins. 2001;44:400–417. doi: 10.1002/prot.1106. [DOI] [PubMed] [Google Scholar]
- 21.Gunner MR, Mao J, Song Y, Kim J. Factors influencing the energetics of electron and proton transfers in proteins. What can be learned from calculations? Biochim Biophys Acta. 2006;1757:942–968. doi: 10.1016/j.bbabio.2006.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanford C. Protein denaturation. Adv Protein Chem. 1968;23:121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- 23.Fersht A. Enzyme structure and mechanism. 2nd ed. New York: W.H. Freeman; 1985. p. 475. [Google Scholar]
- 24.Bartlett GJ, Porter CT, Borkakoti N, Thornton JM. Analysis of catalytic residues in enzyme active sites. J Mol Biol. 2002;324:105–121. doi: 10.1016/s0022-2836(02)01036-7. [DOI] [PubMed] [Google Scholar]
- 25.Harris TK, Turner GJ. Structural basis of perturbed pKa values of catalytic groups in enzyme active sites. IUBMB Life. 2002;53:85–98. doi: 10.1080/15216540211468. [DOI] [PubMed] [Google Scholar]
- 26.Huyghues-Despointes BM, Thurlkill RL, Daily MD, Schell D, Briggs JM, Antosiewicz JM, Pace CN, Scholtz JM. pK values of histidine residues in ribonuclease Sa: effect of salt and net charge. J Mol Biol. 2003;325:1093–1105. doi: 10.1016/s0022-2836(02)01274-3. [DOI] [PubMed] [Google Scholar]
- 27.Laurents DV, Huyghues-Despointes BM, Bruix M, Thurlkill RL, Schell D, Newsom S, Grimsley GR, Shaw KL, Trevino S, Rico M, Briggs JM, Antosiewicz JM, Scholtz JM, Pace CN. Charge–charge interactions are key determinants of the pK values of ionizable groups in ribonuclease Sa (pI = 3.5) and a basic variant (pI = 10.2) J Mol Biol. 2003;325:1077–1092. doi: 10.1016/s0022-2836(02)01273-1. [DOI] [PubMed] [Google Scholar]
- 28.Thurlkill RL, Grimsley GR, Scholtz JM, Pace CN. Hydrogen bonding markedly reduces the pK of buried carboxyl groups in proteins. J Mol Biol. 2006;362:594–604. doi: 10.1016/j.jmb.2006.07.056. [DOI] [PubMed] [Google Scholar]
- 29.Karp DA, Gittis AG, Stahley MR, Fitch CA, Stites WE, Garcia-Moreno EB. High apparent dielectric constant inside a protein reflects structural reorganization coupled to the ionization of an internal Asp. Biophys J. 2007;92:2041–2053. doi: 10.1529/biophysj.106.090266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson DE, Becktel WJ, Dahlquist FW. pH-induced denaturation of proteins: a single salt bridge contributes 3–5 kcal/mol to the free energy of folding of T4 lysozyme. Biochemistry. 1990;29:2403–2408. doi: 10.1021/bi00461a025. [DOI] [PubMed] [Google Scholar]
- 31.Chivers PT, Prehoda KE, Volkman BF, Kim BM, Markley JL, Raines RT. Microscopic pKa values of Escherichia coli thioredoxin. Biochemistry. 1997;36:14985–14991. doi: 10.1021/bi970071j. [DOI] [PubMed] [Google Scholar]
- 32.Dyson HJ, Jeng MF, Tennant LL, Slaby I, Lindell M, Cui DS, Kuprin S, Holmgren A. Effects of buried charged groups on cysteine thiol ionization and reactivity in Escherichia coli thioredoxstructural and functional characterization of mutants of Asp 26 and Lys 57. Biochemistry. 1997;36:2622–2636. doi: 10.1021/bi961801a. [DOI] [PubMed] [Google Scholar]
- 33.Schreiber G, Frisch C, Fersht AR. The role of Glu73 of barnase in catalysis and the binding of barstar. J Mol Biol. 1997;270:111–122. doi: 10.1006/jmbi.1997.1080. [DOI] [PubMed] [Google Scholar]
- 34.Dwyer JJ, Gittis AG, Karp DA, Lattman EE, Spencer DS, Stites WE, Garcia-Moreno EB. High apparent dielectric constants in the interior of a protein reflect water penetration. Biophys J. 2000;79:1610–1620. doi: 10.1016/S0006-3495(00)76411-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bushnell GW, Louie GV, Brayer GD. High-resolution three-dimensional structure of horse heart cytochrome c. J Mol Biol. 1990;214:585–595. doi: 10.1016/0022-2836(90)90200-6. [DOI] [PubMed] [Google Scholar]
- 36.Tishmack PA, Bashford D, Harms E, Van Etten RL. Use of 1H NMR spectroscopy and computer simulations to analyze histidine pKa changes in a protein tyrosine phosphatase: experimental and theoretical determination of electrostatic properties in a small protein. Biochemistry. 1997;36:11984–11994. doi: 10.1021/bi9712448. [DOI] [PubMed] [Google Scholar]
- 37.Pinitglang S, Watts AB, Patel M, Reid JD, Noble MA, Gul S, Bokth A, Naeem A, Patel H, Thomas EW, Steedharan SK, Verma C, Brocklehurst K. A classical enzyme active center motif lacks catalytic competence until modulated electrostatically. Biochemistry. 1997;36:9968–9982. doi: 10.1021/bi9705974. [DOI] [PubMed] [Google Scholar]
- 38.Tolbert BS, Tajc SG, Webb H, Snyder J, Nielsen JE, Miller BL, Basavappa R. The active site cysteine of ubiquitin-conjugating enzymes has a significantly elevated pKa: functional implications. Biochemistry. 2005;44:16385–16391. doi: 10.1021/bi0514459. [DOI] [PubMed] [Google Scholar]
- 39.Lesser GJ, Rose GD. Hydrophobicity of amino acid subgroups in proteins. Proteins. 1990;8:6–13. doi: 10.1002/prot.340080104. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Moreno B, Dwyer JJ, Gittis AG, Lattman EE, Spencer DS, Stites WE. Experimental measurement of the effective dielectric in the hydrophobic core of a protein. Biophys Chem. 1997;64:211–224. doi: 10.1016/s0301-4622(96)02238-7. [DOI] [PubMed] [Google Scholar]
- 41.Kesvatera T, Jonsson B, Thulin E, Linse S. Measurement and modelling of sequence-specific pKa values of lysine residues in calbindin D9k. J Mol Biol. 1996;259:828–839. doi: 10.1006/jmbi.1996.0361. [DOI] [PubMed] [Google Scholar]
- 42.Shaw KL, Grimsley GR, Yakovlev GI, Makarov AA, Pace CN. The effect of net charge on the solubility, activity, and stability of ribonuclease Sa. Protein Sci. 2001;10:1206–1215. doi: 10.1110/ps.440101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pace CN, Hebert EJ, Shaw KL, Schell D, Both V, Krajcikova D, Sevcik J, Wilson KS, Dauter Z, Hartley RW, Grimsley GR. Conformational stability and thermodynamics of folding of ribonucleases Sa, Sa2 and Sa3. J Mol Biol. 1998;279:271–286. doi: 10.1006/jmbi.1998.1760. [DOI] [PubMed] [Google Scholar]
- 44.Greenstein JP, Klemperer FW, Wyman J., Jr Physical chemistry of cystine peptides. J Biol Chem. 1939;129:681–692. [Google Scholar]
- 45.Schaller W, Robertson AD. pH, ionic strength, and temperature dependences of ionization equilibria for the carboxyl groups in turkey ovomucoid third domain. Biochemistry. 1995;34:4714–4723. doi: 10.1021/bi00014a028. [DOI] [PubMed] [Google Scholar]
- 46.Assadi-Porter FM, Fillingame RH. Proton-translocating carboxyl of subunit c of F1Fo H(+)-ATP synthase: the unique environment suggested by the pKa determined by 1H NMR. Biochemistry. 1995;34:16186–16193. doi: 10.1021/bi00049a034. [DOI] [PubMed] [Google Scholar]
- 47.Garner MH, Bogardt RA, Jr, Gurd FR. Determination of the pK values for the alpha-amino groups of human hemoglobin. J Biol Chem. 1975;250:4398–4404. [PubMed] [Google Scholar]


