Abstract
Bacterial microcompartments are supramolecular protein assemblies that function as bacterial organelles by compartmentalizing particular enzymes and metabolic intermediates. The outer shells of these microcompartments are assembled from multiple paralogous structural proteins. Because the paralogs are required to assemble together, their genes are often transcribed together from the same operon, giving rise to a distinctive genomic pattern: multiple, typically small, paralogous proteins encoded in close proximity on the bacterial chromosome. To investigate the generality of this pattern in supramolecular assemblies, we employed a comparative genomics approach to search for protein families that show the same kind of genomic pattern as that exhibited by bacterial microcompartments. The results indicate that a variety of large supramolecular assemblies fit the pattern, including bacterial gas vesicles, bacterial pili, and small heat-shock protein complexes. The search also retrieved several widely distributed protein families of presently unknown function. The proteins from one of these families were characterized experimentally and found to show a behavior indicative of supramolecular assembly. We conclude that cotranscribed paralogs are a common feature of diverse supramolecular assemblies, and a useful genomic signature for discovering new kinds of large protein assemblies from genomic data.
Keywords: supramolecular assembly, bacterial ultrastructure, paralog, homolog, self assembly, carboxysome, bacterial microcompartment
Introduction
Bacterial microcompartments (BMCs) are large (80–150 nm) proteinaceous bodies that encapsulate a series of enzymes carrying out specific metabolic processes in bacteria (reviewed in Refs. 1 and2) [Fig. 1(A,B)]. The carboxysome is the best-studied BMC 6 (reviewed in Ref. 5). In all cyanobacteria and some chemoautotrophs, it encapsulates the enzymes RuBisCO and carbonic anhydrase and, thereby, improves the efficiency of carbon fixation under conditions in which inorganic carbon levels are limiting.7,8 The 1,2-propanediol utilization (Pdu) microcompartment provides another example. In the enteropathic bacterium Salmonella enterica Typhimurium LT2, the Pdu microcompartment is induced in the presence of 1,2-propanediol and metabolizes that compound in the interior.9 The carboxysome and the Pdu microcompartment encapsulate entirely unrelated enzymes, yet their outer shells are similar in key respects. In particular, the major shell proteins from these two microcompartments are homologous to each other.10,11 In addition to the carboxysome and the Pdu microcompartment, other BMCs with related shells but apparently diverse functions have been identified or implicated from genomic sequence data.12,13 The data suggest that these microcompartments are a widespread, but underappreciated subcellular structure in bacteria.
Figure 1.
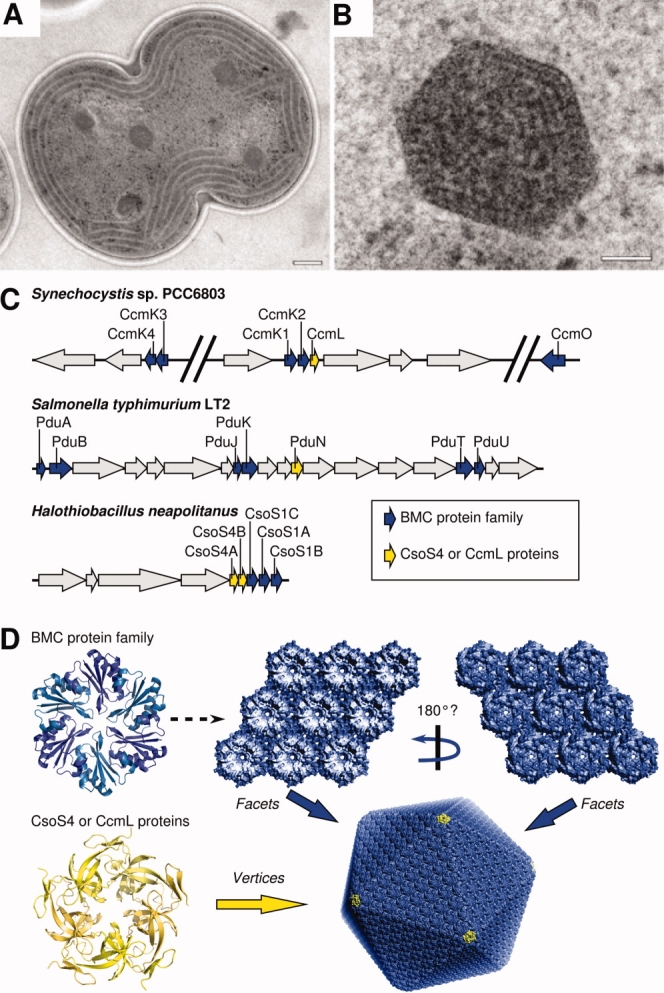
Structure and organization of carboxysomes and related bacterial microcompartments. (A) Carboxysomes visualized by thin-section EM in a dividing cell of the cyanobacterium Synechocystis sp. PCC6803 cell (scale bar, 200 nm). (B) Enlargement of a single carboxysome (scale bar, 50 nm). Panels A and B (courtesy of Wim Vermaas) adapted from Ref. 3. (C) Organization of the microcompartment genes for the carboxysome in Syn. 6803 and in the chemoautotroph H. neapolitanus, and for the Pdu microcompartment in Salmonella typhimurium LT2. Gene families highlighted in the text are colored blue and yellow. (D) Oligomeric structures of proteins from the BMC family (CcmK or CsoS1) and the CcmL/EutN family, and a model for their higher level assembly into a carboxysome, according to Ref. 4. (Figure adapted from Ref. 5).
The shells of BMCs are composed predominantly of a small (∼10 kDa) protein, a few thousand copies of which are assembled together to form a single shell14 (reviewed in Ref. 5). Proteins belonging to this conserved family are referred to here as BMC proteins.15 The diverse microcompartments identified so far share further features. They contain multiple paralogs of the BMC shell protein, typically encoded together in operons alongside the enzymes that carry out the encapsulated processes.10,13 For example, in the carboxysome from H. neapolitanus, there are three BMC proteins (called CsoS1A,B,C) encoded in an operon with genes for the RuBisCO large and small subunits and carbonic anhydrase, along with three other carboxysome proteins (reviewed in Ref. 16); the Synechocystis PCC 6803 genome encodes five BMC proteins [Fig. 1(C)].17 Likewise in the Salmonellapdu operon, there are a total of six distinct BMC genes alongside 15 other genes, several of which are understood to code for enzymes involved in 1,2-propanediol metabolism9 [Fig. 1(C)]. Similarly, in Escherichia coli, the eut operon (which is also present in Salmonella) encodes four BMC protein paralogs, along with enzymes believed to carry out ethanolamine degradation inside the Eut microcompartment.12,18,19 In all cases, there is a clear pattern of multiple paralogous proteins encoded together.
Structural studies are beginning to shed light on the purpose of multiple paralogs in the shells of BMCs. Recent EM tomography studies show that carboxysomes are roughly icosahedral in shape.20,21 Crystal structures of various carboxysome shell proteins have revealed that the conserved BMC-domain proteins form cyclic hexamers, with a tendency to pack side by side to form molecular layers presumed to represent the flat facets of the icosahedral shell.3,4,22 However, differing assembly properties have been noted for some paralogs. The carboxysome shell protein CcmK4 tends to form strips instead of a layer, suggesting that the multiple paralogous proteins in BMC shells might serve specific architectural roles. This would parallel the situation in many viral capsids, where multiple paralogous proteins may be present in different structural environments according to the principles of quasi-equivalence.23–26 Alternatively, the requirement for multiple paralogous proteins might be related to biochemical function. It has been hypothesized based on crystal structures that transport of small molecules across microcompartment shells might occur through the pores down the middle of BMC hexamers.3,22 The diversity of BMC paralogs within a single kind of microcompartment could reflect diverse biochemical functions (e.g., in transport or binding), as suggested by a recent structure of a shell protein from the pdu microcompartment.27 A well-known example of divergence of biochemical function between paralogs in a supramolecular assembly is seen in microtubules, which are composed of paralogous alpha and beta tubulin subunits. The biochemical activities of those two paralogs diverged following duplication of an ancestral FtsZ-like protein, perhaps to support the evolution of motor transport activities in eukaryotes (reviewed in Refs. 28 and29).
Regardless of the reasons for gene duplication and divergence, if paralogous proteins assemble together, they might be expected to be cotranscribed and therefore be encoded near each other in a given prokaryotic genome. Indeed, systematic studies in prokaryotes have shown that, when multiple proteins are involved in stable complexes, they are often encoded in proximity.30 Therefore, in the case of supramolecular assemblies of the type typified by BMCs, two features come into play. First, multiple paralogs are frequently involved. Second, the coassembling structural proteins tend to be encoded by proximal genes. The result is a particular genomic pattern: multiple (typically small) paralogous proteins encoded together. One might expect such a pattern to be somewhat specific for structural assemblies, because though gene duplication and divergence are widespread in other kinds of proteins such as metabolic enzymes, there is generally less selective pressure in those systems for the paralogous genes to remain in proximity.
Studies of prokaryotic ultrastructure have revealed many novel supramolecular structures.31 Discovering novel structures from direct microscopic observation can be critically dependent upon experimental factors, such as whether a microbe in question can be cultured, and whether the structure to be visualized is present under the growth conditions employed. It seems likely therefore that there are other as-yet undiscovered protein-based supramolecular assemblies in nature. In this study, we searched the genomic databases for paralogous proteins that are encoded in close proximity, and are therefore likely to be cotranscribed and coassembled. Among those protein families identified that have already been characterized, the majority are indeed involved in supramolecular assemblies. The uncharacterized families identified therefore constitute predictions of potentially novel supramolecular assemblies. Supportive, preliminary biophysical experiments on one uncharacterized protein family are presented.
Results
To predict protein families forming supramolecular assemblies potentially analogous to the BMC shell, a search was conducted for families with a similar genomic signature to microcompartment shell proteins. Families were defined using the InterPro domain database32 and filtered in a multistep procedure (Fig. 2). Operons were predicted across all archaeal and bacterial sequences from 593 prokaryotic organisms and searched for the desired genomic arrangement of multiple paralogous proteins occurring together [Fig. 1(C)]. Additionally, protein families were filtered for an average length of less than 200 amino acids to obtain proteins that might constitute relatively compact building blocks in larger structures; most microcompartment shell proteins are ∼100 amino acids in length. Families were subsequently filtered to select those with tandem occurrences within an operon in three or more organisms, and with at least three copies in at least one of those organisms, to arrive at a set of families whose analogy to BMC proteins was significant over multiple organisms. Finally, families were ranked according to the average number of paralogous co-occurrences seen in each operon (Supporting Information), and the top 35 were arbitrarily selected for further analysis (Table I)
Figure 2.
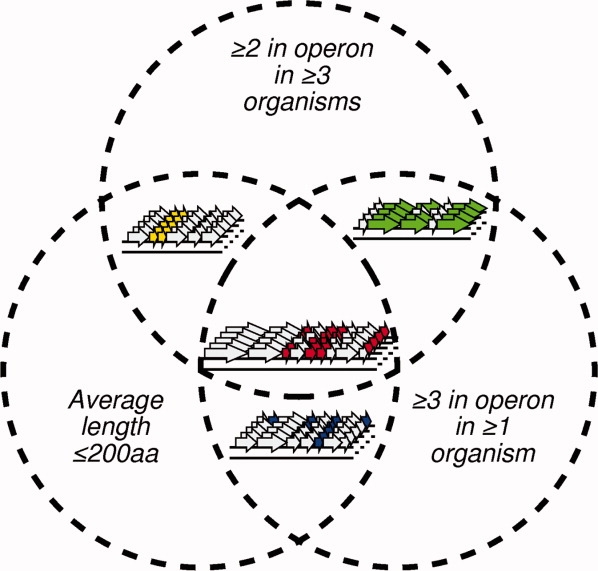
Venn diagram scheme for prediction of putative self-assembling protein families based on genomic context. Each circle is intended to illustrate a criterion used for selection. Proteins were filtered for families that match all three criteria depicted: compactness, multiple members per operon and occurrence of multiple members per operon in three or more organisms. Arrows represent open reading frames (ORFs). Each cluster of ORFs in a row represents a genomic region from one organism. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Table I.
Protein Families Predicted to Form Supramolecular Assemblies Based on Genomic Context
| Protein familya | No. of organismsb | Ave. paralogs per operon | Descriptionc |
|---|---|---|---|
| IPR006728 | 22 | 3.2 | Uncharacterized |
| IPR007966 | 10 | 3.0 | Uncharacterized |
| IPR014994 | 12 | 2.9 | Uncharacterized |
| IPR009482 | 5 | 2.8 | Uncharacterizedd |
| IPR012655 | 9 | 2.7 | Uncharacterized |
| IPR000249 | 104 | 2.6 | Bacterial microcompartments protein3,22,33 |
| IPR012452 | 21 | 2.1 | Uncharacterized |
| IPR010738 | 8 | 2.1 | Uncharacterized |
| IPR012902 | 380 | 2.1 | Prepilin-type cleavage/methylation, N-terminal34 |
| IPR010665 | 10 | 2.0 | Uncharacterized |
| IPR009881 | 17 | 2.0 | Uncharacterized |
| IPR008846 | 11 | 1.9 | Staphylococcus haemolytic peptides |
| IPR001120 | 322 | 1.8 | Prokaryotic N-terminal methylation site34 |
| IPR007670 | 6 | 1.8 | Uncharacterized |
| IPR000638 | 46 | 1.6 | Gas vesicle protein GvpA35 |
| IPR012128 | 28 | 1.6 | Phycobilisome alpha and beta chains36 |
| IPR012661 | 19 | 1.6 | Uncharacterized |
| IPR011747 | 29 | 1.6 | Uncharacterized |
| IPR010351 | 20 | 1.4 | Uncharacterized |
| IPR012495 | 125 | 1.4 | TadE-like34,37 |
| IPR009333 | 25 | 1.3 | Uncharacterized |
| IPR000290 | 24 | 1.3 | Colicin immunity protein/pyocin immunity protein |
| IPR002416 | 199 | 1.3 | Bacterial general secretion pathway protein H34,38 |
| IPR003369 | 428 | 1.3 | Bacterial sec-independent translocation protein mttA/Hcf106 |
| IPR008316 | 19 | 1.3 | Uncharacterized |
| IPR007166 | 13 | 1.3 | Uncharacterized |
| IPR004992 | 98 | 1.2 | Ethanolamine utilization protein EutN/ carboxysome structural protein4 CcmL |
| IPR006817 | 35 | 1.2 | LPP motif |
| IPR003731 | 149 | 1.2 | Dinitrogenase iron-molybdenum cofactor biosynthesis |
| IPR007047 | 98 | 1.2 | Flp/Fap pilin component39 |
| IPR008894 | 33 | 1.2 | WxcM-like, C-terminal |
| IPR002068 | 434 | 1.2 | Heat shock protein Hsp2040,41 |
| IPR010310 | 73 | 1.2 | Uncharacterized |
| IPR012903 | 26 | 1.2 | Uncharacterized |
| IPR010385 | 14 | 1.1 | Uncharacterized |
InterPro domain representing each family.32
Number of sequenced genomes that contain a recognizable member of the indicated protein family.
References are given in cases in which formation of large assemblies has been documented.
Preliminary characterization reported here for this family.
Of these 35 families, 16 have some functional annotation. Of these, only six (IPR008846, IPR000290, IPR003369, IPR006817, IPR003731, and IPR008894) have no evidence in the available literature for supramolecular assembly formation. We cannot rule out the possibility that some of these might actually be involved in forming large assemblies that have not yet been discovered, but at present they constitute false positives in the analysis. As our search criteria were aimed at sensitivity above specificity, it is not unexpected that a number of protein families not involved in supramolecular assemblies would be obtained. The false negatives are distributed toward the lower end of the spectrum in terms of average numbers of paralogs per operon: 1.89, 1.32, 1.30, 1.20, 1.19, and 1.17 versus a mean of 1.78 for the 35 families identified by the search criteria. The other 10 of the 16 characterized families are indeed involved in supramolecular assemblies. This appears to represent a strong enrichment for structural proteins over what would be expected at random, although no attempt was made here to establish how low a number would be expected for a randomly selected set of protein families.
Analysis of computationally identified protein families
Consistent with the design of this study, the annotated family with the highest score (an average of 2.56 paralogs per operon) was the BMC protein family discussed earlier (IPR000249). Of the 538 members of this family identified (see Supporting Information), only 13% (70 proteins) were the sole paralogs in their operon, of which only two cases were the only copies in their organism. Almost two thirds (65%; 348 proteins) occured in operons with at least two other BMC proteins. There are multiple cases with many BMC paralogs per operon: 13 operons have six paralogs, two operons have seven paralogs, and one operon has nine BMC paralogs. It addition, it was surprising to find that the search identified another protein family involved in BMCs. This protein family (IPR004992), which includes proteins known as CcmL, EutN, and CsoS4 in different bacteria, has been suggested to contribute to the vertices of the icosahedral or near-icosahedral microcompartment shell.4 The tendency of this shell protein to appear as multiple paralogs in an operon has hitherto been underappreciated.
The major structural protein from gas vesicles, which are large proteinaceous shells used for buoyancy in prokaryotes, was also identified in our study. The GvpA family (IPR000638) frequently occurs with multiple copies per operon, with 57 of the 96 proteins identified co-occurring together in operons. Of the remaining 39 single occurrences, only 19 are the sole copy in their genome. A small subset of organisms encoding gas vesicles have larger, more complex operon structures. For example, there are a total of 11 GvpA domain proteins in Rhodococcus sp. RHA1 split over seven operons, while Streptomyces avermitilis MA-4680 has three operons encoding three GvpA homologs each. Closer inspection reveals that the majority of cases are split over two proximal operons transcribed in opposite directions, increasing the number of proximal paralogs [Fig. 3(A)]. The structural basis for assembly of GvpA proteins into gas vesicles is not yet understood in detail.
Figure 3.
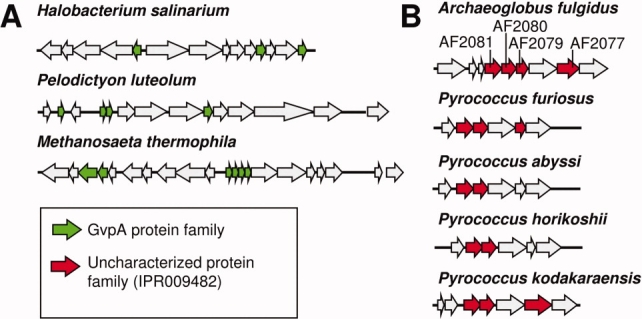
Operon structures for representative protein families identified by genomic context. Panel (A) illustrates the gas vesicle proteins GvpA. Panel (B) illustrates an uncharacterized protein family, IPR009482, which is highlighted in the present study. Arrows represent ORFs. Similar colors indicate homology; unfilled ORFs are not relevant to the analysis. The gene names in A. fulgidus, which were subjected to experimental investigation, are shown. The gene names in the other organisms are (from left to right) P. furiosus: PF0324, PF0325, PF0327; P. abyssi: PAB0981, PAB0982; P. horikoshii: PH0565, PH0564; P. kodakarensis: TK1705, TK1704, TK1702. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
The small heat-shock protein (Hsp) family identified, which includes the alpha-crystallins (IPR002068), is understood to form large assemblies of multiple, different paralogs.40 This family is more widespread (found in 434 organisms) than any others listed in Table I. Although ∼75% (568 of 742) of the members of this family do not co-occur with other paralogous copies in an operon, this nevertheless leaves 150 cases of co-occurrence with another paralog and 24 proteins in operons containing three paralogs.
The phycobiliproteins, represented by the IPR012128 domain, are components of phycobilisomes, which are light-harvesting supramolecular assemblies in cyanobacteria. Biliproteins form dimers of alpha and beta subunits, which assemble further to form large rod-like components of phycobilisomes and subcellar structures that funnel light energy via bound chromophores to a central core.36,42,43 Diverse biliproteins appear to be evolutionarily related; allophycocyanin and phycocyanin were extracted by our study. A total of 28 organisms, all cyanobacteria, contain this protein family, the majority of which (122 proteins of 171) occur together with another paralog.
Finally, our study identifies five domains that are portions of type IV pilins and related proteins: IPR012902, IPR001120, IPR012495, IPR002416, and IPR007047. Type IV pili are long rod-like or filamentous assemblies.44 They are widespread among prokaryotes, where they serve varied roles, especially related to host cell interactions.45 The N-terminal segments of pilin proteins are characterized by two somewhat degenerate methylation consensus sequences, the N-terminal prepilin-type cleavage/methylation motif (IPR012902), and the prokaryotic N-terminal methylation motif (IPR001120). The N-terminal prepilin-type cleavage/methylation site occurs in 2644 proteins split over 380 organisms. Some 205 cases occur with four or more other copies in the same operon. In addition to these widespread sequence motifs, specific pilin-like proteins are also detected here. The IPR002416 domain (which includes the IPR001120 motif noted above as a subdomain) describes the GspH pseudopilin family. Finally, the IPR007047 and IPR012495 domains represent two pilin-related subunits from the tad locus involved in tight, nonspecific adhesion of pathogenic bacteria to surfaces. In both cases, while the proteins occur as single copies the majority of the time, there are nevertheless numerous instances where two or three paralogs occur together.
Of the 35 protein families identified, 19 are completely uncharacterized. These families (IPR006728, IPR007966, IPR014994, IPR009482, IPR012655, IPR012452, IPR010738, IPR010665, IPR009881, IPR007670, IPR012661, IPR011747, IPR010351, IPR009333, IPR008316, IPR007166, IPR010310, IPR012903, and IPR010385) are distributed over a variety of different organisms. Based on an extrapolation from the characterized proteins identified, this set of uncharacterized protein families is likely to be highly enriched in proteins involved in forming novel supramolecular assemblies. We sought to investigate the capacity for self-assembly by one of these uncharacterized families. The family described by the IPR009482 domain was arbitrarily chosen for closer scrutiny. Proteins belonging to this family are found in five hyperthermophilic archaea from our search set, Archaeoglobus fulgidus DSM 4304, Pyrococcus abyssi GE5, Pyrococcus furiosus DSM 3638, Pyrococcus horikoshii OT3, and Pyrococcus kodakarensis KOD1. Figure 3(B) illustrates the arrangement of paralogs from this family in operons. No clear inferences about function could be derived from the other genes encoded in the operons along with these putative structural proteins.
Assembly properties of a previously uncharacterized protein family
To test for the ability of the selected protein family to self-assemble, the four paralogs of the IPR009482 family from Archaeoglobus fulgidus were selected for biophysical characterization and determination of oligomeric state. The genes encoding AF2077, AF2079, AF2080, and AF2081 were cloned, overexpressed in E. coli, and the protein products purified. The four proteins were initially expressed in insoluble form, but after unfolding and refolding from inclusion bodies the target proteins were soluble to varying degrees. AF2079, AF2080, and AF2081 were soluble up to a concentration of 10 mg/mL, while AF2077 remained only marginally soluble. Circular dichroism studies confirmed that all three soluble proteins maintained a similar secondary structure composition, primarily beta sheet [Fig. 4(A)].
Figure 4.
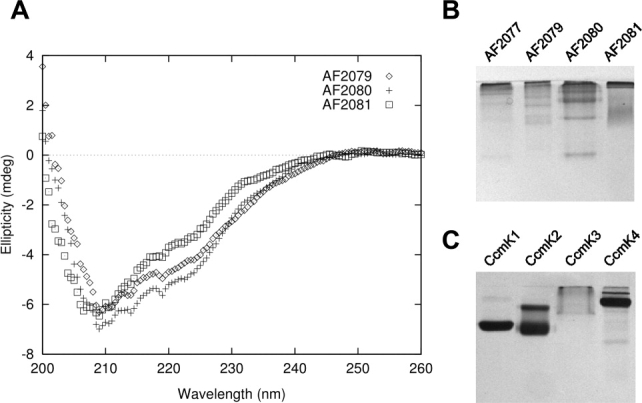
Preliminary biophysical characterization of the IPR009482 protein family. (A) Circular dichroism scans of AF2079, AF2080, and AF2081. (B) A 16% acrylamide native gel of AF2077, AF2079, AF2080, and AF2081 (C) A 12% native gel of carboxysome shell proteins CcmK1, K2, K3, and K4, from the BMC protein family, illustrating similar oligomerization tendencies.
Native PAGE analysis revealed that each protein forms multiple distinct higher-order oligomeric states [Fig. 4(B)]. This behavior was reminiscent of some of the BMC proteins from the carboxysome shell studied earlier [Fig. 4(C), 3]. Determining the stoichiometry of assembly was complicated by the ladder of oligomeric states exhibited by each protein in native gels (Fig. 4). Size exclusion chromatography failed to separate individual species fully, but resultant fractions did show enrichment for different oligomeric states (data not shown). Size-exclusion results indicated that the ladder of oligomeric states ranged from relatively small oligomers to large (>500 kDa) assemblies. Dynamic and static light scattering experiments were consistent with this size range but could not resolve individual species (data not shown). Native PAGE of the size-exclusion fractions showed that they maintained their respective compositions of specific oligomers over a period of at least 6 days, indicating the formation of stable oligomers. In order to estimate the oligomeric state of individual protein species, one protein, AF2081, was analyzed using a Ferguson plot.46,47 This allows an extrapolation of native molecular weight based on the change in mobility versus gel concentration (Fig. 5), without the need to purify separate oligomeric species. The band corresponding to one of the dominant AF2081 oligomeric states was calculated as ∼149 kDa, while the faint, fastest running band was calculated to be 29 kDa. These correspond to hexamer (theoretical mass 147.05 kDa) and monomer (theoretical mass 24.51 kDa), respectively. A pentamer or heptamer could also be within the margin of error, but a hexamer gives an excellent fit. Single native gels of AF2079 and AF2080 and the marginally stable AF2077 revealed a similar pattern of varied oligomeric states [Fig. 4(B)]. The behavior of the proteins from the IPR009482 family is therefore highly suggestive of assembly into high-order structures.
Figure 5.
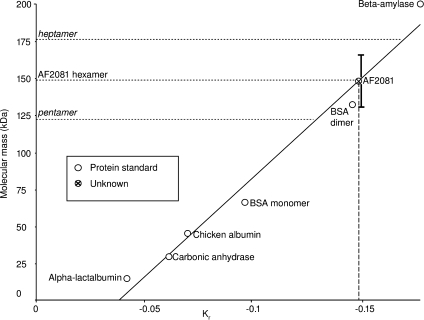
Estimation of oligomeric state for the AF2081 paralog from the IPR009482 protein family, based on native gel electrophoresis and Ferguson plots. The plot shows the slope, Kr, of log(mobility) versus gel concentration (raw data not shown) for a variety of proteins over a range of gel concentrations (see Materials and Methods). Using a series of protein standards of known molecular mass, an approximately linear standard curve is obtained. Molecular masses of standards: alpha-lactalbumin (14 kDa), carbonic anhydrase (29 kDa), chicken albumin (45 kDa), bovine serum albumin (dimer, 132 kDa), and beta-amylase (200 kDa). The dashed vertical line indicates the experimentally determined value of Kr for the AF2081 protein. Its intersection with the linear standard curve is indicated by the crossed circle. The horizontal dotted lines show the molecular masses that would be expected if AF2081 were a pentamer, hexamer, or heptamer. Good agreement is obtained for a hexamer. Error bars are drawn on the AF2081 data point to reflect 1 SD (∼17 kDa) in the estimated MW, based on a least squares analysis of the deviations of the standards from the best fit line.
Further examples
We also searched the literature for other proteins involved in large structures that might have evaded our computational analysis. Sulfur globules provide one case. In sulfur-oxidizing organisms, sulfur is stored in the periplasm for later oxidation in protein-coated sulfur globules.48 In Allochromatium vinosum, the protein coat includes two paralogs, SgpA and SgpB. Although SgpA and SgpB are paralogous, they do not co-occur together in the same operon, thus evading detection by the criteria employed. Intriguingly, these short proteins (∼100 amino acids long) are reported to show some similarity to structural proteins such as keratin, silk fibroin, and plant cell wall proteins.49 Polyhydroxybutyrate (PHB) granules provide another case of proteinaceous encapsulation, in this case for energy storage. These granules serve as storage sites for PHB polymers, which are surrounded by an amphiphilic layer of structural proteins.50 The structural proteins include four paralogs of a protein family referred to as phasins, encoded on separate operons.51 The function of phasins is to control the structure of the PHB granules, but the need for multiple paralogs is unclear.
Discussion
Our initial genomic search identified 35 protein families for which multiple paralogs frequently occur within a single operon. Sixteen of these families have been characterized to some extent, and 10 of those families exhibit evidence for formation of supramolecular assemblies. The set identified therefore appears to be highly enriched in proteins that form large assemblies. These assemblies are of varied geometric types, as discussed later.
The annotated family with the highest average number of paralogs per operon was the BMC protein family, which formed the basis for the search pattern employed. Current data suggest that the divergence of BMC paralogs might be used to achieve both architectural and biochemical specialization. For example, while some of the BMC paralogs are strictly required for forming the Pdu microcompartment shell in Salmonella, others, such as PduT and PduU (T.A. Bobik, unpublished data) appear to have minor structural roles. This suggests architectural specialization. There is also evidence for biochemical specialization between paralogous BMC proteins. Differences in pore structure between CcmK2 and CcmK4 suggest different properties in the passive transport of substrates and products into and out of the carboxysome.3 Variable pore properties have also been observed in the BMC proteins from the Pdu microcompartment,27 while genetic studies in that system also support the idea of biochemical specialization.13
It was surprising that the variously named CcmL/EutN/CsoS4/OrfAB family of proteins—also components of the microcompartment—is identified here. Only a very small fraction of all known protein families were selected by our search, making identification of this distinct family of microcompartment proteins intriguing. The exact role of paralog divergence in this family is unclear. Some members form pentamers that fit at the vertices of an icosahedral shell,4 but some other members form hexamers.4,52 This suggests a probable case of architectural specialization.
Given that microcompartments formed the foundations of our search, it was noteworthy to find that the major structural protein from the unrelated, but somewhat analogous case of gas vesicles was identified. Gas vesicles are large proteinaceous shells found inside certain bacterial cells, where they help maintain buoyancy.35 Their structure is that of a cylinder, capped with cones at each end.53 They are composed primarily of a paralogous family of proteins such as GvpA, GvpJ, and GvpM, some of which occur with additional copies,54 often encoded within two back-to-back, divergently transcribed operons [see Fig. 3(A)]. These proteins assemble into a continuous helical ring that encircles the cylindrical body.53 It has been suggested that various ratios of these paralogs may modulate local shell architecture, such as cylinder radius and tip shape.54
The small Hsp20 heat-shock proteins (or alpha-Hsps) are able to form large oligomeric structures with multiple paralogs. This family, represented by IPR002068, is related to the alpha-crystallins from the mammalian lens, which form homo-oligomeric structures believed to act as chaperones to prevent the aggregation of misfolded proteins.40,55 Ten alpha-Hsps are encoded in the bacterium Bradyrhizobium japonicum, and members of the same subclass have been shown to form functional hetero-oligomers.41 Likewise, in E. coli the paralogs IbpA and IbpB have been shown to be incorporated into a supramolecular structure with a mass in excess of 2 MDa.56 The rationale behind multiple paralogs appears to be primarily related to biochemical function in these cases.
Several pilin proteins and pilin motifs emerge from the analysis. The pili examples show that filamentous and rod-like assemblies tend to exhibit the same kinds of genomic pattern as the shell-like assemblies discussed earlier. The biliprotein family provides another example. Biliproteins form paralogous heterodimers, which in turn form hexamers, in turn forming larger rod-like structures.42,43
The widespread occurrence of multiple paralogs in supramolecular assemblies motivates a consideration of underlying evolutionary events. In the systems discussed here, the paralogous proteins presumably diverged after duplication(s) of a single ancestral protein. We postulate that in most cases this ancestral protein already possessed the ability to form large assemblies. Gene duplication and divergence then provided a route to architectural and functional complexity, which conferred selective advantages in these systems. The apparent generality of this phenomenon gives it predictive utility.
Predictions from genomic analyses provide only a statistical likelihood regarding function, so experimental investigation is essential. Here we report preliminary biophysical results on one novel family of proteins (IPR009482) predicted to be involved in forming large assemblies. The results are highly supportive of the prediction in this case. The four paralogs investigated form assemblies of various sizes when expressed individually, ranging from small oligomers to large aggregates. The paralog that was investigated in the most detail (AF2081) forms predominantly hexameric building blocks. These findings are reminiscent of those for the hexameric BMC proteins from BMCs. Since protein hexamers are fairly uncommon in nature, occurring only about 3% of the time among proteins of known structure,57,58 it is therefore noteworthy that hexamers (cyclic forms of which make good building blocks for large structures) were observed in the family identified by the computational analysis. However, further work will be required to illuminate the natural cellular assembly in more detail and to understand its role in the cell. This is likewise true for the other uncharacterized protein families identified in this study. Follow-up studies on these predictions from genomic context analysis should lead to new biological discoveries.
Materials and Methods
Computational methods
Protein sequences and their encoding gene positions from 593 archaeal and bacterial genomes from the Genome Reviews database59 as of February 26, 2008 were used to predict operons based on contiguous groups of predicted genes separated by no more than 300 base pairs.60 Next, InterPro domains assigned to each protein were taken as domain definitions.32 Initial filtering steps required that the proteins within which the InterPro domains occurred were no more than 200 amino acids in length, and occurred with at least two copies per operon in an least three organisms. Furthermore, at least one of these organisms was required to have three or more paralogs in a single operon. These protein families were further ranked by the average number of occurrences per operon.
Cloning
A. fulgidus genes coding for proteins AF2077, AF2079, AF2080, and AF2081 were amplified directly from A. fulgidus cells lysed by osmotic shock immediately before PCR. Primers were constructed with a BamHI restriction site at the 5′ end and an NcoI restriction site at the 3′ end. Amplified DNA was restricted with BamHI and NcoI as was the pETM-11 vector, providing a TEV protease-cleavable N-terminal his-tag. Vector and insert were ligated overnight at room temperature and transformed into Rosetta cells (Novagen), which were found to provide high expression levels.
Protein expression
Cells were grown to OD between 0.8 and 1.0 at 37°C then induced with 1 mM IPTG. Induction proceeded for 4 h after which cells were pelleted and frozen. All proteins were insoluble. Growth assays at 25°C and 16°C in combination with different IPTG concentrations failed to remedy insoluble expression. Inclusion bodies were purified by lysing cells in 100 mM Tris pH 8.0, 5 mM EDTA, then iteratively centrifuging at 35,000g for 30 min and washing with a succession of buffers: 100 mM Tris pH 8.0, 5 mM EDTA; 100 mM Tris pH 8.0, 0.5% Tergitol, 1% Triton X-100; 100 mM Tris pH 8.0, 1M NaCl. Purified inclusion bodies were resolubilized overnight in 50 mM Tris pH 8.0, 500 mM NaCl, 6M guanidine hydrochloride (GdnHCl), and ∼10 mM dithiothreitol (DTT). Insoluble debris was subsequently pelleted at 35,000g for 30 min in a Sorval RC5C Plus centrifuge, followed by ultracentrifugation in a Beckman Optima LE-80K at 16,0000g for 90 min. Supernatants were chemically reduced with 10 mM DTT, then filtered (0.22 μm) and loaded on a HisTrap column in 50 mM Tris pH 8.0, 500 mM NaCl, 6M GdnHCl. Protein was eluted from the column using 50 mM Tris pH 8.0, 500 mM NaCl, 6M GdnHCl, and 300 mM imidazole. To prevent inadvertant disulfide bond formation, free cysteine residues were alkylated in the dark for 30 min in 50 mM Tris pH 8.0, 500 mM NaCl, 6M GdnHCl, ∼10 mM DTT, and excess (25 mM) iodoacetamide. Proteins were then refolded by dialysis against 50 mM Tris pH 8.0 and 500 mM NaCl with two buffer changes, at least one round of which was overnight. Ultracentrifugation of the refolded protein did not pellet protein assemblies. The finding that the renatured proteins remained in solution supported the conclusion that the proteins were overexpressed in an initially insoluble, misfolded form in the cell, and therefore formed inclusion bodies. Purity was assessed by 10% SDS-PAGE electrophoresis. The CcmK proteins used in Figure 4 were expressed and purified as described in Ref. 3.
Circular dichroism
Circular dichroism data were collected on a JASCO J-715 Spectropolarimeter flushed with liquid nitrogen for 30 min prior to data collection using a 4-s response time, 1 nm bandwidth, 0.5-nm step resolution, 20 nm/min speed, 4+ accumulation, and a scan from 260 to 200 nm.
Ferguson plot analysis of native molecular weights
Ferguson plot analysis was carried out as described previously.46,47 Briefly, native PAGE gels were run at various concentrations of acrylamide on a consistent set of standards and unknowns. Mobility values (R) were calculated for each species and plotted on a log scale against %T across gel concentrations. %C was kept constant using an acrylamide stock of 30%T, 5%C (Sigma), diluting appropriately. The slope of the linear regression of log R against %T was calculated for each species, referred to hereafter as Kr. Standards of known molecular weight and oligomeric state purchased from Sigma-Aldrich were used to construct a standard curve of Kr against molecular weight. Standards used were alpha-lactalbumin (14 kDa), carbonic anhydrase (29 kDa), chicken albumin (45 kDa), bovine serum albumin (monomer, 66 kDa, dimer, 132 kDa), and beta-amylase (200 kDa). The standard curve is approximately linear over the range of standards used.
Acknowledgments
The authors acknowledge Dr. Martin Phillips, Dr. Dan Anderson, Dr. Celia Goulding, and members of the Yeates lab for useful discussions and technical assistance, the Emil Reisler lab for use of their ultracentrifuge, and Dr. Gunter Stier at EMBL, Heidelberg for the pETM-11 plasmid.
References
- 1.Cannon GC, Bradburne CE, Aldrich HC, Baker SH, Heinhorst S, Shively JM. Microcompartments in prokaryotes: carboxysomes and related polyhedra. Appl Environ Microbiol. 2001;67:5351–5361. doi: 10.1128/AEM.67.12.5351-5361.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shively JM, editor. Microbiology Monographs. Vol. 2. Berlin: Springer; 2006. Complex Intracellular Structures in Prokaryotes. [Google Scholar]
- 3.Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO. Protein structures forming the shell of primitive bacterial organelles. Science. 2005;309:936–938. doi: 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO. Atomic-level models of the bacterial carboxysome shell. Science. 2008;319:1083–1086. doi: 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- 5.Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat Rev Microbiol. 2008;6:681–691. doi: 10.1038/nrmicro1913. [DOI] [PubMed] [Google Scholar]
- 6.Shively JM, Ball F, Brown DH, Saunders RE. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science. 1973;182:584–586. doi: 10.1126/science.182.4112.584. [DOI] [PubMed] [Google Scholar]
- 7.Price GD, Badger MR. Isolation and characterization of high CO2-requiring-mutants of the cyanobacterium Synechococcus PCC7942: two phenotypes that accumulate inorganic carbon but are apparently unable to generate CO2 within the carboxysome. Plant Physiol. 1989;91:514–586. doi: 10.1104/pp.91.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buedeker RF, Cannon GC, Kuenen JG, Shively JM. Relations between d-ribulose-1,5-bisphosphate carboxylase, carboxysomes, and CO2 fixing capacity in the obligate chemolithotroph Thiobacillus neapolitanus grown under different limitations in the chemostat. Arch Microbiol. 1980;124:185–189. [Google Scholar]
- 9.Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1, 2-propanediol degradation. J Bacteriol. 1999;181:5967–5975. doi: 10.1128/jb.181.19.5967-5975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannon GC, Heinhorst S, Bradburne CE, Shively JM. Carboxysome genomics: a status report. Funct Plant Biol. 2002;29:175–182. doi: 10.1071/PP01200. [DOI] [PubMed] [Google Scholar]
- 11.Chen P, Andersson DI, Roth JR. The control region of the pdu/cob regulon in Salmonella typhimurium. J Bacteriol. 1994;176:5474–5482. doi: 10.1128/jb.176.17.5474-5482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Penrod JT, Roth JR. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J Bacteriol. 2006;188:2865–2874. doi: 10.1128/JB.188.8.2865-2874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bobik TA. Polyhedral organelles compartmenting bacterial metabolic processes. Appl Microbiol Biotechnol. 2006;70:517–525. doi: 10.1007/s00253-005-0295-0. [DOI] [PubMed] [Google Scholar]
- 14.English RS, Lorbach SC, Qin X, Shively JM. Isolation and characterization of a carboxysome shell gene from Thiobacillus neapolitanus. Mol Microbiol. 1994;12:647–654. doi: 10.1111/j.1365-2958.1994.tb01052.x. [DOI] [PubMed] [Google Scholar]
- 15.Sammut SJ, Finn RD, Bateman A. Pfam 10 years on: 10,000 families and still growing. Brief Bioinf. 2008;9:210–219. doi: 10.1093/bib/bbn010. [DOI] [PubMed] [Google Scholar]
- 16.Heinhorst S, Cannon GC, Shively JM. Complex intracellular structures in prokaryotes. In: Shively JM, editor. Carboxysomes and carboxysome-like inclusions. Berlin: Springer-Verlag; 2006. pp. 141–165. [Google Scholar]
- 17.Price GD, Sültemeyer D, Klughammer B, Ludwig M, Badger MR. The functioning of the CO2 concentrating mechanism in several cyanobacterial strains: a review of general physiological characteristics, genes, proteins, and recent advances. Can J Bot. 1998;76:973–1002. [Google Scholar]
- 18.Kofoid E, Rappleye C, Stojiljkovic I, Roth J. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J Bacteriol. 1999;181:5317–5329. doi: 10.1128/jb.181.17.5317-5329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stojiljkovic I, Baeumler A, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iancu CV, Ding HJ, Morris DM, Dias DP, Gonzales AD, Martino A, Jensen GJ. The structure of isolated Synechococcus strain WH8102 carboxysomes as revealed by electron cryotomography. J Mol Biol. 2007;372:764–773. doi: 10.1016/j.jmb.2007.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmid MF, Paredes AM, Khant HA, Soyer F, Aldrich HC, Chiu W, Shively JM. Structure of Halothiobacillus neapolitanus carboxysomes by cryo-electron tomography. J Mol Biol. 2006;364:526–535. doi: 10.1016/j.jmb.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai Y, Sawaya MR, Cannon GC, Cai F, Williams EB, Heinhorst S, Kerfeld CA, Yeates TO. Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome. PLoS Biol. 2007;5:e144. doi: 10.1371/journal.pbio.0050144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klug A, Caspar DL. The structure of small viruses. Adv Virus Res. 1960;7:225–325. doi: 10.1016/s0065-3527(08)60012-3. [DOI] [PubMed] [Google Scholar]
- 24.Hogle JM, Chow M, Filman DJ. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 25.Rossmann MG, Arnold E, Erickson JW, Frankenberger EA, Griffith JP, Hecht HJ, Johnson JE, Kamer G, Luo M, Mosser AG. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 26.Johnson JE, Speir JA. Quasi-equivalent viruses: a paradigm for protein assemblies. J Mol Biol. 1997;269:665–675. doi: 10.1006/jmbi.1997.1068. [DOI] [PubMed] [Google Scholar]
- 27.Crowley C, Sawaya MR, Bobik TA, Yeates TO. Structure of the PduU shell protein from the Pdu microcompartment of Salmonella. Structure. 2008;16:1324–1332. doi: 10.1016/j.str.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amos LA, van den Ent F, Löwe J. Structural/functional homology between the bacterial and eukaryotic cytoskeletons. Curr Opin Cell Biol. 2004;16:24–31. doi: 10.1016/j.ceb.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 29.Wade RH. Microtubules: an overview. Methods Mol Med. 2007;137:1–16. doi: 10.2119/molecular%20medicine-2006-00038. [DOI] [PubMed] [Google Scholar]
- 30.Huynen M, Snel B, Lathe W, Bork P. Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res. 2000;10:1204–1210. doi: 10.1101/gr.10.8.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jensen GJ, Briegel A. How electron cryotomography is opening a new window onto prokaryotic ultrastructure. Curr Opin Struct Biol. 2007;17:260–267. doi: 10.1016/j.sbi.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 32.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, Copley R, Courcelle E, Das U, Daugherty L, Dibley M, Finn R, Fleischmann W, Gough J, Haft D, Hulo N, Hunter S, Kahn D, Kanapin A, Kejariwal A, Labarga A, Langendijk-Genevaux PS, Lonsdale D, Lopez R, Letunic I, Madera M, Maslen J, McAnulla C, McDowall J, Mistry J, Mitchell A, Nikolskaya AN, Orchard S, Orengo C, Petryszak R, Selengut JD, Sigrist CJ, Thomas PD, Valentin F, Wilson D, Wu CH, Yeats C. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeates TO, Tsai Y, Tanaka S, Sawaya MR, Kerfeld CA. Self-assembly in the carboxysome: a viral capsid-like protein shell in bacterial cells. Biochem Soc Trans. 2007;35:508–11. doi: 10.1042/BST0350508. [DOI] [PubMed] [Google Scholar]
- 34.Hobbs M, Mattick JS. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol Microbiol. 1993;10:233–43. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 35.Walsby AE. Gas vesicles. Microbiol Rev. 1994;58:94–144. doi: 10.1128/mr.58.1.94-144.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas JC, Passaquet C. Characterization of a phycoerythrin without alpha-subunits from a unicellular red alga. J Biol Chem. 1999;274:2472–82. doi: 10.1074/jbc.274.4.2472. [DOI] [PubMed] [Google Scholar]
- 37.Tomich M, Planet PJ, Figurski DH. The tad locus: postcards from the widespread colonization island. Nat Rev Microbiol. 2007;5:363–75. doi: 10.1038/nrmicro1636. [DOI] [PubMed] [Google Scholar]
- 38.Yanez ME, Korotkov KV, Abendroth J, Hol WG. Structure of the minor pseudopilin EpsH from the Type 2 secretion system of Vibrio cholerae. J Mol Biol. 2008;377:91–103. doi: 10.1016/j.jmb.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomich M, Fine DH, Figurski DH. The TadV protein of Actinobacillus actinomycetemcomitans is a novel aspartic acid prepilin peptidase required for maturation of the Flp1 pilin and TadE and TadF pseudopilins. J Bacteriol. 2006;188:6899–914. doi: 10.1128/JB.00690-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narberhaus F. Alpha-crystallin-type heat shock proteins: socializing minichaperones in the context of a multichaperone network. Microbiol Mol Biol Rev. 2002;66:64–93. doi: 10.1128/MMBR.66.1.64-93.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Studer S, Narberhaus F. Chaperone activity and homo- and hetero-oligomer formation of bacterial small heat shock proteins. J Biol Chem. 2000;275:37212–37218. doi: 10.1074/jbc.M004701200. [DOI] [PubMed] [Google Scholar]
- 42.Glazer AN. Phycobilisomes: structure and dynamics. Annu Rev Microbiol. 1982;36:173–198. doi: 10.1146/annurev.mi.36.100182.001133. [DOI] [PubMed] [Google Scholar]
- 43.Schirmer T, Bode W, Huber R, Sidler W, Zuber H. X-ray crystallographic structure of the light-harvesting biliprotein C-phycocyanin from the thermophilic cyanobacterium Mastigocladus laminosus and its resemblance to globin structures. J Mol Biol. 1985;184:257–277. doi: 10.1016/0022-2836(85)90379-1. [DOI] [PubMed] [Google Scholar]
- 44.Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004;2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 45.Andrzejewska J, Lee SK, Olbermann P, Lotzing N, Katzowitsch E, Linz B, Achtman M, Kado CI, Suerbaum S, Josenhans C. Characterization of the pilin ortholog of the Helicobacter pylori type IV cag pathogenicity apparatus, a surface-associated protein expressed during infection. J Bacteriol. 2006;188:5865–5877. doi: 10.1128/JB.00060-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferguson KA. Starch-gel electrophoresis—application to the classification of pituitary proteins and polypeptides. Metab Clin Exp. 1964;13:985–1002. doi: 10.1016/s0026-0495(64)80018-4. [DOI] [PubMed] [Google Scholar]
- 47.Hedrick JL, Smith AJ. Size and charge isomer separation and estimation of molecular weights of proteins by disc gel electrophoresis. Arch Biochem Biophys. 1968;126:155–164. doi: 10.1016/0003-9861(68)90569-9. [DOI] [PubMed] [Google Scholar]
- 48.Prange A, Engelhardt H, Truper HG, Dahl C. The role of the sulfur globule proteins of Allochromatium vinosum: mutagenesis of the sulfur globule protein genes and expression studies by real-time RT-PCR. Arch Microbiol. 2004;182:165–174. doi: 10.1007/s00203-004-0683-3. [DOI] [PubMed] [Google Scholar]
- 49.Pattaragulwanit K, Brune DC, Trüper HG, Dahl C. Molecular genetic evidence for extracytoplasmic localization of sulfur globules in Chromatium vinosum. Arch Microbiol. 1998;169:434–444. doi: 10.1007/s002030050594. [DOI] [PubMed] [Google Scholar]
- 50.Uchino K, Saito T, Gebauer B, Jendrossek D. Isolated poly(3-hydroxybutyrate) (PHB) granules are complex bacterial organelles catalyzing formation of PHB from acetyl coenzyme A (CoA) and degradation of PHB to acetyl-CoA. J Bacteriol. 2007;189:8250–8256. doi: 10.1128/JB.00752-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Potter M, Muller H, Reinecke F, Wieczorek R, Fricke F, Bowien B, Friedrich B, Steinbuchel A. The complex structure of polyhydroxybutyrate (PHB) granules: four orthologous and paralogous phasins occur in Ralstonia eutropha. Microbiology. 2004;150:2301–2311. doi: 10.1099/mic.0.26970-0. [DOI] [PubMed] [Google Scholar]
- 52.Forouhar F, Kuzin A, Seetharaman J, Lee I, Zhou W, Abashidze M, Chen Y, Yong W, Janjua H, Fang Y, Wang D, Cunningham K, Xiao R, Acton TB, Pichersky E, Klessig DF, Porter CW, Montelione GT, Tong L. Functional insights from structural genomics. J Struct Funct Genomics. 2007;8:37–44. doi: 10.1007/s10969-007-9018-3. [DOI] [PubMed] [Google Scholar]
- 53.Offner S, Ziese U, Wanner G, Typke D, Pfeifer F. Structural characteristics of halobacterial gas vesicles. Microbiology. 1998;144(Pt 5):1331–1342. doi: 10.1099/00221287-144-5-1331. [DOI] [PubMed] [Google Scholar]
- 54.Shukla HD, DasSarma S. Complexity of gas vesicle biogenesis in Halobacterium sp. strain NRC-1: identification of five new proteins. J Bacteriol. 2004;186:3182–3186. doi: 10.1128/JB.186.10.3182-3186.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 56.Matuszewska M, Kuczynska-Wisnik D, Laskowska E, Liberek K. The small heat shock protein IbpA of Escherichia coli cooperates with IbpB in stabilization of thermally aggregated proteins in a disaggregation competent state. J Biol Chem. 2005;280:12292–12298. doi: 10.1074/jbc.M412706200. [DOI] [PubMed] [Google Scholar]
- 57.Henrick K, Thornton JM. PQS: a protein quaternary structure file server. Trends Biochem Sci. 1998;23:358–361. doi: 10.1016/s0968-0004(98)01253-5. [DOI] [PubMed] [Google Scholar]
- 58.Levy ED, Erba EB, Robinson CV, Teichmann SA. Assembly reflects evolution of protein complexes. Nature. 2008;453:1262–1265. doi: 10.1038/nature06942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sterk P, Kersey PJ, Apweiler R. Genome reviews: standardizing content and representation of information about complete genomes. OMICS. 2006;10:114–118. doi: 10.1089/omi.2006.10.114. [DOI] [PubMed] [Google Scholar]
- 60.Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]


