Abstract
Carboxysomes are primitive bacterial organelles that function as a part of a carbon concentrating mechanism (CCM) under conditions where inorganic carbon is limiting. The carboxysome enhances the efficiency of cellular carbon fixation by encapsulating together carbonic anhydrase and the CO2-fixing enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO). The carboxysome has a roughly icosahedral shape with an outer shell between 800 and 1500 Å in diameter, which is constructed from a few thousand small protein subunits. In the cyanobacterium Synechocystis sp. PCC 6803, the previous structure determination of two homologous shell protein subunits, CcmK2 and CcmK4, elucidated how the outer shell is formed by the tight packing of CcmK hexamers into a molecular layer. Here we describe the crystal structure of the hexameric shell protein CcmK1, along with structures of mutants of both CcmK1 and CcmK2 lacking their sometimes flexible C-terminal tails. Variations in the way hexamers pack into layers are noted, while sulfate ions bound in pores through the layer provide further support for the hypothesis that the pores serve for transport of substrates and products into and out of the carboxysome. One of the new structures provides a high-resolution (1.3 Å) framework for subsequent computational studies of molecular transport through the pores. Crystal and solution studies of the C-terminal deletion mutants demonstrate the tendency of the terminal segments to participate in protein—protein interactions, thereby providing a clue as to which side of the molecular layer of hexameric shell proteins is likely to face toward the carboxysome interior.
Keywords: bacterial organelles, carboxysome, protein assembly, molecular transport, CO2 fixation
Abbreviations
- 3-PG
3-phosphoglycerate
- CCM
CO2-concentrating mechanism; RuBisCO, ribulose-1,5-bisphosphate carboxylase/oxygenase; RuBP, ribulose-1,5-bisphosphate
- Syn 6803
Synechocystis sp. PCC6803.
Introduction
Bacterial microcompartments are protein-based organelles that serve varied roles in bacteria by sequestering specific groups of enzymes. To date, the best-studied microcompartment is the carboxysome, whose function is to enhance carbon fixation.1–5 Carboxysomes are found in all cyanobacteria and some chemoautotrophic bacteria [Fig. 1(A,B)] (reviewed in Ref.9). The proteinaceous shell of the carboxysome encapsulates the CO2-fixing enzymes ribulose bisphosphate oxygenase/carboxylase (RuBisCO) and carbonic anhydrase.10–12 According to current models (reviewed in Ref.13), bicarbonate from the cytosol of the bacterial cell enters the carboxysome by crossing the protein shell. Bicarbonate is then dehydrated by carbonic anhydrase to form CO2, which is utilized by RuBisCO before it can escape from the carboxysome. By sequestering carbonic anhydrase and RuBisCO together, the carboxysome increases the local concentration of CO2 in order to drive its reaction with ribulose-1,5-bisphosphate (RuBP) at the active site of RuBisCO, an enzyme whose catalytic properties are notoriously poor.14,15
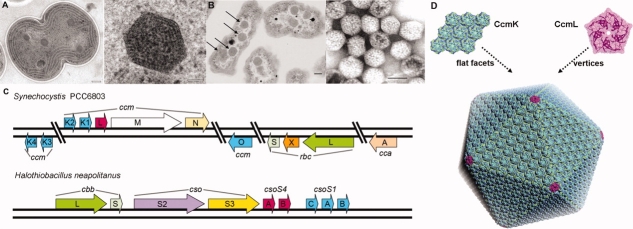
Figure 1.
Transmission electron micrographs showing (A) a section through a dividing cyanobacterial cell (Synechocystis sp. PCC 6803) on the left (scale bar, 200 nm) and an enlargement of a single carboxysome on the right (scale bar, 50 nm), and (B) Halothiobacillus neapolitanus cells on the left with their carboxysomes highlighted by arrows, and purified carboxysomes on the right (scale bars, 100 nm). Their polyhedral shape helps distinguish carboxysomes from other cytoplasmic inclusions. (C) Genomic arrangement of carboxysome-associated proteins. Homologous proteins from two model organisms are shaded and colored similarly. The hexameric (BMC-type) shell proteins are colored in blue. The rbc and cbb genes code for the large and small subunit of RuBisCO. (D) A rough atomic model of a carboxysome constructed from many CcmK hexamers (in light-blue) and 12 CcmL pentamers (in pink) at the icosahedral vertices (triangulation number, T = 75). Panel (A) is courtesy of W. Vermaas, from Ref.6. Panel (B) is courtesy G. Cannon and S. Heinhorst, adapted from Ref.7. Panel (D) is adapted from Ref.8.
Carboxysomes, which were first visualized more than 40 years ago,16–18 can be classified into two types, α and β.9 α-Carboxysomes contain Form 1A RuBisCO and generally have their genes for RuBisCO, carbonic anhydrase, and the shell proteins encoded together in a single operon. β-Carboxysomes, which occur exclusively in cyanobacteria, contain Form 1B RuBisCO and tend to encode their carboxysome proteins in multiple gene clusters. Little is known about the potential functional differences between the two types of carboxysomes, but β-carboxysomes tend to be somewhat larger, typically around 1500 Å in diameter. The small (∼10 kDa) proteins that make up the bulk of the carboxysome shell are homologous in the two types of carboxysomes, though alternate gene names have been retained in the two systems.19 The major shell proteins in the α-carboxysome are encoded by cso-type genes.1,20 In the α-carboxysome from the chemoautotrophic bacterium Halothiobacillus neapolitanus [Fig. 1(C)], which has been studied extensively,1,7,21 the csoS1 genes code for three paralogs of this shell protein (CsoS1ABC). Similarly, the shell proteins in the β-carboxysome are encoded by a series of homologous ccm-type genes.22 In the model cyanobacterium Synechocystis sp. PCC 6803, which is the subject of the present work, five paralogs of the major shell protein are present (CcmK1, CcmK2, CcmK3, CcmK4, and CcmO). The CsoS1 and CcmK proteins belong to the widely distributed BMC (bacterial microcompartment) family of proteins.
Structural studies have been initiated on the carboxysome in an effort to understand in greater detail the mechanistic basis of its cellular function. Recent cryo-electron tomography studies provided important insights into the architecture of carboxysomes by showing that this bacterial microcompartment has a nearly icosahedral shape.23,24 In complementary work, X-ray crystallographic studies have revealed the atomic structures of homologous BMC-type shell proteins from both types of carboxysome, including CcmK2 and CcmK4 from the β-carboxysome6,7 and CsoS1A from the α-carboxysome.7 The various carboxysome shell BMC proteins all form hexamers having a narrow central pore bearing a positive electrostatic potential. Based on the tendency of the hexamers to pack side by side into molecular layers, it has been surmised that these layers represent the flat facets of the shell, and that the narrow pores likely serve for molecular transport.6,7,13 A crystal structure of a BMC-type hexamer from a noncarboxysomal microcompartment—the propanediol utilization or pdu microcompartment, which requires the transport of different molecules across its shell—has a different pore structure.25 Crystal structures of minor carboxysome shell proteins CcmL and CsoS4A, unrelated to the BMC family, were recently elucidated and found to be pentameric.8 The findings from those structural studies led to a rough atomic model of the carboxysome shell in which the pentameric proteins introduce curvature by forming the vertices of the icosahedral microcompartment shell, while the flat facets of the microcompartment are comprised of a large number of hexameric (BMC-type) shell proteins packed into a layer [Fig. 1(D)]. The packing of hexagonal carboxysome shell proteins in a molecular layer appears similar in certain respects to the proposed packing of tetragonal aquaporins in square two-dimensional arrays in the membrane bilayers of lens fibers (reviewed in Ref.26).
Among the five paralogous (BMC-type) proteins encoded by Syn. 6803, CcmK1 and CcmK2 are likely the dominant shell proteins. Direct compositional studies on β-carboxysomes have been difficult, but in the study of another β-carboxysome from Synechococcus PCC7942, inactivation of CcmK1 alone resulted in high CO2 requirement for growth.27 In addition, CcmK1 and CcmK2 are encoded immediately upstream of carboxysome proteins CcmL, CcmM, and CcmN [Fig. 1(C)]; this operon organization suggest that they are likely to play an essential part in carboxysome function. Thus, high-resolution structures of these proteins are important for understanding the properties of the shell, and for computationally modeling molecular transport into and out of the carboxysome.
Obtaining high-resolution structural data on some of the key components of the β-carboxysome shell has been challenging. The previous crystal structure of CcmK2 was limited to 2.8 Å resolution, and refinement was further compromised by crystal growth disorders.6 Some of the difficulties may be attributed to the C-terminal tails on these proteins. The most significant sequence differences between various CcmK paralogs occur in the C-terminal tails [Fig. 2(A)]; CcmK1 is longer than CcmK2 by eight amino acids, while the proteins differ elsewhere at only seven positions. The tails, which are also the regions of greatest structural variation, have been an apparent source of disorder in previous crystal forms.6 That possibility motivated the deletion of the termini in some of the experiments presented here.
Figure 2.
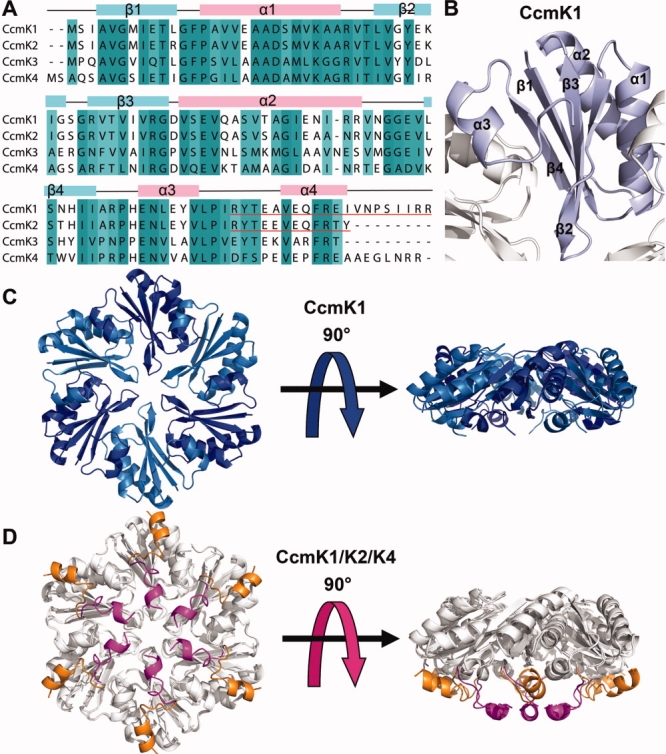
A: A multiple sequence alignment of CcmK shell proteins. Regions of high conservation (>70%) are shaded and the protein secondary structure elements are shown above (pink, α-helices; lightblue, β-strands). The C-terminal deleted regions of CcmK1 and CcmK2 are underlined in red. B: A cartoon diagram of the CcmK1 monomer shaded in light blue. C: Top and side views of a CcmK1 hexamer. D: Top and side views of CcmK2 and CcmK4 superimposed onto CcmK1. Regions where the paralogs are nearly identical are shown in white. The C-termini of CcmK2 and CcmK4 are shown in magenta and orange, respectively. The C-terminus of CcmK1 is disordered. Whether the bottom surface (bearing the C-termini and the concave surface) faces into the carboxysome interior or out to the cytosol has not been established experimentally. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Recent experiments have shed light on the protein–protein interactions that must organize the enzymatic components of the carboxysome in relation to the shell. Two independent studies on β-caboxysomes suggest that the CcmM protein plays a key organizing role.28,29 In one study,29 yeast two hybrid experiments showed direct interactions between CcmM and the CcmK (BMC-type) shell proteins under study in the present work. The atomic level details of those interactions have not been elucidated yet. In fact, the sidedness of the carboxysome shell has not been established by direct experiment. The hexameric structures of the CcmK proteins have been elucidated, but which side faces toward the carboxysome interior—and would therefore be most likely to be involved in protein–protein interactions—is uncertain.7,8
Here we present a series of crystal structures of the CcmK1 and CcmK2 proteins in full length forms and as C-terminal truncations. Comparisons illustrate the structural features that are most conserved, while highlighting potentially relevant structural variations and providing further insights into molecular transport across pores through the hexagonally packed protein layers. The findings also suggest potential roles for the C-terminal tails of the CcmK proteins in higher level interactions in the carboxysome.
Results
Structure of CcmK1
The CcmK1 protein from Synechocystis sp. PCC6803 was expressed in E. coli cells, purified, and crystallized using the vapor diffusion method. Phases for CcmK1 diffraction data were obtained by molecular replacement using a hexamer of CcmK2 (PDB code: 2A1B) as a search model. An atomic model was built and refined at a resolution of 2.0 Å, with final R and Rfree values of 20.3% and 25.0% (Table 1). The final model of CcmK1 consists of a hexamer in the asymmetric unit with 94 of the 111 residues of the protein chain visible in the electron density map [Fig. 2(C)]. The C-terminal region beyond residue 95 was disordered. The C-terminus has been noted to be the region of largest coordinate differences between CcmK paralogs, CcmK2 and CcmK4, elucidated earlier.6 The coordinates of CcmK1 were employed in an earlier study aimed at modeling the assembly of the intact carboxysome,8 but a detailed structural analysis and comparison was not presented. CcmK1 adopts an α/β fold matching the previously determined structures of CcmK2 and CcmK4 [Fig. 2(B)]. The rms deviations between the C-alpha backbone coordinates of CcmK1 and CcmK2 monomers and between CcmK1 and CcmK4 monomers are 0.51 and 0.72 Å, respectively [Fig. 2(D)].
Table I.
Data Collection and Refinement Statistics for CcmK1, CcmK1d, Form 1 and 2 CcmK2d
| Data collection | CcmK1 | CcmK1d | Form 1 CcmK2d | Form 2 CcmK2d |
|---|---|---|---|---|
| PDB ID | 3BN4 | 3DN9 | 3BPI | 3DNC |
| Concentrations (mg/mL) | 4.7 | 13.6 | 35.4 | |
| Space group | C2 | P1 | P6 | |
| Unit cell dimensions (Å, °) | a = 120.9, b = 69.8, c = 78.0 | a = 49.2, b = 49.2, c = 49.2 | a = b = 120.8, c = 29.2 | a = b = 67.4, c = 29.0 |
| α = γ = 90, β = 94.61 | α = 90.65, β = 91.41, γ = 91.43 | α = β = 90, γ = 120 | α = β = 90, γ = 120 | |
| Wavelength (Å) | 0.97944 | 1.000 | 0.979 | 1.5418 |
| Resolution limit (Å) | 60.30–2.00 | 17.61–2.28 | 52.3–1.30 | 58.3–2.05 |
| Rsym (%)a | 12.3 | 10.3 | 7.0 | 15.1 |
| Rsym (%, last shell) | 44.2 | 40.6 | 46.5 | 44.9 |
| I/σ (last shell) | 2.44 | 1.25 | 1.38 | 5.00 |
| Total observations | 230,213 | 26,940 | 191,988 | 31,006 |
| Unique reflections | 43,130 | 16,303 | 58,974 | 4,371 |
| Completeness (%) | 98.8 | 79.8 | 97.4 | 89.2 |
| Completeness (%, last shell) | 97.8 | 42.8 | 99.4 | 99.8 |
| Model refinement | ||||
| Rworkb | 20.3 | 22.2 | 17.1 | 22.2 |
| Rfreec | 25.0 | 27.4 | 22.2 | 27.8 |
| Number of residues (protein/water) | 550/104 | 534/58 | 282/346 | 90/40 |
| Number of protein chains per crystal asymmetric unit | 6 | 6 | 3 | 1 |
| Average B (Å2) (main chain/side chain) | 48.9/54.9 | 33.3/35.3 | 16.1/24.4 | 51.0/53.0 |
| Rmsd bonds (Å) | 0.012 | 0.013 | 0.012 | 0.007 |
| Rmsd angles (°) | 1.37 | 1.093 | 1.42 | 1.022 |
| Rmsd B-values (Å2) (main chain/side chain) | 1.32/2.19 | 1.40/1.48 | 1.13/1.97 | 0.86/0.93 |
Rsym = Σ|I−<1>|2/ΣI2.
Rwork = Σ|Fobs−Fcalc|/ΣFobs.
Rfree = Σ|Fobs−Fcalc|/ΣFobs, where all reflections belong to a test set of 5% randomly selected data.
Structures of C-terminal deletion mutants of CcmK1 and CcmK2
The truncated mutants of CcmK1 and CcmK2 lacking their C-terminal tails were constructed to investigate the effects of these variable regions on the behavior of the proteins, and also to alleviate their problematic effects on obtaining highly ordered crystals. Residues 92–111 and 92–103 were chosen for deletion from CcmK1 and CcmK2, respectively, based on sequence alignments [Fig. 2(A)] and on the C-terminal disorder observed in the CcmK1 structure; the truncated constructs are hereafter referred to as CcmK1d and CcmK2d. The proteins were expressed in E. coli, purified, and crystallized by vapor diffusion. Phases for CcmK1d diffraction data were obtained by molecular replacement using CcmK1 as the search model. An atomic model of CcmK1d was built and refined at a resolution of 2.28 Å, with R and Rfree values of 22.2% and 27.4% (Table 1). The final model consists of a hexamer in the asymmetric unit, with all 91 residues of the complete CcmK1d construct visible in the electron density map. The coordinate deviation between the truncated CcmK1 and the full length CcmK1 was only 0.61 Å over the protein backbone and 1.1 Å over all atoms.
The truncated form of CcmK2 (CcmK2d) crystallized in two different forms, both in space group P6, but with different unit cell lengths. The structures of both forms were obtained by molecular replacement using the structure of CcmK2, omitting the coordinates for residues 92–102. Form 1 crystals diffracted to 1.3 Å resolution and had three protein molecules in the asymmetric unit. A complete atomic model was built and refined with R and Rfree values of 17.1% and 22.2%. Form 2 crystals diffracted to 2.05 Å and had one protein molecule in the asymmetric unit. The final R and Rfree values were 22.2% and 27.8% (Table 1). Both models include all 91 residues of the CcmK2d construct.
Conservation of hexamer structure
The cyclic hexameric arrangement of CcmK subunits is conserved across all the structures reported, including full length and truncated proteins in multiple crystal forms. Complete hexamers from different crystal forms, different constructs, and different paralogs, can be closely superimposed. For example, the backbone rmsd between hexamers of CcmK1 and CcmK2d (Form 1) is 0.57 Å, and the rmsd between hexamers of CcmK2d from the two crystal forms is 0.37 Å. A key feature of the hexamer is a large and somewhat hydrophobic depression on one face, whose appearance is affected by the disposition of the C-termini on that side of the hexamer (and the extent to which the C-termini can be visualized) [Fig. 6(B)]. Variations in the packing of subunits (e.g., by small subunit rotations) within a hexamer would be a possible mechanism for altering the characteristics of the pore through the center of the hexamer, which has been argued to serve a role in molecular transport across the shell. The absence of significant variation in the packing of subunits within hexamers suggests that the properties of the hexameric pores are not modulated by such a mechanism.
Figure 6.
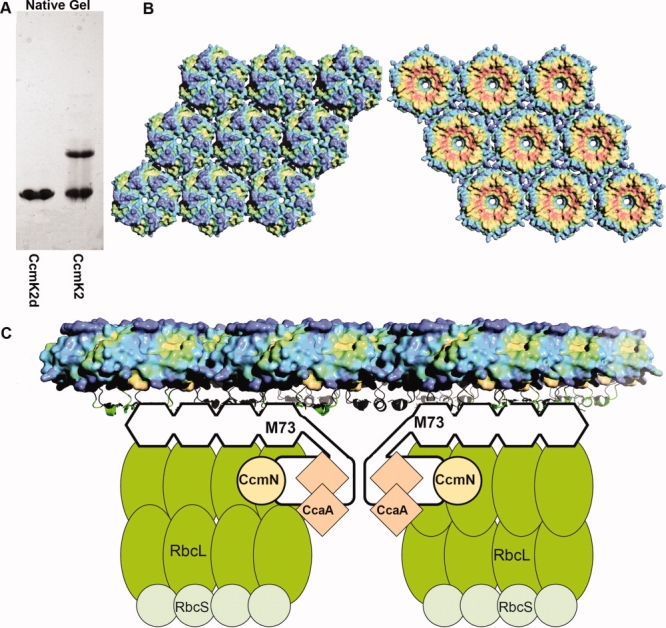
A: A native gel showing the oligomeric state of CcmK2d (left) and CcmK2 (right). B: Hydrophobic character of the two sides of the CcmK2d hexamer layer. The color gradient indicates greater hydrophobicity in red and lower hydrophobicity in blue. The C-terminal tails that are deleted in the CcmK2 construct would be pointing upward in the right side view. C: A model of carboxysome-associated proteins interacting with the shell. The C-terminal side of the hexamer layer (right side view in panel B) is proposed here to face toward the interior of the carboxysome. Possible interactions between the C-terminal regions of CcmM and the shell proteins are indicated. The model incorporates data from recent protein–protein interaction studies.28,29
The structure of Form 1 CcmK2d provides a high resolution view of the hexamer pore. The narrowest point of the pore occurs at the backbone amide nitrogen atom of Ser 39; six copies of that atom fall on a circle of diameter 9.3 Å. Assuming a van der Waals radius of 1.7 Å, the diameter of the space within the pore is 5.9 Å at this point. In CcmK1, the narrowest point of the pore occurs at the terminal oxygen atom of Ser39, and the diameter of the pore is 5.0 Å. At a different position in the hexamer pore, the terminal nitrogen atoms of six Lys36 residues fall on a circle of diameter 17 Å [Supp. Info. Fig. 1(A)]. The lysine residues from neighboring subunits are hydrogen bonded to each other, from the terminal nitrogen atom of one lysine to the backbone carbonyl oxygen of the adjacent lysine, and also to the Glu35 side chain [Supp. Info. Fig. 1(B)]. These lysine residues, which contribute to the positive electrostatic potential in the pore, lie on the same side of the hexamer as the surface depression and the C-terminal tails, and are separated from the narrowest part of the pore by a vertical distance of about 10 Å [Supp. Info. Fig. 1(C)].
Natural substrates and products (i.e., bicarbonate, 3-PG, and RuBP) must cross the shell, but they have not yet been visualized in the pores. However, a sulfate ion was visualized earlier in the CsoS1A hexamer pore from the α-type H. neapolitanus carboxysome.7 In the structure of full length CcmK1 reported here, a sulfate ion is also seen bound in the hexamer pore [Fig. 4(A,C)]. The location of the sulfate ion is consistent with hydrogen bonding to the backbone of Ser39. This is essentially the same position as the sulfate ion observed in CsoS1A,7 which was reported to be hydrogen bonded to the backbone of the corresponding residue in that protein, Gly43.
Figure 4.
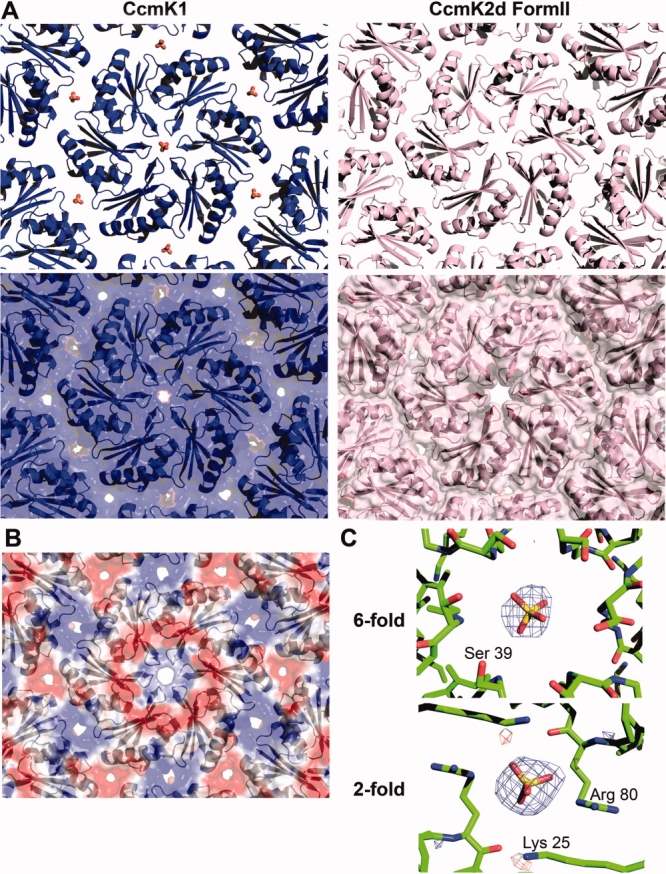
A: Ribbon (top) and space-filling (bottom) diagrams of CcmK1 (left in blue) and Form 2 CcmK2 (right in pink) showing the differences in crystal packing. The spacing of CcmK1 hexamers are wider than Form 2 CcmK2d, and sulfate ions are found in the pores at the sixfold and twofold symmetry axes through the CcmK1 layer. B: The calculated electrostatic potential (blue positive and red negative) in the CcmK1 hexamer layer is positive at the sixfold and twofold axes. C: A difference electron density map (Fobs-Fcalc, αcalc) shown at the sixfold and twofold axes (contoured at 3σ and 4σ, respectively), showing peaks identified as sulfate ions. The views of the molecular layers are the same as in Figure 3.
Molecular layers of carboxysome hexamers
In all of the crystal structures of CcmK1 and CcmK2 hexamers that have been determined, and in CsoS1A as well, the hexamers pack side-by-side to form molecular layers [Fig. 3(A)]. These layers have been postulated to represent the flat facets of the carboxysome shell. The characteristics of the packing in these layers are therefore key to understand the properties of the intact shell. In contrast to the highly conserved arrangement of monomers in individual hexamers, some variation is seen in the packing of hexamers into two-dimensional layers. A difference was noted earlier between the packing of hexamers first visualized in CcmK2 and the particularly tight packing observed in CsoS1A.7 In three of the structures described here (CcmK1, CcmK1d, and Form1 CcmK2d), the packing of hexamers is very similar to the one reported for full length CcmK2. The lateral separation between hexamer centers is very close to 70 Å (see Fig. 3). In contrast, Form 2 CcmK2d has a tighter spacing between hexamers, with a distance of 67.2 Å [Fig. 3(B)]. The key differences between the two distinct packings are illustrated in Figure 4(A). In the packing of CcmK1, CcmK1d, and Form 1 CcmK2d, narrow pores are present at the twofold and threefold axes between hexamers, in addition to the pores through the hexamer centers. Owing to the tighter spacing, pores are present only through the hexamer centers in the Form 2 CcmK2d layer. That situation is reminiscent of the packing visualized earlier in CsoS1A.
Figure 3.
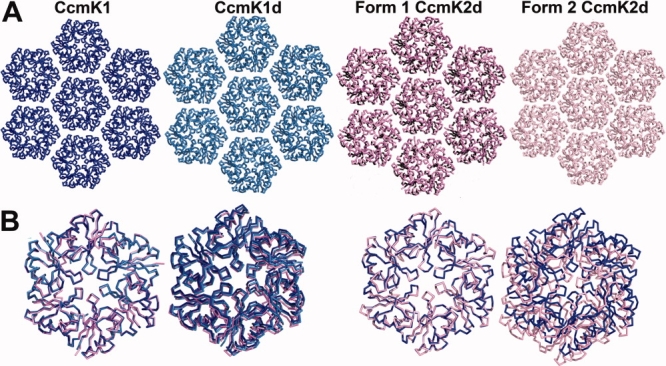
A: Crystal packing of hexamers of CcmK1, CcmK1d, Form 1 CcmK2d, and Form 2 CcmK2d in molecular layers. CcmK1 is blue; CcmK1d is light blue; Form 1 CcmK2d is violet; Form 2 CcmK2d is pink. The packing of Form 2 CcmK2d in a layer is slightly tighter than the other three structures on the left, which are similar to each other. Also, the CcmK1d packing is nearly identical in all directions, despite slight deviations of the unit cell from true hexagonal symmetry. B: One of two adjacent hexamers from CcmK1d, CcmK2, and Forms 1 and 2 CcmK2d is superposed on one of two adjacent CcmK1 hexamers. (Left) The spacings between adjacent hexamers of CcmK1 (blue), CcmK1d (light blue), and Form 1 CcmK2d (violet) are almost identical (70 Å). (Right) The spacing between adjacent hexamers of Form 2 CcmK2d (pink) is closer (67.2 Å) than in the other structures; CcmK1 is shown in blue for comparison. The structures are viewed here from the opposite side of the molecular layer compared to Figure 2(C,D). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
In the structures of CcmK1 and CcmK1d, difference electron density maps suggested possible sulfate ions at two locations in the molecular layer. One site is at the hexagonal pore discussed earlier. The other is at the twofold axis of symmetry where two hexamers meet in the molecular layer [Fig. 4(A,C)]. The sulfate ion there benefits from ionic interactions and potential hydrogen bonds to Arg80 and Lys25. The identification of the bound ligands as sulfate was supported by the atomic refinement, which gave sulfur atom occupancies of 56 and 76% at the sixfold and twofold positions; occupancies under 20% (10 electrons for H2O vs. 50 electrons for sulfate) would have been expected if the features were due to a bound water molecule instead. Furthermore, calculations showed that the electrostatic potential arising from charged protein side chains is positive at both those positions; the calculated potential is negative at the threefold axis, and no sulfate ions were visualized there [Fig. 4(B)]. Taking van der Waals radii into account, the pore at the twofold axis is 3.9 Å across at its narrowest point.
Protein interactions involving the C-terminal tails
In crystals of both CcmK1 and CcmK2 in their full length forms, adjacent molecular layers appear to interact with each other via the C-terminal tails of the protein subunits. The specific atomic interactions cannot be visualized in CcmK1 or CcmK2 layers due to disorder. The missing regions of electron density are particularly prominent in the case of CcmK1, whose C-terminus is longer by eight residues. Although electron density maps are relatively flat, multiple lines of reasoning suggest that the termini must be present in the space between the layers. Interactions between the layers must be present in order for the CcmK1 crystal to be held together in three dimensions (see Fig. 5). The region devoid of electron density is too thin to accommodate another layer of protein molecules, and the presence of the complete amino acid sequence in the purified CcmK1 protein was verified by mass spectrometry (Supp. Info. Fig. 2). Also, deleting the C-termini leads to dramatic changes in the packing of molecular layers (i.e., in crystals of CcmK1d), further underscoring the tendency of the termini to participate in protein–protein interactions.
Figure 5.
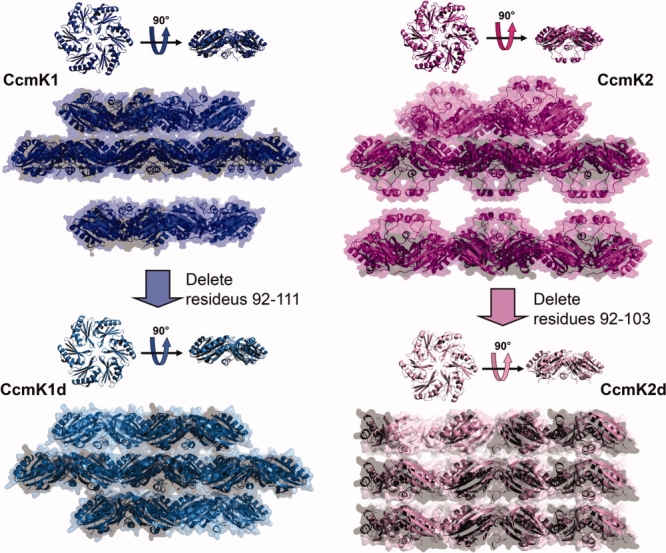
The effects of C-terminal deletion on the molecular layers. The molecular layers of both CcmK1 (blue) and CcmK2 (magenta) adopt face-to-face orientations with space between the C-terminal faces. The molecular layers of both CcmK1d (blue-green) and CcmK2d (pink) adopt face-to-tail orientations with little space between layers. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
When the purified CcmK2 protein was run on a native gel, two distinct oligomeric forms were observed [Fig. 6(A)], whereas only one oligomeric form (the hexamer) was observed when the purified CcmK1 and CcmK4 proteins were run on native gels (data not shown). The two distinct oligomeric forms of CcmK2 were separated by size exclusion chromatography and analyzed by equilibrium sedimentation. The smaller species had a molecular weight consistent with a hexamer. The larger oligomeric species exhibited some molecular weight heterogeneity, consistent with a mixture of a hexameric form and a larger form, with the molecular weight of the larger species being poorly determined but in the range of 9–12 times that of the monomer (data not shown). The molecular weight range, and the absence of other discrete bands besides this one and the hexamer, suggests that the higher molecular weight species is probably a dodecamer comprised of two hexamers. CcmK2d, the truncated form of CcmK2, showed only one oligomeric species on the native gel, corresponding to a hexamer [Fig. 6(A)], again implicating the C-terminal tails in protein–protein interactions.
Discussion
The present work provides additional structural data for understanding the functional mechanisms of the carboxysome. The new structures reiterate the conserved features of the hexameric building blocks that self-assemble to form the carboxysome shell. Across the multiple structures of the CcmK proteins from the β-type carboxysome of Syn 6803 reported here, the arrangement of monomers within a hexameric unit is strongly conserved, so that only minor variations are seen in the side chain conformations on the protein surface and in the central pore through the hexamer. In one of the new structures, a sulfate ion is seen bound in the pore. This substantiates an earlier observation of a sulfate ion in the hexameric pore of a corresponding shell hexamer (Csos1A) from the α-type carboxysome of H. neapolitanus.7 The findings support the view that the pores allow diffusion of small, negatively charged molecules into and out of the carboxysome. Based on the geometric parameters and the presence of a sulfate ion in the hexameric pore, it seems clear that bicarbonate could diffuse through. The positive electrostatic potential of the pore has been discussed previously as a mechanism for enhancing transport of bicarbonate, but not uncharged molecules such as molecular oxygen and carbon dioxide.6,13 Selectivity in transporting bicarbonate over CO2 could be important in retaining CO2 inside the carboxysome after it is generated from bicarbonate by carbonic anhydrase, thereby favoring the reaction of CO2 with RuBisCO. In addition, or alternatively, a preference for allowing bicarbonate rather than O2 to diffuse into the carboxysome would limit the competing, wasteful reaction of O2 with RuBisCO. Efforts to develop mathematical models for how the carboxysome enhances carbon fixation were initiated nearly 20 years ago.30,31 Structures of the shell components reported here and in previous studies should make it possible to incorporate atomic level information into such models. One of the crystals provides a high-resolution structure (1.3 Å), which complements a previous high-resolution structure (1.4 Å) from the α-type carboxysome,7 and which should be valuable for future simulations and computational analyses.
Whether or not the pores through CcmK1 and CcmK2 hexamers can support diffusion of the five-carbon substrate (RuBP) and the three-carbon product (3-PG) of the RuBisCO reaction is not yet known. A smaller pore of 4.3 Å in diameter was reported earlier through the center of the CcmK4 hexamer,6 which is probably a minor component of the shell. Furthermore, there are additional components of the carboxysome whose structures have not been elucidated yet, including CcmN and CcmM in β-type carboxysomes. The possibility that those proteins might play roles in transport cannot be discounted at this time.
Hexamers of the major carboxysome shell proteins show a consistent tendency to further assemble side-by-side into tightly packed molecular layers, but potentially significant variations in packing geometry have been noted.7 That observation is reinforced here. While three of the layer structures observed were very similar to the one reported first for CcmK2,6 one of the structures (Form 2 CcmK2d) showed a particularly tight packing very similar to the one observed in the single structure reported so far from an α-type carboxysome.7 This argues against the idea that the packing variation noted before relates to a difference between the two types of carboxysomes. Whether the packing variations observed are biologically relevant, or whether they represent minor artifacts of crystallization, remains to be established. One possibility is that packing variations could be used to modulate the transport properties of the molecular layer that forms the bulk of the carboxysome shell. Indeed, in the structures exhibiting the looser packing a sulfate ion was visualized at the pore where two hexamers meet. If alterations in subunit packing in the layer are biologically significant, this would parallel the kinds of rearrangements that occur in the maturation of certain viral capsids.32
How the interior of the carboxysome is organized relative to the shell is understood only in rough terms.23,24 Two recent studies have highlighted the role of the CcmM protein in the β-type carboxysome in binding several other carboxysome proteins,28,29 including the CcmK shell proteins.29 The structural details of these interactions have not been illuminated yet. Knowing which regions of the shell proteins interact with other carboxysome proteins, including CcmM and possibly other proteins, would answer key functional questions. In particular, the inside versus outside orientation of the molecular layer of hexamers has not been clarified by earlier structures of the shell proteins,6–8 and has not been addressed by direct experiments. That piece of information is vital to understand the direction in which different molecules diffuse across the pores, and the underlying mechanisms of molecular transport. The studies presented here suggest that the C-terminal tails of the CcmK subunits are likely involved in protein–protein interactions in the native carboxysome that have not been visualized yet [Fig. 6(C)]. For the CcmK2 protein, gel filtration, gel electrophoresis, and genetic truncation experiments show that the C-terminal tails tend to interact with the tails of other hexamers. This was further emphasized by crystallographic results on full length and truncated CcmK1 and CcmK2 proteins, though the interactions observed there are not reflective of the specific interactions that must be present in the context of the native carboxysome. Important interactions could also include the relatively hydrophobic region underlying the C-terminal tails, depending on their conformations. The idea that the C-terminal tails of the major shell proteins are critical elements for interacting with other proteins inside the carboxysome represents a hypothesis to be tested in future work. If the hypothesis is correct, it will provide good evidence that this side of the molecular layer of hexamers faces toward the interior of the carboxysome. That clarification will advance our mechanistic understanding of the carboxysome.
Materials and Methods
Cloning, expression, and purification
The Synechocystis sp. PCC 6803 CcmK1 gene and CcmK2 gene were amplified from Synechocystis sp. PCC 6803 genomic DNA using the following primers:
ccmk1NdeIfwd: 5′-GGGGCATATGTCAATTGCAGTTGGGATGATCGAAACCC-3′,
ccmk1NotIrev: 5′-GGGGGCGGCCGCACGGCGGATGATGGAAGGATTAACGATTT-3′,
ccmk2NdeIfwd: 5′-GGGGCATATGTCCATTGCAGTTGGAATGATCGAAACCC-3′, and
ccmk2NotIrev: 5′-GGGGGCGGCCGCATATGTCCGAAATTGTTCAACTTCTTCGGTATAG-3′ containing NdeI (C∧ATATG) and NotI (GC∧GGCCGC) restriction sites.
PCR products were digested with NdeI and NotI and ligated into the multiple cloning site of pET22b (Novagen), which produces a C-terminal His6 tag to the encoded protein. The sequences of expression vectors were confirmed via plasmid sequencing (DNA Sequencing Facility, University of California, Los Angeles). For expression of recombinant protein, the constructs were transformed into BL21-Gold(DE3) cells (Novagen).
The C-terminal deletion mutants of CcmK1 (CcmK1d) and CcmK2 (CcmK2d) were generated by PCR amplification of the CcmK1 and CcmK2 gene sequences using the following primers:
ccmk1dNdeIfwd: 5′-GGTGGTCATATGTCAATTGCAGTTGGGATGATCGAAACCC-3′,
ccmk1dXhoIrev: 5′-GGTCCTCGAGAATGGGTAGCACATACTCCAGGTTTTCG-3′,
ccmk2dNdeIfwd: 5′-GGTGGTCATATGTCCATTGCAGTTGGAATGATCGAAACCC-3′, and
ccmk2dXhoIrev: 5′-GGTGGTCTCGAGAATGGGGAGGACGTATTCCAGGTTTTC-3′ containing NdeI and XhoI (C∧TCGAG) restrictions sites.
PCR products were digested with NdeI and XhoI and ligated into the multiple cloning site of pET22b (Novagen), which produces a C-terminal His6 tag to the encoded protein. The sequences of expression vectors were confirmed via plasmid sequencing (DNA Sequencing Facility, University of Maine). For expression of recombinant protein, the constructs were transformed into BL21-Gold(DE3) cells (Novagen).
CcmK1, CcmK1d, CcmK2, and CcmK2d expressions were induced by the addition of IPTG to culture grown at 37°C with shaking (225 rpm) until they reached an OD600 reading of 0.8. After addition of IPTG, cell growth continued for 4 h before cells were harvested by centrifugation (5k rpm, 15 min). Cell pellets were resuspended in 20 mM Tris, pH 8.0, 300 mM NaCl, 10% glycerol, 10 mM imidazole, and 1:100 protease inhibitor mixture (Sigma). Resuspended cells were lysed by lysozyme treatment and disrupted with an Ultrasonics Sonicator (model W-220F, Branson). Cell lysates were centrifuged at 30,000g for 30 min and the supernatant was loaded onto a Ni++-NTA column (Qiagen) pre-equilibrated with 20 mM Tris (pH 8.0), 300 mM NaCl, and 10 mM imidazole. The bound proteins were eluted with 300 mM imidazole. The purified proteins were verified by SDS gel electrophoresis. Fractions containing target proteins were then pooled and dialyzed at 4°C overnight against a buffer containing 20 mM Tris (pH 8.0) and 100 mM NaCl. The dialyzed samples of CcmK1, CcmK1d, and CcmK2d were concentrated to a final concentration of 4.7, 13.6, and 35.4 mg/mL, respectively, for crystallization. EDTA was added to a final concentration of 10 mM to the CcmK1 and CcmK1d sample for crystallization.
The dialyzed sample of CcmK2 was concentrated to ∼3 mg/mL and loaded onto a Hi Load 16/60 Superdex75™ (GE Healthcare) size exclusion column to separate large and small species. The fractions containing large CcmK2 and small CcmK2 were verified by SDS gel electrophoresis, fractions from each species pooled separately, and concentrated to 16.5 and 7.7 mg/mL, respectively.
Crystallization of CcmK1, CcmK1d, and CcmK2d
Diffraction quality protein crystals of the CcmK1 protein were grown using the hanging drop vapor diffusion method, in 0.1M MES pH 6.0, 15% MPD, and 0.1M lithium sulfate at room temperature. Crystals were observed within a few days. CcmK1 crystals belong to space group C2 with unit cell dimensions a = 120.9 Å, b = 69.8 Å, c = 78.0 Å α = γ = 90.00°, β = 94.61. Similarly, CcmK1d protein crystals were grown using the hanging drop vapor diffusion method, in 0.1M MES, pH 6.5, 10% dioxane, 1.6M ammonium sulfate at room temperature. CcmK1d crystals belong to space group P1 with unit cell dimensions a = 49.2 Å, b = 49.2 Å, c = 49.2 Å, α = 90.65°, β = 91.41°, γ = 91.43°. Although the unit cell had edges of nearly identical length, the unit cell angles are unequal and the data could not be reduced in any higher symmetry space group. CcmK2d protein crystals were grown in 0.1M CHES, pH 9.5, 0.15M NaCl, 1.26M ammonium sulfate. CcmK2d (Form I) crystal belong to space group P6 with unit cell dimensions a = b = 120.8 Å, c = 29.2 Å, α = β = 90°, γ = 120°, and three CcmK2d monomers in the asymmetric unit. CcmK2d (Form II) crystal belong to space group P6 with unit cell dimensions a = b = 67.4 Å, c = 29.0 Å, α = β = 90°, γ = 120°, and one CcmK2d monomer in the asymmetric unit.
CcmK1 structure determination and refinement
X-ray diffraction data were collected at the Advanced Photon Source beamline 24-ID-C using an ADSC quantum 315 CCD area detector. Data were collected at 100 K from a single crystal that was cryo-protected by a quick dip in a solution containing 80% reservoir and 20% MPD. Data were processed using DENZO/SCALEPACK.33 The data set was strongly anisotropic, with diffraction limits of 2.0 Å along the a* direction, 2.0 Å along the b* direction, and 3.0 Å along the c* direction. For this reason, data were truncated that fell outside an ellipse centered at the reciprocal lattice origin and having vertices at 1/2, 1/2, and 1/3 Å−1 along a*, b*, and c*, respectively. The data were scaled anisotropically using the procedure described in Strong et al., 2006 then used for refinement with REFMAC.35 The structure was solved readily by molecular replacement using the CcmK2 hexamer (PDB code = 2A1B) as a search model. The six subunits comprising the asymmetric unit were positioned using the program Phaser.36 Initially, the six molecules in asymmetric unit were refined using tight sixfold symmetric NCS restraints. Later, the grouping was broken into two separate threefold symmetric groups with tight restraints for main chain atoms and loose restraints for side chain atoms. Throughout the refinement, TLS parameters were refined to model anisotropic disorder in the hexamer. The model was refined to (Rwork = 20.3% and Rfree = 25.0%) using REFMAC35 and visualized using COOT.37
The geometric quality of the model was assessed with the following structure validation tools: ERRAT,38 PROCHECK,39 and WHATIF.40 PROCHECK reported 90.6% of the residues fall in the most favored region of the Ramachandran plot and the remaining residues (9.4%) were in additionally allowed regions. ERRAT reported an overall quality factor of 86.6%. Images of protein structures were created using PyMOL.41
CcmK1d structure determination and refinement
X-ray diffraction data were collected at the Advanced Light Source beamline 8.2.2 using an ADSC Quantum 315 CCD are detector. Data were collected from a single crystal cryo-protected with a solution containing 65% reservoir and 35% glycerol and processed as described above. The structure was solved readily by molecular replacement using the CcmK1 hexamer as a search model. The six subunits comprising the asymmetric unit were positioned using Phaser. The model was refined to (Rwork = 22.2% and Rfree = 27.4%) as described above using NCS restraints and TLS parameters. PROCHECK reported 91.8% of the residues fall in the most favored region of the Ramachandran plot and the remaining residues (8.2%) were in additionally allowed regions. ERRAT reported an overall quality factor of 88.8%.
CcmK2d Form I structure determination and refinement
X-ray diffraction data were collected at the Advanced Photon Source beamline 24-ID-C using an ADSC quantum 315 CCD area detector. Data were collected and processed as described earlier. The structure was solved readily by molecular replacement using the CcmK2 monomer (PDB: 2A1B) as a search model. Based on the mutant sequence, C-terminal sequence of the CcmK2 coordinate was trimmed to residue 91. The three subunits comprising the asymmetric were positioned using Phaser. The model was refined to (Rwork = 17.1% and Rfree = 22.2%). PROCHECK reported 87.9% of the residues in the most favored region of the Ramachandran plot and the remainder in additionally allowed regions. ERRAT reported an overall quality factor of 83.3% for the three molecules in the asymmetric unit. Comparatively, this ERRAT score was low relative to the scores from other carboxysome shell structures, and therefore warranted further investigation. The lowest scoring region of the molecule corresponded to the C-terminal His-tag; removal of its coordinates improved the score to 88.0%. After examination of a simulated annealing omit map, we concluded that the His-tag was modeled correctly, and the low scores were probably due to its artificial attachment. Low scores from other surface residues are probably due to the fact that the coordinates used for ERRAT calculation were removed from context of the biological hexamer. The score was further improved to 91% when the three monomers were examined in the context of neighboring monomers from two other hexamers.
CcmK2d Form II structure determination and refinement
X-ray diffraction data were collected using Rigaku FR-D rotating anode X-ray generator (CuKα) equipped with a Raxis-IV++ imaging plate detection at the University of California, Los Angeles. Data were collected and processed as described earlier. The structure was solved readily by molecular replacement using the CcmK2 monomer (PDB: 2A1B) as a search model. Based on the mutant sequence, C-terminal sequence of the CcmK2 coordinate was trimmed to residue 91. The one subunit comprising the asymmetric unit was positioned using Phaser. TLS parameters were refined for anisotropic disorder of the monomer, and the model was refined to (Rwork = 22.2% and Rfree = 27.8%). PROCHECK reported 94.7% of the residues in the most favored region of the Ramachandran plot and the remaining residues in additionally allowed regions. ERRAT reported an overall quality factor of 88.0%. The score was further improved to 94.7% when one monomer was included with neighboring monomers of a hexamer.
Acknowledgments
The authors thank Duilio Cascio for crystallographic assistance, Julian Whitelegge for mass spectrometry assistance, and Cheryl Kerfeld, Sabine Heinhorst, and Gordon Cannon for helpful discussions.
References
- 1.Cannon GC, Bradburne CE, Aldrich HC, Baker SH, Heinhorst S, Shively JM. Microcompartments in prokaryotes: carboxysomes and related polyhedra. Appl Environ Microbiol. 2001;67:5351–5361. doi: 10.1128/AEM.67.12.5351-5361.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shively JM, English RS, Baker SH, Cannon GC. Carbon cycling: the prokaryotic contribution. Curr Opin Microbiol. 2001;4:301–306. doi: 10.1016/s1369-5274(00)00207-1. [DOI] [PubMed] [Google Scholar]
- 3.Badger MR, Price GD. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J Exp Bot. 2003;54:609–622. doi: 10.1093/jxb/erg076. [DOI] [PubMed] [Google Scholar]
- 4.Badger MR, Price GD, Long BM, Woodger FJ. The environmental plasticity and ecological genomics of the cyanobacterial CO2 concentrating mechanism. J Exp Bot. 2006;57:249–265. doi: 10.1093/jxb/eri286. [DOI] [PubMed] [Google Scholar]
- 5.Heinhorst S, Cannon GC, Shively JM. Carboxysomes and carboxysome-like inclusions. In: Shively JM, editor. Complex intracellular structures in prokaryotes. Berlin: Springer-Verlag; 2006. pp. 141–165. [Google Scholar]
- 6.Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO. Protein structures forming the shell of primitive bacterial organelles. Science. 2000;309:936–938. doi: 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- 7.Tsai Y, Sawaya MR, Cannon GC, Cai F, Williams EB, Heinhorst S, Kerfeld CA, Yeates TO. Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome. PLoS Biol. 2007;5:1345–1354. doi: 10.1371/journal.pbio.0050144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO. Atomic-level models of the bacterial carboxysome shell. Science. 2008;319:1083–1086. doi: 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- 9.Badger MR, Hanson DT, Price GD. Evolution and diversity of CO2 concentrating mechanisms in cyanobacteria. Funct Plant Biol. 2002;29:161–173. doi: 10.1071/PP01213. [DOI] [PubMed] [Google Scholar]
- 10.Shively JM, Ball F, Brown DH, Saunders RE. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science. 1973;182:584–586. doi: 10.1126/science.182.4112.584. [DOI] [PubMed] [Google Scholar]
- 11.Price GD, Coleman JR, Badger MR. Association of carbonic anhydrase activity with carboxysomes isolated from the Cyanobacterium Synechococcus PCC7942. Plant Physiol. 1992;100:784–793. doi: 10.1104/pp.100.2.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker SH, Williams DS, Aldrich HC, Gambrell AC, Shively JM. Identification and localization of the carboxysome peptide Csos3 and its corresponding gene in Thiobacillus neapolitanus. Arch Microbiol. 2000;173:278–283. doi: 10.1007/s002030000141. [DOI] [PubMed] [Google Scholar]
- 13.Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat Rev Microbiol. 2008;6:681–691. doi: 10.1038/nrmicro1913. [DOI] [PubMed] [Google Scholar]
- 14.Cleland WW, Andrews JT, Gutteridge S, Hartman FC, Lorimer GH. Mechanism of RubisCO: the carbamate as general base. Chem Rev. 1998;98:549–561. doi: 10.1021/cr970010r. [DOI] [PubMed] [Google Scholar]
- 15.Tabita FR. Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a different perspective. Photosynth Res. 1999;60:1–28. [Google Scholar]
- 16.Drews G, Niklowitz W. Beiträge zur cytologie der blaualgen. II. zentroplasma und granulare einschlüsse von Phormidium uncinatum. Arch Mikrobiol. 1956;24:147–162. [PubMed] [Google Scholar]
- 17.Gantt E, Conti SF. Ultrastructure of blue-green algae. J Bacteriol. 1969;97:1486–1493. doi: 10.1128/jb.97.3.1486-1493.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shively JM, Ball FL, Kline BW. Electron microscopy of the carboxysomes (polyhedral bodies) of Thiobacillus neapolitanus. J Bacteriol. 1973;116:1405–1411. doi: 10.1128/jb.116.3.1405-1411.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cannon GC, Heinhorst S, Bradburne CE, Shively JM. Carboxysome genomics: a status report. Funct Plant Biol. 2002;29:175–182. doi: 10.1071/PP01200. [DOI] [PubMed] [Google Scholar]
- 20.English RS, Lorbach SC, Qin X, Shively JM. Isolation and characterization of a carboxysome shell gene from Thiobacillus neapolitanus. Mol Microbiol. 1994;12:647–654. doi: 10.1111/j.1365-2958.1994.tb01052.x. [DOI] [PubMed] [Google Scholar]
- 21.Shively JM, Bradburne CE, Aldrich HC, Bobik TA, Mehlman JL, Jin S, Baker SH. Sequence homologs of the carboxysomal polypepide CsoS1 of the thiobacilli are present in cyanobacteria and enteric bacteria that form carboysomes-polyhedral bodies. Can J Bot. 1998;76:906–916. [Google Scholar]
- 22.Price GD, Badger MR. Evidence for the role of carboxysomes in the cyanobacterial CO2 concentrating mechanism. Can J Bot. 1991;69:963–973. [Google Scholar]
- 23.Schmid MF, Paredes AM, Khant HA, Soyer F, Aldrich HC, Chiu W, Shively JM. Structure of Halothiobacillus neapolitanus carboxysomes by cryo-electron tomography. J Mol Biol. 2006;364:526–535. doi: 10.1016/j.jmb.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iancu CV, Ding HJ, Morris DM, Dias DP, Gonzales AD, Martino A, Jensen GJ. The structure of isolated Synechococcus strain WH8102 carboxysomes as revealed by electron cryotomography. J Mol Biol. 2007;372:764–773. doi: 10.1016/j.jmb.2007.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crowley C, Sawaya MR, Bobik TA, Yeates TO. Structure of the PduU shell protein from the Pdu microcompartment of Salmonella. Structure. 2008;16:1324–1332. doi: 10.1016/j.str.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engel A, Fujiyoshi Y, Gonen T, Walz T. Junction-forming aquaporins. Curr Opin Struct Biol. 2008;18:229–235. doi: 10.1016/j.sbi.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price GD, Howitt SM, Harrison K, Badger MR. Analysis of a genomic DNA region from the cyanobacterium Synechococcus sp. strain PCC7942 involved in carboxysome assembly and function. J Bacteriol. 1993;75:2871–2879. doi: 10.1128/jb.175.10.2871-2879.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Long BM, Badger MR, Whitney SM, Price GD. Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple Rubisco complexes with carboxysomal proteins CcmM and CcaA. J Biol Chem. 2007;282:29323–29335. doi: 10.1074/jbc.M703896200. [DOI] [PubMed] [Google Scholar]
- 29.Cot SS, So AK, Espie GS. A multiprotein bicarbonate dehydration complex essential to carboxysome function in cyanobacteria. J Bacteriol. 2008;190:936–945. doi: 10.1128/JB.01283-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reinhold L, Zviman M, Kaplan A. A quantitative model for inorganic carbon fluxes and photosynthesis in cyanobacteria. Plant Physiol Biochem. 1989;27:945–954. [Google Scholar]
- 31.Reinhold L, Kosloff R, Kaplan A. A model for inorganic carbon fluxes and photosynthesis in cyanobacterial carboxysomes. Can J Bot. 1991;69:984–988. [Google Scholar]
- 32.Conway JF, Wikoff WR, Cheng N, Duda RL, Hendrix RW, Johnson JE, Steven AC. Virus maturation involving large subunit rotations and local refolding. Science. 2001;292:744–748. doi: 10.1126/science.1058069. [DOI] [PubMed] [Google Scholar]
- 33.Otwinowsk Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 36.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 40.Vriend G, Sander C. Quality control of protein models: directional atomic contact analysis. J Appl Cryst. 1993;26:47–60. [Google Scholar]
- 41.DeLano WL. The PyMOL user's manual. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]