Abstract
The muscleblind-like (MBNL) proteins 1, 2, and 3, which contain four CCCH zinc finger motifs (ZF1–4), are involved in the differentiation of muscle inclusion by controlling the splicing patterns of several pre-mRNAs. Especially, MBNL1 plays a crucial role in myotonic dystrophy. The CCCH zinc finger is a sequence motif found in many RNA binding proteins and is suggested to play an important role in the recognition of RNA molecules. Here, we solved the solution structures of both tandem zinc finger (TZF) motifs, TZF12 (comprising ZF1 and ZF2) and TZF34 (ZF3 and ZF4), in MBNL2 from Homo sapiens. In TZF12 of MBNL2, ZF1 and ZF2 adopt a similar fold, as reported previously for the CCCH-type zinc fingers in the TIS11d protein. The linker between ZF1 and ZF2 in MBNL2 forms an antiparallel β-sheet with the N-terminal extension of ZF1. Furthermore, ZF1 and ZF2 in MBNL2 interact with each other through hydrophobic interactions. Consequently, TZF12 forms a single, compact global fold, where ZF1 and ZF2 are approximately symmetrical about the C2 axis. The structure of the second tandem zinc finger (TZF34) in MBNL2 is similar to that of TZF12. This novel three-dimensional structure of the TZF domains in MBNL2 provides a basis for functional studies of the CCCH-type zinc finger motifs in the MBNL protein family.
Keywords: NMR, solution structure, CCCH-type zinc finger motif, muscleblind-like (MBNL), myotonic dystrophy (DM)
Introduction
The human muscleblind-like (MBNL) 1, 2, and 3 proteins promote the inclusion or exclusion of specific exons on the corresponding pre-mRNAs by antagonizing the activities of the CUG-BP and ETR-3-like factors (CELF proteins), which are bound to distinct intronic sites.1,2 Reflecting the importance of the MBNL protein family, highly conserved homologues of MBNL are found in many species, ranging from worms and insects to mammals.2,3
MBNL2 was identified as an RNA binding protein that colocalizes with integrin α3 mRNA in the cytoplasm,4 and MBNL1 is known to be pathologically related to myotonic dystrophy (DM).1,5–8 DM is the most common form of muscular dystrophy affecting adults. Especially, DM1, which accounts for ∼98% of the cases, is caused by the repetitive expansion of a trinucleotide (CTG) in the 3′-untranslated region of the DM1 protein kinase (DMPK) gene.9,10 In DM1 cells, the transcripts containing the expanded RNA repeats form an unusual hairpin secondary structure and accumulate in nuclear foci.5,6 The MBNL1 protein exhibits RNA-binding activity toward the transcripts with the expanded trinucleotide repeats and is sequestered on the CUG repeats in the pre-mRNAs. This sequestering consequently causes the loss of MBNL1 function, leading to the DM pathogenesis.5,11,12
All three human MBNL proteins contain four CCCH zinc finger motifs (ZF1–4), and the amino acid sequences spanning ZF1–ZF2 and ZF3–ZF4 are very well conserved among the MBNL proteins, respectively. (Besides the zinc finger motifs, the linker lengths for ZF1–ZF2 and ZF3–ZF4 are identical among the MBNL protein family members (14 residues for ZF1–ZF2, and 16 residues for ZF3–ZF4) [Fig. 1(B)].) In addition, the N-terminal extension of ZF1 includes the highly conserved sequence (R/K) KWLTLEV immediately preceding the first Cys ligand residue of ZF1. Moreover, within each member of the MBNL family, ZF1 and ZF3 share sequence homology with each other, and have identical spacing between the Cys and His ligand residues (CX7CX6CX3H). This is also the case for ZF2 and ZF4 (CX7CX4CX3H). Thus, it is considered that ZF1-ZF2 and ZF3-ZF4 in the MBNL proteins constitute novel tandem CCCH zinc finger (TZF) motifs (CX7CX6CX3H–CX7CX4CX3H), referred to as TZF12 and TZF34, respectively.7 The mRNA-binding activity of the MBNL family reportedly resides in the two highly conserved TZF domains (TZF12 and TZF34) [Fig. 1(A)].11,12
Figure 1.
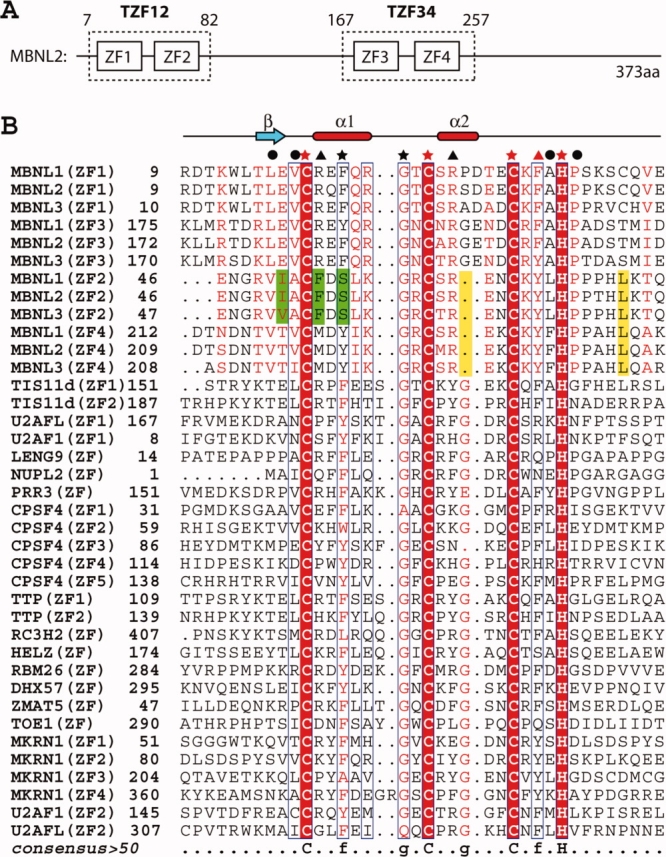
(A) Schematic diagram of the CCCH-type zinc finger motifs contained in human MBNL2. (B) Multiple sequence alignment of CCCH-type zinc finger motifs from selected human proteins. The three Cys and one His ligand residues involved in zinc coordination are marked by red stars. Other strongly conserved residues in CCCH-type zinc fingers are marked by black stars. Residues involved in hydrophobic interactions within the TZF domains are marked by black circles. Residues under the black and red triangles are the amino acid residues with different (aromatic and positively charged residues) and the same properties as compared with the residues at the corresponding positions of the CCCH zinc finger in TIS11d, respectively. Characteristic residues or positions from ZF2 only and from ZF2 and ZF4 are colored green and yellow, respectively. Secondary structure elements are depicted above the aligned sequences. The sequence alignment output was created with the ESPript program.13 [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
The CCCH-type zinc finger is an evolutionarily conserved motif found in a number of proteins that perform diverse RNA-processing functions, and the structure of the tandem CCCH-type zinc finger domain of the TIS11d protein in the complex with the AU-rich element 5′-UUAUUUAUU-3′ was solved.14 In the case of TZF in the TIS11d protein, each finger adopts minimal regular secondary structure, with a short α-helix between the first and second zinc ligand residues and a 310 helix between the second and third zinc ligand residues. The two CCCH zinc fingers fold independently, and the linker between the two CCCH zinc finger domains is flexible in the free state. Upon binding to the target RNA, the two domains of TIS11d are arranged almost parallel to one another and bind to adjacent 5′-UAUU-3′ subsites. Several intercalative stacking interactions between the conserved aromatic side chains and the RNA bases were observed upon the recognition of the single-stranded RNA molecule. In this manner, the spatial relationship between the two zinc finger domains becomes fixed, without direct interactions between these two domains.14
Unlike the TIS11d protein, MBNL2 was described originally as a double-stranded RNA-binding protein, and it can bind to expanded CUG repeats that form an extended hairpin in vitro.6 Therefore, the RNA recognition mode by TZFs in the MBNL family seems to be quite distinct from that observed in the TIS11d protein. To elucidate the mechanism of the accumulation of MBNL proteins on the expanded CUG repeats, structural information about the zinc finger motifs of the MBNL proteins is necessary. In this study, we solved the solution structures of both tandem zinc finger motifs TZF12 (ZF1 and ZF2) and TZF34 (ZF3 and ZF4) in MBNL2 from Homo sapiens, and we discuss the structural differences of the TZFs between the MBNL2 and TIS11d proteins.
Results
Resonance assignments
Backbone sequential and side-chain resonance assignments of the 13C- and 15N-doubly labeled samples were achieved with standard 2D and 3D triple resonance NMR experiments (for details, see the “Materials and Methods” section). The assignments of the backbone resonances and the nonlabile side-chain resonances of TZF12 were almost complete, except for the amide protons of Val8, Asp10, Ser28, Asn47, and His74 as well as the entire side-chain of Pro7, Hδ and Hγ of Arg9, and Hζ of Phe36 and Phe54. Using 15N-edited NOESY and HSQC spectra, all of the side-chain NH2 resonances of the Asn/Gln residues were assigned, except for Gln21 and Gln44 of TZF12. For TZF34, the proton assignments were complete, except for Ser205, the amide protons of Thr206, Met207, and Ile208 as well as the side-chain NH2 resonance of Gln253. For all of the X-Pro bonds in TZF12 and TZF34, the trans conformation was confirmed independently by the intense X (Hα)-Pro (Hδ) sequential NOESY cross peaks, and by the 13Cβ and 13Cγ chemical shift differences.15 The assignments were confirmed by tracing the dNN,dαN, and dβN connectivities in NOESY experiments, following the original methodology of Wüthrich.16
Zinc coordination
The zinc coordination by the Cys residues was confirmed by the chemical shifts of their Cβ atoms, which are consistent with the sulfur groups coordinating zinc ions.17 Furthermore, the 2D 1H-15N long-range HMQC spectrum18 of the TZF domains in the presence of the zinc ions clearly showed two of the 15N resonances, corresponding to Nɛ2 of His38 and His70 in TZF12, and His201 and His235 in TZF34 shifted downfield as a result of zinc coordination, indicating that these His residues are in the ɛ tautomeric state, with the Nδ1 protonated. The other His residues are protonated at the Nɛ1 positions.
The experiments described above indicated that eight amino acids, six Cys and two His, are used in the zinc ion coordination. Correspondingly, for the TZF12 domain, an initial structure calculation based on the NOE restraints revealed that Cys19, Cys27, Cys34, and His38 are clustered and oriented to coordinate one zinc ion. Similarly, Cys53, Cys61, Cys66, and His70 are in the proximity of another zinc ion. For the TZF34 domain, residues Cys182, Cys190, Cys197, and His201 are coordinated with one zinc ion, while residues Cys218, Cys226, Cys231, and His235 are coordinated with another. Therefore, we decided to determine the structures of these TZF12 and TZF34 domains, on the basis of the coordination information.
Structure calculation
The statistics regarding the quality and precision of the final 20 best conformers [Figs. 2(A) and 3(A)] that represent the solution structures of the TZF12 and TZF34 domains of MBNL2 are summarized in Table I. The structures are well-defined and show excellent agreement with the experimental data. Among the 2662/2547 (for TZF12/TZF34) cross peaks that had been identified in the 15N- and 13C-edited 3D NOESY spectra, 99%/99% were assigned by the program CYANA2.0,19 resulting in 1244/1339 nonredundant distance restraints. Almost 16.6/14.7 NOE distances restraints per residue, including 390/388 long-range distances restraints as well as the restraints for the coordination with zinc ions (described in the “Materials and Methods” section), were used in the final structure calculations with CYANA2.0. The precision of the structure is characterized by RMSD values to the mean coordinates of 0.35/0.37 Å for the backbone and 0.99/0.96 Å for all heavy atoms [see Fig. 2(A)]. Finally, the quality of the structure is also reflected by the fact that 73.8%/76.7% of the (φ,ψ) backbone torsion angle pairs were found in the most favored regions and 25.5%/22.6% were in the additionally allowed regions of the Ramachandran plot, according to the program PROCHECK-NMR.20
Figure 2.
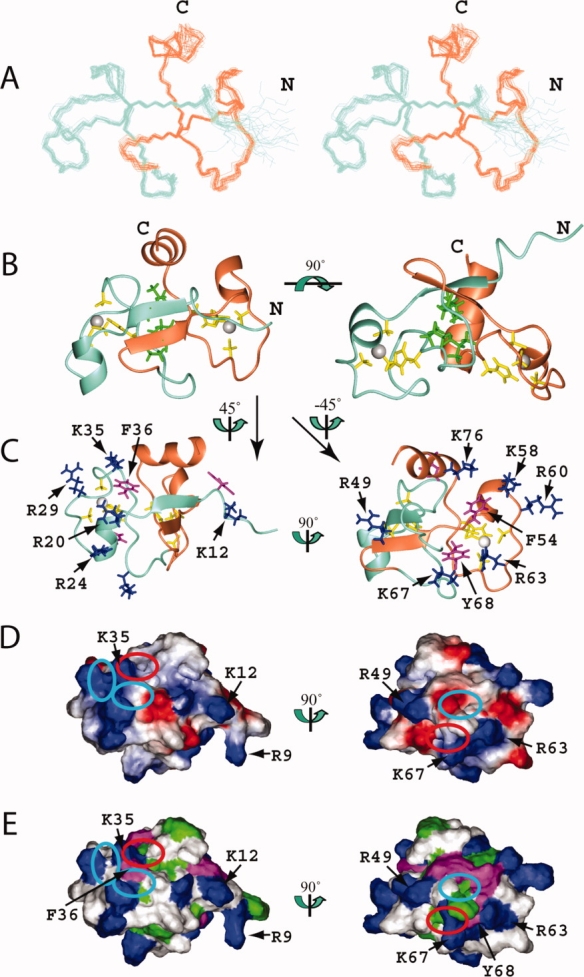
Solution structure of the TZF12 domain in human MBNL2. (A) Stereo-view of the best 20 structures of the TZF12 domain (residues Val8-Asn82). ZF1 and ZF2 of TZF12 are colored aquamarine and coral, respectively. (B) Ribbon presentation of the lowest energy structure of the TZF12 domain. The colors for the backbones of ZF1 and ZF2 are the same as in A. The side chains of the ligand residues and the zinc ions are shown in yellow and gray, respectively. The side chains of the hydrophobic residues that constitute the hydrophobic core of TZF12 are shown in green. The ribbon diagram in the right panel is rotated horizontally by 90° from that on the left. (C) Ribbon diagrams of TZF12 with the side chains of the positively charged and aromatic residues of ZF1 (left panel) and ZF2 (right panel), which are shown in blue and magenta, respectively. (D) Electrostatic surface potential of the TZF12 domain. The blue and red colors represent positive and negative electrostatic surface potential, respectively. (E) Hydrophobic and positively charged surface residues of the TZF12 domain. In C, D, and E, the ZF1 and ZF2 surface orientations (left and right panel) were obtained by 45° and −45° vertical rotations of the ribbon structure on the left in B. The ribbon diagrams (C), the electrostatic potential (D), and the hydrophobic surfaces (E) have the same orientation. The cyan and red ellipses on the surfaces (D, E) of TZF12 mark the pockets formed by the conserved residues. The red ellipse marks the pocket that results from the formation of a compact global fold with the tandem zinc fingers. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Figure 3.
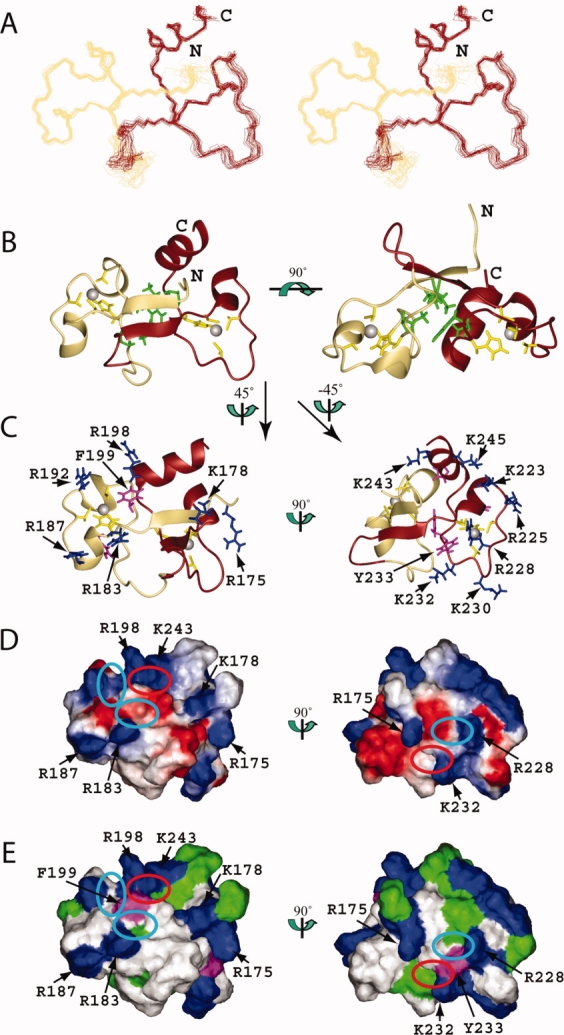
Solution structure of the TZF34 domain in human MBNL2. (A) Stereo-view of the best 20 structures of the TZF34 domain (residues Leu174-Ala247). ZF3 and ZF4 of TZF34 are colored khaki and brown, respectively. (B) Ribbon presentation of the lowest energy structure of the TZF34 domain. The colors for the backbones of ZF3 and ZF4 are the same as in A. The side chains of the ligand residues and the zinc ions are shown in yellow and grey, respectively. The side chains of the hydrophobic residues that constitute the hydrophobic core of TZF34 are shown in green. The ribbon diagram in the right panel is rotated horizontally by 90° from that on the left. (C) Ribbon diagrams of TZF34 with the side chains of the positively charged and aromatic residues of ZF3 (left panel) and ZF4 (right panel), which are shown in blue and magenta, respectively. (D) Electrostatic surface potential of the TZF34 domain. The blue and red colors represent positive and negative electrostatic surface potential, respectively. (E) Hydrophobic and positively charged surface residues of the TZF34 domain. In C, D, and E, the ZF3 and ZF4 surface orientations (left and right panel) were obtained by 45° and −45° vertical rotations of the left ribbon structure on the left in B. The ribbon diagrams (C), the electrostatic potential (D) and the hydrophobic surfaces (E) have the same orientation. The cyan and red ellipses on the surfaces (D, E) of TZF34 mark the pockets formed by the conserved residues. The red ellipse marks the pocket that results from the formation of a compact global fold with the tandem zinc fingers. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Table I.
Statistics of the 20 Final Solution Structures of the TZF12 and TZF34 Domains From Human MBNL2
| TZF12 | TZF34 | |
|---|---|---|
| NMR distance and dihedral angle restraints | ||
| Distance restraints | ||
| Total NOE | 1244 | 1339 |
| Intraresidue | 337 | 406 |
| Interresidue | ||
| Sequential (|i − j| = 1) | 279 | 316 |
| Medium range (1 < |i − j| < 5) | 238 | 229 |
| Long range (|i − j| ≥ 5) | 390 | 388 |
| Zinc-related restraints (upper limit, lower limit) | 26/26 | 26/26 |
| φ/ψ dihedral angle restraints (TALOS) | 60/48 | 42/39 |
| χ1/χ2 dihedral angle restraints | 19/8 | 4/4 |
| CYANA target function (Å2) | 0.026 | 0.13 |
| Structure statistics | ||
| Maximal NOE distance restraint violation (Å) | 0.03 | 0.13 |
| Maximal dihedral angle restraint violation (Å) | 1.2 | 5.0 |
| Ramachandran plot statistics (%)a | ||
| Residues in most favored regions | 73.8 | 76.7 |
| Residues in additional allowed regions | 25.5 | 22.6 |
| Residues in generously allowed regions | 0.7 | 0.7 |
| Residues in disallowed regions | 0.1 | 0.0 |
| Average pairwise rmsd (Å)b | ||
| Backbone | 0.35 | 0.37 |
| Heavy | 0.99 | 0.96 |
Values calculated for residues 11–82 of TZF12 and 173–248 for TZF34.
Values calculated for residues 15–40 and 49–71 of TZF12 and residues 178–202 and 214–237 of TZF34.
Tertiary structure of ZF1 and ZF2 in the TZF12 domain
Using structural data obtained from NMR spectroscopy, we determined the solution structures of the TZF12 and TZF34 domains of human MBNL2. The 20 conformers with the lowest target function values that represent the solution structures of TZF12 and TZF34 are superimposed in Figures 2 and 3, respectively. The TZF12 domain comprises two CCCH-type zinc finger motifs (ZF1: Val8-Val45; ZF2: Glu46-Asn82). The tertiary structure of the TZF12 domain forms a compact global fold, and each of the zinc fingers (ZF1 and ZF2) independently adopts the typical CCCH-type structure, binding one zinc ion through the aforementioned three Cys and one His ligand residues (Cys19, Cys27, Cys34, and His38 for ZF1; Cys53, Cys61, Cys66, and His70 for ZF2). The His ligand residues in each zinc finger are coordinated to zinc via their Nɛ2 atom. The χ1 torsion angles for all of the Cys ligand residues are close to 180°. The χ1 values of the His ligand residues are −60°. There is one short α-helix (Arg20-Arg24) immediately after the first Cys ligand residue and another short α-helix (Ser30-Glu34) between the second and third Cys ligand residues in ZF1 [Fig. 2(B)]. Correspondingly, there is a short α-helix (Phe54-Lys58) just after the first Cys ligand residue in ZF2. However, the α-helix between the second and third Cys ligand residues is missing in ZF2 [Figs. 2(B) and 4(B,C)].
Figure 4.
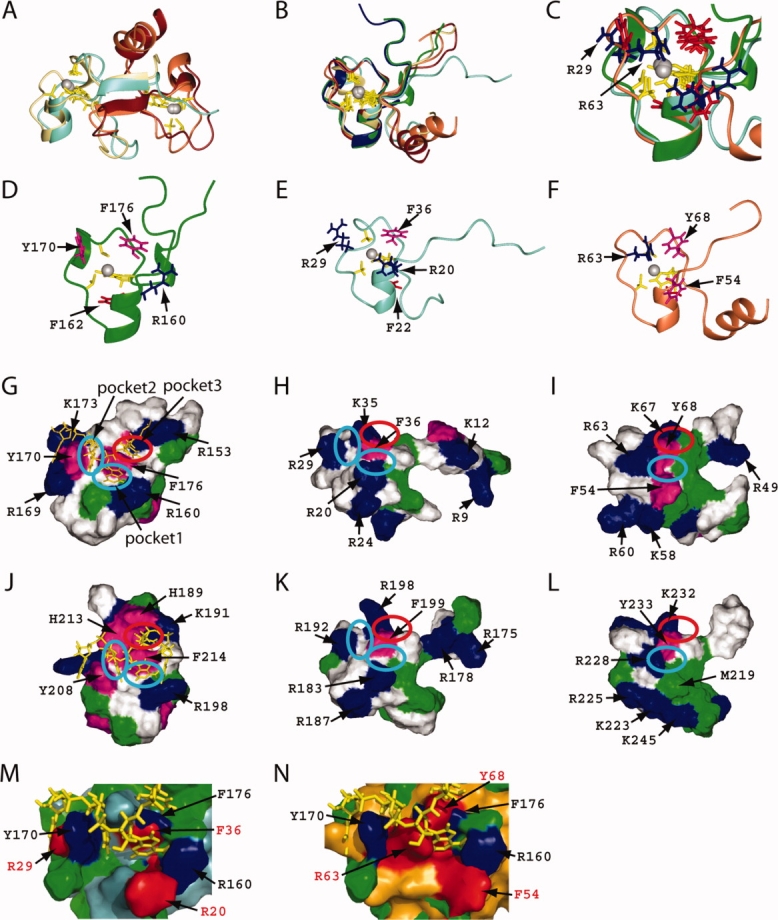
Comparison of the CCCH-type zinc finger motif structures of TIS11d and MBNL2. (A) Superposition of the ribbon diagrams of the TZF12 and TZF34 domains of MBNL2 with the ligand residue side-chains and zinc ions. (B) Superposition of the ribbon diagrams of ZF1 and ZF2 of TIS11d and ZF1-4 of MBNL2 with the ligand residue side-chains and zinc ions. ZF1 and ZF2 of TIS11d are shown in green and blue, respectively. The colors of ZF1-ZF4 of MBNL2 are the same as in Figures 2 and 3. (C) Expanded view of the pocket region with the side chains of the conserved residues. The colors of the side chains are the same as in D–F. (D–F) Ribbon presentations with the side chains of the conserved positively charged and aromatic residues for ZF1 of TIS11d (D), ZF1 of MBNL2 (E), and ZF2 of MBNL2 (F). (G–L) Surface representations showing hydrophobic and positively charged residues of the ZF1-RNA complex of TIS11d (G), ZF1 of MBNL2 (H), ZF2 of MBNL2 (I), the ZF2-RNA complex of TIS11d (J), ZF3 of MBNL2 (K), and ZF4 of MBNL2 (L). The RNA is shown in yellow in (G) and (J). Positively charged residues are colored blue, and aliphatic and aromatic hydrophobic residues are in green and magenta, respectively. The cyan and red ellipses mark pockets 1-2 and pocket 3, respectively. (M, N) Surface superposition of the ZF1 complexed with RNA in TIS11d with ZF1 (M) and ZF2 (N) of the TZF12 in MBNL2. The surfaces of the ZF1 of TIS11d, and of ZF1 and ZF2 of TZF12 in MBNL2 are colored green, light blue, and orange, respectively. The conserved residues of the ZF1 in TIS11d and the TZF12 in MBNL2 with the red annotations are colored blue and red, respectively. The RNA is depicted by yellow sticks. Figures M and N were generated by PyMOL program (DeLano Scientific LLC). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Hydrophobic interactions between ZF1 and ZF2 in the TZF12 domain
In TZF12, the two CCCH-type zinc finger motifs interact with each other through hydrophobic interactions [Fig. 2(A,B)]. The N-terminal extension of ZF1 (Thr15-Glu17) and the linker between ZF1 and ZF2, that is, the N-terminal extension of ZF2 (Arg49-Ile51), form an antiparallel β-sheet. This β-sheet involves both ZF1 and ZF2, which are aligned with a C2 symmetry axis. Simultaneously, the C-terminal extended region of ZF1 (Ala37-Pro39) and that of ZF2 (Leu69-Pro71), which both have the His ligand residue in the middle, are closely aligned in an antiparallel manner [Fig. 2(A,B)]. These C-terminal extended regions (down) and the N-terminal strands in the β-sheet (up) are “weaved” to form the hydrophobic core of the compact global fold of TZF12 [Fig. 2(B)]. The hydrophobic residues (Leu16, Val18, Ala37, and Pro39 from ZF1; Val50, Ala52, Leu69, and Pro71 from ZF2) that compose the hydrophobic core are all well conserved in the MBNL proteins [Fig. 1(B)].
Furthermore, the additional α-helix (spanning residues His74-Asn82) at the C-terminal end of ZF2 folds back to stabilize the TZF domain. Especially, Leu 75 at the C-terminus of the α-helix is well conserved in ZF2 and ZF4, and it contacts the other Leu residue in the LEV box (Leu16) preceding the first Cys ligand residue of ZF1. Consequently, the relative positions of ZF1 and ZF2 are fixed, and the two zinc ions are located ∼20 Å apart [Fig. 2(B)].
Tertiary structure of TZF34 of MBNL2
TZF34 has 46.2% sequence identity and 24.4% additional strong similarity to TZF12. Inevitably, TZF34 adopts a similar solution structure to that of TZF12 (see Fig. 3). The secondary elements in TZF34 are as follows [Fig. 3(B)]. There is one short α-helix (Arg183-Arg187) immediately after the first Cys ligand residue and one α-helical turn (G193-Asp196) between the second and third Cys ligand residues in ZF3, and there is one short α-helix (Met219-Lys223) just after the first Cys ligand residue and one long α-helix (Ala238-Gln248) at the C-terminal end in ZF4. The interactions between ZF3 and ZF4 are similar to those between ZF1 and ZF2, as described for TZF12. Namely, the N-terminal strand of ZF3 (Lys178-Glu180) and that of ZF4 (Thr214-Thr216) form an antiparallel β-sheet structure. TZF34 has similar hydrophobic interactions between ZF3 and ZF4 [Fig. 3(A,B)]. The solution structures of TZF12 and TZF34 are quite similar, with corresponding secondary structure elements and the same tertiary structures [Figs. 2, 3, and 4(A)].
Discussion
RNA recognition by the CCCH-type zinc finger of TIS11d
TIS11d contains a pair of CCCH-type zinc finger motifs of the CX8CX5CX3H type, with an 18-residue linker. The linker between the two zinc fingers of TIS11d is flexible in the RNA-free state. Upon binding to the target RNA, the spatial relationship between the two zinc fingers becomes fixed.14
Both zinc fingers of TIS11d have three pockets that accommodate three bases of the RNA molecule. These pockets are arranged around the well-conserved, key aromatic amino acid residue, located two residue positions after the third Cys ligand residue (position 0). In addition, another aromatic amino acid residue, located two residues after the second Cys ligand residue (position 1), and the Arg residue following the first Cys ligand residue (position 2) are involved in the formation of the first and second pockets, respectively.
Moreover, the N-terminal conserved (R/K)YKTEL extensions of the zinc fingers of TIS11d intimately contact the segment between the third Cys ligand residue and the fourth His ligand residues to form the third pocket, with the key aromatic amino acid for the recognition of one base moiety of RNA.14
CCCH-type zinc fingers in MBNL2
As described earlier, among the MBNL protein family members, the amino acid sequences of ZF1-ZF2 and ZF3-ZF4 are very well conserved. ZF1 and ZF3, and ZF2 and ZF4 belong to the CX7CX6CX3H and CX7CX4CX3H types, respectively. A superposition of the CCCH-type zinc finger motifs of TIS11d and the four zinc finger motifs of MBNL2 [Fig. 4(B,C)] reveals that the zinc fingers of TIS11d are closer to ZF1 and ZF3 than to ZF2 and ZF4. Although an aromatic residue resides at position 1 in the two zinc fingers of TIS11d, positively charged Arg residues occupy the corresponding positions in all four zinc fingers of the MBNL proteins [Fig. 4(G–L)]. Despite the replacement, we could find the pocket (pocket 1) between the key aromatic amino acid residue (position 0) and the Arg residue (position 1) for ZF1 and ZF3 [Fig. 4(H,K)]. On the other hand, in the case of ZF2 and ZF4, the loop between the second and third Cys ligand residues lacks one turn of the α-helix, as compared to ZF1 and ZF3. Accordingly, the side chain of the Arg residue, which is conserved at position1 among all of the CCCH zinc fingers in MBNL2, interacts with the key aromatic amino acid residue (position 0), and we could not find an obvious pocket between the two residues [Fig. 4(C,F,I,L)].
The positively charged residue at position 2 of TIS11d is conserved for ZF1 and ZF3 in the MBNL proteins. In contrast, in the cases of ZF2 and ZF4, position 2 is occupied by Phe and Met residues, respectively. Regardless of the type of amino acid residue at position 2, we could observe the pocket between the key aromatic amino acid residue and the position 2 residues for all four CCCH-type zinc fingers of MBNL2.
A detailed examination of the amino acid sequences of the zinc finger motifs in the MBNL proteins revealed that ZF2 is unique among them [Fig. 1(B)]. Namely, the position two residues preceding the first Cys ligand residue is occupied by Glu or Thr in ZF1, ZF3 and ZF4, but by a hydrophobic amino acid in ZF2. Furthermore, in ZF1, ZF3 and ZF4, the third position from the first Cys ligand residue is occupied by an aromatic amino acid that stacks with the His ligand residue to stabilize the zinc finger fold. In the case of ZF2, however, a Ser residue is located at this position. Interestingly, this replacement does not cause an obvious structural change in ZF2 [Fig. 1(B)].
Tertiary structure comparison with the TZF domain of TIS11d
In the RNA complex structure of the TZF of TIS11d, the two zinc fingers of TIS11d bind to adjacent 5′-UAUU-3′ subsites on the single-stranded RNA in almost a parallel arrangement, and face the RNA molecule in a similar direction. However, in the TZF12 domain of MBNL2, the two zinc finger motifs are fixed through hydrophobic interactions, and they exhibit a 180° difference in orientation because of their C2 symmetry (see Fig. 5). Thus, it is impossible to superimpose the TZF domain of TIS11d in the complex with RNA and the TZF domains of MBNL2 in the free state.
Figure 5.
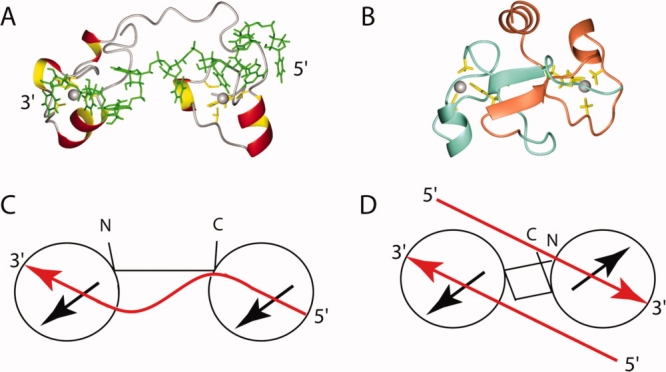
(A) Ribbon diagram presentation of the TZF complex of TIS11d, where the side chains of the ligand residues are shown in yellow, the zinc ions are gray, and the RNA molecule is green, respectively. (B) Ribbon diagram presentation of TZF12 of MBNL2, with the side chains of the ligand residues and zinc ions. The colors are the same as in Figure 2. (C) and (D) Cartoon representations of the TZF of TIS11d and the TZF12 of MBNL2, respectively. Each circle represents a CCCH-type zinc finger motif, and the black arrows represent the zinc finger orientation corresponding to the direction of the first helix in each of the domains. The red arrows in (C) indicate the 5′–3′ direction of the RNA strand in the TZF complex in TIS11d, and those in (D) indicate the direction of the putative accommodated RNA on the TZF12 of MBNL2. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
In contrast to TIS11d, the N-terminal extensions of the CCCH-type zinc fingers in MBNL2 (the LEV box for ZF1 and ZF3, as well as the VXh (hydrophobic) box for ZF2 and ZF4, where X stands for any amino acid) are located far away from their own CCCH zinc finger bodies. Instead, both boxes are involved in the formation of the “inter-zinc-finger” β-sheet in the TZF domains. Moreover, the hHP box, just located at the fourth His ligand residue, is also characteristic of the MBNL proteins, and the hHP box of ZF1 and that of ZF2 interact with each other. These two boxes contain the key residues contributing to the hydrophobic interactions of the TZF domains, as described earlier.
A comparison of the amino acid sequences among several CCCH-type ZF domains, which constitute the tandem ZF domains, revealed that only the fifth ZF domain in the CPSF4 protein contains the Pro residues just after the fourth His ligand residue. However, even for the fifth ZF domain of CPSF4, the segment corresponding to the LEV or Vxh box in the ZF domains of MBNL2 has the RVI sequence. Thus, it seems unusual that the fifth ZF domain of the CPSF4 forms the same structure as those of the TZF domains in MBNL2, as the first position of the segment must be occupied by a hydrophobic amino acid residue for the formation of the characteristic TZF structure.
RNA binding site formed by the two consecutive CCCH-type zinc fingers in TZF
As described above, we can predict the RNA binding site on each of the ZF domains of MBNL2, on the basis of the structural data for the TIS11d complex [Figs. 2(D,E), 3(D,E), and 4(G,J,M,N)]. In addition, we identified the putative RNA binding sites that are formed by the interactions between the two ZF domains.
The additional C-terminal helix plays an important role in stabilizing the TZF12 structure, as described in the Results. At the same time, the α-helix is involved in the formation of the third pocket on ZF1, with the conserved key aromatic residue (position 0) in ZF1. Namely, the third pocket at ZF1 of TZF12 is formed by Lys35, Phe36 from ZF1, and Gln78 residue from the α-helix at the C-terminal end of ZF2 [Figs. 2(D,E) and 4(H,I)]. In addition, the pocket corresponding to the third pocket observed in TIS11d is formed on ZF2 by Lys67, Tyr68 from ZF2, and Gln44 in the linker region (Ser42-Asn47) between ZF1 and ZF2 [Figs. 2(D,E) and 4(H,I)]. This implies that each of the CCCH-type zinc fingers in TZF12 requires amino acid residues from the other zinc finger domain as well as its own key aromatic amino acid residues for the formation of the binding pocket. This also holds true for the TZF34 domain [Figs. 3(D,E) and 4(H,I)].
Furthermore, the positively charged residues located in the N-terminal segments of ZF1 and ZF3 are close to the binding surface of the other CCCH zinc finger motif in the TZF domains (ZF2 and ZF4), respectively. They constitute the positively charged patch with the residue (K/R) three positions before the third Cys ligand residue and the residue (K/R) following the third Cys ligand residue in ZF2 and ZF4. This patch may provide an additional region for their partner interactions with RNA molecules [Figs. 2(C–E) and 3(C–E)].
Intriguingly, as the linker between the two zinc finger motifs of the TZF domain is not flexible, the two predicted binding sites are fixed on opposite sides of the compact global fold of TZF12 and TZF34 in antiparallel directions, as mentioned above (see Fig. 5). This suggests that each of the ZF domains in the TZF domain may work independently, and thus they could bind the two single-stranded RNAs, which are aligned in opposite directions to each other. The double-stranded RNA with pyrimidine–pyrimidine mismatches is reportedly the target of MBNL proteins.11,21 Thus, we propose the hypothesis that, upon binding to the TZF domain, the double-stranded structure becomes untwined, and each of the single-stranded RNAs is recognized by the TZF domain [Fig. 5(D)]. Whether the TZF domain binds to single-stranded RNA in a similar manner as TZF of TIS11d does or binds to double-stranded RNA in a hypothetical manner or both remains to be determined by future structural experiments.
Materials and Methods
NMR sample preparation
Protein samples of the TZF12 and TZF34 domains of human MBNL2 were produced by using an E. coli based cell-free protein synthesis system.22 For the structure determination, the uniformly 13C- and 15N-labeled proteins (about 1.1 mM) were prepared in a solution consisting of 20 mM2H-Tris-HCl buffer, 100 mM NaCl, 1 mM dithiothreitol (DTT), 0.02% (w/v) NaN3, 0.05 mM ZnCl2, 1 mM IDA, and 10% (v/v) 2H2O at pH 7.0. The engineered TZF12 and TZF34 regions span residues Pro7-Asn82 and Ser167-Ala257 of human MBNL2, respectively.
NMR spectroscopy
All NMR experiments were carried out at 298 K on Bruker AVANCE spectrometers equipped with triple axes gradients, operating at 600 and 800 MHz, with the 13C- and 15N-doubly labeled samples in Shigemi susceptibility-matched tubes. The 1H, 15N, and 13C chemical shifts were referenced to the frequency of the 2H lock resonance of water, and the 1H chemical shift for water was determined by measuring the difference between referencing on water and referencing on DSS in a very dilute DSS solution, under spectrometer conditions identical to those used for the measurements of the TZF domains. A series of 2D and 3D standard triple resonance NMR experiments were recorded.23 2D [1H,15N]-HSQC, and 3D HNCO, HN(CA)CO, HNCA, HN(CO)CA, HNCACB, and CBCA(CO)NH spectra were used for sequence-specific backbone assignments. Side-chain assignments were obtained using 2D [1H,13C]-HSQC, and 3D HBHA(CO)NH, H(CCCO)NH, (H)CC(CO)NH, HCCH-COSY, HCCH-TOCSY, and (H)CCH-TOCSY spectra. 3D 15N- and 13C-edited NOESY-HSQC spectra were recorded with an 80 ms mixing time for collecting NOE restraints. These 3D NOESY spectra were also used to check the chemical shift assignments for consistency. The tautomeric state of the histidine ring was determined with a 2D heteronuclear multiple quantum coherence (1H-15N HMQC) experiment optimized for histidine side-chains.18 All NMR data were processed with the NMRPipe program.24 Linear prediction and zero filling were applied in the indirect dimensions to achieve better resolution. The programs NMRView25 and Kujira26 were used for NMR spectra analysis.
Structure calculations
NOE data from 15N and 13C-edited 3D NOESY spectra were used for the structure calculations. The NOESY peak lists were generated automatically with manual checking, and the peak volumes were determined by the automatic integration function of NMRView.25 The three-dimensional structure was determined by combined automated NOESY cross peak assignment.27 and structure calculation with torsion angle dynamics,28 implemented in the CYANA program.19 The standard CYANA protocol of seven iterative cycles of NOE assignment and structure calculation, followed by a final structure calculation, was applied. Dihedral angle restraints for φ and ψ were obtained from the main chain and 13Cβ chemical shift values, using the program TALOS.29 In the final refinement stage, distance restraints were added for Zn-Sγ (2.25-2.35 Å) and Zn-Nɛ2 (1.95-2.05 Å)30,31 and for the distances between the four zinc coordinating atoms, to ensure tetrahedral zinc coordination geometry. Experimental data and structural statistics are summarized in Table I. The quality of the solution structure was evaluated using PROCHECK-NMR.20 Structural figures were prepared with the MOLMOL program.32
Protein data bank accession numbers
The best 20 structures calculated by CYANA of the TZF12 and ZTF34 domains of human MBNL2 have been deposited in the Protein Data Bank (PDB entries 2YXI and 2E5S, respectively).
Acknowledgments
The authors are grateful to Yasuko Tomo, Masaomi Ikari, and Kazuharu Hanada for their help in sample preparation.
Glossary
Abbreviations
- HSQC
heteronuclear single quantum coherence
- MBNL
muscleblind-like
- MBNL2
MBNL protein 2
- NOE
nuclear Overhauser effect
- NOESY
NOE spectroscopy
References
- 1.Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. Embo J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pascual M, Vicente M, Monferrer L, Artero R. The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation. 2006;74:65–80. doi: 10.1111/j.1432-0436.2006.00060.x. [DOI] [PubMed] [Google Scholar]
- 3.Begemann G, Paricio N, Artero R, Kiss I, Perez-Alonso M, Mlodzik M. Muscleblind, a gene required for photoreceptor differentiation in Drosophila, encodes novel nuclear Cys3His-type zinc-finger-containing proteins. Development. 1997;124:4321–4331. doi: 10.1242/dev.124.21.4321. [DOI] [PubMed] [Google Scholar]
- 4.de Andrade GR, Jansen RP. Moving with Muscleblind. Nat Cell Biol. 2005;7:1155–1156. doi: 10.1038/ncb1205-1055. [DOI] [PubMed] [Google Scholar]
- 5.Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. Embo J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, Henderson D, Schalling M, Swanson MS, Thornton CA. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum Mol Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- 7.Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, Brook JD. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum Mol Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- 8.Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 9.Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T, Sohn R, Zemelman B, Snell RG, Rundle SA, Crow S, Davies J, Shelbourne P, Buxton J, Jones C, Juvonen V, Johnson K, Harper PS, Shaw DJ, Houseman DE. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 10.Mahadevan M, Tsilfidis C, Sabourin L, Shutler G, Amemiya C, Jansen G, Neville C, Narang M, Barcelo J, O'Hoy K, Leblond S, Earle-Macdonald J, de Jong PJ, Wieringa B, Korneluk RG. Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- 11.Warf MB, Berglund JA. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007;13:2238–2251. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan Y, Compton SA, Sobczak K, Stenberg MG, Thornton CA, Griffith JD, Swanson MS. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007;35:5474–5486. doi: 10.1093/nar/gkm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 14.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol. 2004;11:257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 15.Schubert M, Labudde D, Oschkinat H, Schmieder P. A software tool for the prediction of Xaa-Pro peptide bond conformations in proteins based on 13C chemical shift statistics. J Biomol NMR. 2002;24:149–154. doi: 10.1023/a:1020997118364. [DOI] [PubMed] [Google Scholar]
- 16.Wüthrich K. NMR of proteins and nucleic acids. New York: Wiley; 1986. [Google Scholar]
- 17.Lee MS, Palmer AG, 3rd, Wright PE. Relationship between 1H and 13C NMR chemical shifts and the secondary and tertiary structure of a zinc finger peptide. J Biomol NMR. 1992;2:307–322. doi: 10.1007/BF01874810. [DOI] [PubMed] [Google Scholar]
- 18.Pelton JG, Torchia DA, Meadow ND, Roseman S. Tautomeric states of the active-site histidines of phosphorylated and unphosphorylated IIIGlc, a signal-transducing protein from Escherichia coli, using two-dimensional heteronuclear NMR techniques. Protein Sci. 1993;2:543–558. doi: 10.1002/pro.5560020406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Güntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 20.Laskowski RA, Rullmann JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 21.Goers ES, Voelker RB, Gates DP, Berglund JA. RNA binding specificity of Drosophila muscleblind. Biochemistry. 2008;47:7284–7294. doi: 10.1021/bi702252d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kigawa T, Yabuki T, Matsuda N, Matsuda T, Nakajima R, Tanaka A, Yokoyama S. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J Struct Funct Genomics. 2004;5:63–68. doi: 10.1023/B:JSFG.0000029204.57846.7d. [DOI] [PubMed] [Google Scholar]
- 23.Clore GM, Gronenborn AM. Multidimensional heteronuclear nuclear magnetic resonance of proteins. Methods Enzymol. 1994;239:349–363. doi: 10.1016/s0076-6879(94)39013-4. [DOI] [PubMed] [Google Scholar]
- 24.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 25.Johnson BA, Blevins RA. NMR view: a computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi N, Iwahara J, Koshiba S, Tomizawa T, Tochio N, Güntert P, Kigawa T, Yokoyama S. KUJIRA, a package of integrated modules for systematic and interactive analysis of NMR data directed to high-throughput NMR structure studies. J Biomol NMR. 2007;39:31–52. doi: 10.1007/s10858-007-9175-5. [DOI] [PubMed] [Google Scholar]
- 27.Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J Biomol NMR. 2002;24:171–189. doi: 10.1023/a:1021614115432. [DOI] [PubMed] [Google Scholar]
- 28.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 29.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 30.Summers MF, Henderson LE, Chance MR, Bess JW, Jr, South TL, Blake PR, Sagi I, Perez-Alvarado G, Sowder RC, III, Hare DR, Arthur LO. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1992;1:563–574. doi: 10.1002/pro.5560010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang B, Alam SL, Meyer HH, Payne M, Stemmler TL, Davis DR, Sundquist WI. Structure and ubiquitin interactions of the conserved zinc finger domain of Npl4. J Biol Chem. 2003;278:20225–20234. doi: 10.1074/jbc.M300459200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]


