Abstract
Susceptibility of methionine residues to oxidation is a significant issue of protein therapeutics. Methionine oxidation may limit the product's clinical efficacy or stability. We have studied kinetics of methionine oxidation in the Fc portion of the human IgG2 and its impact on the interaction with FcRn and Protein A. Our results confirm previously published observations for IgG1 that two analogous solvent-exposed methionine residues in IgG2, Met 252 and Met 428, oxidize more readily than the other methionine residue, Met 358, which is buried inside the Fc. Met 397, which is not present in IgG1 but in IgG2, oxidizes at similar rate as Met 358. Oxidation of two labile methionines, Met 252 and Met 428, weakens the binding of the intact antibody with Protein A and FcRn, two natural protein binding partners. Both of these binding partners share the same binding site on the Fc. Additionally, our results shows that Protein A may serve as a convenient and inexpensive surrogate for FcRn binding measurements.
Keywords: methionine oxidation, human IgG2 antibody, protein A chromatography, protein A binding, FcRn binding
Introduction
Therapeutic proteins can undergo a variety of degradation processes, including aggregation, deamidation, isomerization, oxidation, disulfide bond scrambling, and truncation. These events can occur during fermentation, purification, formulation, manufacturing, and storage. The product's clinical utility or shelf-life can be compromised if degradation adversely affects the biological activity of the molecule or induces immunogenicity. One of the most common modifications is oxidation of labile amino acid side chains, which is induced by reactive oxygen species.1 Although protein oxidation can occur at cysteine, tryptophan, lysine and other amino acids,2–4 methionine is often the most susceptible residue to oxidation.5,6 The most common product of methionine oxidation is methionine sulfoxide, which is more polar and less hydrophobic than methionine. Susceptible methionines are typically located on the surface of the protein and exposed to the solvent.7–10 Met oxidation can have adverse effects on proteins, including decreased stability,5,11–16 and decreased biological activity.5,8,12,17–20
The immunoglobulin gamma (IgG) monoclonal antibody (mAb) has emerged as one of the most promising drug class in the biopharmaceutical industry. More than 20 antibody drug products have been approved by the FDA for human therapeutic use across many clinical settings.21,22 The Fab portion of antibodies is responsible for drug specificity, whereas the Fc portion is responsible for the effector functions and modulation of the half-life of the antibody.23,24
The Fc portion is a homodimer composed of the C-terminus of the heavy chain, including a short hinge region, the CH2, and CH3 domains. The sequences of CH2 and CH3 domains in human IgG1 and IgG2, two widely used therapeutic mAb subtypes, are shown in Figure 1(A). The Fc regions of human IgG1 and IgG2 are nearly identical, with sequence homology as high as 97%. The human IgG2 Fc has four methionines, Met 252, Met 358, Met 397, and Met 428. Valine replaces methionine at position 397 in human IgG1 Fc. Met 252 is located in the CH2 domain, whereas the other methionines are located in the CH3 domain. The crystal structure of human IgG1 Fc is shown in Figure 1(B). Met 252 is located at the end of a short α-helix structure, which is a part of the CH2-CH3 interface. Met 358 is located in a short α-helix, whereas Met 397 is presumably located in a β-sheet since Val occupies this position in the human IgG1 Fc. These two methionines are located in the CH3-CH3 interface between the two Fc polypeptide chains. Similar to Met 252, Met 428 is located at the CH2-CH3 interface, which is the consensus binding site that interacts with several natural proteins including Staphylococcal Protein A,25 and FcRn.26
Figure 1.
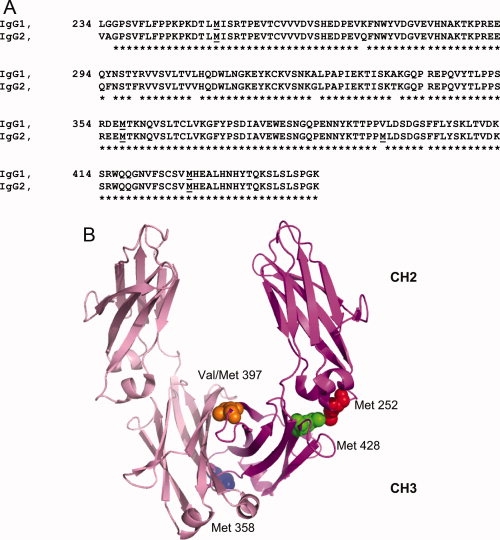
A: Alignment of the amino acid sequences of human IgG1 and IgG2 Fc. Residues are numbered using the EU numbering system. Methionine residues are underlined. B: The cartoon representation of the Fc. The crystal structure of human IgG1 Fc (pdb code:1FC1) was used to prepare the figure. Met 252 was colored red. Met 358 was colored blue. Val 397, which is equivalent to Met 397 in human IgG2, was colored orange and Met 428 was colored green. The sphere presentations of these residues are only shown in one peptide chain of the Fc. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]:
Protein A is a 42 kDa membrane protein in the cell wall of Staphylococcus aureus, consisting of five homologous domains.27 Binding studies between Protein A and antibodies have shown that all five domains of Protein A bind to the IgG Fc domain.28 Because of the high specificity, Protein A affinity chromatography is a common technique for the purification of antibody or Fc fusion proteins.
FcRn mediates transport of maternal IgG across the placenta in humans, thereby conferring humoral immunity to the newborn against antigens encountered by the mother.29 In addition, FcRn protects IgG from degradation by diverting endogenous IgG from lysosomes.30 IgGs that bind to FcRn in endosomes at acidic pH (pH 6–6.5) after fluid phase endocytosis are recycled back to the cell surface where they are released at the neutral pH (pH ∼ 7.4).31 In contrast, IgGs that do not bind to FcRn are degraded in the lysosome. The half-life of IgG is reduced from 5 days to about 1 day in FcRn-deficient mice.32 The relative longer half-life is one of the major advantages of antibody therapeutics.33
Several studies on methionine oxidation in the Fc have been reported. Shen et al.34 studied the susceptibility of methionines in the humanized monoclonal IgG1 antibody HER2 by incubation with tert-butyl hydroperoxide (TBHP). Chumsae et al.35 reported that Met 252 and Met 428 are the two most susceptible methionines, based on thermal stability studies on a fully human IgG1 antibody. Nearly one third of Met 258 and Met 432 were spontaneously oxidized after incubation in the formulation buffer at 25°C for 12 months. Liu et al.36 reported that detectable changes in secondary and tertiary structures have been observed by CD and NMR upon oxidation of Met 252 and Met 428. The melting temperature of the CH2 domain was significantly reduced, and the aggregation and deamidation rates at Asn 286 and Asn 315 were increased. Although extensive biochemical and biophysical characterizations have been performed on methionine oxidation in the Fc region, the potential changes in the binding of natural partners, such as Protein A and FcRn, have not been studied.
In our studies, we evaluated the impact of the oxidation of methionine residues in the Fc region on human IgG2 binding to Protein A and FcRn. The similarity in the impact of methionine oxidation on the binding of these two molecules to the antibody, and the implication of potential changes in the antibody serum half-life will be discussed.
Results and Discussion
Oxidation susceptibility of methionines in human IgG2 Fc
The relative oxidation rate for each methionine in human IgG2 Fc was determined by the forced oxidation study. The antibody was incubated with 1% TBHP for various time periods. The reaction was stopped by diluting the sample 100 fold into ice-cold acetate buffer, quickly followed by diafiltration to remove the TBHP. The oxidized samples were subjected to the peptide mapping analysis. In this study, a protein concentration of 0.4 mM was used, whereas the TBHP concentration was at 110 mM, resulting in a TBHP to antibody molar ratio of ∼270. A nonreduced Lys-C peptide map was performed to monitor the oxidation reaction. An expanded overlay of peptide maps of the untreated sample and the treated sample that has been incubated with TBHP for 4 h is shown in Figure 2. In the peptide map of the untreated sample, four methionine containing peptides, D249-K288/C321-K322, G341-K360, T393-K409, and N361-K370/S415-K439 (“/” represents one disulfide bond), were identified by the MS/MS analysis. After the incubation with TBHP for 4 h, peak intensities of these peptides were reduced and +16 Da species eluting earlier were observed. The MS/MS analysis shows that the +16 Da species are the oxidized forms, with methionines being oxidized to the sulfoxide. The peak intensities of the nonoxidized forms were continuously decreased as the incubation time increased, whereas the peak intensities of the oxidized forms were increased. Chemical modifications other than methionine oxidation were not observed in the peptide mapping analysis.
Figure 2.
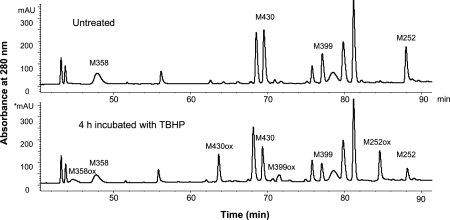
Peptide maps for nonreduced Lys-C digests for the untreated IgG2 and treated IgG2 that has been incubated with TBHP for 4 h.
Figure 3(A) shows the time course of the oxidation reaction for each methionine from time zero to 6 h. The reaction kinetics was fitted to an exponential decay equation to obtain the oxidation rate constants. The result (R2 > 0.99) indicates that the oxidation is a pseudo-first order reaction. Three different oxidation rates were observed. Met 252 oxidizes with the fastest rate at 0.19 h−1. Met 428 oxidizes with the second fastest rate at 0.09 h−1, which is about half of the Met 252 oxidation rate. Met 358 and Met 397 have the slowest oxidation rates, which are 0.05 h−1 and 0.04 h−1, respectively.
Figure 3.
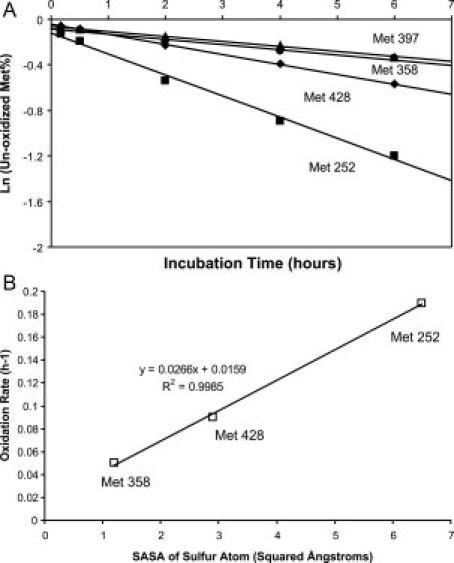
A: Kinetics of methionine oxidation. Oxidation rates of each methionine (square for Met 252, diamond for Met 428, circle for Met 358, and triangle for Met 397) were determined from the slopes of the linear fits through the data. B: The correlation between oxidation rates and solvent accessible surface area of sulfur atom for each methionine.
The solvent accessible surface areas (SASAs) of the sulfur atoms in the Met 252, Met 358, and Met 428 are 6.5, 1.2, and 2.9 Å2, respectively.37 These values are calculated from the crystal structure of IgG1 Fc (1FC1), since the structure of human IgG2 Fc is not available. The SASA data for Met 397 is not available, since IgG1 Fc has Valine at position 397. The observed oxidation rates correlate well with the SASAs of the sulfur atoms [Fig. 3(B)]. The sulfur atom of Met 358 is deeply buried inside of the CH3-CH3 interface between two Fc peptide chains, so it has a slower oxidation rate compared with Met 252, which has a sulfur atom that is largely exposed to solvent. Data obtained from this study is in general agreement with what has been reported previously for recombinant monoclonal antibodies. Shen et al.34 reported that Met 358 oxidizes much slower than Met 252 and Met 428 in TBHP-treated rhμMab HER2, a humanized IgG1 antibody. Liu et al.36 observed that Met 252 oxidizes two-fold faster than Met 428 in the E.coli expressed human IgG1 Fc treated by H2O2. Our results support the contention that Met 252 and Met 428 are more exposed to the solvent than Met 358 and Met 397, and are therefore more susceptible to oxidation.
Methionine oxidation in Fc decreases the binding affinity to protein A
Protein A affinity chromatography is a well established technique for antibody purification.38,39 In the typical “bind and elute” mode, Protein A tightly binds the Fc portion of the antibody under neutral pH conditions (pH 7–8), while impurities are washed away, after which the acidic pH buffer (pH 3–4) is quickly introduced to release the antibody from the Protein A column. To exploit potential differences in affinity between oxidized variants of IgG2 antibody and Protein A, we have developed a novel method to separate antibody structural variants using pH gradient elution. In this application, a pH-gradient is generated by mixing a neutral pH buffer and an acidic pH buffer to elute the IgGs bound to the Protein A column. The untreated and TBHP treated samples from the forced oxidation were analyzed by this technique, and the chromatograms are shown in Figure 4. The untreated IgG2 was resolved into two peaks, a minor prepeak A and a main peak. The level of prepeak A in the untreated IgG2 is 12.1%. After 2 h of incubation with TBHP, the prepeak A was increased to 67.7%. Prepeak B and prepeak C, which elute earlier than the prepeak A, appeared after incubation with TBHP for 6 and 24 h, respectively. No new peaks were observed from incubation times exceeding 24 h. The pH gradient was superimposed in Figure 4. A small fraction of eluent was collected every 2 min and was measured by pH meter. The elution pH for the main peak, prepeak A, prepeak B, and prepeak C were determined to be 4.32, 4.50, 4.62, and 4.95, respectively. The differences of the pHs for these peaks to elute are quite small, averaging 0.2 pH unit apart.
Figure 4.
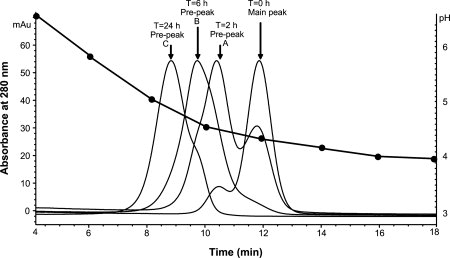
pH-gradient Protein A chromatograms of an IgG2 antibody treated by TBHP for various time periods. The pH gradient was superimposed. Each filled circle represents the pH value of the eluent collected in every 2 min.
To characterize the prepeaks and the main peak species, nonreduced Lys-C peptide mapping analysis was performed. Prepeak A and the main peak were collected from the untreated IgG2 sample and were further concentrated and buffer-exchanged. Prepeak B and prepeak C were prepared by buffer-exchanging samples after incubation with TBHP for 6 and 24 h. The percentages of oxidation for each methionine in IgG2 antibody were quantified by the peak areas under the nonoxidized and oxidized peaks in the peptide maps, as shown in Table I. Our experiments show that the main peak of IgG2 has a low level of oxidation of all methionine residues ranging from 2.9 to 5.4%. This could be attributed to the manufacturing process or the artifact of sample handling. In the prepeak A, two methionines, Met 252 and Met 428, show 46.5 and 30% oxidation, respectively. Interestingly, the other two methionines, Met 358 and Met 397, have virtually the same low oxidation level as in the main peak. No significant differences for other chemical modifications were observed for these two samples by peptide mapping analysis. In prepeak B, the oxidation levels in Met 252 and Met 428 are 69.7 and 43.4%, respectively. The other two buried methionines, Met 358 and Met 397, were partially oxidized. In prepeak C, Met 252 and Met 428 were nearly fully oxidized, reaching 95.2 and 82.0%, respectively. The buried methionines also reached high level of oxidation. Our data show that the multiple prepeaks observed in the pH-gradient Protein A chromatography result from oxidation of the methionine residues, specifically Met 252 and Met 428, in both Fc peptide chains.
Table I.
Oxidation Level of Four Fc Methionines in the Prepeaks and Main Peak of a Human IgG2 Antibody Separated by pH-Gradient Protein A Chromatography
| % Met 252 | % Met 358 | % Met 397 | % Met 428 | |
|---|---|---|---|---|
| Main Peak | 2.9 | 5.4 | 3.4 | 4.6 |
| Prepeak A | 46.5 | 5.3 | 3.0 | 30.0 |
| Prepeak B | 69.7 | 28.3 | 23.2 | 43.4 |
| Prepeak C | 95.2 | 63.9 | 66.7 | 82.0 |
To quantitatively assess the impact of Fc methionine oxidation on Protein A binding, a Surface Plasmon Resonance (SPR) binding experiment was performed on three samples with different oxidation levels, which include the main peak, prepeak A and prepeak C. The main peak has virtually no methionine oxidation. Prepeak A has partial oxidation of Met 252 and Met 428, whereas prepeak C has nearly full oxidation of Met 252 and Met 428 plus partial oxidation of Met 358 and Met 397. Samples with five concentrations ranging from 666 to 8.2 nM were analyzed on the Biacore chip that contains immobilized Protein A. The sensorgrams shown in Figure 5(A) demonstrate different binding kinetics with Protein A for the main peak fraction, prepeak A, and prepeak C. The association and dissociation rates fitted from these curves are shown in Table II. The prepeak A associates with Protein A slower than the main peak, and prepeak C associates with Protein A even slower than prepeak A. For the dissociation of the antibody and Protein A complex, prepeak A dissociates three-fold faster than the main peak and prepeak C showed three-fold faster dissociation rate than the prepeak A. The dissociation equilibrium constants (KD) were calculated from the association and dissociate rates (ka and kd) as shown in Table II. Prepeak A (KD = 3 nM) has lower affinity than the main peak fraction (KD = 1 nM), whereas the prepeak C, which is nearly fully oxidized at Met 252 and Met 428, has the lowest affinity (KD = 10 nM). Since no differences were found in the oxidation level of Met 358 and Met 397 in main peak and prepeak A, it is likely that only Met 252 and Met 428 are responsible for the change of binding kinetics with Protein A, thereby affecting the binding affinity. The results demonstrate that oxidation of Met 252 and Met 428 reduces the affinity of the antibody with Protein A. Our data are in general agreement with the SPR analyses of a human IgG1 antibody.40 Full oxidation of Met 252 and Met 428 resulted in about two-fold increase in the association rate and 10-fold increase in the dissociation rate of the antibody and Protein A, which led to a 4.2-fold increase in the dissociation equilibrium constant (KD).
Figure 5.
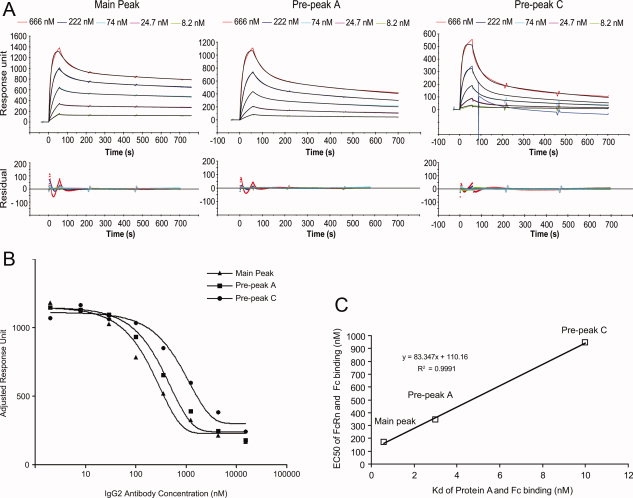
A: The sensorgrams (colored lines) of the main peak, prepeak A, and prepeak C binding with Protein A immobilized on the Biacore chip. In each panel, the fitted curves are colored black. A spike was observed in the sensorgram of injecting 222 nM prepeak C to the Biacore chip probably due to an air bubble. The curve of this sensorgram was fitted before the spike and the residuals weren't shown after the spike. B: The competition binding curves of the main peak, prepeak A, and prepeak C with FcRn. C: The correlation between Kd of Fc and Protein A and the EC50 value of Fc and FcRn for main peak, prepeak A, and prepeak C. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]:
Table II.
Association Rate, Dissociation Rate, and Dissociation Equilibrium Constant of Main Peak, Prepeak A, and Prepeak C Binding with Protein A Measured by Biacore Binding Experiment
| kon (1/M*s) with Protein A | koff (1/s) with Protein A | Kd (nM) with Protein A | EC50 (nM) with FcRn | |
|---|---|---|---|---|
| Main Peak | 5.35 ± 0.06 e5 | 3.36 ± 0.18 e-4 | 0.6 | 170 (134–216) |
| Prepeak A | 3.76 ± 0.09 e5 | 1.02 ± 0.03 e-3 | 3.0 | 347 (312–386) |
| Prepeak C | 3.25 ± 0.24 e5 | 3.18 ± 0.09 e-3 | 10.0 | 947 (590–1520) |
The values represent mean ± standard error calculated from the BIAevaluation software. EC50 of main peak, prepeak A, and prepeak C binding with FcRn measured by a Biacore competitive assay is also included with 95% confidence intervals shown in the brackets.
The interaction between the IgG1 Fc and Protein A has been studied in detail by X-ray crystallography and 13C NMR.41,42 The interaction between Protein A and Fc are primarily hydrophobic interactions, involving many residues from the interface between the CH2 and CH3 domains.41 One key hydrophobic interaction between IgGs and Protein A involves Met 252. Met 252, which is located within a short α-helical structure of the CH2 domain, is a highly conserved residue in the CH2-CH3 interface. This residue makes a very close contact with two residues in Protein A, Phe 124 and Gln 129. The distances between the sulfur atom in Met 252 and several atoms (one nitrogen and four carbon atoms) in Phe 124 in Protein A are all less than 4 Å.37 An electron rich oxygen added to the sulfur is very likely to disturb this binding pocket. Met 428 is located within the CH3 domain and is another conserved residue in the CH2-CH3 interface. Although Met 428 is not directly involved in the binding, it is in the proximity of Met 252 and His 435, which are two critical residues in the interaction between Protein A and the Fc. The side chains of Met 252, His 435 and Met 428 are tightly packed against each other. His 435 interacts with a histidine (His 137) on Protein A28 at low pH (pH 3), generating electrostatic repulsion and overcoming the hydrophobic interaction between the Fc and Protein A. Addition of oxygen to Met 428 is likely to generate unfavorable interactions and affect these residues, resulting in decreased affinity between Protein A and Fc. Met 358 and Met 397 are not close to the binding sites, so they likely have minimal impact on the binding between the Fc and Protein A.
Methionine oxidation in Fc decreases the binding affinity of FcRn
The same set of samples was also evaluated for their binding affinity to the FcRn by a competition binding assay. A fixed amount of human FcRn was incubated with various amounts of samples for one hour before injection over the Biacore chip containing immobilized human Fc. A decreased binding response with increasing concentrations of antibody indicated that antibody binds to FcRn in solution, which blocks FcRn from binding to the immobilized human Fc surface. The competitive binding curves for main peak fraction, prepeak A and prepeak C shown in Figure 5(B) illustrate that they bind to FcRn with different affinities. The EC50 values and the 95% confidence intervals for antibody binding with FcRn calculated from the binding curves are shown in Table II. The EC50 value of the prepeak A is twice of the EC50 value of the main peak. Extensive methionine oxidation induced by TBHP in prepeak C increases the EC50 value by five-fold. These results indicate that oxidation of Fc methionines significantly decreases the affinity between the antibody and FcRn.
The FcRn interaction site on IgG has been mapped by X-ray crystallography and many mutation studies.26,43–51 The sharp pH dependence of FcRn/Fc interaction is attributed to the titration of two conserved histidines in human Fc.45 In addition to the charge interactions, the center of the Fc/FcRn interface includes a core of hydrophobic residues including Met 252, which is surrounded by several key residues, such as Leu 251 and Ile 253, which play critical roles in the hydrophobic interactions between Fc and FcRn. The NMR signal of Leu 251 disappears upon the oxidation of Met 252, suggesting that it underwent a structural change.36 Oxidation of Met 252 is likely to disturb these hydrophobic interactions. Met 252 also makes direct contacts with Pro 134 and Glu 135 in FcRn. The distances between each pair are all close to 4 Å, calculated from the co-crystal structure of human IgG1 Fc and Rat FcRn. Although Met 428 is not directly located in the binding pocket, it interacts with His 435, one of the two histidine residues involved in the pH-dependent binding.45 HSQC NMR signal of His 435 also disappeared upon the oxidation of Met 428.36 Oxidation of Met 428 may perturb the charge interaction between Fc and FcRn.
As FcRn is responsible for the extended persistence of IgG in the serum, modulation of the FcRn-IgG interaction has become an area of active research focused on extending the pharmacokinetics of antibodies by engineering the FcRn/Fc interface.31 Several studies have demonstrated a correlation between the binding affinity of IgG mAbs to FcRn and their serum half-lives in mice and primates.48–52 Since oxidation of Met 252 and Met 428 have been shown to decrease the FcRn binding affinity at pH 6 while maintaining the dissociation at neutral pH, the oxidized IgGs may display attenuated pharmacokinetics. Our results warrant a further study to explore whether methionine oxidation in the Fc changes the serum half-life of human IgGs. Careful consideration must be taken to eliminate other factors such as other biochemical and biophysical differences in the antibodies tested.
Similarity in the protein A/Fc and FcRn/Fc interactions
The Protein A/Fc and FcRn/Fc crystal structures reveal that both Protein A and FcRn bind to the interface of the CH2 and CH3 domains, a versatile region of the Fc that also binds to several other natural proteins, such as Protein G and rheumatoid factor.53,54 The residues in the Fc involved in the hydrophobic interactions and charge interactions with Protein A and FcRn are nearly identical, including three patches on IgG Fc, Met 252 to Ser 254, Leu 309 to Gln and Asn 434 to Tyr 436. Although Protein A doesn't form salt bridges with the Fc, resulting in less sharp pH-dependence, the interaction with the Fc is still dependent upon titrating two key histidine residues, His 310 and His 435, in human IgG Fc.
The binding equilibrium constant (Kd) of Protein A and Fc correlates well with the EC50 value of FcRn and Fc for samples with different oxidation levels [Fig. 5(C)]. The partially oxidized IgG2 antibody (prepeak A) has a five-fold decrease of Protein A binding affinity and two-fold decrease of FcRn binding affinity compared with nonoxidized antibody (main peak). The fully oxidized antibody (prepeak C) has a three-fold decrease of Protein A binding affinity and also has a three-fold decrease of FcRn binding affinity compared with partially oxidized antibody (prepeak A). These observations demonstrate that the methionine oxidation in Fc has similar impact to the binding affinity with both Protein A and FcRn. Since the Protein A mimics the interaction between Fc and FcRn, the Protein A binding can potentially serve as a convenient and inexpensive surrogate for the FcRn binding to assess functionality of the therapeutic IgGs.
Materials and Methods
Materials
The recombinant monoclonal antibodies used in this study were expressed in Chinese Hamster Ovary cells and purified using conventional manufacturing process steps. The antibodies were prepared in 10 mM sodium acetate, pH 5.2. TBHP solution (70%) was purchased from Alfa Aesar. All other reagents were purchased from Sigma (St. Louis, MO) unless otherwise specified.
pH-gradient protein A chromatography
The POROS A/20 Protein A column was purchased from Applied Biosystems (Foster City, CA). The chromatographic system is an Agilent 1100 HPLC equipped with a diode-array detector, autosampler, and micro-flow cell (Agilent, Palo Alto, CA). UV absorbance was monitored at 280 nm. The buffers are (A) 20 mM Tris, 150 mM NaCl, pH 7.0 ± 0.1 and (B) 20 mM Acetate, 150 mM NaCl, pH 3.1 ± 0.1. The antibody was injected on the Protein A column after it was equilibrated at 0% B for at least 20 min. A linear gradient from 0 to 35% B was run over 22 min. The flow rate was 2 mL/min. The column was kept at room temperature. The antibody was diluted to 2 mg/mL before the injection and 100 μg of antibody was injected into the column. Roughly 1 mL of eluent was collected offline in every 2 min and was measured by pH meter.
Forced oxidation by TBHP
The stock TBHP solution (∼70%) was diluted to 10% with 10 mM sodium acetate buffer (pH 5.2). An aliquot of 10% TBHP solution was added to the antibody solution (70 mg/mL) with a volume ratio of 1:9 to make the final TBHP concentration at 1%. The mixture was incubated at 37°C. During the incubation, several aliquots were removed from the mixture at various time intervals. The aliquots were diluted 100-fold into ice-cold acetate buffer (10 mM sodium acetate buffer, pH 5.2). The solutions were quickly buffer exchanged three times with the same acetate buffer using a Vivaspin 20 (membrane cut-off at 10,000 MW, Vivascience).
Lys-C enzymatic digestion
The concentration of the antibody samples was diluted to 13 mg/mL with 8M guanidine hydrochloride, 0.5M Na2HPO4, 0.5M NaH2PO4, 20 mM NEM to a total volume of 100 μL. The solution was then placed at 37°C for 2 h to denature the antibody. Lys-C digestion was performed for 24 h at 37°C by mixing 15 μL denatured antibody with 85 μL digestion buffer (8M urea, 0.2M phosphate, 40 mM NH2OH, pH 7.0), 90 μL H2O and 10 μg Lys-C (Waco Chemicals). The digests were quenched with 15 μL of 5% TFA and stored at −70°C.
LC/MS of Lys-C digests
The Lys-C digests were separated on a Jupiter C18 column (250 mm × 2.0 mm, Phenomenex, CA) using a two step linear gradient from 2 to 22% B over 38 min and then from 22 to 42% over 80 min. Solvent A was 0.1% TFA in water, and solvent B contained 0.1% TFA, 90% acetonitrile (Burdick & Jackson, MI) and 10% water. The column temperature was maintained at 50°C. The flow rate was 0.2 mL/min, and a total of 50 μg of protein digest was injected onto the column for analysis. Agilent 1100 HPLC equipped with a diode-array detector, autosampler and binary pumps (Agilent, Palo Alto, CA) was used to separate the peptides. The UV wavelength was set at 215 nm. The HPLC was directly coupled to a Finnigan LCQ Deca ion trap mass spectrometer (Thermo Electron, San Jose, CA) equipped with an electrospray ionization source. The spray voltage was 4.5 kV, and the capillary temperature was 250°C. The fragmentation mass spectra were obtained using ion trap collision energies of 35%. Peptides were identified automatically by Mass Analyzer, a software program written by Zhongqi Zhang in Amgen. The software was employed to correlate the experimental tandem mass spectra against theoretical tandem mass spectra generated from the known antibody amino acid sequence for peptide identification.
Protein A binding assay
BIACore 3000, sensor chip SA, Glycine pH 1.5 and HBS-EP buffer, were from BIACore, Inc. (Piscataway, NJ). Protein A was from GE Healthcare (Piscataway, NJ) and the Protein A biotinylation was done by the Amgen Analytical and Formulation Sciences Department. Biotinylated Protein A was diluted to 10 μg/mL in HBS-EP buffer. The sensor chip surface was prepared by injecting 5 μL biotinylated Protein A over flow cell 1, 2, 3, and 4 at 50 μL/min. The bound protein on the sensor chip SA generated a signal of about 900 resonance units (RU). The Protein A binding assay was carried out on the immobilized Protein A surface. Samples were diluted in HBS-EP buffer using a three-fold serial dilution scheme from 666 to 8.2 nM, and then injected over the specific flow cell for each sample. For each cycle, the samples were allowed to associate with Protein A for 1 min and dissociate for 10 min at a flow rate of 100 μL/min, then the surface was regenerated by glycine buffer. For calculation of the association rate constant (ka), dissociation rate constant (kd) and equilibrium dissociation constant (KD), the BIAevaluation 2.1 software package was used to fit the binding curves using the 1:1 binding model with mass transport limitations.
FcRn binding assay
BIACore 3000, sensor chip CM5, surfactant P-20, and amine coupling kit, were from BIACore, Inc. (Piscataway, NJ). Phosphate-buffered saline (PBS, 1×, no calcium chloride, no magnesium chloride) was from Gibco. Bovine serum albumin (BSA, fraction V, IgG free) was from Sigma. Human Fc and human FcRn were expressed by Amgen Protein Science Department. Immobilization of human Fc to a CM5 sensor chip surface was performed according to the manufacturer's instructions. Briefly, carboxyl groups on the sensor chip surfaces were activated by injecting 150 μL of a mixture containing 0.2M-ethyl-N′-(dimethylaminopropyl) carbodiimide and 0.05MN-hydroxysuccinimide. Human Fc was diluted in 10 mM sodium acetate, pH 4.0 at 65 μg/mL and injected over the activated chip surface at 10 μL/min for 12 min. Excess reactive groups on the surfaces were deactivated by injecting 60 μL of 1M ethanolamine. The final immobilized level was ∼8000 RU. A blank reference surface was also prepared on the sensor chips by activation and deactivation without injection of protein. Binding assays were carried out on the immobilized human Fc surface. huFcRn (10 nM) was incubated with various amounts of samples in sample buffer (5 mM sodium acetate, 150 mM NaCl, pH 5.5, 0.1 mg/mL BSA, 0.005% P20) for 1 h before injection over the immobilized HuFc surface for 60 min at 2 μL/min. Following the sample injection, bound FcRn was allowed to dissociate from the Fc surface in PBS, pH 7.4 containing 0.005% P20 for 2 min. 100% FcRn binding signal was determined in the absence of antibody. A decreased binding response with increasing concentrations of antibody indicated that antibody was binding to FcRn in solution, blocking FcRn from binding to the immobilized human Fc surface. The EC50 values for antibody binding with 10 nM huFcRn were calculated by plotting the binding signal versus antibody concentrations, and using the one site competition model in GraphPad Prism.
Conclusions
Instability of methionines is a significant challenge in the development of protein therapeutics. Methionine oxidation has been shown to decrease bioactivity and stability, which may limit the product's clinical application or shelf-life. In this article, we have studied the methionine oxidation kinetics in the human IgG2 Fc and its impact on Protein A and FcRn binding. Met 252 is more readily oxidized than Met 428. Both of these residues are more susceptible to oxidation than the other two methionines in the Fc of human IgG2 antibody. The oxidation rates have excellent correlation with the SASA of the sulfur atom for each methionine. Our results further indicate that oxidation of Met 252 and Met 428 weakens the binding of the intact antibody with Protein A and FcRn, two natural proteins that share the same binding site on the Fc.
Methionine oxidation in the Fc domain is not expected to affect the potency of the antibody since the binding site is located in the CDRs within the Fab portion. However, we demonstrated that Fc Met oxidation, especially at Met 252 and Met 428, may have an impact on the biological functions of the human monoclonal IgG 2 antibody, since the decrease of FcRn binding affinity may translate to a decrease in the half-life of the IgG antibody.
Acknowledgments
The authors thank Ming Li, Anuradha Dixit, and Kelly Oliner for valuable assistance in performing the FcRn binding experiments. They also thank Drew Kelner and Sam Guhan for comments on the manuscript.
Glossary
Abbreviations:
- FcRn
neonatal Fc receptor
- IgG
immunoglobulin gamma
- Met
methionine
- mAb
monoclonal antibody
- NEM
N-ethylmaleimide
- SASA
solvent accessible surface area
- SPR
surface plasmon resonance
- TBHP
tert-butyl hydroperoxide
References
- 1.Shacter E. Quantification and significance of protein oxidation in biological samples. Drug Metab Rev. 2000;32:307–326. doi: 10.1081/dmr-100102336. [DOI] [PubMed] [Google Scholar]
- 2.Scislowski PW, Foster AR, Fuller MF. Regulation of oxidative degradation of l-lysine in rat liver mitochondria. Biochem J. 1994;300(Pt 3):887–891. doi: 10.1042/bj3000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hovorka S, Schoneich C. Oxidative degradation of pharmaceuticals: theory, mechanisms and inhibition. J Pharm Sci. 2001;90:253–269. doi: 10.1002/1520-6017(200103)90:3<253::aid-jps1>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 4.Slavica A, Dib I, Nidetzky B. Single-site oxidation, cysteine 108 to cysteine sulfinic acid, in D-amino acid oxidase from Trigonopsis variabilis and its structural and functional consequences. Appl Environ Microbiol. 2005;71:8061–8068. doi: 10.1128/AEM.71.12.8061-8068.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu HS, Fausset PR, Narhi LO, Horan T, Shinagawa K, Shimamoto G, Boone TC. Chemical modification and site-directed mutagenesis of methionine residues in recombinant human granulocyte colony-stimulating factor: effect on stability and biological activity. Arch Biochem Biophys. 1999;362:1–11. doi: 10.1006/abbi.1998.1022. [DOI] [PubMed] [Google Scholar]
- 6.Stadtman ER, Moskovitz J, Berlett BS, Levine RL. Cyclic oxidation and reduction of protein methionine residues is an important antioxidant mechanism. Mol Cell Biochem. 2002;234–235:3–9. [PubMed] [Google Scholar]
- 7.Frelinger AL, III, Zull JE. The role of the methionine residues in the structure and function of parathyroid hormone. Arch Biochem Biophys. 1986;244:641–649. doi: 10.1016/0003-9861(86)90632-6. [DOI] [PubMed] [Google Scholar]
- 8.Teh LC, Murphy LJ, Huq NL, Surus AS, Friesen HG, Lazarus L, Chapman GE. Methionine oxidation in human growth hormone and human chorionic somatomammotropin. Effects on receptor binding and biological activities. J Biol Chem. 1987;262:6472–6477. [PubMed] [Google Scholar]
- 9.Sasaoki K, Hiroshima T, Kusumoto S, Nishi K. Oxidation of methionine residues of recombinant human interleukin 2 in aqueous solutions. Chem Pharm Bull (Tokyo) 1989;37:2160–2164. doi: 10.1248/cpb.37.2160. [DOI] [PubMed] [Google Scholar]
- 10.Lam XM, Yang JY, Cleland JL. Antioxidants for prevention of methionine oxidation in recombinant monoclonal antibody HER2. J Pharm Sci. 1997;86:1250–1255. doi: 10.1021/js970143s. [DOI] [PubMed] [Google Scholar]
- 11.Gao J, Yin DH, Yao Y, Sun H, Qin Z, Schoneich C, Williams TD, Squier TC. Loss of conformational stability in calmodulin upon methionine oxidation. Biophys J. 1998;74:1115–1134. doi: 10.1016/S0006-3495(98)77830-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu JL, Lu KV, Eris T, Katta V, Westcott KR, Narhi LO, Lu HS. In vitro methionine oxidation of recombinant human leptin. Pharm Res. 1998;15:632–640. doi: 10.1023/a:1011998331254. [DOI] [PubMed] [Google Scholar]
- 13.Uversky VN, Yamin G, Souillac PO, Goers J, Glaser CB, Fink AL. Methionine oxidation inhibits fibrillation of human alpha-synuclein in vitro. FEBS Lett. 2002;517:239–244. doi: 10.1016/s0014-5793(02)02638-8. [DOI] [PubMed] [Google Scholar]
- 14.Wood MJ, Becvar LA, Prieto JH, Melacini G, Komives EA. NMR structures reveal how oxidation inactivates thrombomodulin. Biochemistry. 2003;42:11932–11942. doi: 10.1021/bi034646q. [DOI] [PubMed] [Google Scholar]
- 15.Chen B, Mayer MU, Squier TC. Structural uncoupling between opposing domains of oxidized calmodulin underlies the enhanced binding affinity and inhibition of the plasma membrane Ca-ATPase. Biochemistry. 2005;44:4737–4747. doi: 10.1021/bi0474113. [DOI] [PubMed] [Google Scholar]
- 16.Chugha P, Sage HJ, Oas TG. Methionine oxidation of monomeric lambda repressor: the denatured state ensemble under nondenaturing conditions. Protein Sci. 2006;15:533–542. doi: 10.1110/ps.051856406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brot N, Weissbach H. Biochemistry and physiological role of methionine sulfoxide residues in proteins. Arch Biochem Biophys. 1983;223:271–281. doi: 10.1016/0003-9861(83)90592-1. [DOI] [PubMed] [Google Scholar]
- 18.Glaser CB, et al. Oxidation of a specific methionine in thrombomodulin by activated neutrophil products blocks cofactor activity. A potential rapid mechanism for modulation of coagulation. J Clin Invest. 1992;90:2565–2573. doi: 10.1172/JCI116151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dow LK, Changela A, Hefner HE, Churchill ME. Oxidation of a critical methionine modulates DNA binding of the Drosophila melanogaster high mobility group protein, HMG-D. FEBS Lett. 1997;414:514–520. doi: 10.1016/s0014-5793(97)01059-4. [DOI] [PubMed] [Google Scholar]
- 20.Kornfelt T, Persson E, Palm L. Oxidation of methionine residues in coagulation factor VIIa. Arch Biochem Biophys. 1999;363:43–54. doi: 10.1006/abbi.1998.1071. [DOI] [PubMed] [Google Scholar]
- 21.Brekke OH, Sandlie I. Therapeutic antibodies for human diseases at the dawn of the twenty-first century. Nat Rev Drug Discov. 2003;2:52–62. doi: 10.1038/nrd984. [DOI] [PubMed] [Google Scholar]
- 22.Maggon K. Monoclonal antibody “gold rush”. Curr Med Chem. 2007;14:1978–1987. doi: 10.2174/092986707781368504. [DOI] [PubMed] [Google Scholar]
- 23.Burton DR. Immunoglobulin G: functional sites. Mol Immunol. 1985;22:161–206. doi: 10.1016/0161-5890(85)90151-8. [DOI] [PubMed] [Google Scholar]
- 24.Carter PJ. Potent antibody therapeutics by design. Nat Rev. 2006;6:343–357. doi: 10.1038/nri1837. [DOI] [PubMed] [Google Scholar]
- 25.Sjodahl J. Structural studies on the four repetitive Fc-binding regions in protein A from Staphylococcus aureus. Eur J Biochem. 1977;78:471–490. doi: 10.1111/j.1432-1033.1977.tb11760.x. [DOI] [PubMed] [Google Scholar]
- 26.Burmeister WP, Huber AH, Bjorkman PJ. Crystal structure of the complex of rat neonatal Fc receptor with Fc. Nature. 1994;372:379–383. doi: 10.1038/372379a0. [DOI] [PubMed] [Google Scholar]
- 27.Langone JJ. Protein A of Staphylococcus aureus and related immunoglobulin receptors produced by streptococci and pneumonococci. Adv Immunol. 1982;32:157–252. [PubMed] [Google Scholar]
- 28.Moks T, Abrahmsen L, Nilsson B, Hellman U, Sjoquist J, Uhlen M. Staphylococcal protein A consists of five IgG-binding domains. Eur J Biochem. 1986;156:637–643. doi: 10.1111/j.1432-1033.1986.tb09625.x. [DOI] [PubMed] [Google Scholar]
- 29.Simister NE, Mostov KE. An Fc receptor structurally related to MHC class I antigens. Nature. 1989;337:184–187. doi: 10.1038/337184a0. [DOI] [PubMed] [Google Scholar]
- 30.Ghetie V, Ward ES. Multiple roles for the major histocompatibility complex class I-related receptor FcRn. Annu Rev Immunol. 2000;18:739–766. doi: 10.1146/annurev.immunol.18.1.739. [DOI] [PubMed] [Google Scholar]
- 31.Roopenian DC, Akilesh S. FcRn: the neonatal Fc receptor comes of age. Nat Rev. 2007;7:715–725. doi: 10.1038/nri2155. [DOI] [PubMed] [Google Scholar]
- 32.Israel EJ, Wilsker DF, Hayes KC, Schoenfeld D, Simister NE. Increased clearance of IgG in mice that lack beta 2-microglobulpossible protective role of FcRn. Immunology. 1996;89:573–578. doi: 10.1046/j.1365-2567.1996.d01-775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lobo ED, Hansen RJ, Balthasar JP. Antibody pharmacokinetics and pharmacodynamics. J Pharm Sci. 2004;93:2645–2668. doi: 10.1002/jps.20178. [DOI] [PubMed] [Google Scholar]
- 34.Shen FJ, Kwong MY, Keck RG, Harris RJ. The application of tert-butylhydroperoxide oxidation to study sites of potential methionine oxidation in a recombinant antibody. Tech Protein Chem. 1996;7:275–284. [Google Scholar]
- 35.Chumsae C, Gaza-Bulseco G, Sun J, Liu H. Comparison of methionine oxidation in thermal stability and chemically stressed samples of a fully human monoclonal antibody. J Chromatogr. 2007;850:285–294. doi: 10.1016/j.jchromb.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 36.Liu D, Ren D, Huang H, Dankberg J, Rosenfeld R, Cocco MJ, Li L, Brems DN, Remmele RL., Jr Structure and stability changes of human IgG1 Fc as a consequence of methionine oxidation. Biochemistry. 2008;47:5088–5100. doi: 10.1021/bi702238b. [DOI] [PubMed] [Google Scholar]
- 37.Vriend G, Krause R. WHAT IF. 2007. [accessed March 2008]. Available at http://swift.cmbi.kun.nl/WIWWWI/
- 38.Hober S, Nord K, Linhult M. Protein A chromatography for antibody purification. J Chromatogr. 2007;848:40–47. doi: 10.1016/j.jchromb.2006.09.030. [DOI] [PubMed] [Google Scholar]
- 39.Shukla AA, Hubbard B, Tressel T, Guhan S, Low D. Downstream processing of monoclonal antibodies—application of platform approaches. J Chromatogr. 2007;848:28–39. doi: 10.1016/j.jchromb.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 40.Gaza-Bulseco G, Faldu S, Hurkmans K, Chumsae C, Liu H. Effect of methionine oxidation of a recombinant monoclonal antibody on the binding affinity to protein A and protein G. J Chromatogr. 2008;870:55–62. doi: 10.1016/j.jchromb.2008.05.045. [DOI] [PubMed] [Google Scholar]
- 41.Deisenhofer J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9-and 2.8-A resolution. Biochemistry. 1981;20:2361–2370. [PubMed] [Google Scholar]
- 42.Kato K, Gouda H, Takaha W, Yoshino A, Matsunaga C, Arata Y. 13C NMR study of the mode of interaction in solution of the B fragment of staphylococcal protein A and the Fc fragments of mouse immunoglobulin G. FEBS Lett. 1993;328:49–54. doi: 10.1016/0014-5793(93)80963-u. [DOI] [PubMed] [Google Scholar]
- 43.Martin WL, Bjorkman PJ. Characterization of the 2:1 complex between the class I MHC-related Fc receptor and its Fc ligand in solution. Biochemistry. 1999;38:12639–12647. doi: 10.1021/bi9913505. [DOI] [PubMed] [Google Scholar]
- 44.West AP, Jr, Bjorkman PJ. Crystal structure and immunoglobulin G binding properties of the human major histocompatibility complex-related Fc receptor. Biochemistry. 2000;39:9698–9708. doi: 10.1021/bi000749m. [DOI] [PubMed] [Google Scholar]
- 45.Martin WL, West AP, Jr, Gan L, Bjorkman PJ. Crystal structure at 2.8 A of an FcRn/heterodimeric Fc complex: mechanism of pH-dependent binding. Mol Cell. 2001;7:867–877. doi: 10.1016/s1097-2765(01)00230-1. [DOI] [PubMed] [Google Scholar]
- 46.Shields RL, Namenuk AK, Hong K, Meng YG, Rae J, Briggs J, Xie D, Lai J, Stadlen A, Li B, Fox JA, Presta LG. High resolution mapping of the binding site on human IgG1 for Fc gamma RI, Fc gamma RII, Fc gamma RIII, and FcRn and design of IgG1 variants with improved binding to the Fc gamma R. J Biol Chem. 2001;276:6591–6604. doi: 10.1074/jbc.M009483200. [DOI] [PubMed] [Google Scholar]
- 47.Dall'Acqua WF, Woods RM, Ward ES, Palaszynski SR, Patel NK, Brewah YA, Wu H, Kiener PA, Langermann S. Increasing the affinity of a human IgG1 for the neonatal Fc receptor: biological consequences. J Immunol. 2002;169:5171–5180. doi: 10.4049/jimmunol.169.9.5171. [DOI] [PubMed] [Google Scholar]
- 48.Hinton PR, Johlfs MG, Xiong JM, Hanestad K, Ong KC, Bullock C, Keller S, Tang MT, Tso JY, Vasquez M, Tsurushita N. Engineered human IgG antibodies with longer serum half-lives in primates. J Biol Chem. 2004;279:6213–6216. doi: 10.1074/jbc.C300470200. [DOI] [PubMed] [Google Scholar]
- 49.Vaccaro C, Zhou J, Ober RJ, Ward ES. Engineering the Fc region of immunoglobulin G to modulate in vivo antibody levels. Nat Biotechnol. 2005;23:1283–1288. doi: 10.1038/nbt1143. [DOI] [PubMed] [Google Scholar]
- 50.Dall'Acqua WF, Kiener PA, Wu H. Properties of human IgG1s engineered for enhanced binding to the neonatal Fc receptor (FcRn) J Biol Chem. 2006;281:23514–23524. doi: 10.1074/jbc.M604292200. [DOI] [PubMed] [Google Scholar]
- 51.Hinton PR, Xiong JM, Johlfs MG, Tang MT, Keller S, Tsurushita N. An engineered human IgG1 antibody with longer serum half-life. J Immunol. 2006;176:346–356. doi: 10.4049/jimmunol.176.1.346. [DOI] [PubMed] [Google Scholar]
- 52.Petkova SB, Akilesh S, Sproule TJ, Christianson GJ, Al Khabbaz H, Brown AC, Presta LG, Meng YG, Roopenian DC. Enhanced half-life of genetically engineered human IgG1 antibodies in a humanized FcRn mouse model: potential application in humorally mediated autoimmune disease. Int Immunol. 2006;18:1759–1769. doi: 10.1093/intimm/dxl110. [DOI] [PubMed] [Google Scholar]
- 53.Sauer-Eriksson AE, Kleywegt GJ, Uhlen M, Jones TA. Crystal structure of the C2 fragment of streptococcal protein G in complex with the Fc domain of human IgG. Structure. 1995;3:265–278. doi: 10.1016/s0969-2126(01)00157-5. [DOI] [PubMed] [Google Scholar]
- 54.Corper AL, Sohi MK, Bonagura VR, Steinitz M, Jefferis R, Feinstein A, Beale D, Taussig MJ, Sutton BJ. Structure of human IgM rheumatoid factor Fab bound to its autoantigen IgG Fc reveals a novel topology of antibody-antigen interaction. Nat Struct Biol. 1997;4:374–381. doi: 10.1038/nsb0597-374. [DOI] [PubMed] [Google Scholar]


