Abstract
The protein folding process is often in vitro rate-limited by slow cis-trans proline isomerization steps. Importantly, the rate of this process in vivo is accelerated by prolyl isomerases (PPIases). The archetypal PPIase is the human cyclophilin 18 (Cyp18 or CypA), and Arg 55 has been demonstrated to play a crucial role when studying short peptide substrates in the catalytic action of Cyp18 by stabilizing the transition state of isomerization. However, in this study we show that a R55A mutant of Cyp18 is as efficient as the wild type to accelerate the refolding reaction of human carbonic anhydrase II (HCA II). Thus, it is evident that the active-site located Arg 55 is not required for catalysis of the rate-limiting prolyl cis-trans isomerization steps during the folding of a protein substrate as HCA II. Nevertheless, catalysis of cis-trans proline isomerization in HCA II occurs in the active-site of Cyp18, since binding of the inhibitor cyclosporin A abolishes rate acceleration of the refolding reaction. Obviously, the catalytic mechanisms of Cyp18 can differ when acting upon a simple model peptide, four residues long, with easily accessible Pro residues compared with a large protein molecule undergoing folding with partly or completely buried Pro residues. In the latter case, the isomerization kinetics are significantly slower and simpler mechanistic factors such as desolvation and/or strain might operate during folding-assisted catalysis, since binding to the hydrophobic active site is still a prerequisite for catalysis.
Keywords: cis-trans proline isomerization, cyclophilin 18, prolyl isomerases, human carbonic anhydrase II
Introduction
The protein folding process in vitro is often rate-limited by slow cis-trans proline isomerization steps. Importantly, the rate of this process in vivo is accelerated by prolyl isomerases (PPIases). The archetypal PPIase is the human cyclophilin 18 (Cyp18 or CypA) for which the structure-function relationship has been comprehensively investigated.1 Cyp18 has also been used as a model enzyme for studies of the linkage between dynamics and catalysis.2–4 In the catalytic action, Arg 55 has been demonstrated to play a critical role when using peptide substrates. In this study we show, however, that a R55A mutant is as effective as the wild-type enzyme in catalyzing the cis-trans proline isomerization on a folding protein substrate (human carbonic anhydrase II; HCA II).
Despite the large amount of data that have been accumulated from studies on the action of Cyp18 on peptide substrates, the molecular mechanism has not been definitely resolved. In the favored mechanism, Arg 55 plays a crucial role by lowering the rotational barrier of the prolyl bond. This is accomplished by stabilization of the lone electron pair at the prolyl imide nitrogen in transition state by a hydrogen bond from the guanidino group of Arg 55.1,5–7
In the crystal structure of Cyp18 with bound peptide substrate molecules, interactions of the guanidino group of Arg with the imide nitrogen of proline in the substrate have been shown.6,7 Support for this mechanism also stems from the almost complete inactivation of Cyp18 by a R55A mutation (0.1% remaining activity).8 Moreover, the backbone nitrogen of Arg 55 from NMR chemical shift and relaxation measurements has been observed to undergo conformational exchange coupled to the isomerization and binding process.2 This motion with the substrate of Arg 55 leading to stronger hydrogen bond interactions in transition state is also supported by simulation studies.9–12 To gain insight into the catalytic mechanism, small model peptide substrates have usually been employed and not folding proteins that are the natural substrates in the cell. Therefore, we engineered the R55A variant and used the folding protein HCA II as a substrate. HCA II contains two cis-peptidyl-Pro bonds (Pro 30 and Pro 202) out of 17 Pro residues in the native state,13 and we have previously shown that refolding of the protein is rate-limited by cis-trans proline isomerization.14 Moreover, the refolding reaction has been demonstrated to be accelerated by wild-type Cyp18.14–17
Results and Discussion
To verify the reduced prolyl isomerise activity on a peptide substrate (succinyl-Ala-Leu-Pro-Phe-4-nitroanilide) caused by the R55A mutation in human Cyp18, we initially measured the activity for Cyp18 (wild-type) and Cyp18R55A in the commonly used protease-coupled assay. We observed that the R55A mutation gave rise to loss of the prolyl isomerise activity and the mutant showed the same kinetics as the uncatalyzed reaction [Fig. 1(A)]. On the contrary, Cyp18 efficiently catalyzed rapid isomerization of the peptide substrate, which was almost completed within the time of manual mixing.
Figure 1.
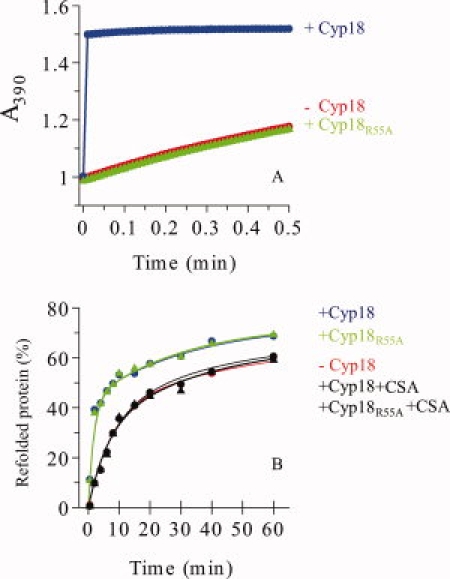
(A) Prolyl isomerase activity of Cyp18 and Cyp18R55A in the protease-coupled assay on the peptide substrate succinyl-Ala-Leu-Pro-Phe-4-nitroanilide as measured by the increase in A390 nm after the coupled hydrolysis by chymotrypsin. The uncatalyzed reaction (•: red), cis → trans isomerization catalyzed by Cyp18 (•: blue), and Cyp18R55A (▴: green). (B) Time courses of reactivation of unfolded HCA IIpwt in the presence and absence of Cyp18 variants. The kinetic data were best fitted to two exponential terms and the parameters are given in Table I. Uncatalyzed (•: red), with Cyp18 (•: blue), with Cyp18R55A (▴: green), with Cyp18 + CsA (▴: black), and with Cyp18R55A+ CsA (•: black).
To compare the prolyl isomerise activity between Cyp18 and Cyp18R55A in a proline rate-limiting protein folding reaction, we used denatured HCA IIpwt as a substrate protein. HCA IIpwt is a pseudowild type with a C206S mutation that has indistinguishable folding and functional properties to those of the wild type.18
The HCA II variants were denatured for 1 h in 5M GuHCl to reach cis-trans equilibrium, and the Cyp18 variants were added to the dilution buffer used to initiate refolding. The halftime of the total reactivation process of denatured HCA IIpwt was lowered to ∼3 min in the presence of Cyp18 as compared to 10 min for the uncatalyzed refolding reaction in agreement with previous data.14 Interestingly, the refolding kinetics of HCA IIpwt in the presence of Cyp18R55A followed identical reactivation kinetics as in the presence of Cyp18 [Fig. 1(B); Table I)]. Thus, it is clear that the active-site located Arg 55 is not required for catalysis of the rate-limiting prolyl cis-trans isomerization steps during the folding of a protein substrate such as HCA II.
Table I.
Kinetic Data for Reactivation of Denatured HCA IIpwt With and Without Assistance of Cyp18 Variants
| Protein | k1 (min−1) | A1 | k2 (min−1) | A2 | t1/2a (min) |
|---|---|---|---|---|---|
| HCA IIpwt | 0.14 | 0.38 | 0.020 | 0.30 | 10 |
| HCA IIpwt + Cyp18 | 0.56 | 0.43 | 0.029 | 0.31 | 2.8 |
| HCA IIpwt + Cyp18R55A | 0.55 | 0.43 | 0.032 | 0.31 | 2.9 |
| HCA IIpwt + Cyp18+CsA | 0.14 | 0.34 | 0.028 | 0.33 | 9.9 |
| HCA IIpwt + Cyp18R55A+CsA | 0.16 | 0.31 | 0.026 | 0.36 | 10 |
The halftime of the total reactivation process.
Of course, a protein folding reaction with in this case involvement of cis-trans isomerization of 17 Pro residues is much more complicated than the corresponding isomerization of a single Pro residue in a small peptide. In the peptide reaction, Cyp18 is operating on the millisecond time scale (kcat > 600 s−1; Ref.1), whereas the catalyzed folding reaction is proceeding on the minute time scale (k1 = 0.56 min−1; Table I). Therefore, the reaction mechanisms are not necessarily the same for a simple peptide substrate and a much larger protein substrate. Addition of the inhibitor CsA stops the rate acceleration of the reactivation of HCA II for both Cyp18 and Cyp18R55A [Fig. 1(B)(B)]. Consequently, binding of the protein substrate to the hydrophobic active-site located at the surface of Cyp18 is also important for Cyp18R55A for catalysis. However, in a folding protein the accessibility of substrate Pro residues for the active-site located residue Arg 55 can rapidly be limited because of the initial hydrophobic collapse hiding hydrophobic residues (like Pro) in the interior of the protein. For HCA II, it has also been shown that a fraction of the Pro residues very rapidly becomes buried for Cyp18 during the initiation of the folding process.15,17 Therefore, the catalytic effect of binding to the hydrophobic active-site region for a large protein substrate such as the 29 kDa HCA II might be limited to desolvation and/or strain, mechanisms that previously have also been proposed to explain the prolyl isomerase activity.19,20 In this context, it is interesting to note that the yeast mitochondrial cyclophilin Cpr3 with a homologous R73A mutation did not affect the prolyl-isomerase activity on a mutated form of T1 RNase but lacked catalytic activity on peptide substrates.21
Proline cis-trans isomerization has important functional consequences during protein folding, misfolding, and regulation of isoformic conformations. Recently, proline isomerization gained considerable interest in the protein misfolding and amyloidogenesis arena. It was demonstrated that proline isomerization governed the population of an amyloidogenic intermediate in β-2-microglobulin misfolding22,23 and proline isomerization controlled the phosphorylation of Thr 668-Pro 669, mediated by Pin1, in amyloid precursor protein that facilitates proteolytic cleavage that affords the Aβ-peptides in Alzheimer's disease.24 Furthermore, prefibrillar tetrameric assemblies of the protein stefin B (from the cystatin superfamily) have been shown to be controlled by proline isomerization that mediated extensive intermolecular contacts of domain swapped dimers.25 Recently, we hypothesized that proline isomerization governs alternate supermolecular folding of tetrameric functional conformational isoforms of a thermophilic alcohol dehydrogenase in a process reminiscent of conformational variations of yeast prion strains.26 Notably, all these aforementioned conformational transitions occur within large protein molecules.
To conclude, the catalytic role of Arg 55 in Cyp18 should not be generalized from its action toward simple model peptides to more complicated in vivo substrates such as a folding protein molecule. This does, however, not exclude involvement of Arg 55 toward protein substrates, since for instance Cyp18 has been shown to bind to a short exposed proline-containing loop of the HIV-1 CA protein, where Arg 55 probably facilitates cis-trans isomerization by anchoring the proline oxygen and activating the proline imide of the isomeric peptide bond.27 It should be pointed out, on the other hand, that it is different to act on exposed protein loops in the native state of a protein than to interact with partly exposed or buried prolines in a protein during folding.
Materials and Methods
Cloning, production, and purification of Cyp18 and Cyp18R55A
The human cyclophilin A (Cyp18) gene was isolated from a human lymphocyte cDNA library (CD4 positive) in the cloning vector lambda gt11 from Clontech and cloned into the expression vector, pACA.28 Site-directed mutagenesis and expression of Cyp18 and Cyp18R55A in Escherichia coli as well as purification followed the procedure described by Zydowsky et al.8 with some modifications. Thus, Dnase I (a spatula tip) treatment of the cell solution was performed prior to the protamine sulfate precipitation of nucleic acids. The buffers in the chromatographic steps were complemented with 0.5 mM DTT. In the final gel filtration step, Sephadex G-50 Fine was used and protein-containing fractions were pooled and concentrated. The purity of the prepared Cyp18 and Cyp18R55A proteins were analyzed by SDS-PAGE. Ten micrograms of the Cyp18 variants were applied in each lane and stained with Commassie Brilliant Blue R-250. No significant impurity bands could thereby be detected.
Prolyl isomerase activity in the peptide assay
The PPIase assay was performed in a coupled assay with chymotrypsin essentially as described by Scholz et al.21 This assay is based on monitoring the time course of the chymotryptic cleavage of the 4-nitroanilide bond of N-succinylated tetrapeptide-4-nitroanilides as substrates, which is kinetically coupled to the cis → trans isomerization of the peptidylprolyl bond of the peptide. The chromogenic peptide substrate succinyl-Ala-Leu-Pro-Phe-4-nitroanilide was used and the release of the nitroanilide moiety was followed by an increase in absorbance at 390 nm (Varian, Cary 100 Bio UV–visible spectrophotometer)). In the assay, Cyp18 or Cyp18R55A (final concentrations of 3–7 nM) were added to a 600 μM solution of α-chymotrypsin (Fluka) buffered with 0.1M Tris-HCl, pH 8.0. The activity measurement was initiated by adding a 7.8 mM solution of the chromogenic peptide succinyl-Ala-Leu-Pro-Phe-4-nitroanilide, dissolved in trifluroethanol that additionally contained 0.45M LiCl. The peptide solution was thereby diluted 100 times yielding a final concentration of 78 μM. PPIase inhibition was performed by addition of 50 nM of cyclosporin A (CsA), dissolved in ethanol, prior to addition of the peptide substrate in the assay.
Reactivation kinetics of denatured HCA II variants with and without Cyp18 variants
A pseudo wild-type variant of HCA II (HCA IIpwt) with a C206S mutation was used in these measurements. HCA IIpwt has been shown to have indistinguishable folding and functional properties when compared with the wild-type enzyme and was constructed as described earlier.18
HCA IIpwt was denatured in 5.0M GuHCl for 1 h and renatured by dilution to 0.3M GuHCl at 21°C. The measurements of the regain in CO2 hydration activity during refolding were performed as described previously.14 The concentration of HCA IIpwt during refolding was 0.83 μM and the concentration of Cyp18 and Cyp18R55A was 8.3 μM, and when applicable CsA was added to a concentration of 25 μM. All solutions were buffered with 0.1M Tris-H2SO4, pH 7.5. The rate constants and amplitudes were calculated using a nonlinear least squares fit program (TableCurve, Jandel Scientific). The data were fitted to the double exponential zero intercept equation:
where y is the refolding yield in %, t denotes the time in minutes, A1 and A2 denote the amplitudes of the first and second phase, respectively, and k1 and k2 are the rate constants for the first and second phases.
References
- 1.Fanghänel J, Fischer G. Insights into the catalytic mechanism of peptidyl prolyl cis/trans isomerases. Front Biosci. 2004;9:3453–3478. doi: 10.2741/1494. [DOI] [PubMed] [Google Scholar]
- 2.Eisenmesser EZ, Bosco DA, Akke M, Kern D. Enzyme dynamics during catalysis. Science. 2002;295:1520–1523. doi: 10.1126/science.1066176. [DOI] [PubMed] [Google Scholar]
- 3.Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf-Watz M, Bosco DA, Skalicky JJ, Kay LE, Kern D. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438:117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 4.Kern D, Eisenmesser EZ, Wolf-Watz M. Enzyme dynamics during catalysis measured by NMR spectroscopy. Methods Enzymol. 2005;394:507–524. doi: 10.1016/S0076-6879(05)94021-4. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Y, Chen Y, Schutkowski M, Fischer G, Ke H. Cyclophilin A complexed with a fragment of HIV-1 gag proteinsights into HIV-1 infectious activity. Structure. 1997;5:139–146. doi: 10.1016/s0969-2126(97)00172-x. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Y, Ke H. Crystal structure implies that cyclophilin predominantly catalyzes the trans to cis isomerization. Biochemistry. 1996;35:7356–7361. doi: 10.1021/bi9602775. [DOI] [PubMed] [Google Scholar]
- 7.Zhao Y, Ke H. Mechanistic implication of crystal structures of the cyclophilin-dipeptide complexes. Biochemistry. 1996;35:7362–7368. doi: 10.1021/bi960278x. [DOI] [PubMed] [Google Scholar]
- 8.Zydowsky LD, Etzkorn FA, Chang HY, Ferguson SB, Stolz LA, Ho SI, Walsh CT. Active site mutants of human cyclophilin A separate peptidyl-prolyl isomerase activity from cyclosporin A binding and calcineurin inhibition. Protein Sci. 1992;1:1092–1099. doi: 10.1002/pro.5560010903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hur S, Bruice TC. The mechanism of cis-trans isomerization of prolyl peptides by cyclophilin. J Am Chem Soc. 2002;124:7303–7313. doi: 10.1021/ja020222s. [DOI] [PubMed] [Google Scholar]
- 10.Li G, Cui Q. What is so special about Arg 55 in the catalysis of cyclophilin A? insights from hybrid QM/MM simulations. J Am Chem Soc. 2003;125:15028–15038. doi: 10.1021/ja0367851. [DOI] [PubMed] [Google Scholar]
- 11.Agarwal PK, Geist A, Gorin A. Protein dynamics and enzymatic catalysis: investigating the peptidyl-prolyl cis-trans isomerization activity of cyclophilin A. Biochemistry. 2004;43:10605–10618. doi: 10.1021/bi0495228. [DOI] [PubMed] [Google Scholar]
- 12.Trzesniak D, van Gunsteren WF. Catalytic mechanism of cyclophilin as observed in molecular dynamics simulations: pathway prediction and reconciliation of X-ray crystallographic and NMR solution data. Protein Sci. 2006;15:2544–2551. doi: 10.1110/ps.062356406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Håkansson K, Carlsson M, Svensson LA, Liljas A. Structure of native and apo carbonic anhydrase II and structure of some of its anion-ligand complexes. J Mol Biol. 1992;227:1192–1204. doi: 10.1016/0022-2836(92)90531-n. [DOI] [PubMed] [Google Scholar]
- 14.Fransson C, Freskgård P-O, Herbertsson H, Johansson A, Jonasson P, Mårtensson L-G, Svensson M, Jonsson B-H, Carlsson U. Cis-trans isomerization is rate-determining in the reactivation of denatured human carbonic anhydrase II as evidenced by proline isomerase. FEBS Lett. 1992;296:90–94. doi: 10.1016/0014-5793(92)80410-i. [DOI] [PubMed] [Google Scholar]
- 15.Freskgård P-O, Bergenhem N, Jonsson B-H, Svensson M, Carlsson U. Isomerase and chaperone activity of prolyl isomerase in the folding of carbonic anhydrase. Science. 1992;258:466–468. doi: 10.1126/science.1357751. [DOI] [PubMed] [Google Scholar]
- 16.Kern G, Kern D, Schmid FX, Fischer G. Reassessment of the putative chaperone function of prolyl-cis/trans-isomerases. FEBS Lett. 1994;348:145–148. doi: 10.1016/0014-5793(94)00591-5. [DOI] [PubMed] [Google Scholar]
- 17.Kern G, Kern D, Schmid FX, Fischer G. A kinetic analysis of the folding of human carbonic anhydrase II and its catalysis by cyclophilin. J Biol Chem. 1995;270:740–745. doi: 10.1074/jbc.270.2.740. [DOI] [PubMed] [Google Scholar]
- 18.Mårtensson L-G, Jonsson B-H, Freskgård PO, Kihlgren A, Svensson M, Carlsson U. Characterization of folding intermediates of human carbonic anhydrase II: probing substructure by chemical labeling of SH groups introduced by site-directed mutagenesis. Biochemistry. 1993;32:224–231. doi: 10.1021/bi00052a029. [DOI] [PubMed] [Google Scholar]
- 19.Harrison RK, Stein RL. Mechanistic studies of peptidyl prolyl cis-trans isomerase: evidence for catalysis by distortion. Biochemistry. 1990;29:1684–1689. doi: 10.1021/bi00459a003. [DOI] [PubMed] [Google Scholar]
- 20.Radzicka A, Wolfenden R. Analogues of intermediates in the action of pig kidney prolidase. Biochemistry. 1991;30:4160–4164. doi: 10.1021/bi00231a008. [DOI] [PubMed] [Google Scholar]
- 21.Scholz C, Schindler T, Dolinski K, Heitman J, Schmid FX. Cyclophilin active site mutants have native prolyl isomerase activity with a protein substrate. FEBS Lett. 1997;414:69–73. doi: 10.1016/s0014-5793(97)00979-4. [DOI] [PubMed] [Google Scholar]
- 22.Jahn TR, Parker MJ, Homans SW, Radford SE. Amyloid formation under physiological conditions proceeds via a native-like folding intermediate. Nat Struct Mol Biol. 2006;13:195–201. doi: 10.1038/nsmb1058. [DOI] [PubMed] [Google Scholar]
- 23.Eakin CM, Berman AJ, Miranker AD. A native to amyloidogenic transition regulated by a backbone trigger. Nat Struct Mol Biol. 2006;13:202–208. doi: 10.1038/nsmb1068. [DOI] [PubMed] [Google Scholar]
- 24.Pastorino L, Sun A, Lu PJ, Zhou XZ, Balastik M, Finn G, Wulf G, Lim J, Li SH, Li X, Xia W, Nicholson LK, Lu KP. The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-beta production. Nature. 2006;440:528–534. doi: 10.1038/nature04543. [DOI] [PubMed] [Google Scholar]
- 25.Jenko Kokalj S, Guncar G, Stern I, Morgan G, Rabzelj S, Kenig M, Staniforth RA, Waltho JP, Zerovnik E, Turk D. Essential role of proline isomerization in stefin B tetramer formation. J Mol Biol. 2007;366:1569–1579. doi: 10.1016/j.jmb.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 26.Mishra R, Olofsson L, Karlsson M, Carlsson U, Nicholls IA, Hammarström P. A conformationally isoformic thermophilic protein with high kinetic unfolding barriers. Cell Mol Life Sci. 2008;65:827–839. doi: 10.1007/s00018-008-7517-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howard BR, Vajdos FF, Li S, Sundquist WI, Hill CP. Structural insights into the catalytic mechanism of cyclophilin A. Nat Struct Biol. 2003;10:475–481. doi: 10.1038/nsb927. [DOI] [PubMed] [Google Scholar]
- 28.Nair SK, Calderone TL, Christanson DW, Fierke C. Altering the mouth of a hydrophobic pocket. Structure and kinetics of human carbonic anhydrase II mutants at residue Val-121. J Biol Chem. 1991;266:17320–17325. [PubMed] [Google Scholar]


