Abstract
OSR1 (oxidative stress-responsive-1) and SPAK (Ste20/Sps1-related proline/alanine-rich kinase) belong to the GCK-VI subfamily of Ste20 group kinases. OSR1 and SPAK are key regulators of NKCCs (Na+/K+/2Cl− cotransporters) and activated by WNK family members (with-no-lysine kinase), mutations of which are known to cause Gordon syndrome, an autosomal dominant form of inherited hypertension. The crystal structure of OSR1 kinase domain has been solved at 2.25 Å. OSR1 forms a domain-swapped dimer in an inactive conformation, in which P+1 loop and αEF helix are swapped between dimer-related monomers. Structural alignment with nonswapped Ste20 TAO2 kinase indicates that the integrity of chemical interactions in the kinase domain is well preserved in the domain-swapped interfaces. The OSR1 kinase domain has now been added to a growing list of domain-swapped protein kinases recently reported, suggesting that the domain-swapping event provides an additional layer of complexity in regulating protein kinase activity.
Keywords: OSR1, STE20 kinase family, domain swapping, crystallography
Introduction
A large emerging group of protein kinases with complex and diverse functions is the Ste20 kinase family, which was identified as a prototypical mitogen-activated protein kinase kinase kinase kinase (MAP4K) involved in the yeast pheromone-induced mating pathway.1–3 Ste20 kinases regulate a wide range of fundamental cellular processes such as cell cycle, apoptosis, development, growth, stress responses, cellular volume sensing, and regulation.4–7 Based upon phylogenetic relationships, Ste20 kinases have been further divided into the p21-activated kinases (PAKs) and the germinal center kinases (GCKs). The PAKs bind to GTP-liganded forms of the Rho family small G proteins Rac and Cdc42 and regulate cytoskeletal organization and cell motility.8 The GCKs often function as MAP4Ks activating c-Jun kinases (JNKs) or p38 MAP kinases but have been shown to directly phosphorylate ion transporters.9 OSR1 (oxidative stress-responsive-1) and SPAK (Ste20/Sps1-related proline/alanine-rich kinase) have been grouped in the GCK-VI subfamily of Ste20 kinases3 and share 96% sequence identity in their kinase domains. OSR1 was originally identified and named because of its sequence similarity to SOK1 (Ste20/oxidant stress responsive kinase-1).10 OSR1 is widely expressed but highest in heart and skeletal muscle.10 The distribution of OSR1 at the organ/tissue level largely overlaps with SPAK.4,7 OSR1 and SPAK have been suggested to be coupled to cellular events such as cell differentiation, cytoskeleton rearrangement, cell proliferation, and transformation.3,11–15 In addition, extensive biochemical and physiological studies demonstrate that OSR1 and SPAK are also involved in the regulation of ion homeostasis and volume control in mammalian cells. For example, during hyperosmotic stress, OSR1 and SPAK interact with and activate NKCC1 (Na+/K+/2Cl− Cotransporter-1), a ubiquitous ion carrier that plays essential roles in epithelial transport, ion homeostasis in the central nervous system, and also cell-volume regulation.4,5,16,17 More interestingly, OSR1 and SPAK are phosphorylated and activated by WNK1 (with-no-lysine kinase 1) and WNK4, mutations of which are known to cause Gordon syndrome.18–21 Gordon syndrome, also known as PHAII (pseudohypoaldosteronism type II), is an autosomal dominant form of inherited hypertension and hyperkalemia.22 Like other members of the GCK branch of the Ste20 kinases, OSR1 and SPAK harbor an N-terminal kinase catalytic domain and two conserved C-terminal regulatory regions, named PF1 and PF2 domains [Fig. 1(A)]. OSR1 is a 58-kDa protein of 527 amino acids. OSR1 has a 17-residues insertion prior to the kinase domain, whereas SPAK contains an upstream stretch of 74 residues. Previous studies suggest that the PF1 domain, located immediately C-terminal to the catalytic domain, is required for catalytic activity.13 The PF2 domain, also known as CCT (conserved carboxy-terminal) domain, is involved in recognizing its substrates as well as its activators. The CCT domain of OSR1 and SPAK interacts with an RFXV (Arg-Phe-Xaa-Val) motif present in their substrate NKCC1 and activators WNK1/WNK4.20,23,24 Recently, the crystal structure of the CCT domain of OSR1 kinase in complex with a peptide containing an RFXV motif derived from WNK4 was reported, providing the molecular mechanism by which the CCT domain recognizes its activators and substrates.25 Despite the recent progress in biochemical studies of OSR1 and SPAK, however, the lack of structural information about the core kinase domain limits our view of the elaborate regulatory mechanism. It remains to be determined how the kinase domains of OSR1 and SPAK are coupled to their regulatory domains and what the unique features of their kinase domains are. Here, we present the crystal structure of human OSR1 kinase domain complexed with MgAMP-PNP, which revealed a novel mode of dimerization characterized by domain-swapping of the activation segment as well as the αEF helix.
Figure 1.
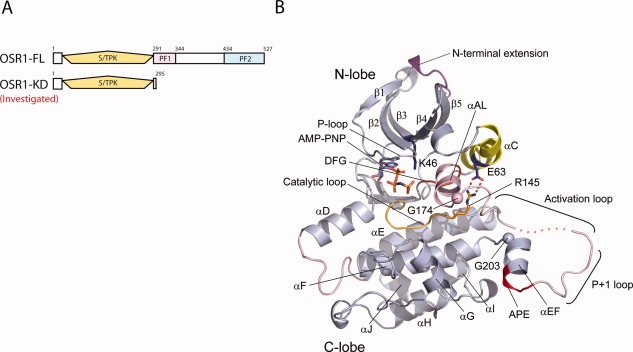
The monomeric structure of OSR1 kinase domain. (A) Organization of OSR1. The upper diagram represents domain organization of full-length OSR1 kinase. The lower diagram indicates the construct, which was investigated for crystallographic studies. The Serine/Threonine protein kinase (S/TPK) domain is represented in yellow. (B) The monomeric structure of OSR1 kinase domain. Note that there are four molecules in asymmetric unit and one subunit was displayed. The disordered region of activation loop is shown in dotted line.
Results and Discussion
Structure determination of OSR1 kinase domain
As initial efforts to crystallize recombinant full–length 527–residue human OSR1 were not successful, we screened shorter constructs for solubility. A construct encoding residues 1–295 of human OSR1, which includes the kinase domain and a short N-terminal tail (OSR1-KD) produced soluble protein. The purified OSR1-KD was crystallized as a binary complex with Mg-AMPPNP and yielded diffraction data to 2.25 Å resolution (see Materials and Methods). OSR1-KD crystallizes in space group P212121 with four polypeptide chains in the asymmetric unit. The structure was solved by molecular replacement utilizing the structures of the checkpoint kinase 1 (Chk1; PDB ID 1NVR; Ref.26) and the Ste20 TAO2 kinase (TAO2; PDB ID 1U5R; Ref.27) as N-terminal and C-terminal lobe search models, respectively. The final refined structure has a conventional R factor of 0.23 and R free of 0.26 with reasonable stereochemistry (Table I). In general, clear interpretable electron density is observed for the entire kinase domain, except for the first five residues from Met1 to Ser5 in the N-terminal lobe and about 10 residues spanning the central part of the activation loop from Asp176 to Phe186 in the C-terminal lobe. The four monomers in the asymmetric unit form two tight dimers. The individual polypeptides are quite similar, with an average root mean square deviation (rmsd) of 0.35 Å.
Table I.
Crystallographic Data Collection and Refinement Statistics
| OSR1KD-AMP-PNP complex | |
|---|---|
| Data collection | |
| Wavelength (Å) | 0.9789 |
| Space group | P212121 |
| Cell dimensions (Å) | |
| a | 74.2 |
| b | 104.4 |
| c | 162.7 |
| Resolution (Å) | 50–2.25 |
| Unique reflections | 59432 |
| Rsym (outer shell)a | 0.082(0.45) |
| I/σ (outer shell) | 23.2(2.9) |
| Multiplicity (outer shell) | 5.5(4.4) |
| Completeness (%) (outer shell) | 98.2(91.9) |
| Refinement statistics | |
| Resolution (Å) | 29.74–2.25 |
| Number of reflections in refinement | 56330 |
| Rwork/Rfree (%)b | 23.4/26.7 |
| Rmsd bond length (Å)c | 0.016 |
| Rmsd bond angle (°)c | 1.665 |
| Average B values (Å2) | 38.6 |
| Ramachandran plot regions (%) | |
| Most favored regions | 91.0 |
| Additional allowed regions | 7.5 |
| Generously allowed regions | 1.4 |
| Disallowed regions | 0.0 |
Rsym = ∑|Iavg − Ij|/∑Ij.
Rfactor = ∑|Fo − Fc|/∑Fo, where Fo and Fc are observed and calculated structure factors, respectively, Rfree was calculated from a randomly chosen 5% of reflections excluded from the refinement, and Rfactor was calculated from the remaining 95% of reflections.
Rmsd is the root mean square deviation from ideal geometry.
Architecture of the monomeric OSR1 kinase domain
The crystal structure of monomeric OSR1-KD displays the canonical bilobal protein kinase fold [Fig. 1(B)]. The N-terminal lobe has the five-stranded antiparallel β-sheet (β1–β5) and αC. The N-terminal extension possesses 17 residues, visible starting at residue 6, and packs between β2 and β5 at the top of the protein. A single-turn helix corresponding to αB of protein kinase A (PKA)28 is present between and β3 and αC. The C-terminal lobe possesses the standard helices αD, αE, αF, αG, αH, and αI, as well as a short seven-residue helix αJ. Compared with TAO2, OSR1 has a seven-residue insertion between αD and αE (Ala109 to Ser115) that elongates αD one turn, and a second six-residue insertion in the linker between αG and αH (Gly250 to Glu 255) that lacks secondary structure. These two inserts form close contacts with each other. The activation segment that houses the activating phosphorylation site (Thr185) adopts a unique structure. At its N-terminus, a two-turn helix forms between Gly166 and Gly174. Residues Asp176 to Thr185 are disordered. Then the P+1 recognition site loop and αEF helix surprisingly extend away from the kinase core. The crystals formed in the presence of AMPPNP, which is bound in the canonical nucleotide binding site between the two lobes, contacting the glycine-rich phosphate binding loop (GXGXFG). Despite the presence of AMPPNP, the OSR1-KD adopts a clearly inactive conformation, according to several structural cues. Most apparent, a hallmark ion pair of active kinases between a conserved lysine in strand β3 (Lys46) and a conserved glutamate in helix C (Glu63) (“K-E ion pair”)29,30 is not formed, as discussed later [Fig. 1(B)].
OSR1 kinase is a domain-swapped dimer
Domain swapping is a process for making dimers of proteins by exchanging identical structural elements, while maintaining chemical interactions observed in monomeric forms.31 OSR1-KD forms dimers in which the P+1 recognition loop and αEF (Phe186 to Gly203) is domain-swapped (Fig. 2). The hinge points for the domain swap are Gly174 at the C-terminus of αAL in the activation segment and Gly203 [Fig. 1(B)]. Tyr204, which is a conserved residue involved in phosphate binding (TAO2 crystal structure), occupies its normal position. Residues in the activation segment from Gly174 to Val187 (including Asn180 to Thr185 that are missing in the electron density) are involved by crossing between the two subunits of the dimer. The electron density over the intersubunit region is very clear from Phe186 to Asp209, justifying the domain-swap interpretation (see Fig. 3). In addition, the omit map over the central part of intersubunit region ranging from Met198 to Gly203 was generated by simulated annealing refinement with program Phenix and was in full agreement with the OSR1 kinase structure (see Supp. Info. Figs. A and B).32 Also, a structural alignment of dimeric OSR1-KD with TAO2 revealed that P+1 loop and αEF helix derived from the adjacent molecule overlays with the corresponding residues TAO2 (rmsd = 0.18) and makes the same chemical interactions, thus satisfying the definition of domain swapping (see Fig. 4). Two interactions are particularly striking. One concerns a buried ion pair between the glutamate in the conserved APE sequence at the beginning of αEF, and a conserved arginine between αH and αI.33 In OSR1-KD, this ion pair is in the domain-swapped interface with Arg279 forming an ion pair with Glu196′ (′ denotes opposite subunit) [Fig. 5(A)]. The second is a cation-π interaction between Lys148 Nɛ and Trp192′ [Fig. 5(B)]. A tabulation of cation-π interactions34 has revealed that they are present more often in domain-swapped interfaces (95%) than in other interfaces (80%),35 although the reason for this is unclear.
Figure 2.
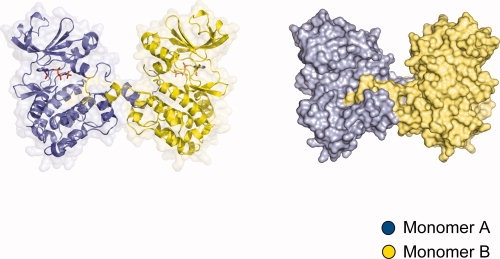
The structure of domain-swapped OSR1 kinase domain dimer. Front view of domain-swapped dimer. One subunit is represented in blue and the other dimer-related subunit is in yellow. The left model is rendered in cartoon and the right model represents surface diagram of domain-swapped dimer.
Figure 3.
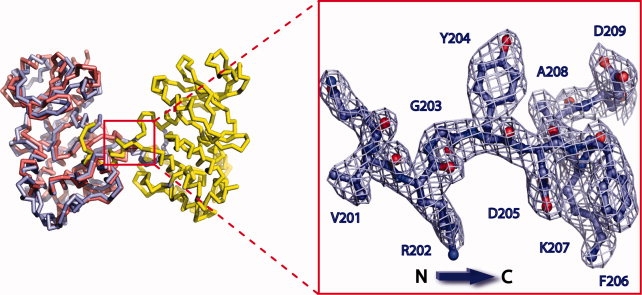
Electron density of intersubunit region in OSR1 kinase domain. Electron density from residues (V201 to D209) spanning intersubunit region between αEF and αF helices.
Figure 4.
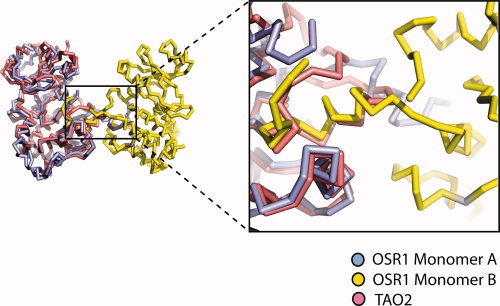
Structural alignment of TAO2 with OSR1 domain-swapped dimer. Non-swapped Ste20 TAO2 kinase (PDB:1U5R) was aligned with one subunit of domain-swapped dimer of OSR1.
Figure 5.
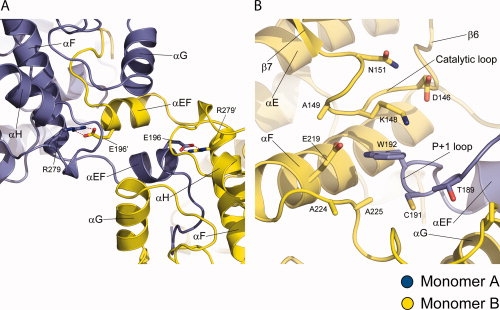
Domain-swapped interface in OSR1 kinase domain. (A) Intersubunit interface between αEF and αF. Polar interaction between R279 and E196′ is represented in red dotted line. Each subunit is rendered in blue and yellow. (B) Intersubunit cation-π interaction between E219 and W192. E219 is from αF and W219 is from P+1 loop of the other subunit.
OSR1-KD is in an inactive conformer
With numerous structures of active and inactive kinases available, structural cues have been identified as hallmarks of active kinases. The N-terminal lobe K-E ion pair noted above is one.29,30 Lys46 in β3 and Glu63 in helix αC are separated by about 12 Å [Fig. 1(B)]. Moreover, helix αC is displaced outward by a helix, αAL, formed of activation loop sequences (Val167 to Gly174). Similar mechanisms of inactivation occur in CDK2, WNK1, and in MEK1/2.29,36,37 In OSR1, both Lys46 and Glu63 are well-pinned down in alternative interactions. Ly46 forms an ion pair with Asp164 (in the DFG motif). Glu63 forms an ion pair with Arg145, which is in the catalytic loop (conserved RD motif) that binds phosphothreonine in active kinases. Second, in active kinases, the activation loop or activation segment contains a β-strand, β9, which forms a β-ribbon with strand β6–β9 ribbon.38–40 These residues are formed into αAL in OSR1 [Fig. 1(B)]. This helix is shared with CDK2, MEK1/2, and WNK1. A third hallmark of active kinases is the presence of a hydrogen bonding network in the active site between two conserved residues in the catalytic loop, Asp151 [TAO2 numbering as in Fig. 6(A)] and Lys153, and Thr185 in the P+1 loop at the C-terminus of activation loop, thereby forming the “D-K-T catalytic triad”27,28,41 [Fig. 6(A)]. The lysine residue (Lys148 in OSR1) is the same as that forming the cation-π interaction discussed earlier. In OSR1-KD, the P+1 loop is derived from the opposite subunit of the domain-swapped dimer, Thr189′ has weak electron density and is displaced from its expected active position, and the D-K-T catalytic triad is not formed [Fig. 6(B)]. This mechanism of inactivation occurs in a very large number of kinases in the CMGC and STE groups of kinases.41 In addition, a formation of hydrophobic spine characteristic of active protein kinase is disrupted (over Tyr78, Met67, Phe165, and His144), with DFG Phe165 flipping out of the active site.42,43
Figure 6.
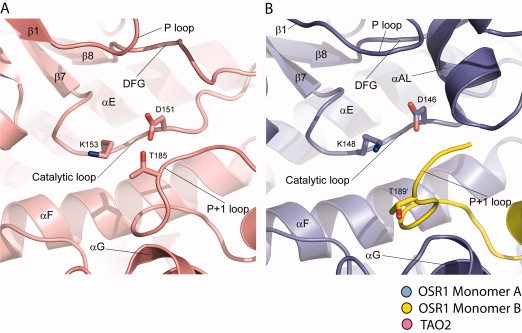
D-K-T catalytic triad in TAO2 and OSR1. (A) D-K-T catalytic triad in TAO2 kinase. (B) D-K-T catalytic triad in domain-swapped OSR1 kinase.
Comparison of OSR1 with other domain-swapped kinases
Recently, CHK2 and DAPK3 in the CAMK group, and SLK, LOK, and STK10 in the STE group have been reported to be domain-swapped (referred to by authors as “activation segment exchange”).44,45 There are similarities and differences between OSR1 and the other domain swapped kinases. In Figure 7, OSR is compared with SLK (PDB; 2JFL). The hinges for the domain swap are similar but not identical. In OSR1, the first hinge is at the C-terminus of αAL (in the activation loop) [Fig. 7(A)]. In SLK, strand β9 is formed in the activation segment, and the hinge directly follows it [Fig. 7(B)]. The orientations of the domain-swapped segment are different in each dimer, giving rise to slightly different subunit orientations.45 The most obvious difference between OSR1 and the SLK is αAL, rather than strand β9. Thus, OSR1 appears more “structurally inactive” than SLK. OSR1 is five residues longer here, which may explain the ability to form αAL. On the other hand, SLK, as well as the other domain-swapped kinases45 do not form the D-K-T catalytic triad discussed earlier. Thus, we conclude all of the kinases listed above are also “structurally inactive.” SLK and the others make many of the same domain-swapped contacts as does OSR1, especially the buried ion pair discussed earlier.
Figure 7.
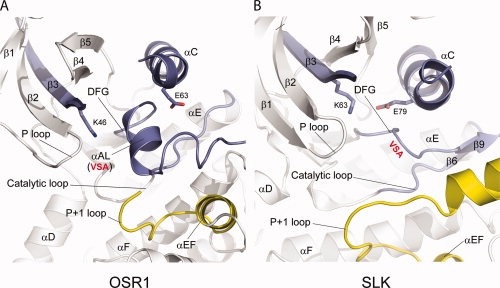
Activation segment in OSR1 and SLK. (A) Activation segment and its neighboring region in OSR1. β9 strand is converted into αAL in domain-swapped OSR1 kinase domain. (B) Activation segment and its neighboring region in SLK. In the doubly phosphorylated SLK (PDB: 2JFL), β9 strand is intact and interacts with β6 in its sheet formation.
Sequence alignment in our hands did not give any clear answer concerning what these proteins have in common that allows them to display domain swapping (see Fig. 8). The domain-swapped kinases have the cation-π interaction (between Lys148 and Trp192′). However, this interaction occurs in all Ste20 kinases, even those that are not swapped. The activation loops, on the other hand, do contain greater similarity to each other. The known domain-swapped Ste20 kinases share the same VSA sequence from strand β9, even OSR1 which has no strand β9. The sequence RXXF is also conserved. The phenylalanine is visible in OSR1 and stabilizes the P+1 loop. In SLK, although not in OSR1-KD, the arginine is visible and forms a buried ion-dipole interaction with an asparagine in the opposite subunit. However, it is really unclear what role this might have for triggering the domain swap event.
Figure 8.
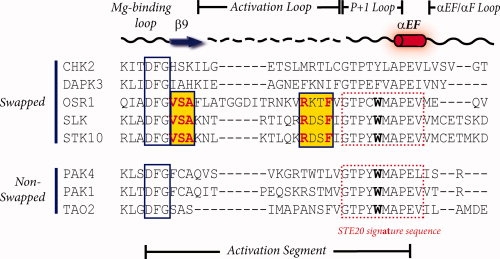
Sequence alignment of activation segments in swapped and nonswapped protein kinases. The sequences of activation segments from swapped and nonswapped protein kinases were aligned. The activation segment encompasses DFG Mg binding motif, β9 strand, activation loop followed by P+1 loop, αEF helix. Only among the domain-swapped Ste20 kinases, conserved residues are found in activation loop, although their function is not clear.
Discussion
What is the function of the domain swapping in these kinases? The idea that has been put forward most strongly is that the organization may allow for trans-autophosphorylation.45 This may well be the case. In OSR1, the available data show that kinase dead OSR1 is not phosphorylated by wild-type WNK1.20 This result supports the idea that trans-autophosphorylation is occurring. On the other hand, the kinase activity of WNK1 is required in cells and in vitro for its activation of OSR1, casting some doubt on the autophosphorylation scheme.20 OSR1 requires sequences outside the kinase domain for activity and its activation,13 and thus clearly there are events occurring that cannot be garnered from knowledge of the kinase domain alone. We are intrigued by the added layers of control potentially conferred by the dimers. The dimers are inactive. Apparently, conformational changes induced by interactions of substrate at remote sites are required to induce an active conformation. This assures that only the correct substrates will be phosphorylated, assuring pathway specificity. Further, the phosphorylation sites are trapped in the dimer interface, where they are inaccessible for “processing,” either by kinases or phosphatases. As discussed earlier, WNK1 and WNK4, which phosphorylate OSR1, contains a docking motif similar to OSR1 substrates, and thus likely also bind to the remote PF2 domains.21 Interactions with the correct processing enzymes, as with substrates, are likely to induce long range conformational changes that release these phosphorylation sites from the interface. Substrate or processing enzyme docking induced conformational changes occur in MAPKs46 and in MAP2Ks (Min et al., submitted for publication). We look forward to more structures of larger fragments and complexes of OSR1 to truly understand its regulation at a molecular level.
Materials and Methods
Cloning, protein expression, and purification
The kinase domain of human OSR1 (residues1–295) was cloned into a pHisParallel vector containing an N-terminal His6 tag and TEV protease cleavage site.47 Transformed Rosetta 2 (DE3) cells (Novagen) were grown in Luria-Bertani medium containing 100 μg/mL ampicillin overnight at 37°C. Twenty milliliters of starter culture was used to inoculate 1 L of LB medium containing 3% ethanol.48 Cells were grown 37°C to an OD600 of 0.7–0.8, then induced with 1 mM isopropyl-thio-galactopyranoside (IPTG), and grown for 16 h at 30°C. Cells were harvested by centrifugation and stored at −80°C. Cell pellets were resuspended in lysis buffer [50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 10 mM imidazole] containing protease inhibitors. Following lysis by sonication, material was sedimented at 35,000g for 1 h. The supernatant was loaded onto a fast-chelating Sepharose column precharged with 0.1 mM NiSO4 and the target protein was eluted with 250 mM imidazole. The eluted fractions containing the human OSR1 kinase domain (residues1–295) were pooled and digested by TEV protease at 16°C overnight to remove N-terminal His6 tag. The reaction mixture was then loaded unto Ni-NTA agarose (Qiagen) to remove the His6 tag and the undigested OSR1. The protein was applied to MonoQ-HR5/5 column (Amersham Pharmacia) and then Superdex 75 16/60 for further purification. By gel filtration, the OSR1 ran as a monomer, eluting at 65 mL. The purified protein was then exchanged to storage buffer (20 mM Tris pH 8.0, 50 mM NaCl, and 2 mM DTT) and concentrated to a final concentration of 10 mg/mL.
Crystallization and data collection
Initial crystallization trials of human OSR1-KD (residues1–295) utilized Crystal Screen I & II (Hampton Research, CA) and other kits, using the hanging-drop vapor diffusion method with 10 mg/mL OSR1-KD in storage buffer, at 16°C. Crystallization trials were also conducted with nucleotide analogs. Following extensive optimization trials, successful condition was then replicated using PEG3350. Crystals were grown by mixing 1.5 μL of protein solution with 1.5 μL of well solution containing 20% (w/v) PEG 3350, 200 mM tri-Lithium citrate tetrahydrate, only in the presence of AMP-PNP. Prior to crystallization, 1 mM AMP-PNP and 5 mM MgCl2 were added to purified protein solution at low protein concentration (2 mg/mL) and the mixture was then concentrated to 12 mg/mL. Crystals appeared about 3 days after setup and grew to dimensions 50 × 60 × 300 in 7 days. Crystals were cryoprotected prior to low-temperature data collection, by transferring them stepwise into a stabilizing solution containing 25% glycerol, 23% (w/v) PEG 3350, 200 mM tri-Lithium citrate tetrahydrate. Crystals were flash frozen by immersion in liquid isopropanol. Diffraction data to 2.25 Å sets were collected for the OSR1 kinase domain/AMP-PNP binary complex at APS beamline 19BM from a single crystal at 100 K. Diffraction data were indexed, integrated, and scaled using HKL2000 programs.49
Structure determination and refinement
Crystals of human OSR1 kinase domain (residues 1–295) complexed with AMP-PNP belong to space group P212121, with 4 molecules per asymmetric unit. The structure of the human OSR1 kinase domain was solved by molecular replacement with the program Phaser.50,51 For the molecular replacement, the checkpoint kinase Chk1 (PDB code: 1NVR) and Ste20 TAO2 kinase (PDB code: 1U5R) were utilized as search models for N-terminal lobe and C-terminal lobe of OSR1 kinase domain, respectively. Density modification combined with solvent flattening and NCS averaging in the CCP4 package produced clear and interpretable map.52 Iterative cycles of rigid-body refinement and restrained refinement with translation-liberation-screw (TLS) using the program REFMAC553 were intervened by manual rebuilding of the model utilizing program Coot.54
Coordinates for the structure of OSR1 kinase domain (1–295) have been deposited in the Protein Data Bank under accession code 3DAK.
Acknowledgments
The authors thank the members of the Goldsmith lab for helpful discussion and Diana Thomchick for help with diffraction data collection and structure refinement. The authors are grateful to the staff at Argonne National Laboratory for help with Synchrotron data collection. Use of the Argonne National Laboratory Structural Biology Center beamlines at the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Biological and Environmental Research, under Contract No. W-31-109-ENG-38.
References
- 1.Leberer E, Dignard D, Harcus D, Thomas DY, Whiteway M. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein beta gamma subunits to downstream signalling components. EMBO J. 1992;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramer SW, Davis RW. A dominant truncation allele identifies a gene, STE20, that encodes a putative protein kinase necessary for mating in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1993;90:452–456. doi: 10.1073/pnas.90.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- 4.Piechotta K, Garbarini N, England R, Delpire E. Characterization of the interaction of the stress kinase SPAK with the Na+-K+-2Cl-cotransporter in the nervous system: evidence for a scaffolding role of the kinase. J Biol Chem. 2003;278:52848–52856. doi: 10.1074/jbc.M309436200. [DOI] [PubMed] [Google Scholar]
- 5.Strange K, Denton J, Nehrke K. Ste20-type kinases: evolutionarily conserved regulators of ion transport and cell volume. Physiology (Bethesda) 2006;21:61–68. doi: 10.1152/physiol.00139.2005. [DOI] [PubMed] [Google Scholar]
- 6.Flatman PW. Cotransporters, WNKs and hypertension: important leads from the study of monogenetic disorders of blood pressure regulation. Clin Sci (Lond) 2007;112:203–216. doi: 10.1042/CS20060225. [DOI] [PubMed] [Google Scholar]
- 7.Delpire E, Gagnon KB. SPAK and OSR1: STE20 kinases involved in the regulation of ion homoeostasis and volume control in mammalian cells. Biochem J. 2008;409:321–331. doi: 10.1042/BJ20071324. [DOI] [PubMed] [Google Scholar]
- 8.Bokoch GM. Biology of the p21-activated kinases. Annu Rev Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- 9.Denton J, Nehrke K, Yin X, Morrison R, Strange K. GCK-3, a newly identified Ste20 kinase, binds to and regulates the activity of a cell cycle-dependent ClC anion channel. J Gen Physiol. 2005;125:113–125. doi: 10.1085/jgp.200409215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamari M, Daigo Y, Nakamura Y. Isolation and characterization of a novel serine threonine kinase gene on chromosome 3p22-21.3. J Hum Genet. 1999;44:116–120. doi: 10.1007/s100380050121. [DOI] [PubMed] [Google Scholar]
- 11.Johnston AM, Naselli G, Gonez LJ, Martin RM, Harrison LC, DeAizpurua HJ. SPAK, a STE20/SPS1-related kinase that activates the p38 pathway. Oncogene. 2000;19:4290–4297. doi: 10.1038/sj.onc.1203784. [DOI] [PubMed] [Google Scholar]
- 12.Tsutsumi T, Ushiro H, Kosaka T, Kayahara T, Nakano K. Proline-and alanine-rich Ste20-related kinase associates with F-actin and translocates from the cytosol to cytoskeleton upon cellular stresses. J Biol Chem. 2000;275:9157–9162. doi: 10.1074/jbc.275.13.9157. [DOI] [PubMed] [Google Scholar]
- 13.Chen W, Yazicioglu M, Cobb MH. Characterization of OSR1, a member of the mammalian Ste20p/germinal center kinase subfamily. J Biol Chem. 2004;279:11129–11136. doi: 10.1074/jbc.M313562200. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Hu J, Vita R, Sun B, Tabata H, Altman A. SPAK kinase is a substrate and target of PKCtheta in T-cell receptor-induced AP-1 activation pathway. EMBO J. 2004;23:1112–1122. doi: 10.1038/sj.emboj.7600125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delpire E, Gagnon KB. SPAK and OSR1, key kinases involved in the regulation of chloride transport. Acta Physiol (Oxf) 2006;187:103–113. doi: 10.1111/j.1748-1716.2006.01565.x. [DOI] [PubMed] [Google Scholar]
- 16.Piechotta K, Lu J, Delpire E. Cation chloride cotransporters interact with the stress-related kinases Ste20-related proline-alanine-rich kinase (SPAK) and oxidative stress response 1 (OSR1) J Biol Chem. 2002;277:50812–50819. doi: 10.1074/jbc.M208108200. [DOI] [PubMed] [Google Scholar]
- 17.Dowd BF, Forbush B. PASK (proline-alanine-rich STE20-related kinase), a regulatory kinase of the Na-K-Cl cotransporter (NKCC1) J Biol Chem. 2003;278:27347–27353. doi: 10.1074/jbc.M301899200. [DOI] [PubMed] [Google Scholar]
- 18.Moriguchi T, Urushiyama S, Hisamoto N, Iemura S, Uchida S, Natsume T, Matsumoto K, Shibuya H. WNK1 regulates phosphorylation of cation-chloride-coupled cotransporters via the STE20-related kinases, SPAK and OSR1. J Biol Chem. 2005;280:42685–42693. doi: 10.1074/jbc.M510042200. [DOI] [PubMed] [Google Scholar]
- 19.Vitari AC, Deak M, Morrice NA, Alessi DR. The WNK1 and WNK4 protein kinases that are mutated in Gordon's hypertension syndrome phosphorylate and activate SPAK and OSR1 protein kinases. Biochem J. 2005;391:17–24. doi: 10.1042/BJ20051180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anselmo AN, Earnest S, Chen W, Juang YC, Kim SC, Zhao Y, Cobb MH. WNK1 and OSR1 regulate the Na+, K+, 2Cl-cotransporter in HeLa cells. Proc Natl Acad Sci USA. 2006;103:10883–10888. doi: 10.1073/pnas.0604607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zagorska A, et al. Regulation of activity and localization of the WNK1 protein kinase by hyperosmotic stress. J Cell Biol. 2007;176:89–100. doi: 10.1083/jcb.200605093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson FH, et al. Human hypertension caused by mutations in WNK kinases. Science. 2001;293:1107–1112. doi: 10.1126/science.1062844. [DOI] [PubMed] [Google Scholar]
- 23.Vitari AC, Thastrup J, Rafiqi FH, Deak M, Morrice NA, Karlsson HK, Alessi DR. Functional interactions of the SPAK/OSR1 kinases with their upstream activator WNK1 and downstream substrate NKCC1. Biochem J. 2006;397:223–231. doi: 10.1042/BJ20060220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delpire E, Gagnon KB. Genome-wide analysis of SPAK/OSR1 binding motifs. Physiol Genomics. 2007;28:223–231. doi: 10.1152/physiolgenomics.00173.2006. [DOI] [PubMed] [Google Scholar]
- 25.Villa F, Goebel J, Rafiqi FH, Deak M, Thastrup J, Alessi DR, van Aalten DM. Structural insights into the recognition of substrates and activators by the OSR1 kinase. EMBO Rep. 2007;8:839–845. doi: 10.1038/sj.embor.7401048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao B, Bower MJ, McDevitt PJ, Zhao H, Davis ST, Johanson KO, Green SM, Concha NO, Zhou BB. Structural basis for Chk1 inhibition by UCN-01. J Biol Chem. 2002;277:46609–46615. doi: 10.1074/jbc.M201233200. [DOI] [PubMed] [Google Scholar]
- 27.Zhou T, Raman M, Gao Y, Earnest S, Chen Z, Machius M, Cobb MH, Goldsmith EJ. Crystal structure of the TAO2 kinase domaactivation and specificity of a Ste20p MAP3K. Structure. 2004;12:1891–1900. doi: 10.1016/j.str.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 28.Knighton DR, Zheng JH, Ten Eyck LF, Ashford VA, Xuong NH, Taylor SS, Sowadski JM. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 29.De Bondt HL, Rosenblatt J, Jancarik J, Jones HD, Morgan DO, Kim SH. Crystal structure of cyclin-dependent kinase 2. Nature. 1993;363:595–602. doi: 10.1038/363595a0. [DOI] [PubMed] [Google Scholar]
- 30.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, Eisenberg D. 3D domain swapping: as domains continue to swap. Protein Sci. 2002;11:1285–1299. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung L-W, Read RJ, Adams PD. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor SS, Radzio-Andzelm E. Three protein kinase structures define a common motif. Structure. 1994;2:345–355. doi: 10.1016/s0969-2126(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 34.Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proc Natl Acad Sci USA. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dehouck Y, Biot C, Gilis D, Kwasigroch JM, Rooman M. Sequence-structure signals of 3D domain swapping in proteins. J Mol Biol. 2003;330:1215–1225. doi: 10.1016/s0022-2836(03)00614-4. [DOI] [PubMed] [Google Scholar]
- 36.Min X, Lee B-H, Cobb MH, Goldsmith EJ. Crystal structure of the kinase domain of WNK1, a kinase that causes a hereditary form of hypertension. Structure. 2004;12:1303–1311. doi: 10.1016/j.str.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 37.Ohren JF, et al. Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat Struct Mol Biol. 2004;11:1192–1197. doi: 10.1038/nsmb859. [DOI] [PubMed] [Google Scholar]
- 38.Johnson LN, Noble MEM, Owen DJ. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 39.Johnson LN, Lowe ED, Noble MEM, Owen DJ. The structural basis for substrate recognition and control by protein kinases. FEBS Lett. 1998;430:1–11. doi: 10.1016/s0014-5793(98)00606-1. [DOI] [PubMed] [Google Scholar]
- 40.Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 41.Goldsmith EJ, Akella R, Min X, Zhou T, Humphreys JM. Substrate and docking interactions in serine/threonine protein kinases. Chem Rev. 2007;107:5065–5081. doi: 10.1021/cr068221w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci USA. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kornev AP, Taylor SS, Ten Eyck LF. A helix scaffold for the assembly of active protein kinases. Proc Natl Acad Sci USA. 2008;105:14377–14382. doi: 10.1073/pnas.0807988105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliver AW, Paul A, Boxall KJ, Barrie SE, Aherne GW, Garrett MD, Mittnacht S, Pearl LH. Trans-activation of the DNA-damage signalling protein kinase Chk2 by T-loop exchange. EMBO J. 2006;25:3179–3190. doi: 10.1038/sj.emboj.7601209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pike AC, Rellos P, Niesen FH, Turnbull A, Oliver AW, Parker SA, Turk BE, Pearl LH, Knapp S. Activation segment dimerization: a mechanism for kinase autophosphorylation of non-consensus sites. EMBO J. 2008;27:704–714. doi: 10.1038/emboj.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou T, Sun L, Humphreys J, Goldsmith EJ. Docking interactions induce exposure of activation loop in the MAP kinase ERK2. Structure. 2006;14:1011–1019. doi: 10.1016/j.str.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 47.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 48.Thomas JG, Baneyx F. Divergent effects of chaperone overexpression and ethanol supplementation on inclusion body formation in recombinant Escherichia coli. Protein Expr Purif. 1997;11:289–296. doi: 10.1006/prep.1997.0796. [DOI] [PubMed] [Google Scholar]
- 49.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW, editor; Sweet RM, editor. Methods in enzymology. Vol. 276. San Diego, CA: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 50.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 51.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.The CCP4 suite: programs for protein crystallography. Collaborative Computational Project, Number 4. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 53.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]


