Summary
The mammalian AP-endonuclease (APE1) repairs abasic (AP) sites and strand breaks with 3' blocks in the genome that are formed both endogenously and as intermediates during base excision repair (BER). APE1 has an unrelated activity as a redox activator (and named Ref-1) for several trans-acting factors. In order to identify whether any of the seven cysteine residues in human APE1 (hAPE1) affects its enzymatic function, we substituted these singly or multipully with serine. The repair activity is not affected in any of the mutants except those with C99S mutation. The Ser99-containing mutants lost affinity for DNA and its activity was inhibited by 10 mM Mg2+. However, the Ser99 mutant has normal activity in 2 mM Mg2+. Using crystallographic data and molecular dynamic (MD) simulation, we have provided a mechanistic basis for the altered properties of the C99S mutant. We earlier predicted that Mg2+ with potential binding sites A and B, bound at the B-site of WT APE1-substrate complex and moves to the A-site after cleavage occurs, as observed in the crystal structure. The APE1-substrate complex is stabilized by a H-bond between His309 and the AP-site in the C99S mutant. We now show that this bond is broken to destabilize the complex in the absence of the Mg2+. This effect due to the mutation of Cys99, ~16Å from the active site, on the DNA binding and activity is surprising.
Mg2+ at the B-site promotes stabilization of the C99S mutant complex. At higher Mg2+ concentration the A-site is also filled causing the B-site Mg2+ to shift together with the AP-site. At the same time, the H-bond between His309 and the AP-site shifts towards the 5' site of DNA. These shifts could explain the lower activity of the C99S mutant at higher [Mg2+]. The unexpected involvement of Cys99 in APE1’s substrate binding and catalysis provides an example of involvement of a residue far from the active site.
Keywords: Human APE1, Ref-1, AP-endonuclease, DNA base excision repair, DNA binding
Introduction
The genomes of all organisms are being continuously damaged due to both spontaneous chemical reactions and exposure to endogenous genotoxic agents, in particular, reactive oxygen species (ROS) that are generated as by-products of respiration. 103 to 105 DNA lesions are estimated to be induced in a genome per day including oxidized bases, AP-sites and DNA single-strand breaks 1,2. Unrepaired AP-sites and their cleavage products are cytotoxic and mutagenic 3. AP-endonucleases (APEs), responsible for repair of the AP-sites and 3' end cleaning at single-strand breaks, are highly conserved among various species. APEs belong to two families, Xth and Nfo, whose prototypes are exonuclease III (Xth) and endonuclease IV (Nfo) in E. coli 4–6. Mammalian cells express only one active APE, APE1, an ortholog of E. coli Xth with an unconserved N-terminal extension 4,7. All APEs have two intrinsic activities in DNA repair. They act as an endonuclease in cleaving AP-sites to generate 3'OH and 5' phosphodeoxyribose termini. These also act as a 3' phosphodiesterase/exonuclease to remove 3' blocking phosphodeoxyribose or its fragments generated during strand breaks, and by ROS or by DNA glycosylases that excise oxidized bases in the first step of BER 4. Thus APE’s key function is to produce a 3'OH terminus which serves as a primer for repair synthesis 8.
The mammalian APE1 was independently discovered as redox-factor 1 (Ref-1), involved in activation shown to many critical trans-acting proteins including C-Jun and p53 9–12. Cys65 (Cys64 in mouse APE1), initially identified as the reductant for such activation, was subsequently shown to be dispensable for this function 13,14. APE1/Ref-1 was also identified as a component of the nCaRE-bound protein complexes 15,16. We have recently shown that Lys6 or Lys7 of APE1 is acetylated by the histone acetyltransferase p300 that enhances APE1’s binding to nCaREs 17. APE1 was earlier shown to be essential for embryo development and no APE1-null mammalian cell could be established 13. Our subsequent studies established essentiality of APE1 in somatic cells which is due to both acetylation-dependent functions and DNA repair activity 9.
The X-ray crystallographic structure of mammalian/human APE1 (hAPE1) has provided significant insight into the molecular mechanism of AP-site recognition and cleavage by APE1 18,19. The phosphodiester bond cleavage by APE1 requires Mg2+, like other DNase I family members. We proposed earlier that this divalent metal ion shuttles between two metal binding sites, A- and B in hAPE1 20. The kinetic and binding studies of WT and mutant APE1 show that product dissociation is Mg2+ concentration dependent 21. Arg156 and Tyr128 are the predominant residues bound to the minor groove of AP-site containing DNA for stabilizing the enzyme-DNA complex 22. Several amino acid residues in hAPE1 have been identified by biochemical and structural studies that are required for phosphodiester cleavage at the AP-site 4,18,21,23–26. The critical residues required for phosphodiester bond cleavage and Mg2+ coordination are Glu96, Asp210 and His309 all of which are conserved in the Xth family members. His309 forms an H-bond with the AP-phosphate and is thus critical for substrate/product binding 20,21,24,27. APE1 contains several highly conserved sequence motifs, including 94LQETK98, in which Glu96 is involved in coordinating Mg2+ 23.
Redox regulation by APE1 via one or more of 7 Cys residue(s) has been investigated for more than a decade. Cys65, Cys93, and Cys310 have been implicated in its redox activity 13,29. Interestingly, some Cys residues are located close to the catalytic core domain for APE activity. These include C93 and C99 which are adjacent to the metal coordinating domain, and C310 which is adjacent to the active site H309. However, there have been no systematic studies to elucidate the possible role of the Cys residues in catalysis. This study is an effort to identify the active site for redox activation; we individually mutated all 7 Cys residues in hAPE1 to Ser, and tested for a possible linkage between the redox function and AP-site cleavage activity. Our results indicate that all Cys residues are dispensable for the AP-endonuclease or phosphodiesterase activity. However, unexpectedly, the C99S mutant lost intrinsic affinity for the AP-DNA substrate in the absence of Mg2+, and its AP-endonuclease activity, comparable to the WT APE1 at low Mg2+ concentration, is strongly inhibited at high Mg2+ concentration. This unusual behaviour of the Cys99 mutant can be explained by molecular dynamic (MD) simulation analysis based on our previously proposed moving metal mechanism20.
Results
Cys Residues in APE1 are Dispensable for AP-endonuclease Activity
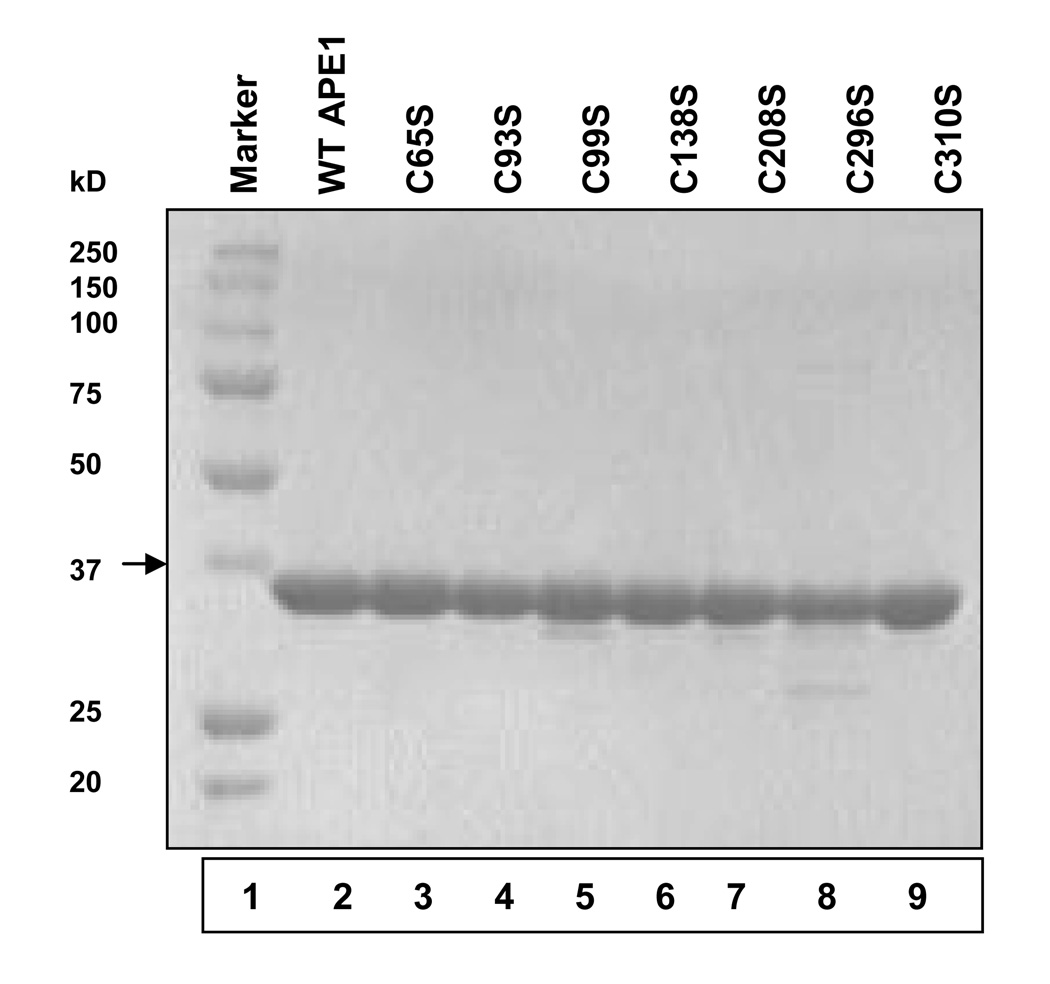
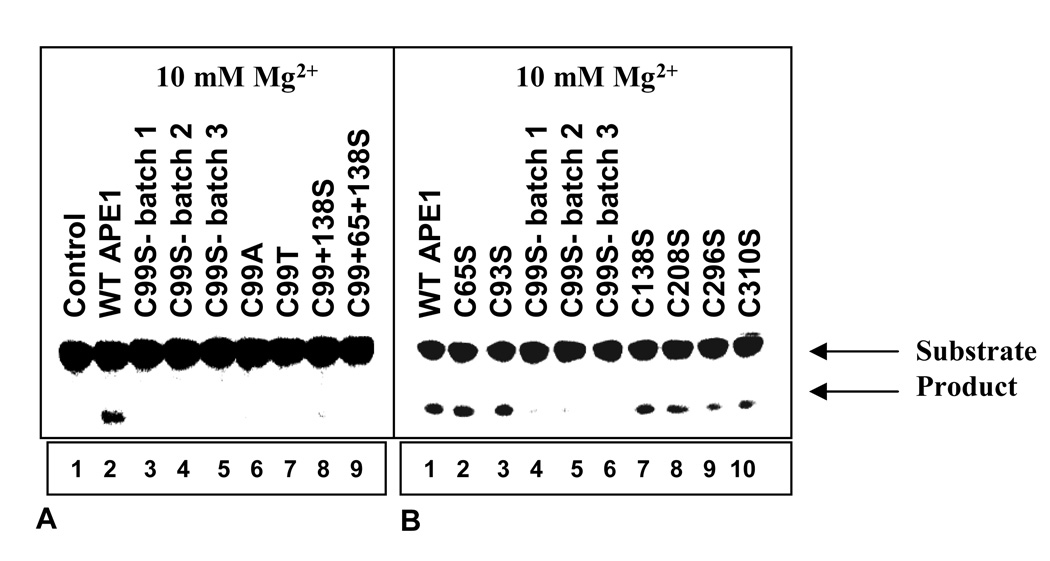
WT APE1 and its Cys mutants were expressed as His-tag fusion polypeptides as described in Materials and Methods. After affinity purification and cleavage of the His-tag, we purified the untagged proteins to near homogeneity by FPLC using Heparin-Sepharose (Fig.1). To test whether the mutation affected APE1’s activity, we used a 43-mer duplex oligo containing the stable AP-site analog tetrahydrofuran (THF) as position 31 30. Our standard assay buffer contained 10 mM Mg2+, and we ensured a linear dependence of activity with respect to the amount of enzyme and time. We observed that all Cys mutants except those with C99S mutation were at least as active as the WT APE1 (Fig. 2b). We concluded that the Cys residues, including those reported to be involved in Ref-1 function are dispensable for APE1’s repair activity. However, the AP-endonuclease activity of the C99S mutant as well as of the double mutants C138S/C99S and C65S/C99S was strongly inhibited in the presence of 10 mM Mg2+. Furthermore, the loss of activity due to Cys99 mutation was independent of Cys substitution with Ser, Ala or Thr (Fig. 2a). Finally, in order to confirm that the activity loss due to Cys99 was not an artefact, we produced several independent batches of the C99S mutant all of which behaved identically.
Figure 1.
Purification of WT and mutant (Cys→Ser) APE1 proteins. Coomassie blue staining of purified WT APE1 and Ser mutants (0.5 µg protein) after SDS-PAGE (10% polyacrylamide). Lane 1, protein marker; lane 2, WT APE1; lane 3, C65S; lane 4, C93S; lane 5, C99S; lane 6, C138S; lane 7, C208S; lane 8, C296S; and lane 9, C310S.
Figure 2.
Mg2+- dependent AP-endonuclease activity of the WT and mutant (Cys→Ser, Ala and Thr) APE1 proteins. The 43-mer, THF-containing oligonucleotide was incubated with 33 pM APEs for 3 min at 37° C. AP-endonuclease activity measured using a buffer containing 10 mM Mg2+. (a) Lane 1, no protein; lane 2, WT APE1; lanes 3–5, three different batches of C99S mutant; lane 6, C99A mutant; lane 7, C99T mutant; lane 8, C99S/C138S double mutant; lane 9, C99S/C65S/C138S triple mutant. (b) Lane 1, WT APE1; lanes 2, C65S mutant; lane 3, C93S mutant; lanes 4–6, three different batches of C99S mutant; lane 7, C138S mutant; lane 8, C208S mutant; lane 9, C296S mutant; lane 10, C310S mutant.
The Cys99 APE1 Mutant is Inhibited by High [Mg2+]
Further studies revealed that the C99S mutant was active in the presence of low Mg2+ concentration. In fact, the activity of Cys99 mutants was comparable to that of the WT and other Cys mutants in 0.5 or 2 mM Mg2+ (Supplementary Fig. 1) while only these Cys99 mutants were strongly inhibited at 5 mM and higher Mg2+ concentration.
Loss of Substrate DNA Binding due to C99S Mutation
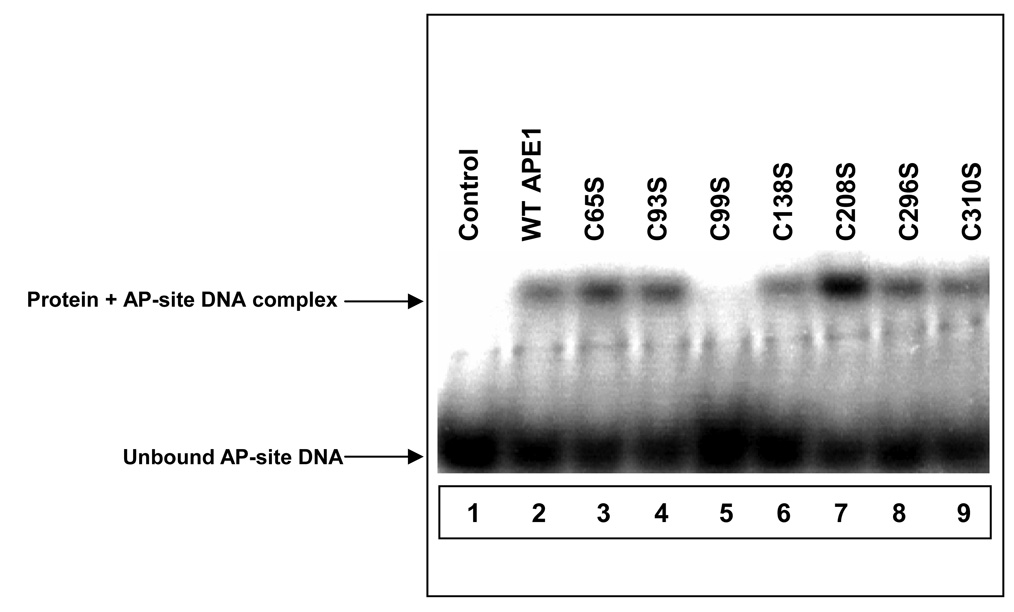
APE1 has significant affinity for the AP-DNA substrate in the absence of Mg2+ and the structure of APE1-substrate oligo complex without a divalent metal ion was solved earlier by X-ray crystallography 19. We used EMSA to confirm binding of WT APE1 to the AP-site in the absence of Mg2+ (to prevent cleavage of the bound substrate). However, unlike WT and other Ser mutant polypeptides, the C99S mutant did not bind to the AP-site, (Fig. 3). This suggested that, unlike other Cys residues, Cys99 in hAPE1 is directly or indirectly involved in contact with the substrate AP-site.
Figure 3.
EMSA of THF-DNA binding by WT APE1 and mutants. Lane 1, no protein; lane 2, WT APE1; lane 3, C65S mutant; lane 4, C93S mutant; lane 5, C99S mutant; lane 6, C138S mutant; lane 7, C208S mutant; lane 8, C296S mutant; and lane 9, C310S mutant. The experiments have been repeated at least four times. Other details are given in Materials and Methods.
Fluorescence Emission Spectra of WT and C99S Mutant APE1
Intrinsic tryptophan (Trp) fluorescence emission spectra were generated for the WT and C99S APE1. The fluorescence output for the mutant was less than that of the WT protein without any change in the emission spectra (Supplementary Fig. 2). Similar decrease in emission intensity without a change in spectra was earlier observed for the K98R mutant APE1 which we had interpreted to be due to an increase in collisional quenching 23.
Kinetic Parameters for WT vs. C99S APE1
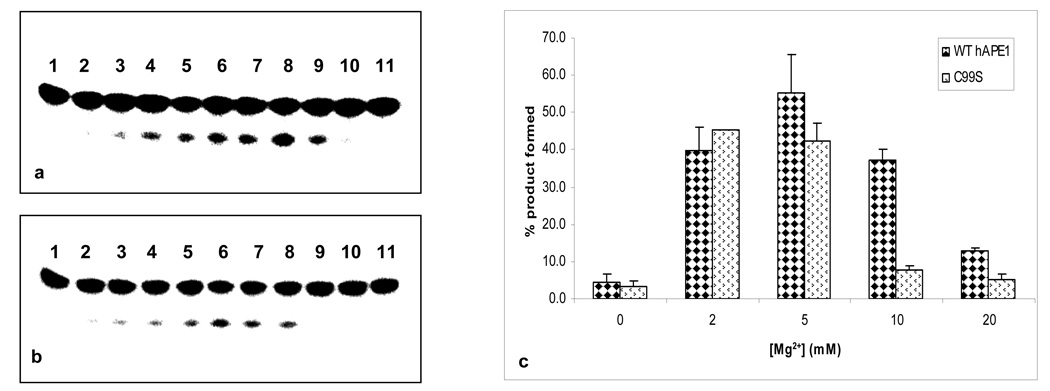
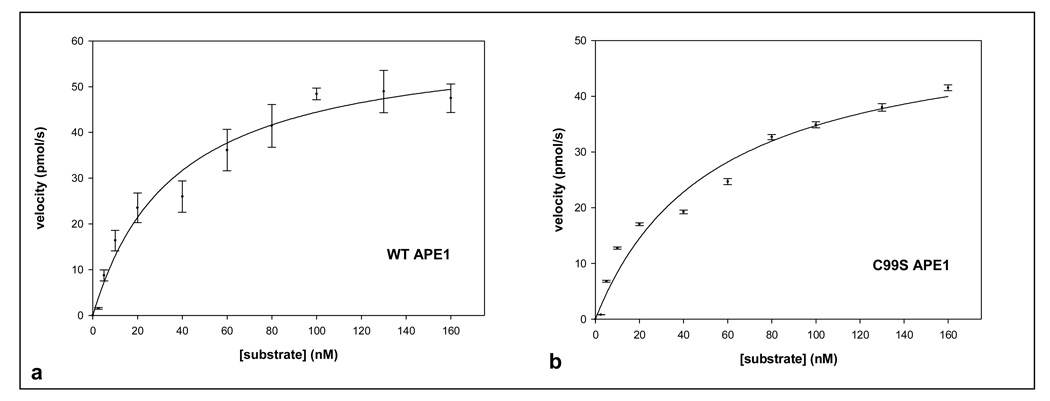
To further probe the effect of the C99S mutation on APE1’s enzymatic activity, we determined the kinetic parameters of purified WT and mutant enzymes. Increasing Mg2+ concentration to 10 mM inhibited product formation by 5.4-fold in the mutant as compared for the WT APE1 (Fig. 4c). At 20 mM Mg2+, the product formation with WT APE1 was inhibited 4.2-fold and with the C99S mutant ~14-fold relative to the activity of the WT protein in 5 mM Mg2+ (Fig. 4c). Fig. 5 shows that the Km of the C99S mutant in 2 mM Mg2+ was slightly higher (53.6 nM), than of the WT enzyme (36.8 nM), which is comparable to the published values 23,31. However, the kcat values of the mutant and WT proteins were comparable in 2 mM Mg2+ (Table 1).
Figure 4.
Effect of Mg2+ on AP-endonuclease activity of (a) WT and (b) C99S mutant APE1. Lane 1, control; lanes 2–11, 0, 0.1, 0.5, 1, 2, 3, 5, 10, 20, and 40 mM Mg2+ respectively. Lane 2 also contains 0.1 mM EDTA. (c) Effect of Mg2+ on the activity of WT and Ser99 mutants.
Figure 5.
Reaction kinetics of WT and C99S mutant APE1 (33 pM) in presence of 2 mM [Mg2+]. (a) WT APE1 and (b) C99S mutant proteins were incubated with 0–160 nM of THF-containing oligonucleotide at 37° C for 3 min. Other details are described in Materials and Methods. Data represent mean values of four independent experiments.
Table 1.
Kinetic parameters for AP-endonuclease activity of wild-type and C99S mutant APE1 proteins in presence of 2 mM Mg2+
| Km (nM) | kcat (s−1) | kcat /Km(nM−1 s−1) | |
|---|---|---|---|
| WT APE1 | 36.8±7.6 | 3.36±0.23 | 0.091 |
| C99S APE1 | 53.6±13.1 | 2.91±0.28 | 0.054 |
Molecular Dynamic Simulation
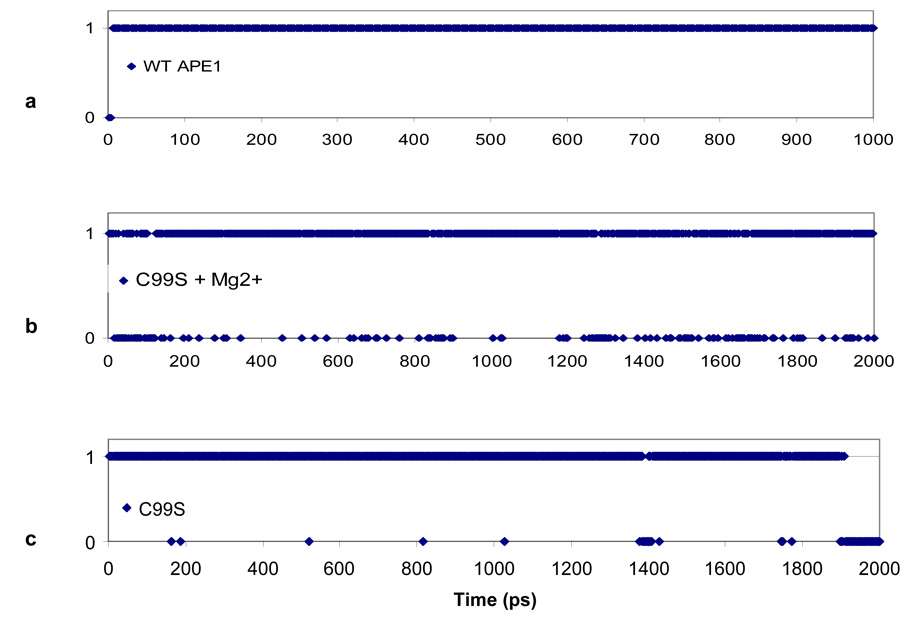
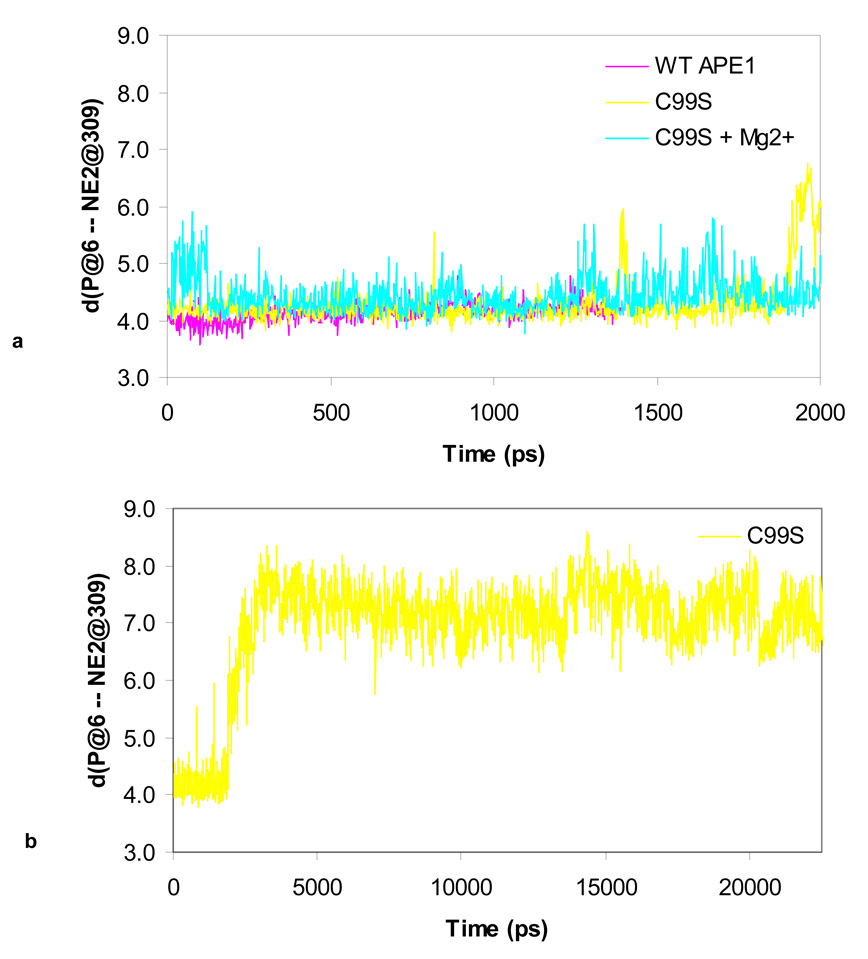
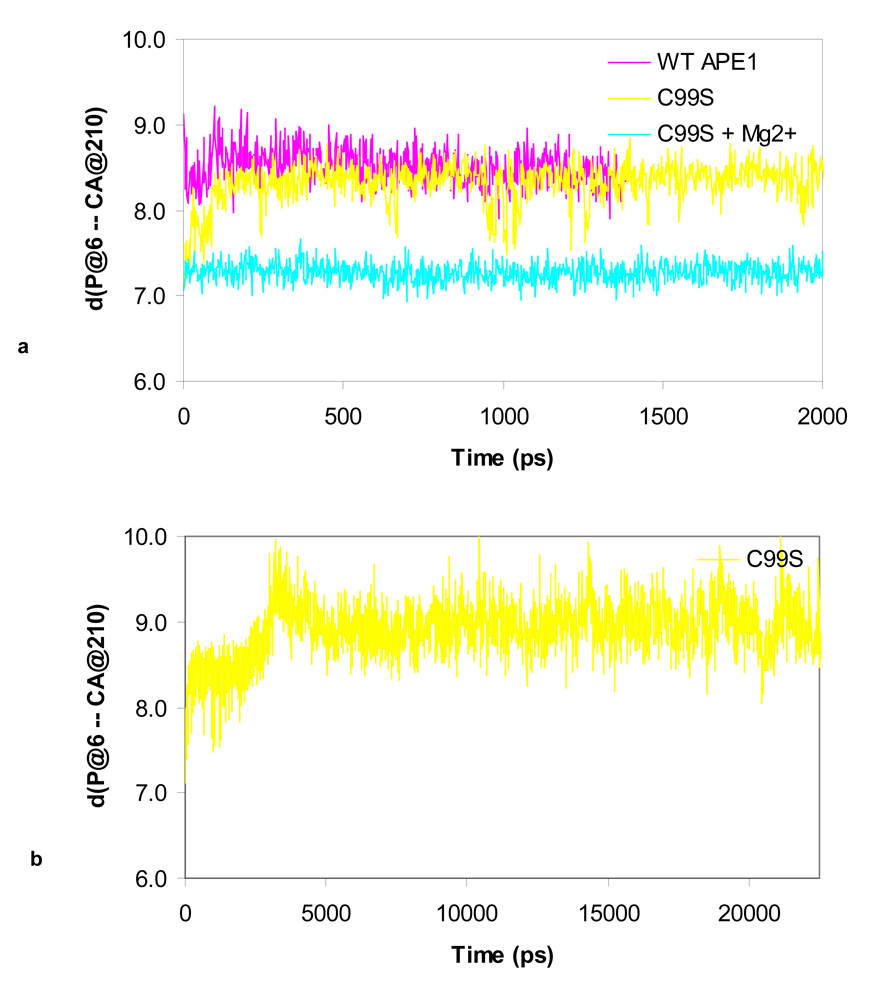
The impact of the C99S mutation on the DNA binding and endonuclease activity of APE1 was quite surprising in view of the fact that Cys99 is ~16Å away from the active site. We then performed MD-simulation analysis to examine the effect of this mutation on the structure and dynamics of hAPE1 as our previous work on simulation for WT hAPE1 led us to postulate a “moving ion mechanism” 20. We predicted that in the pre-cleavage complex of APE1 and DNA, the divalent metal ion is present at the buried B-site, where it is coordinated by Asp210, Glu96, Asn212 and AP-phosphate. We also showed that two H-bonds are stably formed between Asp283 and His309 and between His309 and the AP-phosphate oxygen. Here we compare the presence of the H-bond between His309 and the AP-site in the WT and C99S APE1 during a 2 ns dynamics simulation. Fig. 6 shows whether this H-bond is formed (1) or broken (0), while Fig. 7 shows the distances among the atoms NE2 of His309 and P of the AP-site. The H-bond in the WT enzyme is stable, and independent of the presence of Mg2+ (Fig. 6a and Fig. 7a). For the C99S mutant this H-bond between His309 and AP-site is metastable in the presence of Mg2+ (Fig. 6b), and in the absence of Mg2+ this bond is completely broken at ~ 1800 ps (Fig. 6c, Fig. 7a & b). At this time, the AP-site moves away, increasing the distance from ~ 4.2 Å to 8 Å between the atoms at NE2 of His309 and the P on the AP-site (Fig. 7). At the same time, the binding to DNA weakens as the distance to the CA atom of Asp210 increases (Fig. 8b). Adding Mg2+ to the C99S mutant changes the dynamics of the AP-site along with rest of the DNA. As shown in Fig. 8a, Mg2+ pulls the AP-site down by ~ 1 Å into the active site pocket towards Asp210.
Figure 6.
Kinetics of H-bond formation/breakage between NE2 of H309 and O1P of the AP-site phosphate for (a) WT APE1, (b) C99S mutant with Mg2+ at the B-site, and (c) C99S APE1 mutant. The value is 1 or 0 when H-bond is formed or not respectively.
Figure 7.
Time course of the distance between atoms NE2 on the His309 and P on the AP-site for (a) WT APE1, C99S and C99S + Mg2+ simulations, and (b) C99S APE1 mutant simulation.
Figure 8.
Typical time course for the distance between atoms CA of Asp210 and P of the AP-site for (a) WT APE1, C99S and C99S + Mg2+ simulations, and (b) C99S APE1 mutant simulation.
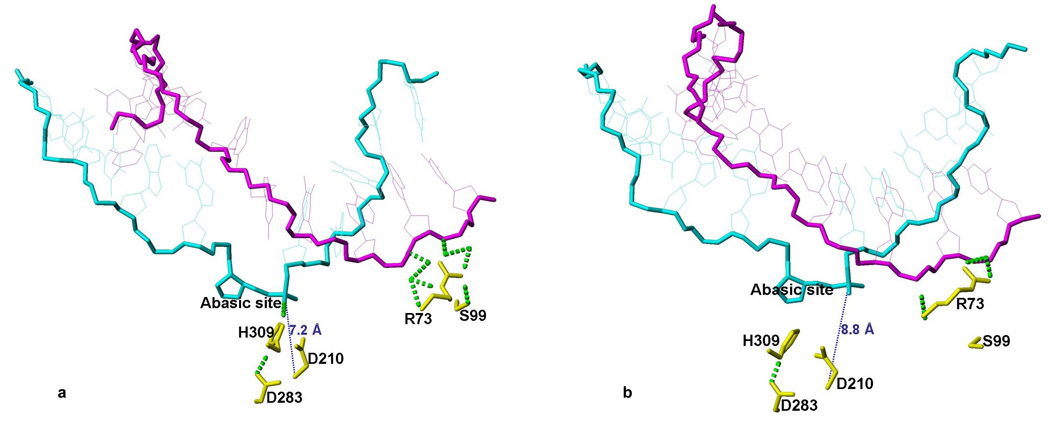
Cys99 in WT APE1 does not interact with DNA at all as we had shown earlier in several independent simulations 20. The shortest H-bonds to the complimentary strand in DNA are formed with the Arg73 side chain. However, in several independent simulations with C99S mutation, Arg73 forms more H-bonds to DNA phosphate compared to WT APE1, and Ser99 directly forms an H-bond to the DNA or to Arg73. These additional H-bonds pull the complementary strand closer to Arg73 and Ser99. The change in the relative position of the complimentary strand is transmitted via the DNA back bone and weakens the H-bond between the AP-site and His309. Fig. 9 shows two snapshots of simulation with the C99S mutant at 1 ps and at 22 ns. At the beginning, there are 7 H-bonds between Arg73 and the DNA, one H-bond is also formed between Arg73 and Ser99 and towards the end, only 3 H-bonds are formed between Arg73 and DNA, which is similar to the situation with WT APE1.
Figure 9.
Snapshots of the C99S mutant simulation at the beginning after 1 ps (panel a) and at the end after 22500 ps (panel b). Only the DNA and key residues that are important for this study in the C99S mutant protein are shown. The AP-site containing DNA strand is shown in cyan and the complementary strand in magenta. The side chains of key residues of the C99S mutant are shown in yellow. Dotted green and blue lines represent H-bonds and the distance is indicated.
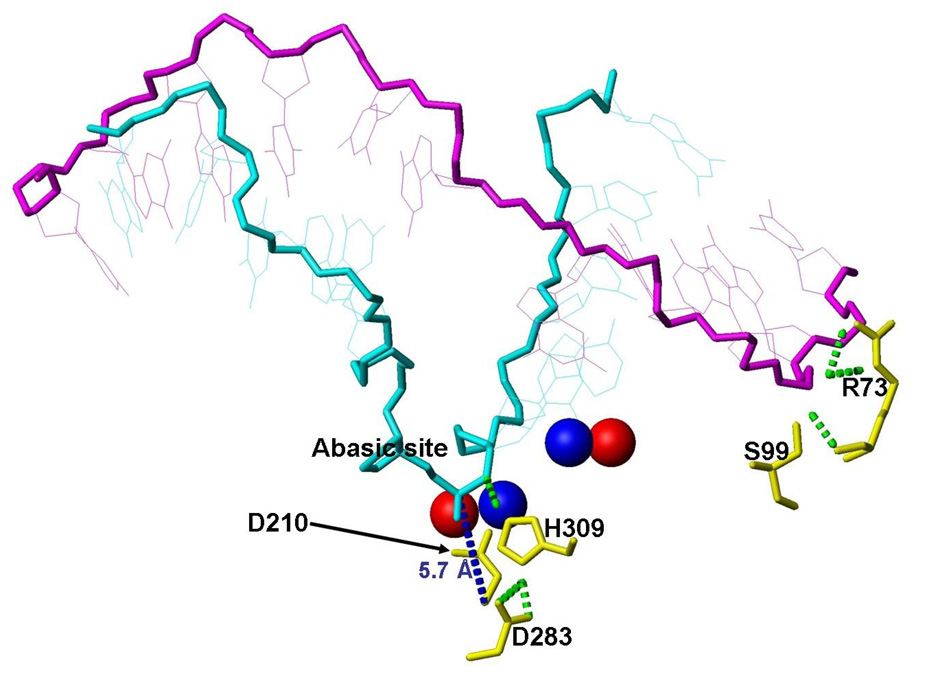
Our earlier study with WTAPE1 showed that Mg2+ located at the A- or B-site in WT APE1 were 5.7 Å apart in the unperturbed position20. However, at higher [Mg2+] when both sites are filled, two Mg2+ ions move apart and the distance increases to 7.7 Å after 500 ps. Analogous simulation with the C99S mutant shows further increase of the inter-metal distance to 9.6 Å (Fig. 10). The final positions of Mg2+ are shown in red spheres, and the unperturbed positions of the A- and B-sites are shown in blue spheres. In this study and our previous simulations, Mg2+ at the B-site was coordinated by the phosphate oxygen of the AP-site and Asp210. Accordingly, the AP-site and the P are also displaced by 2.6 Å. The AP-site is still bound at the active site as indicated by its distance of 5.7 Å from the CA-atom of Asp210. However, the substrate stabilizing H-bond is now between His309 and O3'. It is shifted towards the 5' end of the DNA. At the end of the simulation the relative positions of the active site constituents were significantly disturbed.
Figure 10.
Snapshots of simulation of the C99S mutant after 500 ps with two Mg2+ ions in the active site. The positions of key DNA residues and Mg2+ are shown. The final Mg2+ positions are shown in red and for comparison the starting positions, which are identical to the unperturbed pre-cleavage A- and B-site, are shown in blue. Important H-bonds are shown in dotted green lines.
Taken together, our simulations show that the Ser99 mutation introduces stronger interaction between the DNA, Arg73 and Ser99 sites. This tension propagates via H-bonds between the DNA strands along the backbone to the AP-site. In the absence of Mg2+, this weakens and finally breaks the AP-site stabilizing H-bond between the H309 to an AP-site phosphate oxygen. The Cys99Ser mutation thus has the similar effect as the His309 to Asn309 mutation also breaks the H-bond to the phosphate oxygen and releases the substrate at higher rate than the WT enzyme 21. In the presence of Mg2+, the tension, which originates from the pull of Arg73 and Ser99 site is partially compensated by the interaction with Mg2+. Thus the H-bond between His309 and an AP-site phosphate oxygen is weakened; thus the inter-nuclear distance fluctuates and eliminating the interaction. Nevertheless, at higher Mg2+ concentration, when both metal binding sites are expected to be filled, the enzymatic activity decreases. This is consistent with our earlier finding that having two metal ions in the active site caused the metal at the B-site to move about 2.6 Å and the inter-nuclear distance increased from 5.7 Å (unperturbed A to B-site distance) to 7.7 Å in WTAPE120. In simulation with the Ser99 mutant and two Mg2+ ions at the active site, this distance increased even further to 9.6 Å. This alters the active site geometry to the point where the phosphodiester bond cleavage activity is strongly suppressed while the substrate is still bound in the active site.
Discussion
The mammalian APE1 is an extremely unusual DNA repair protein because of its second unrelated activity in transcriptional regulation. The situation turned out to be more complex with the discovery of two regulatory activities in APE1/Ref-1, one in reductive activation of some transcription factors 11,32, and the other in acetylation-dependant modulation of other trans-acting factors 17. We have shown that the second regulatory function is essential for survival, in addition to the DNA repair activity 9. However, essentiality of the repair activity was unexpected because the lack of APEs including APE1’s orthologs in E. coli and yeast does not cause cell death 33,34.
The Cys residues in mammalian APE1 became important after the discovery of redox activation (Ref-1) function of this protein. It is interesting to note that among the 7 Cys residues in hAPE1, Cys208 and Cys310 are completely conserved among all APEs while Cys99 and Cys93 and Cys296 are conserved only among metazoan APEs. It is also intriguing that Cys208, Cys310 and Cys99 are close to the key residues Asp210, His309 and Glu96 respectively, which are involved in catalysis and metal binding for the AP-endonuclease activity. Furthermore, an earlier study suggested direct involvement of Cys310 in the reaction chemistry of hAPE1 29. Although Cys residues have not been implicated in the catalytic steps of other APEs, we decided to directly assess their role in hAPE1 by characterizing site-specific mutations. Our results clearly show that at low [Mg2+], substitution of individual Cys with Ser for all 7 Cys residues did not affect enzyme turnover in regards to either endonuclease or phosphodiesterase activity. In fact, Ser138 and Ser208 mutants may have slightly higher specific activity than the WT enzyme. However, the aberrant behaviour of the Ser99 mutant both in regards to its affinity loss for the substrate in the absence of Mg2+ was surprising. A small reduction in quantum yield without a change in the emission spectra of intrinsic Trp fluorescence suggested only a subtle change in the tertiary structure of hAPE1 due to the C99S mutation. To gain more definitive insight about the structural perturbation which caused this unusual behaviour, we exploited to MD modeling and extended our previous simulation studies.
Earlier we have shown that APE1’s binding to the substrate/product DNA via H-bonds are necessary for efficient functioning of the BER pathway 35. The present study shows the importance of the AP-site stabilizing H-bond from His309 to the AP-site phosphate oxygen. Weakening of this H-bond in the C99S mutant reduces the affinity of the AP-site DNA in absence of the Mg2+. At low Mg2+ concentration this interaction is partially restored. The reduced affinity of the C99S mutant for AP-site containing DNA was specifically addressed in MD simulations. Being so far from the active site (~ 16Å), Cys99 indirectly affects the relative position of the AP-site which is bound in the active site to H309. We postulate that in the Ser99 mutant Mg2+ at high concentration occupies both A and B-sites resulting in inhibition of the endonuclease activity as we had proposed earlier 20. MD simulation gave further indication that the H-bond between the active site H309 and the AP-site is metastable in the C99S mutant in the absence of Mg2+ as compared to the WT enzyme. In the absence of the metal ion, once the H-bond is broken, the AP-site containing DNA moves away resulting in increase in distance between NE2 of H309 and P on the AP-site. This also leads to an increase in distance between CA atom of Asp210 and DNA binding weakens. The presence of Mg2+ at the B-site changes the dynamics of the AP-site forcing it downwards to the active site towards Asp210.
In summary, we have identified Cys99 in hAPE1 as a critical residue modulating APE’s function for ensuring optimum activity. Substitution of this residue alters the enzyme’s ability to recognize the AP-site substrate in the absence of Mg2+. Further studies with this mutant protein may shed light on metal-dependent substrate recognition and processing in the BER pathway.
Materials and Methods
Purification of Wild-Type and Mutant APE1s
The coding sequence for full-length APE1 was inserted in the pET15b vector (Novagen) at Nde I/Xho I sites for expression of APE1 in E. coli. Other single site-directed mutations at all 7 Cys amino acid positions including Cys65Ser, Cys93Ser, Cys99Ser, Cys138Ser, Cys208Ser, Cys296Ser and Cys310Ser, were created in the APE1 cDNA by following manufacturer’s guidelines of site-directed mutagenesis kit (QuickChange II XL, Stratagene). The DNA sequence of the WT APE1 and mutant cDNAs were confirmed by UTMB's recombinant DNA laboratory (RDL).
WT APE1 and the Ser mutants were purified as previously described 23 with slight modifications. After transforming with the pET15b-based APE1 expression plasmids E. coli BL21 (DE3) cultures (1L in L-broth) were grown to 0.6 OD at 600nm. APE1 expression was then induced with 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 16° C for 14–16 h 23. After collecting the cells in suspension buffer containing 20 mM Tris (pH 8.0) and 0.5 M NaCl, these were sonicated and centrifuged. The supernatant were loaded on to Ni-NTA (Qiagen) column (3 mL) which was first washed with 18 mL sonication buffer containing 20 mM imidazole. WT and mutant APE1s were then eluted with 10 mL sonication buffer containing 200 mM imidazole, and the eluate was dialyzed vs. 20 mM Tris-Cl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 1 mM dithiotreitol (DTT) and 10% glycerol. The poly His-tags in the proteins were cleaved by overnight incubation at 4° C with thrombin. The APE1 polypeptides were finally purified to near homogeneity by FPLC in an 1 mL SP-Sepharose column (LCC-500 PLUS; Pharmacia) and the final preparations were dialyzed vs. 20 mM Tris (pH 8.0), 300 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 50% glycerol, and stored at −20° C.
AP-endonuclease Assay
A 43-mer oligonucleotide containing the AP-site analog tetrahydrofuran (THF) at nt 31 (Midland Corp), was prepared as described previously 23,39. The oligo was 5'-end-labeled with [γ-32P]ATP using T4 polynucleotide kinase. After annealing to the complementary strand with A opposite THF, the duplex oligo was purified from the free nucleotides by gel filtration column (Clontech, Chroma Spin TE 10).
The duplex THF-containing oligo was incubated with recombinant WT APE1 or Cys mutants at 37° C for 3 min during which the reaction rate was linear in a 15 µL reaction mixture containing 50 mM Tris–HCl (pH 8.5), 50 mM KCl, 1 mM DTT, 0.1m MEDTA and 100 µg/mL BSA and various concentration of MgCl2. After stopping the reaction with 10 µL 80% formamide/40 mM NaOH containing 0.05% xylene cyanol, followed by heating at 95° C for 5 min, the samples were kept on ice until denaturing gel electrophoresis in 20% polyacrylamide containing 8 M urea to separate the substrate oligo from the cleaved product. After drying the gels, the radioactivity was quantitated by phosphoimager analysis in a Storm system (Molecular Dynamics). The enzyme kinetics data were fitted by nonlinear least-squares regression to obtain Vmax and Km using the Michaelis-Menten equation using Sigma plot software. The kinetic parameters Km and kcat were calculated after incubating 33 pM enzyme at 37° C for 3 min with various substrate concentration.
Electrophoretic Mobility Shift Analysis (EMSA)
The 5′-[γ32P]-labeled 43-mer THF-oligo (about 25 pM) was incubated with 5 ng of WT or mutant APE1 in the EMSA-binding buffer containing 40 mM HEPES, 50 mM KCl, 1 mM DTT, 2 mM EDTA for 15 min at 25° C followed by electrophoresis in nondenaturing 6% polyacrylamide gel containing 7 mM Tris-HCl (pH 8), 3.3 mM sodium acetate, 1 mM EDTA at 4° C for 1 h at 150 V. The gels were processed as described earlier 35.
Fluorescence Spectroscopy
The fluorescence emission spectra with 0.5 µM of WT and C99S mutant APE1 proteins in PBS buffer were acquired using an excitation wavelength of 295 nm in a Shimadzu RF-5301PC fluorimeter. An average of four repetitive scans were considered 35.
Molecular Dynamic Simulation
Molecular dynamic (MD) simulations were done with the AMBER9 program 40 with complexes of WT APE1, uncleaved AP-site containing DNA substrate and the cofactor Mg2+. Models for the WT APE1, Cys99→Ser99 mutant and the DNA were generated from the crystal structure 1DE8 19. The 11 bp DNA oligo has flipped out and AP-site bound to the WT APE1. Active site His309 was protonated in all models and succeeding simulation to account for its role in stabilizing the substrate/product. It was shown experimentally that H309N mutation abolishes endonuclease activity and increases substrate release rate much faster21,24. In previous work we showed that His309 is only forming the stabilizing H-bond to the AP-site oxygen when it is protonated 20. For simulations with the cofactor Mg2+ we placed the Mg2+ ions at the A and B-site positions 20,25.
Na+ ions were added to all complexes to achieve electrostatic neutrality and then soaked in TIP3 water box 41. The dimensions of the water box were chosen to be 10 Å beyond any complex atom. These models were first energy minimized over 2000 steps using standard procedure with the Sander program of the AMBER9 distribution 40. Dynamic simulations over several ns followed the minimization. The parameters for the MD-simulations were constant pressure and temperature, periodic boundary conditions and the ff03 force field 42. Finally, analysis of the trajectories and visualizations were done with the programs CARNAL of the AMBER6 distribution and MOLMOL 43.
Supplementary Material
Mg2+ dependent AP-endonuclease activity of the WT and mutant APE1 proteins. The 43-mer, THF-containing oligonucleotide was incubated with 33 pM APEs for 3min at 37° C. AP-endonuclease activity of WT and mutant APE1 proteins in the presence of (a) 0.5 mM Mg2+, (b) 2 mM Mg2+, and (c) 10 mM Mg2+.
Fluorescence emission spectra of the PBS buffer, WT (continuous line) and C99S mutant APE1 (dashed line) indicating intrinsic tryptophan fluorescence. The excitation wavelength was 295 nm. The spectra represent an average of four individual experiments.
Acknowledgements
This research was supported by USPHS grants RO1 CA53791 and RO1 ES08457, P30 ES006676 (to SM), RO1 CA98664 (to TI), and US DOE grant DE-FG02-04ER63826 (to WEB). We thank Drs. T. Hazra, and M. Hegde at UTMB, and Dr. G. Tell at University of Udine for helpful discussions during the study. We thank Dr. R. Chattopadhyay, University of Virginia for his technical guidance. We acknowledge Dr. R. Roy of Georgetown University for critically reviewing the manuscript. We also thank Drs. T. Wood of the Recombinant DNA Laboratory for DNA sequencing and J. Navarro for providing fluorescence spectroscopy facility at UTMB. Ms. W. Smith’s secretarial assistance is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 2.Mitra S, Izumi T, Boldogh I, Bhakat KK, Chattopadhyay R, Szczesny B. Intracellular trafficking and regulation of mammalian AP-endonuclease 1 (APE1), an essential DNA repair protein. DNA Repair (Amst) 2007;6:461–469. doi: 10.1016/j.dnarep.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 3.Doetsch PW, Cunningham RP. The enzymology of apurinic/apyrimidinic endonucleases. Mutat Res. 1990;236:173–201. doi: 10.1016/0921-8777(90)90004-o. [DOI] [PubMed] [Google Scholar]
- 4.Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 5.Johnson RE, Torres-Ramos CA, Izumi T, Mitra S, Prakash S, Prakash L. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 1998;12:3137–3143. doi: 10.1101/gad.12.19.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ribar B, Izumi T, Mitra S. The major role of human AP-endonuclease homolog Apn2 in repair of abasic sites in Schizosaccharomyces pombe. Nucleic Acids Res. 2004;32:115–126. doi: 10.1093/nar/gkh151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitra S, Boldogh I, Izumi T, Hazra TK. Complexities of the DNA base excision repair pathway for repair of oxidative DNA damage. Environ Mol Mutagen. 2001;38:180–190. doi: 10.1002/em.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson SH. Mammalian base excision repair and DNA polymerase beta. Mutat Res. 1998;407:203–215. doi: 10.1016/s0921-8777(98)00002-0. [DOI] [PubMed] [Google Scholar]
- 9.Izumi T, Brown DB, Naidu CV, Bhakat KK, Macinnes MA, Saito H, Chen DJ, Mitra S. Two essential but distinct functions of the mammalian abasic endonuclease. Proc Natl Acad Sci U S A. 2005;102:5739–5743. doi: 10.1073/pnas.0500986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans AR, Limp-Foster M, Kelley MR. Going APE over ref-1. Mutat Res. 2000;461:83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- 11.Xanthoudakis S, Miao G, Wang F, Pan YC, Curran T. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tell G, Damante G, Caldwell D, Kelley MR. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxid Redox Signal. 2005;7:367–384. doi: 10.1089/ars.2005.7.367. [DOI] [PubMed] [Google Scholar]
- 13.Walker LJ, Robson CN, Black E, Gillespie D, Hickson ID. Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol Cell Biol. 1993;13:5370–5376. doi: 10.1128/mcb.13.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ordway JM, Eberhart D, Curran T. Cysteine 64 of Ref-1 is not essential for redox regulation of AP-1 DNA binding. Mol Cell Biol. 2003;23:4257–4266. doi: 10.1128/MCB.23.12.4257-4266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okazaki T, Chung U, Nishishita T, Ebisu S, Usuda S, Mishiro S, Xanthoudakis S, Igarashi T, Ogata E. A redox factor protein, ref1, is involved in negative gene regulation by extracellular calcium. J Biol Chem. 1994;269:27855–27862. [PubMed] [Google Scholar]
- 16.Fuchs S, Philippe J, Corvol P, Pinet F. Implication of Ref-1 in the repression of renin gene transcription by intracellular calcium. J Hypertens. 2003;21:327–335. doi: 10.1097/00004872-200302000-00024. [DOI] [PubMed] [Google Scholar]
- 17.Bhakat KK, Izumi T, Yang SH, Hazra TK, Mitra S. Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. EMBO J. 2003;22:6299–6309. doi: 10.1093/emboj/cdg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorman MA, Morera S, Rothwell DG, de La Fortelle E, Mol CD, Tainer JA, Hickson ID, Freemont PS. The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J. 1997;16:6548–6558. doi: 10.1093/emboj/16.21.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected] Nature. 2000a;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 20.Oezguen N, Schein CH, Peddi SR, Power TD, Izumi T, Braun W. A "moving metal mechanism" for substrate cleavage by the DNA repair endonuclease APE-1. Proteins. 2007;68:313–323. doi: 10.1002/prot.21397. [DOI] [PubMed] [Google Scholar]
- 21.Masuda Y, Bennett RAO, Demple B. Rapid dissociation of human apurinic endonuclease (Ape1) from incised DNA induced by magnesium. J. Biol. Chem. 1998a;273:30360–30365. doi: 10.1074/jbc.273.46.30360. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen LH, Barsky D, Erzberger JP, Wilson DM., 3rd Mapping the protein-DNA interface and the metal-binding site of the major human apurinic/apyrimidinic endonuclease. J Mol Biol. 2000;298:447–459. doi: 10.1006/jmbi.2000.3653. [DOI] [PubMed] [Google Scholar]
- 23.Izumi T, Malecki J, Chaudhry MA, Weinfeld M, Hill JH, Lee JC, Mitra S. Intragenic suppression of an active site mutation in the human apurinic/apyrimidinic endonuclease. J Mol Biol. 1999;287:47–57. doi: 10.1006/jmbi.1999.2573. [DOI] [PubMed] [Google Scholar]
- 24.Masuda Y, Bennett RAO, Demple B. Dynamics of the interaction of human apurinic endonuclease (Ape1) with its substrate and product. J. Biol. Chem. 1998b;273:30352–30359. doi: 10.1074/jbc.273.46.30352. [DOI] [PubMed] [Google Scholar]
- 25.Beernink PT, Segelke BW, Hadi MZ, Erzberger JP, Wilson DM, III, Rupp B. Two divalent metal ions in the active site of a new crystal form of human apurinic/apyrimidinic endonuclease, Ape1: implications for the catalytic mechanism. J. Mol. Biol. 2001;307:1023–1034. doi: 10.1006/jmbi.2001.4529. [DOI] [PubMed] [Google Scholar]
- 26.Mol CD, Hosfield DJ, Tainer JA. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3' ends justify the means. Mutat Res. 2000b;460:211–229. doi: 10.1016/s0921-8777(00)00028-8. [DOI] [PubMed] [Google Scholar]
- 27.Rothwell DG, Hang B, Gorman MA, Freemont PS, Singer B, Hickson ID. Substitution of Asp-210 in HAP1 (APE/Ref-1) eliminates endonuclease activity but stabilises substrate binding. Nucleic Acids Res. 2000;28:2207–2213. doi: 10.1093/nar/28.11.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schein CH, Ozgun N, Izumi T, Braun W. Total sequence decomposition distinguishes functional modules, "molegos" in apurinic/apyrimidinic endonucleases. BMC Bioinformatics. 2002;3:37. doi: 10.1186/1471-2105-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelley MR, Parsons SH. Redox regulation of the DNA repair function of the human AP endonuclease Ape1/ref-1. Antioxid Redox Signal. 2001;3:671–683. doi: 10.1089/15230860152543014. [DOI] [PubMed] [Google Scholar]
- 30.Barzilay G, Mol CD, Robson CN, Walker LJ, Cunningham RP, Tainer JA, Hickson ID. Identification of critical active-site residues in the multifunctional human DNA repair enzyme HAP1. Nat Struct Biol. 1995;2:561–568. doi: 10.1038/nsb0795-561. [DOI] [PubMed] [Google Scholar]
- 31.Wilson DM, 3rd, Takeshita M, Grollman AP, Demple B. Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J Biol Chem. 1995;270:16002–16007. doi: 10.1074/jbc.270.27.16002. [DOI] [PubMed] [Google Scholar]
- 32.Xanthoudakis S, Curran T. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J. 1992;11:653–665. doi: 10.1002/j.1460-2075.1992.tb05097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guillet M, Boiteux S. Origin of endogenous DNA abasic sites in Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:8386–8394. doi: 10.1128/MCB.23.22.8386-8394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saporito SM, Gedenk M, Cunningham RP. Role of exonuclease III and endonuclease IV in repair of pyrimidine dimers initiated by bacteriophage T4 pyrimidine dimer-DNA glycosylase. J Bacteriol. 1989;171:2542–2546. doi: 10.1128/jb.171.5.2542-2546.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Izumi T, Schein CH, Oezguen N, Feng Y, Braun W. Effects of backbone contacts 3' to the abasic site on the cleavage and the product binding by human apurinic/apyrimidinic endonuclease (APE1) Biochemistry. 2004;43:684–689. doi: 10.1021/bi0346190. [DOI] [PubMed] [Google Scholar]
- 36.Kostrewa D, Winkler FK. Mg2+ binding to the active site of EcoRV endonuclease: a crystallographic study of complexes with substrate and product DNA at 2 A resolution. Biochemistry. 1995;34:683–696. doi: 10.1021/bi00002a036. [DOI] [PubMed] [Google Scholar]
- 37.Thomas MP, Brady RL, Halford SE, Sessions RB, Baldwin GS. Structural analysis of a mutational hot-spot in the EcoRV restriction endonuclease: a catalytic role for a main chain carbonyl group. Nucleic Acids Res. 1999;27:3438–3445. doi: 10.1093/nar/27.17.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chattopadhyay R, Wiederhold L, Szczesny B, Boldogh I, Hazra TK, Izumi T, Mitra S. Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells. Nucleic Acids Res. 2006;34:2067–2076. doi: 10.1093/nar/gkl177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramana CV, Boldogh I, Izumi T, Mitra S. Activation of apurinic/apyrimidinic endonuclease in human cells by reactive oxygen species and its correlation with their adaptive response to genotoxicity of free radicals. Proc Natl Acad Sci U S A. 1998;95:5061–5066. doi: 10.1073/pnas.95.9.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comp Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jorgensen WL, Chandrasekhar J, Madura JD. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 42.Ponder JW, Case DA. Force fields for protein simulations. Adv Protein Chem. 2003;66:27–85. doi: 10.1016/s0065-3233(03)66002-x. [DOI] [PubMed] [Google Scholar]
- 43.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mg2+ dependent AP-endonuclease activity of the WT and mutant APE1 proteins. The 43-mer, THF-containing oligonucleotide was incubated with 33 pM APEs for 3min at 37° C. AP-endonuclease activity of WT and mutant APE1 proteins in the presence of (a) 0.5 mM Mg2+, (b) 2 mM Mg2+, and (c) 10 mM Mg2+.
Fluorescence emission spectra of the PBS buffer, WT (continuous line) and C99S mutant APE1 (dashed line) indicating intrinsic tryptophan fluorescence. The excitation wavelength was 295 nm. The spectra represent an average of four individual experiments.