Abstract
Objectives
The aim of the present study was to purify and clone the trypsinogen isoforms from the guinea pig pancreas and characterize their activation properties.
Methods
Trypsinogens from pancreatic homogenates were isolated by ecotin-affinity chromatography, followed by cation-exchange chromatography. Activation of trypsinogens was tested with enteropeptidase, cathepsin B and trypsin. Complementary DNAs for pre-trypsinogens were cloned from total RNA after reverse transcription and PCR-amplification.
Results
Purification of trypsinogens yielded a single peak with an N-terminal amino-acid sequence of LPIDD. Cloning of pre-trypsinogen cDNAs revealed two distinct but nearly identical isoforms. At the amino acid level, the only difference between the two isoforms is an Ala/Ser change at position 15 within the signal peptide. Thus, both cDNA variants give rise to the same mature trypsinogen upon secretion. Guinea pig trypsinogen is readily activated by enteropeptidase and cathepsin B, but exhibits essentially no autoactivation, under conditions where human cationic and anionic trypsinogens rapidly autoactivate.
Conclusions
The observations suggest that multiple trypsinogen isoforms and their ability to autoactivate are not required universally for normal digestive physiology in mammals. Furthermore, the inability of guinea pig trypsinogen to undergo autoactivation suggests that this species might be more resistant to pancreatitis than humans, where increased autoactivation of cationic trypsinogen mutants has been linked to hereditary pancreatitis.
Keywords: zymogen activation, enteropeptidase, cathepsin B, autoactivation, trypsinogen isoforms, ecotin
INTRODUCTION
The guinea pig pancreas is a classic model of exocrine glands, which was used to elucidate the basic principles of regulated enzyme secretion (e.g. 1, 2). More recently, the guinea pig pancreas has gained popularity as a model system for the study of transport processes of the excretory duct system (3, 4). Although early studies using one or two-dimensional gel electrophoresis identified a trypsinogen in the guinea pig pancreas (5, 6), the number of isoforms and their amino-acid or cDNA sequence has not been determined. Furthermore, activation of guinea pig trypsinogens and activity of guinea pig trypsins has never been studied either.
Trypsin plays an important initiator role in experimental acute pancreatitis. A number of animal models of acute pancreatitis have been established, which include the secretagouge-induced edematous (7), the L-arginine-induced necrotizing (8–10), and the bile acid-induced necrotizing and hemorrhagic (11, 12) types. These models are well-characterized in the rat and mouse, however, only a few studies utilized guinea pigs (13, 14).
Trypsin also contributes to the pathogenesis of chronic pancreatitis in humans. The human pancreas expresses three isoforms of trypsinogen, commonly referred to as cationic trypsinogen, anionic trypsinogen and mesotrypsinogen. These isoenzymes are encoded by separate genes, the PRSS1 (protease, serine, 1), PRSS2 and PRSS3 genes (15). Relatively recent genetic studies identified several variants in the PRSS1 gene that are associated with hereditary or sporadic idiopathic chronic pancreatitis (16, 17). The majority of the disease-causing PRSS1 variants stimulate autoactivation of cationic trypsinogen in vitro and most likely cause premature intrapancreatic trypsin activation in vivo (17). Variants in the PRSS2 or PRSS3 genes have not been found in association with chronic pancreatitis, however, the recently identified p.G191R PRSS2 variant, which undergoes rapid autodegradation, was shown to protect against chronic pancreatitis (18). The central role of trypsin in pancreatitis pathogenesis is also underscored by the finding that loss-of-function mutations in the SPINK1 gene, that encodes the physiological inhibitor of trypsin, are also associated with chronic pancreatitis (19). Furthermore, loss-of-function mutations in the trypsin-degrading enzyme chymotrypsin C (CTRC) were found in association with chronic pancreatitis (20).
Here we report the cDNA cloning and purification of trypsinogen from the guinea pig pancreas. We found that the guinea pig pancreas secretes a single major trypsinogen isoform, which is encoded by two distinct mRNA species. Surprisingly, autoactivation of the guinea pig trypsinogen was minimal, suggesting that autoactivation is relatively unimportant for physiological zymogen activation in this species. The lack of autoactivation also suggests that this species might be less prone to the development of pancreatitis.
MATERIALS AND METHODS
Materials
Whole pancreata from Abyssinian guinea pigs were purchased from Rockland Immunochemicals (Gilbertsville, PA). The pancreata were flash frozen in liquid nitrogen, shipped on dry ice and stored at −80 °C. RNAqueous™ Phenol-free total RNA Isolation kit was from Ambion Inc. (Austin, TX), SMART™ RACE cDNA Amplification Kit from Clontech Laboratories Inc. (Mountain View, CA), Zero Blunt TOPO PCR Cloning Kit from Invitrogen Corp. (Carlsbad, CA) and N-CBZ-Gly-Pro-Arg-p-nitroanilide from Sigma-Aldrich (St. Louis, MO). Human trypsinogens were expressed in Escherichia coli and affinity-purified as described previously (21–24). Recombinant human pro-enteropeptidase was from R&D Systems (Minneapolis, MN). Human pro-enteropeptidase (0.07 mg/mL stock solution) was activated with 50 nM human cationic trypsin in 0.1 M Tris-HCl (pH 8.0), 10 mM CaCl2 and 2 mg/ml bovine serum albumin (final concentrations) for 30 min at room temperature. Human recombinant cathepsin B was a generous gift from Paul M. Steed (Research Department, Novartis Pharmaceuticals, Summit, New Jersey). Before use, cathepsin B was activated with 1 mM dithiothreitol (final concentration) for 30 min on ice.
Purification of trypsinogen
Guinea pig pancreatic tissue (5 g) was homogenized in 0.1 M Tris-HCl, pH 8.0 using a rotor-stator tissue homogenizer (T 25, IKA Works) at 11,000 – 13,000 rpm and the homogenate was centrifuged for 8 min at 13,000 rpm, at 4 °C. The supernatant was further clarified by a second round of centrifugation, and then loaded onto a 2 mL ecotin-affinity column. The column was washed with 20 mM Tris–HCl (pH 8.0), 0.2 M NaCl and elution was performed with 50 mM HCl. The ecotin-eluate (~1 mg protein in 3–4 mL volume) was directly loaded onto a Mono S HR 5/5 strong cation-exchange column equilibrated with 20 mM Na-acetate (pH 5.0). The Mono S column was developed with a linear gradient of 0–0.5 M NaCl. Concentrations of guinea pig trypsinogen preparations were estimated from UV absorbance at 280 nm, using a theoretical extinction coefficient of 37,650 M−1 cm−1 (http://ca.expasy.org/tools/protparam.html).
Complementary DNA cloning
Total RNA isolation was performed from 30 mg homogenized guinea pig pancreas by the RNAqueous™ Phenol-free total RNA Isolation kit modified with an additional phenol-chloroform extraction. First-strand cDNA was synthesized using the SMART™ RACE cDNA Amplification Kit with 2 µg RNA. For the 5' RACE reaction, first-strand synthesis was primed with an oligo(dT) primer. The reverse transcriptase used in the reaction exhibits terminal transferase activity and adds a series of dC residues to the 3' end of the cDNA. An oligonucleotide (SMART IIA) present in the reaction mixture anneals to the dC stretch and serves as template for further extension of the cDNA. The end result of the reaction is a full length cDNA with an additional SMART sequence at the 5' end. For the 3' RACE, the mRNA is reverse transcribed with an oligo(dT) primer which anneals to the polyA tail and also incorporates the SMART sequence at its 5' end. Guinea pig trypsinogen cDNA was PCR-amplified in 2 reactions from the first-strand 5' and 3' RACE cDNAs using a gene-specific primer and a Universal primer that annealed against the SMART sequence incorporated into the first-strand cDNA either at the 5' or 3' ends. The gene specific GP1 primers annealed with relatively low stringency to the region around the conserved catalytic Ser200 (see Fig 1); the GP1 sense primer (5`-TGT CAG GGA GAC TCT GGT GGC CCA GTT GTC TGC-3`) corresponded to codons 196–206, and the GP1 antisense primer (5`-GCA GAC AAC TGG GCC ACC AGA GTC TCC CTG-3') corresponded to codons 197–206 (Fig 1). These primers were previously designed against the corresponding region of the Bothrops jararaca trypsinogen (GenBank #AF190273) and used successfully in other cloning projects in our laboratory. PCR products were purified and directly subcloned into the pCR-Blunt II-TOPO vector using the Zero Blunt TOPO PCR Cloning Kit (Invitrogen). Fifteen clones from the 5' RACE reaction and 3 clones from the 3' RACE reaction were sequenced. The initial sequence information indicated that 2 almost identical cDNA species were cloned. On the basis of the initial sequencing results, new trypsinogen-specific primers were designed. The GP2 sense primer (5'- AGC TAC AGC AGC AGC ACC CTG AAC -3') anneals to codons 98–105, and the GP2 antisense primer (5'- GAT CTG TCC AGG ATA GGC GGA CTG -3') anneals to codons 172–179 (Fig 1). Using the GP2 primers and the Universal primer additional trypsinogen-specific amplicons were generated from the first-strand 5' RACE and 3' RACE cDNA and subcloned into the pCR-Blunt II-TOPO vector. Five clones from the 5' RACE reaction and 4 clones from the 3' RACE reaction were sequenced. Finally, the full length cDNA sequences of the 2 guinea pig pre-trypsinogen isoforms were reconstructed from overlapping fragments. The guinea pig pre-trypsinogen cDNA sequences have been deposited to GenBank under accession numbers EF371801 and EF371802.
Figure 1.
Complementary DNA and deduced amino acid sequence of guinea pig pre-trypsinogens. Amino-acid numbering starts with the initiator methionine codon. Isoform 1 is shown, with the nucleotide differences present in isoform 2 indicated by superscripts. Note that Ala15 becomes Ser15 in isoform 2, whereas Lys92 remains unchanged. The sequences where the GP1 and GP2 primers anneal are underlined. The activation peptide is highlighted in gray.
Assay of trypsin activity
Trypsin activity was determined using the synthetic chromogenic substrate N-CBZ-Gly-Pro-Arg-p-nitroanilide. Kinetics of the chromophore release was followed at 405 nm in 0.1 M Tris-HCl (pH 8.0), 1 mM CaCl2, at 22 °C using a Spectramax Plus 384 microplate reader (Molecular Devices Corp.).
Trypsinogen activation with enteropeptidase
2 µM (final concentration) trypsinogen was incubated with 28 ng/mL human enteropeptidase holoenzyme (final concentration) at 37 °C, in 0.1 M Tris-HCl, pH 8.0, 2 mg/mL bovine serum albumin and 1 mM CaCl2 in a final volume of 100 µL. Aliquots of 2 µL were withdrawn from reaction mixtures at the indicated times, and trypsin activity was determined with the synthetic substrate N-CBZ-Gly-Pro-Arg-p-nitroanilide.
Trypsinogen activation with human cathepsin B
Trypsinogens were activated at 2 µM concentration with human cathepsin B (90 µg/mL final concentration) at 37 °C in 0.1 M Na-acetate buffer (pH 4.0) in the presence of 1 mM dithiothreitol, 2 mg/mL bovine serum albumin, 1 mM K-EDTA, and 0.3 mM benzamidine; as described previously (25). Aliquots (2 µL) were withdrawn at indicated times and trypsin activity was measured on the synthetic substrate.
Trypsinogen autoactivation
Trypsin-mediated trypsinogen activation was measured at 37 °C, using 2 µM trypsinogen, 1 mM CaCl2, 0.1 M Tris-HCl (pH 8.0), 2 mg/mL bovine serum albumin and 200 nM trypsin initial concentrations.
Trypsin autolysis
Trypsinogens (final concentration 2 µM) were activated to trypsin with enteropeptidase for 60 min at 37 °C in the presence of 1 mM CaCl2. Autocatalytic inactivation of trypsins was then followed at 37 °C after addition of K-EDTA (pH 8.0) to a final concentration of 2 mM. At indicated time points aliquots (2 µL) were withdrawn and residual trypsin activity was measured on the synthetic substrate.
RESULTS
Purification of guinea pig trypsinogens
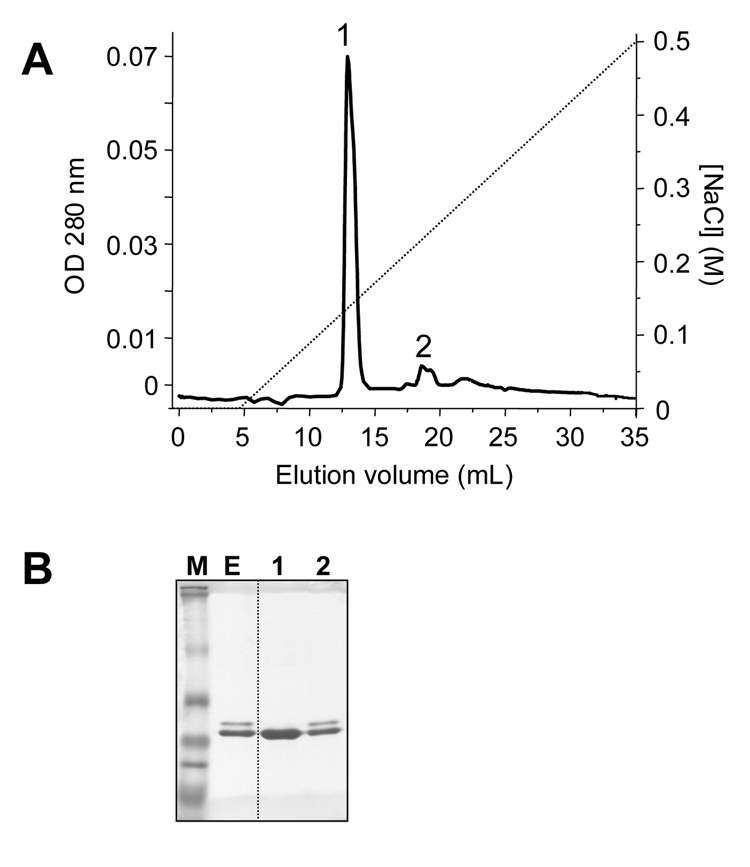
Tissue extracts from the guinea pig pancreas were applied to an ecotin-affinity column (26). Ecotin is a pan-serine-protease inhibitor, which binds to pancreatic serine proteases (trypsins, chymotrypsins, elastases) and their zymogen forms. The flow-through of the ecotin-column was analyzed for the presence of trypsin activity after activation with enteropeptidase. The complete absence of trypsin activity indicated that all trypsinogen isoforms were bound to the ecotin column. Elution was performed under acidic conditions, and SDS-PAGE analysis revealed two closely migrating bands, a lighter upper band and a strong lower band, indicating that at least 2 different proteins were present in the ecotin eluate (Fig 2). To resolve these proteins, the ecotin eluate was loaded onto a Mono S cation-exchange column equilibrated with 20 mM Na-acetate (pH 5.0) and eluted with a NaCl gradient (0–0.5 M). A single major peak was eluted at approximately 0.15 M NaCl concentration (Fig 2A), which was identified as trypsinogen on the basis of its enzymatic activity after activation with enteropeptidase. In addition, a small, broader peak was eluted at approximately 0.22 M NaCl, which exhibited both trypsin and chymotrypsin activity upon activation with enteropeptidase or trypsin, respectively. Although not shown, the number of peaks eluted did not change when the NaCl gradient was extended to 1.0 M. Aliquots of fractions were analyzed by SDS-PAGE and Coomassie blue staining. The first, main peak showed only a single band, whereas the second peak contained two bands, one of which co-migrated with the trypsinogen band recovered in the first peak (Fig 2B). N-terminal sequencing of the first, major trypsinogen peak revealed a single, homogeneous sequence of Leu-Pro-Ile-Asp-Asp, suggesting that this peak contains a single trypsinogen isoform. This trypsinogen isoform was novel, as the N-terminal sequence determined was distinct from the known trypsinogen sequences found in protein databases. The second peak contained a mixture of two different N-terminal sequences, a trypsinogen N terminus identical to the one found in the first, major peak and a chymotrypsinogen-like N terminal sequence of Cys-Gly-Val-Pro-Ala. Therefore, the enzyme activity assays and N terminal sequencing are in agreement demonstrating that the smaller Mono S peak contains a mixture of a trypsinogen and a chymotrypsinogen. The identical N-terminal sequences of the trypsinogens recovered in the first and second peaks suggested that the two peaks contained the same isoform and the small second trypsinogen peak represented an oligomeric or aggregate form of guinea pig trypsinogen. This conclusion was also supported by comparative MS/MS peptide sequencing of trypsinogens from the two peaks (see below).
Figure 2.
Purification of the guinea pig trypsinogen. (A) Mono S ion-exchange chromatography of the guinea pig trypsinogens eluted from the ecotin affinity column. See text for details. The Mono S column was developed with a 0–0.5 M gradient of NaCl at a flow rate of 1 mL/min. (B) SDS-PAGE analysis of the ecotin-eluate (E, 50 µL loaded; ~15 µg protein) and Mono S peaks 1 and 2 (100 µL from the 1 mL fraction loaded; ~15 µg protein). Samples were precipitated with trichloroacetic acid (10 % final concentration); solubilized in reducing Laemmli sample buffer, heat-denatured, electrophoresed on 13 % minigels, and stained with Coomassie blue. MultiMark multi-colored standards (Invitrogen) were used as molecular weight markers (M).
Complementary DNA cloning of guinea pig trypsinogens
To determine the complete cDNA sequence of the guinea pig trypsinogen(s), we utilized a PCR-based cloning method after 5' and 3' RACE, as described in Experimental Procedures. The assembled cDNA sequences were essentially identical with the exception of 2 positions (Fig 1). Thus, at c.43 (in codon 15) a G/T variation was observed, whereas at c.276 (in codon 92) an A/G variation was found. The variant with c.43G and c.276A was arbitrarily designated as isoform 1 and the variant with c.43T and c.276G as isoform 2. The sequences of the 2 isoforms have been deposited with GenBank under accession numbers EF371801 and EF371802. The relative amounts of the two isoforms at the mRNA level appeared to be comparable, as judged from the isoform distribution of the sequenced 5' RACE clones. The G/T variation at codon 15 affects the encoded amino acid, which is predicted either as Ala (GCC) or Ser (TCC). In contrast, the G/A variation at position 92 has no affect at the protein level as both AAG and AAA codons encode Lys. Therefore, the deduced amino-acid sequences of the 2 isoforms differ only in amino-acid 15, which is either Ala or Ser. Position 15 is the last amino-acid of the secretory signal peptide, which is removed upon entry into the endoplasmic reticulum. The mature trypsinogens resulting from the two variants are thus identical, and the guinea pig pancreas secretes a single trypsinogen isoform.
The guinea pig pre-trypsinogens are composed of 246 amino acid residues, and the 231 amino-acid long mature trypsinogen has a predicted molecular weight of 24,378.5 Da. The guinea pig trypsinogen is cationic in character, with a theoretical pI value of 8.0 (http://ca.expasy.org/tools/protparam.html). The N-terminal 15 amino acids form the signal peptide, which is followed by the 8 amino-acid long trypsinogen activation peptide including the highly conserved tetra-aspartate motif (Fig 1). Importantly, the predicted and experimentally determined N-terminal sequences of guinea pig trypsinogen are in perfect agreement. The molecule is stabilized by 6 disulfide bridges, characteristic of most vertebrate trypsinogens. Acidic determinants of the calcium-binding loop are also fully conserved (Glu75, Glu82 and Glu85). Similarly to human mesotrypsinogen, position 79 is Lys in the guinea pig trypsinogen, instead of the more typical Glu found in other mammalian trypsinogens. The functional role of this side-chain is unclear, but the Glu79→Lys mutation (E79K) in human cationic trypsinogen seems to be associated with chronic pancreatitis (27). It is interesting to note that the otherwise highly conserved Arg122 is replaced with a Lys in the guinea pig trypsinogen. Arg122 is a trypsin sensitive site, which is believed to be important for autocatalytic degradation of bovine, rat and human trypsins (28, 29). Mutation of Arg122 to His causes hereditary chronic pancreatitis in humans (16). Relative to human trypsinogens, there is a low ratio of charged side-chains in the guinea pig trypsinogen (Arg, Lys, Asp and Glu residues; 11.3 % in guinea pig trypsinogen versus 18.1 % in both human trypsinogens), and a higher abundance of Ser residues (14.3 % in guinea pig trypsinogen versus 8.6 % and 10.3 % in human cationic and anionic trypsinogens, respectively). The guinea pig trypsinogen exhibits the strongest similarity to the bovine cationic trypsinogen (82 % identity), whereas the identity with human cationic and anionic trypsinogens are 76 % and 75 %, respectively.
MS/MS peptide sequencing of purified guinea pig trypsinogen
To obtain independent sequence information from the purified guinea pig trypsinogen, a sample was subjected to tandem MS peptide sequencing. After electrophoresis on a 15 % SDS-PAGE, the trypsinogen band was excised and digested in-gel with sequencing grade modified porcine trypsin (Promega Corp., Madison, WI). The resulting peptide mixture was then analyzed by LC-MS/MS (ProtTech, Inc.; Eagleville, PA). The following tryptic peptides were sequenced. LPIDDDDK (from Leu16 to Lys23); VSEGSEQFITASK (from Val80 to Lys92); HPSYSSSTLNNDIMLIK (from His96 to Lys112); and LASAANLNSK (from Leu113 to Lys122). Although this approach yielded only about 20 % coverage of the protein sequence; the peptides sequenced were in perfect agreement with the amino-acid sequence predicted from the cloned cDNA. The trypsinogen band from the small, second Mono S peak was also subjected to MS/MS analysis and the exact same tryptic fragments were identified, confirming that this peak contained the same isoform of trypsinogen as the first, main Mono S peak.
Activation of guinea pig trypsinogen
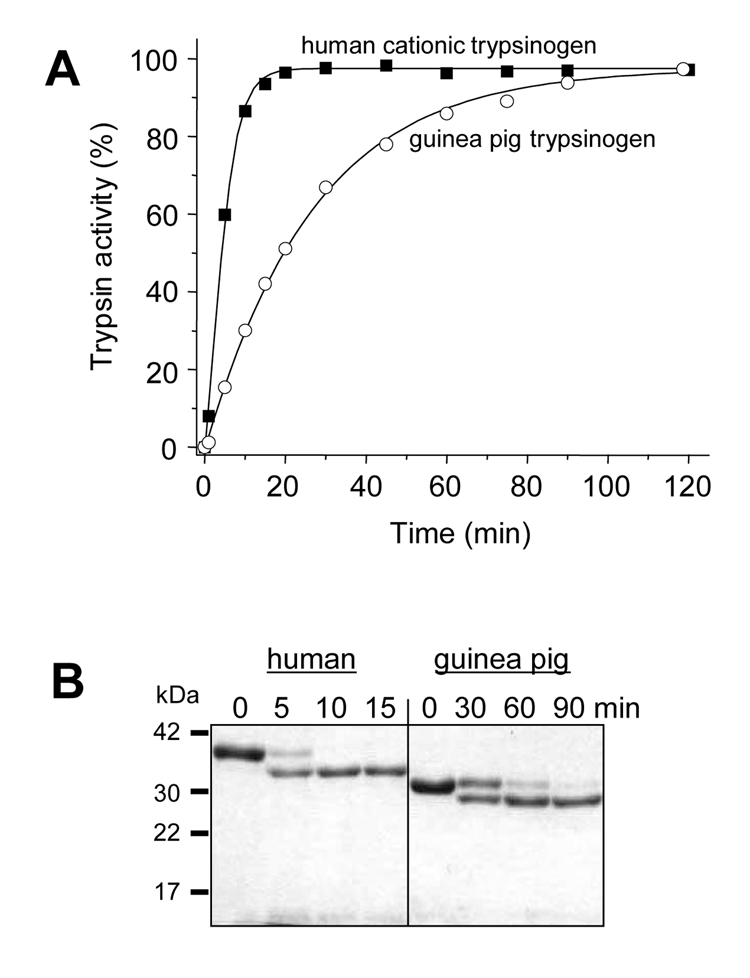
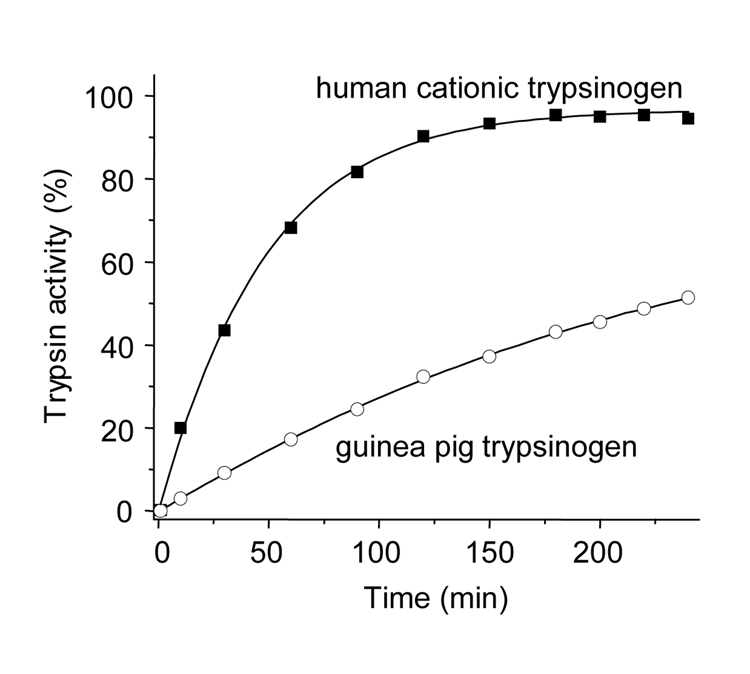
Activation characteristics and catalytic properties of guinea pig trypsinogen were compared to those of human anionic and cationic trypsinogens, which comprise 90–95 % of trypsinogens in humans. The physiological activator enteropeptidase readily activated guinea pig trypsinogen at pH 8.0, although at a rate that was slower than activation of human cationic trypsinogen (Fig 3). Similarly, activation of guinea pig trypsinogen by the pathological activator cathepsin B at pH 4.0 was slower, but still significant, relative to human cationic trypsinogen (Fig 4). Because we used human enteropeptidase and cathepsin B preparations in our experiments, the observed differences in activation rates between the human and guinea pig trypsinogens should be interpreted cautiously. The important conclusion of the experiments in Fig 3 and Fig 4 is that both enteropeptidase and cathepsin B can activate guinea pig trypsinogen under certain conditions.
Figure 3.
Activation of trypsinogen by enteropeptidase. (A) 2 µM (final concentration) of guinea pig trypsinogen (open circles) and human cationic trypsinogen (solid squares) were incubated with 28 ng/mL enteropeptidase (final concentration) at 37 °C, in 0.1 M Tris-HCl (pH 8.0), 2 mg/mL bovine serum albumin and 1 mM CaCl2 in a final volume of 100 µL. Aliquots of 2 µl were withdrawn from reaction mixtures at the indicated times, and trypsin activity was determined with the synthetic substrate N-CBZ-Gly-Pro-Arg-p-nitroanilide. Trypsin activity was expressed as a percentage of full activity. (B) Samples (100 µL) were precipitated with 10 % trichloroacetic acid (final concentration) at the indicated times and analyzed by reducing SDS/PAGE (13%) and Coomassie Blue staining.
Figure 4.
Activation of trypsinogen by cathepsin B. Guinea pig trypsinogen (open circles) and human cationic trypsinogen (solid squares) were activated at 2 µM concentration with human cathepsin B (90 µg/mL) at 37 °C in 0.1 M sodium acetate buffer (pH 4.0) in the presence of 1 mM dithiothreitol, 2 mg/mL bovine serum albumin, 1 mM K-EDTA and 0.3 mM benzamidine. Aliquots (2 µL) were withdrawn at indicated times and trypsin activity was measured on the synthetic substrate.
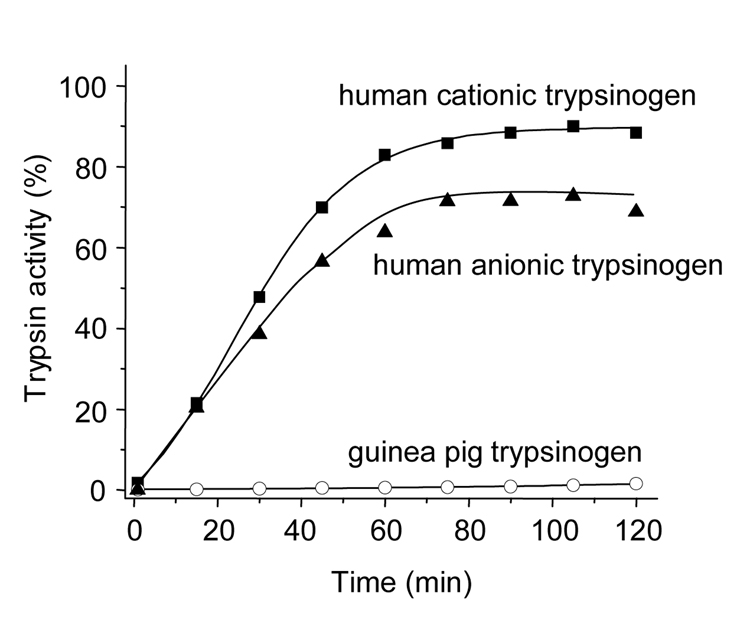
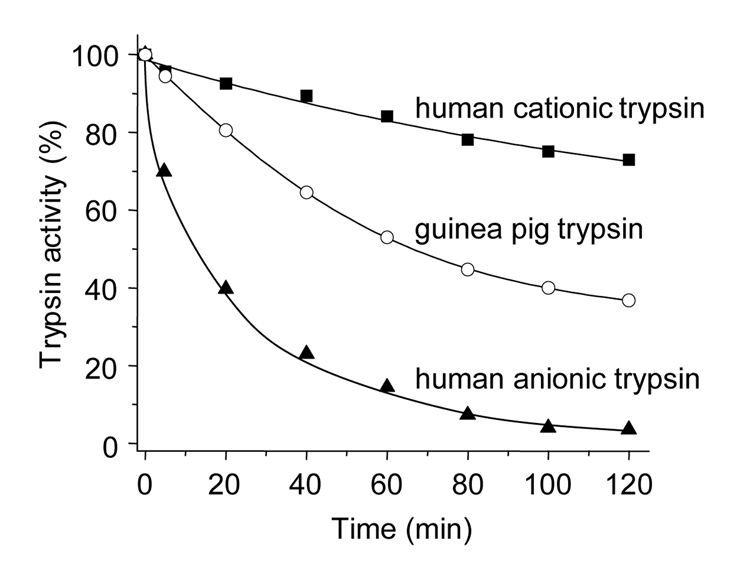
In contrast, a completely different picture emerged when autoactivation of guinea pig trypsinogens and human trypsinogens were compared. Human anionic and cationic trypsinogens autoactivated rapidly, whereas guinea pig trypsinogen exhibited essentially no autoactivation (Fig 5).
Figure 5.
Trypsin-mediated trypsinogen activation (autoactivation). Guinea pig trypsinogen (open circles), human cationic trypsinogen (solid squares) and human anionic trypsinogen (solid triangels) were activated with their respective trypsins. The reactions were carried out at 37 °C, using 2 µM trypsinogen, 1 mM CaCl2, 0.1 M Tris-HCl (pH 8.0), and 200 nM trypsin initial concentrations. Although not shown, increasing the CaCl2 concentration to 10 mM had no measurable stimulatory effect on the autoactivation of guinea pig trypsinogen in this time range.
Catalytic activity and autolysis of guinea pig trypsin
Catalytic parameters of guinea pig trypsin were determined on the peptide substrate N-CBZ-Gly-Pro-Arg-p-nitroanilide. The kcat and KM values were similar to those of human trypsins, indicating that lack of autoactivation (see above) was not due to a catalytic defect in guinea pig trypsin (Table 1).
Table 1.
Kinetic parameters of guinea pig trypsin and human cationic trypsin on the synthetic substrate N-CBZ-Gly-Pro-Arg-p-nitroanilide.
| kcat (s−1) | KM (µM) | kcat/KM (M−1·s−1) | |
|---|---|---|---|
| guinea pig | 115.6 | 14.6 | 7.9 × 106 |
| human cationic | 85.3 | 17.6 | 4.8 × 106 |
Autocatalytic degradation (autolysis) of guinea pig trypsin was also measured. Fig 6 demonstrates that at pH 8.0, in the absence of Ca2+, the guinea pig trypsin underwent relatively slow autolysis at an intermediate rate between those of the two human trypsins. As observed previously, human cationic trypsin was resistant to autolysis, whereas anionic trypsin rapidly autodegraded (22–24, 30, 31).
Figure 6.
Autocatalytic degradation (autolysis) of trypsin. Guinea pig trypsinogen (open circles), human cationic trypsinogen (solid squares), and human anionic trypsinogen (solid triangles) were activated at 2 µM concentration with enteropeptidase for 60 min at 37 °C in the presence of 1 mM CaCl2. Autocatalytic inactivation of trypsins was then followed at 37 °C after addition of K-EDTA (pH 8.0) to a final concentration of 2 mM. Residual activities were expressed as percentage of the full trypsin activity measured after enteropeptidase activation.
DISCUSSION
The present study offers two interesting observations for discussion. First, the guinea pig pancreas secretes a single major trypsinogen isoform, which is somewhat atypical, as most vertebrate species express multiple trypsinogen isoforms. Second, compared to the human trypsinogens, the guinea pig trypsinogen is defective in autoactivation.
The trypsinogen genes are intercalated between the beta T-cell receptor genes in humans, the mouse and the chicken (32–35). This unusual location might explain the tendency of the trypsinogen genes to undergo significant rearrangements during speciation, which includes duplications with the formation of new genes and pseudogenes as well as gene losses. All species sequenced to date contain multiple trypsinogen genes in their genome, of which only a limited number are expressed at the protein level (36). In humans, 9 genomic trypsinogen sequences were identified, but only 3 isoforms are expressed to measurable protein levels (13). The mouse genome contains 20 trypsinogen genes, but the normal mouse pancreas secretes only 4 trypsinogen isoforms (M. S.-T., unpublished data). The finding that the guinea pig pancreas secretes a single major isoform indicates that multiple isoforms are not an obligatory requirement for digestion, but they probably confer species-specific adaptive advantages. The guinea pig genome has not been sequenced yet, but it is very likely that it also contains multiple trypsinogen genes. This assumption is supported by the cloning of the 2 nearly identical cDNA forms, indicating the presence of at least 2 recently duplicated genes.
Although we posit that the guinea pig pancreas expresses 2 different mRNAs for pre-trypsinogen, which result in the expression of a single mature trypsinogen, there are caveats to this conclusion. First, the biochemical approach utilized here may not detect isoforms present in low quantities (1 % or less of total trypsinogens). Second, the cDNA cloning approach used may not identify isoforms in which the cDNA sequence around the catalytic Ser is less conserved, or isoforms present in very low amounts. Finally, hormonal stimulation of the pancreas may result in the expression of additional trypsinogen isoforms, which have not been found in this study.
Autoactivation of trypsinogen is believed to facilitate physiological zymogen activation in the duodenum. During autoactivation trypsin activates trypsinogen to trypsin, resulting in a self-amplifying reaction. Mutations in the human cationic trypsinogen that cause acceleration of autoactivation in vitro have been associated with hereditary pancreatitis, suggesting that autoactivation may be important for premature trypsinogen activation during pancreatitis, at least in humans (16, 17). The inability of the guinea pig trypsinogen to autoactivate indicates that this mode of trypsinogen activation is not universally required for digestion. Therefore, the strong tendency for autoactivation observed with the human trypsinogens may indicate a positive evolutionary selection. The relatively high abundance of surface exposed charged side chains (Glu, Asp, Lys, Arg) in the human trypsinogens and their absence in the guinea pig trypsinogen suggests that efficient autoactivation requires interactions associated with these strongly polar amino acids. Finally, the inability of the guinea pig trypsinogen to undergo autoactivation also suggests that this species might be more resistant to the development of pancreatitis, which would explain the scarcity of experimental models of acute pancreatitis using the guinea pig (13, 14).
Acknowledgments
This work was supported by NIH grant DK058088 to M. S.-T and a scholarship from the Rosztoczy Foundation to B. O.
Contributor Information
Béla Ózsvári, Department of Molecular and Cell Biology, Boston University Goldman School of Dental Medicine, Boston, Massachusetts; and 1st Department of Medicine, University of Szeged, Faculty of Medicine, Szeged, Hungary.
Péter Hegyi, 1st Department of Medicine, University of Szeged, Faculty of Medicine, Szeged, Hungary.
Miklós Sahin-Tóth, Department of Molecular and Cell Biology, Boston University Goldman School of Dental Medicine, Boston, Massachusetts.
REFERENCES
- 1.Scheele GA, Palade GE. Studies on the guinea pig pancreas. Parallel discharge of exocrine enzyme activities. J Biol Chem. 1975;250:2660–2670. [PubMed] [Google Scholar]
- 2.Tartakoff AM, Jamieson JD, Scheele GA, et al. Studies on the pancreas of the guinea pig. Parallel processing and discharge of exocrine proteins. J Biol Chem. 1975;250:2671–2677. [PubMed] [Google Scholar]
- 3.Case RM, Argent BE. Pancreatic duct secretion: control and mechanisms of transport. In: Go VLW, DiMagno EP, et al., editors. The Pancreas: Biology, Pathobiology, and Disease. New York: Raven Press; 1993. pp. 301–350. [Google Scholar]
- 4.Hegyi P, Rakonczay Z., Jr The inhibitory pathways of pancreatic ductal bicarbonate secretion. Int J Biochem Cell Biol. 2007;39:25–30. doi: 10.1016/j.biocel.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 5.Tartakoff A, Greene LJ, Palade GE. Studies on the guinea pig pancreas. Fractionation and partial characterization of exocrine proteins. J Biol Chem. 1974;249:7420–7431. [PubMed] [Google Scholar]
- 6.Scheele GA. Two-dimensional gel analysis of soluble proteins. Charaterization of guinea pig exocrine pancreatic proteins. J Biol Chem. 1975;250:5375–5385. [PubMed] [Google Scholar]
- 7.Lampel M, Kern HF. Acute interstitial pancreatitis in the rat induced by excessive doses of a pancreatic secretagogue. Virchows Arch A Pathol Anat Histol. 1977;373:97–117. doi: 10.1007/BF00432156. [DOI] [PubMed] [Google Scholar]
- 8.Mizunuma T, Kawamura S, Kishino Y. Effects of injecting excess arginine on rat pancreas. J Nutr. 1984;114:467–471. doi: 10.1093/jn/114.3.467. [DOI] [PubMed] [Google Scholar]
- 9.Hegyi P, Rakonczay Z, Jr, Sari R, et al. L-arginine-induced experimental pancreatitis. World J Gastroenterol. 2004;15:2003–2009. doi: 10.3748/wjg.v10.i14.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawra R, Sharif R, Phillips P, et al. Development of a new mouse model of acute pancreatitis induced by administration of L-arginine. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1009–G1018. doi: 10.1152/ajpgi.00167.2006. [DOI] [PubMed] [Google Scholar]
- 11.Aho HJ, Koskensalo SM, Nevalainen TJ. Experimental pancreatitis in the rat. Sodium taurocholate-induced acute haemorrhagic pancreatitis. Scand J Gastroenterol. 1980;15:411–416. doi: 10.3109/00365528009181493. [DOI] [PubMed] [Google Scholar]
- 12.Laukkarinen JM, Van Acker GJ, Weiss ER, et al. A mouse model of acute biliary pancreatitis induced by retrograde pancreatic duct infusion of Na-taurocholate. Gut. 2007;56:1590–1598. doi: 10.1136/gut.2007.124230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harell D, Orda R, Wiznitzer T, et al. Proteolytic proenzymes in the pancreas in the course of experimental bile-induced pancreatitis in the guinea pig. Digestion. 1978;18:394–401. doi: 10.1159/000198225. [DOI] [PubMed] [Google Scholar]
- 14.Frick TW, Hailemariam S, Heitz PU, et al. Acute hypercalcemia induces acinar cell necrosis and intraductal protein precipitation in the pancreas of cats and guinea pigs. Gastroenterology. 1990;98:1675–1681. doi: 10.1016/0016-5085(90)91106-g. [DOI] [PubMed] [Google Scholar]
- 15.Chen JM, Férec C. Trypsinogen genes: evolution. In: Cooper DN, editor. Nature encyclopedia of the human genome. London: Macmillan; 2003. pp. 645–650. [Google Scholar]
- 16.Whitcomb DC, Gorry MC, Preston RA, et al. Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene. Nat Genet. 1996;14:141–145. doi: 10.1038/ng1096-141. [DOI] [PubMed] [Google Scholar]
- 17.Teich N, Rosendahl J, Tóth M, et al. Mutations of human cationic trypsinogen (PRSS1) and chronic pancreatitis. Hum Mutat. 2006;27:721–730. doi: 10.1002/humu.20343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Witt H, Sahin-Tóth M, Landt O, et al. A degradation-sensitive anionic trypsinogen (PRSS2) variant protects against chronic pancreatitis. Nat Genet. 2006;38:668–673. doi: 10.1038/ng1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Witt H, Luck W, Hennies HC, et al. Mutations in the gene encoding the serine protease inhibitor, Kazal type 1 are associated with chronic pancreatitis. Nat Genet. 2000;25:213–216. doi: 10.1038/76088. [DOI] [PubMed] [Google Scholar]
- 20.Rosendahl J, Witt H, Szmola R, et al. Chymotrypsin C (CTRC) variants that diminish activity or secretion are associated with chronic pancreatitis. Nat Genet. 2008;40:78–82. doi: 10.1038/ng.2007.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahin-Tóth M. Human cationic trypsinogen. Role of Asn-21 in zymogen activation and implications in hereditary pancreatitis. J Biol Chem. 2000;275:22750–22755. doi: 10.1074/jbc.M002943200. [DOI] [PubMed] [Google Scholar]
- 22.Sahin-Tóth M, Tóth M. Gain-of-function mutations associated with hereditary pancreatitis enhance autoactivation of human cationic trypsinogen. Biochem Biophys Res Commun. 2000;278:286–289. doi: 10.1006/bbrc.2000.3797. [DOI] [PubMed] [Google Scholar]
- 23.Szilágyi L, Kénesi E, Katona G, et al. Comparative in vitro studies on native and recombinant human cationic trypsins. Cathepsin B is a possible pathological activator of trypsinogen in pancreatitis. J Biol Chem. 2001;276:24574–24580. doi: 10.1074/jbc.M011374200. [DOI] [PubMed] [Google Scholar]
- 24.Kukor Z, Tóth M, Sahin-Tóth M. Human anionic trypsinogen. Properties of autocatalytic activation and degradation and implications in pancreatic diseases. Eur J Biochem. 2003;270:2047–2058. doi: 10.1046/j.1432-1033.2003.03581.x. [DOI] [PubMed] [Google Scholar]
- 25.Kukor Z, Mayerle J, Kruger B, et al. Presence of cathepsin B in the human pancreatic secretory pathway and its role in trypsinogen activation during hereditary pancreatitis. J Biol Chem. 2002;277 doi: 10.1074/jbc.M200878200. 21389-212396. [DOI] [PubMed] [Google Scholar]
- 26.Lengyel Z, Pál G, Sahin-Tóth M. Affinity purification of recombinant trypsinogen using immobilized ecotin. Protein Expr Purif. 1998;12:291–294. doi: 10.1006/prep.1997.0837. [DOI] [PubMed] [Google Scholar]
- 27.Teich N, Le Maréchal C, Kukor Z, et al. Interaction between trypsinogen isoforms in genetically determined pancreatitis: mutation E79K in cationic trypsin (PRSS1) causes increased transactivation of anionic trypsinogen (PRSS2) Hum Mutat. 2004;23:22–31. doi: 10.1002/humu.10285. [DOI] [PubMed] [Google Scholar]
- 28.Várallyay E, Pál G, Patthy A, et al. Two mutations in rat trypsin confer resistance against autolysis. Biochem Biophys Res Commun. 1998;243:56–60. doi: 10.1006/bbrc.1997.8058. [DOI] [PubMed] [Google Scholar]
- 29.Kukor Z, Tóth M, Pál G, et al. Human cationic trypsinogen. Arg117 is the reactive site of an inhibitory surface loop that controls spontaneous zymogen activation. J Biol Chem. 2002;277:6111–6117. doi: 10.1074/jbc.M110959200. [DOI] [PubMed] [Google Scholar]
- 30.Mallory PA, Travis J. Human pancreatic enzymes. Characterization of anionic human trypsin. Biochemistry. 1973;12:2847–2851. doi: 10.1021/bi00739a011. [DOI] [PubMed] [Google Scholar]
- 31.Colomb E, Guy O, Deprez P, et al. The two human trypsinogens: catalytic properties of the corresponding trypsins. Biochim Biophys Acta. 1978;525:186–193. doi: 10.1016/0005-2744(78)90213-9. [DOI] [PubMed] [Google Scholar]
- 32.Hood L, Rowen L, Koop BF. Human and mouse T-cell receptor loci: Genomics, evolution, diversity, and serendipity. Ann NY Acad Sci. 1995;758:390–412. doi: 10.1111/j.1749-6632.1995.tb24844.x. [DOI] [PubMed] [Google Scholar]
- 33.Wang K, Gan L, Lee I, et al. Isolation and characterization of the chicken trypsinogen gene family. Biochem J. 1995;307:471–479. doi: 10.1042/bj3070471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rowen L, Koop BF, Hood L. The complete 685-kilobase DNA sequence of the human beta T cell receptor locus. Science. 1996;272:1755–1762. doi: 10.1126/science.272.5269.1755. [DOI] [PubMed] [Google Scholar]
- 35.Glusman G, Rowen L, Lee I, et al. Comparative genomics of the human and mouse T cell receptor loci. Immunity. 2001;15:337–349. doi: 10.1016/s1074-7613(01)00200-x. [DOI] [PubMed] [Google Scholar]
- 36.Roach JC, Wang K, Gan L, et al. The molecular evolution of the vertebrate trypsinogens. J Mol Evol. 1997;45:640–652. doi: 10.1007/pl00006268. [DOI] [PubMed] [Google Scholar]