Abstract
Background
Pancreatic intraepithelial neoplasia (PanIN) is a precursor to invasive ductal adenocarcinoma of the pancreas. Observations made in genetically engineered mouse models suggest that the acinar/centroacinar compartment can give rise to ductal neoplasia. In order to integrate findings in mice and men, we examined human acinar cells, acinar-ductal metaplasia (ADM) lesions and PanINs for KRAS2 gene mutations.
Methods
Surgically resected pancreata were screened for foci of ADM with or without an associated PanIN lesion. Stromal cells, acinar cells, ADMs, and PanINs, were separately isolated using laser capture microdissection. KRAS2 status was analyzed using genomic DNA isolated from the microdissected tissue.
Results
Twelve of these 31 foci of ADM occurred in isolation, while 19 were in the same lobules as a PanIN lesion. All 31 microdissected foci of acinar cells were KRAS2 gene wild-type, as were all 12 isolated ADM lesions lacking an associated PanIN. KRAS2 gene mutations were present in 14 of 19 (74%) PanIN lesions, and in 12 of the 19 (63%) foci of ADM associated with these PanINs. All ADM lesions with a KRAS2 gene mutation harbored the identical KRAS2 gene mutation found in their associated PanIN lesions.
Conclusions
Ductal neoplasms of the human pancreas, as defined by KRAS2 gene mutations, do not appear to arise from acinar cells. Isolated AMD lesions are genetically distinct from those associated with PanINs, and the latter may represent retrograde extension of the neoplastic PanIN cells, or less likely are PanIN precursor lesions.
Introduction
Pancreatic intraepithelial neoplasia (PanIN) has been recognized morphologically for over a century, but the biological significance of these lesions has only recently been defined (1–3). In 1976, Cubilla and Fitzgerald described histologically distinct proliferative lesions in the pancreatic ducts and ductules adjacent to infiltrating exocrine pancreatic cancer. They showed that these lesions were more common in pancreata with an invasive carcinoma than those without carcinoma (2). PanINs are currently classified into three grades, PanIN-1, −2 and −3, based on the degree of architectural and cytological atypia (3). A number of histopathological and clinical studies have provided strong evidence that PanINs in the pancreas can progress to invasive carcinoma (4). In addition, molecular genetic analyses have demonstrated that almost all of the genetic alterations identified in infiltrating ductal adenocarcinomas of the pancreas can also be identified in PanINs, and the prevalence of these genetic alterations in PanINs increases in parallel with morphologic progression from PanIN-1 to PanIN-3 to invasive carcinoma (3). Based on these studies, a genetic progression model has been established for PanINs, and PanINs are now recognized as one of the precursors to invasive adenocarcinoma of the pancreas (5). In addition, these studies have demonstrated that KRAS2 gene mutations are one of earliest genetic alterations in pancreatic neoplasia, approximately 45% of early PanIN lesions, and 90% of infiltrating pancreatic ductal adenocarcinomas harbor KRAS2 gene mutations (6–11). Therefore, KRAS2 gene mutations provide a tool to study the origins of human pancreatic neoplasia.
A number of animal models have recently been generated which recapitulate the morphologic progression of human pancreatic ductal adenocarcinoma, and these models have been used to define the populations of cells that can give rise to invasive pancreatic adenocarcinoma (12). Not surprisingly, most of these models are driven by mutant KRAS (13, 14). The expression of mutant KRAS in ductal epithelium under the control of cytokeratin 19 promoter failed to produce PanINs or pancreatic ductal adenocarcinoma (15), however, selective expression of endogenous mutant KRAS in acinar/centroacinar cells during early embryonic development produces a full range of mouse PanINs (mPanINs) and invasive pancreatic ductal adenocarcinoma (16–18). In addition, ADM is a prominent component of many of these genetically engineered mouse models (17–19), and in some models ADM appears to precede the appearance of mouse PanIN lesions (17). The genetically engineered mouse models, therefore, suggest the possibility of an acinar/centroacinar origin of pancreatic adenocarcinoma, with progression to ADM, mPanINs and eventually invasive carcinoma.
In humans, Brune et al. and Detlefsen et al. have observed that PanIN lesions are frequently associated with lobulocentric atrophy and ADM (20, 21). In addition, it has been demonstrated that these ADM structures contain both acinar and duct cell phenotypes (19). These observations in human tissues, when taken together with the findings in genetically engineered mouse models, suggest the possibility that human “ductal” neoplasia may arise prior to the PanIN lesion, perhaps even in acinar cells. (22). In order to integrate findings in mice and men, we examined human acinar cells, ADM lesions and PanINs for KRAS2 gene mutations to determine if KRAS2 gene mutations occur before the development of PanINs in human pancreata.
Materials and Methods
Case selection
We obtained appropriate institutional approval for all experiments involving human subjects. All pancreatoduodenectomy (Whipple) resections in 2006 (approximately 240 cases) were microscopically reviewed for the presence of either PanIN with associated ADM, or ADM in isolation. Excluded were any cases that contained invasive carcinoma on the original slide or in the approximately 60 microns cut for microdissection. PanINs were identified as a microscopic papillary or flat noninvasive epithelial neoplasms arising in a pancreatic duct, composed of cuboidal to columnar cells with varying amounts of mucin and degrees of cytologic and architecture atypia (23). PanINs were further graded into PanIN-1, PanIN-2 and PanIN-3 lesions based on the degree of cytologic and architectural atypia (23). A similar classification system has been developed for precursor lesions in genetically engineered mouse models (12). Because we were interested in the earliest events in human pancreatic neoplasia, the PanIN lesions selected for analysis in this study were mostly PanIN-1 lesions, which are low-grade, flat or papillary epithelial lesions composed of tall columnar cells with basally located nuclei and abundant supranuclear mucin (Figure 1A and C).
Figure 1.
ADM with (A,C,E) and without (B,D,F) PanINs. A. H&E staining (40x) showing central PanIN lesion (PanIN), ADM (ADM) and acinar cells (AC) in an acinar lobule. B. H&E staining (40X) showing ADM only lesions that are composed of normal duct, metaplastic acinar structures (ADM) and normal acinar cells (AC). C. Higher resolution (100X) of the ADM showing that metaplastic cells contain abundant intracytoplasm mucin, resembling ductal epithelium in the PanIN. D. Higher magnification (100X) of the ADM showing that metaplastic cells are cuboidal cells with minimal mucin. E. Higher resolution (200X) of the interface between the ADM and normal acinar cells. Note that some metaplastic structures are composed of both acinar cells (arrows) and duct-like cells. F. Higher resolution (200X) of the interface between the ADM and normal acinar cells. Note that some metaplastic structures are composed of both acinar cells (arrows) and duct-like cells. Arrows in E and F identify residual acinar cells in partially metaplastic acini.
Tissue procurement and processing
Twelve 5-µm serial sections were cut from formalin-fixed, paraffin-embedded tissue blocks and placed on UV-irradiated membrane-coated slides (Carl Zeiss Microimaging Inc, Thornwood, NY). Slides were stained with hematoxylin and eosin (H&E) using a modified “H&E Staining for PALM Laser Capture” protocol. Briefly, the paraffin was first removed by semi-melting it at 65° C on a heat block for 1 minute and then dipping it in xylene for 1 minute. The slides were re-hydrated with 100%, 96%, and 70% ethanol consecutively for 1 minute each. Nuclei were stained with Hematoxylin (Sigma-Aldrich, St. Louis, MO) for 10 minutes, and the cytoplasm was stained with Eosin (Sigma-Aldrich) for 5 minutes.
Stromal cells, mature acinar cells immediately adjacent to ADM, ADM lesions, and associated PanIN lesions within a single lobule were each separately microdissected in that order using a laser capture microdissection technique from PALM Technologies (Bernried, Germany). Dissected tissues were catapulted into adhesive caps (Carl Zeiss Microimaging, Inc) by defocused laser pulses. Representative photographs before and after microdissection of the ADM and PanIN are shown in Figure 2.
Figure 2.
Representative photographs taken before and after microdissection of ADM lesions with associated PanINs. Photographs before (A) and after (B) microdissection of the PanIN. Representative photographs taken before (C) and after (D) microdissection of the ADM (ADM) in MP16. Note that dissection of the ADM was performed first to prevent potential contamination by the PanIN.
DNA extraction and PCR amplification of KRAS2
Genomic DNA was extracted from the microdissected tissues using QIAamp DNA Micro Kit from Qiagen (Valencia, CA). DNA concentrations were measured using Pico Green® (Topac Inc., Cohasset, MA), ranging from 0.1 to 1 ng/µl. Isolated DNA (3 µl) was subjected to PCR amplification of the region of KRAS2 gene containing codons 12 and 13. The forward and reverse primers were 5’-GAGTTTGTATTAAAAGGTACTGGTGGA-3’ and 5’-TGGATCATATTCGTCCACAAAA-3’, respectively. Amplifications were performed by initial denaturation at 94°C for 2 min followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 30 s and extension at 72°C for 30 min, followed by 7 min of final extension at 72°C, using PCR SuperMix from Invitrogen (Carlsbad, CA). Due to low DNA concentrations for most of the specimens, a second round, 35-cycle nested PCR was followed using a nested PCR primer pair, 5’-AAGGTACTGGTGGAGTATTTGATAGTG-3’ (forward) and 5’-GATCATATTCGTCCACAAAATGA-3’ (reverse). These PCR products were then purified using Qiagen PCR purification kit, and sequenced using the BigDye Terminator 3.1 Cycle Sequencing Kit and an ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA).
LigAmp analysis of KRAS2 gene mutations
LigAmp analyses for KRAS2 gene mutations were also performed to supplement the KRAS2 gene sequencing with a significantly more sensitive method for detecting mutations. LigAmp analysis of KRAS2 gene mutations has been previously described in detail (24, 25). To simultaneously determine mutant and wild-type KRAS2, we performed 2 reactions, each including a wild-type and one mutant (GAT or GTT) upstream oligonucleotide in addition to taqMan probes for wild-type (16S rDNA probe) and mutant (LacZ probe) DNA in the reactions.
Immunohistochemical (IHC) labeling for Ki-67
Five-µm sections were prepared from formalin-fixed, paraffin-embedded tissue blocks, and immunolabeled for Ki-67 using a BenchMark XT automated stainer (Ventana, Tucson, AZ) after steam-mediated antigen retrieval. Approximately 300–500 cells were counted depending on the size of the lesions. Percentages of positively labeled cells in the PanIN associated and isolated ADM lesions were calculated and compared using Student T test.
TUNEL assay
Five-µm tissue sections were first deparaffinized in fresh xylene, and followed by rehydration in 95%, 85%, 75% and 50% ethanol and by fixation in 4% paraformaldehyde. The sections were then washed twice with PBS, permeabilized with proteinase K, rinsed in PBS, and refixed with 4% paraformaldehyde. Apoptotic cells in the PanIN associated and isolated ADM lesions were stained using The DeadEnd™ Colorimetric TUNEL System from Promega (Madison, WI ) according to the manufacturer's protocol, using mouse spleen and small intestine as positive and negative controls.
Results
KRAS2 gene status in acinar cells
To address these questions, KRAS2 gene mutations were analyzed because these mutations are one of the earliest known mutations to occur in human PanINs, and because most of the genetically engineered mouse models that have suggested an acinar cell origin of pancreatic cancer are driven by mutant KRAS. Thirty-one foci of acinar cells associated with an ADM and/or a PanIN lesion were sequenced and all 31 were KRAS2 gene wild-type, both by standard sequencing and by LigAmp (Table 1). The LigAmp technique is an ultrasensitive point mutation detection strategy capable of detecting low levels of point mutant DNA in a dominant population of wild-type molecules [1:10,000, (24, 25)]. This finding suggests that acinar cells in human pancreata are unlikely “the cell of origin” for human pancreatic ductal adenocarcinoma, at least as defined by a clonal KRAS2 gene mutation. Similarly, all 31 foci of microdissected stromal cells were KRAS2 gene wild-type by both techniques.
Table 1.
KRAS2 mutations in acinar ductal metaplasia with (Group I) or without associated PanINs (Group II)
| Group | Case# | Age | Sex | Diagnosis | Stroma | Acinar cells | Metaplasia | PanIN | |
|---|---|---|---|---|---|---|---|---|---|
| Group I | IA | MP1 | 80’s | M | Endocrine neoplasm | WT | WT | GAT | GAT |
| Leisons with | MP4 | 50’s | F | Endocrine neoplasm | WT | WT | GTT* | GTT | |
| an associated | MP5 | 80’s | F | Cholangiocarcinoma | WT | WT | GAT | GAT | |
| PanIN | MP6 | 60’s | F | Ampullary adenocarcinoma | WT | WT | GAT* | GAT* | |
| MP8 | 50’s | M | Chronic pancreatitis | WT | WT | GTT* | GTT | ||
| MP9 | 70’s | M | Duodenal adenocarcinoma | WT | WT | GTT* | GTT* | ||
| MP11 | 60’s | F | Duodenal adenocarinoma | WT | WT | GTT | GTT | ||
| MP13 | 40’s | F | Endocrine neoplasm | WT | WT | GAT | GAT | ||
| MP14 | 70’s | F | IPMN | WT | WT | GAT* | GAT | ||
| MP16 | 40’s | F | Serous cystadenoma | WT | WT | GAT+GTT | GTT | ||
| MP17 | 30’s | F | Endocrine neoplasm | WT | WT | GTT* | GTT* | ||
| MP19 | 50’s | M | Endocrine neoplasm | WT | WT | GTT* | GTT | ||
| IB | MP3 | 60’s | M | Ductal adenocarcinoma | WT | WT | WT | GTT* | |
| MP15 | 60’s | F | Ductal adenocarcinoma | WT | WT | WT | CGT | ||
| IC | MP2 | 80’s | M | Ductal adenocarcinoma | WT | WT | WT | WT | |
| MP7 | 60’s | F | Ampullary adenocarcinoma | WT | WT | WT | WT | ||
| MP10 | 60’s | F | Ampullary adenocarcinoma | WT | WT | WT | WT | ||
| MP12 | 60’s | M | Serous cystadenoma | WT | WT | WT | WT | ||
| MP18 | 60’s | M | Ampullary adenocarcinoma | WT | WT | WT | WT | ||
| Group II | M1 | 60’s | M | Chronic pancreatitis | WT | WT | WT | ||
| Lesions | M2 | 60’s | M | Ductal adenocarcinoma | WT | WT | WT | ||
| without | M3 | 50’s | M | Serous cystadenoma | WT | WT | WT | ||
| an associated | M4 | 50’s | M | Chronic pancreatitis | WT | WT | WT | ||
| PanIN | M5 | 60’s | M | Chronic pancreatitis | WT | WT | WT | ||
| M6 | 60’s | M | Chronic pancreatitis | WT | WT | WT | |||
| M7 | 60’s | M | Chronic pancreatitis | WT | WT | WT | |||
| M8 | 70’s | M | Serous cystadenoma | WT | WT | WT | |||
| M9 | 50’s | F | Serous cystadenoma | WT | WT | WT | |||
| M10 | 30’s | F | Endocrine neoplasm | WT | WT | WT | |||
| M11 | 60’s | M | Ampullary adenocarcinoma | WT | WT | WT | |||
| M12 | 60’s | M | IPMN | WT | WT | WT | |||
Case numbers and mutations are shown in bold. WT: wild type GGT codons. Age is provided as decades of life to maintain patient privacy.
Asterisk indicates cases that were wildtype by DNA sequencing, but where a mutation was found using LigAmp.
Initial Characterization of ADM lesions
ADM is characterized by the abnormal transformation of a mature acinar cell to a cell with ductal differentiation (26). ADM lesions initially appear as collections of acinar cells which have lost some of their normal apical granularity and polarization. These cells then gradually acquire features of ductal differentiation as they become cuboidal to columnar and express mucin (Figure 1). These metaplastic cells may focally involve a lobule, or a lobule of acinar cells may be completely replaced by cells with ductal differentiation. At the interface between foci of complete metaplasia and normal acini, the ADM structures may contain both cells with acinar differentiation and cells with ductal differentiation (Figure 1E and Figure 1F).
In this study, foci of ADM associated with a PanIN lesion were analyzed separately from foci of ADM not associated with a PanIN lesion. In the latter cases, the ducts draining the metaplastic lobules were histologically normal, and the metaplasia lesions typically contained little or no mucin (Figure 1B, 1D, and 1F). Once we identified an isolated ADM lesion, we cut and stained an additional 12 sections (approximately 60 microns) for laser capture microdissection. These confirmed the absence of co-existing PanIN in the planes of section examined, although we cannot exclude the possibility of co-existing PanIN in other regions not examined.
To test whether the two types of ADMs varied in proliferation rates, we performed IHC for Ki67, which demonstrated that both ADM lesions (with and without an associated PanIN lesion) have low proliferative indices, however ADMs associated with a PanIN have a slightly higher proliferation rate (2.8% vs. 1.6%, p=0.047). To assess if either lesion had high rates of apoptosis, we performed TUNEL staining, which was essentially negative in both lesions, with only extremely rare positive cells.
KRAS2 status in isolated ADM lesions
Twelve ADM lesions involving lobules without an associated PanIN in the same lobule were examined (Table 1, Group II, M1-M12). The metaplastic acinar cells in the cases selected were small, cuboidal duct-like epithelium with no mucinous cytoplasm (see Figure 1B, 1D and 1F, Materials and Methods). The twelve lesions were selected from 9 surgically resected pancreata. Neither PanIN lesions nor pancreatic invasive carcinomas were present in the same slides in order to avoid contamination by these lesions/cancers.
Sequencing the KRAS2 gene in these 12 lesions revealed that all 12 were KRAS2 wild-type. No mutations were found in the pure ADMs. As expected, the acinar and stromal cells in these cases were also KRAS2 gene wild-type. Testing the pure ADM lesions for the possibility of small numbers of mutation containing cells using LigAmp confirmed that they were KRAS2 gene wild-type.
KRAS2 gene status in ADM lesions associated with PanINs
Nineteen ADM lesions associated with a PanIN lesion were microdissected from 18 surgically resected pancreata (designated as MP1 to MP19 in Table 1). The non-neoplastic stromal cells, histologically normal acinar cells, foci of ADM, and PanIN lesions from each of these 19 foci were separately microdissected (Figure 2) and then analyzed for activating KRAS2 gene mutations by DNA sequencing and by LigAmp. Codon 12 KRAS2 gene mutations were detected in 14 out of 19 (74%) PanIN lesions (Table 1, Groups IA and IB). These mutations include GGT to GTT (8/14), GAT (5/14) and CGT (1/14). Ten of the 14 KRAS2 gene mutations were detected by DNA sequencing and an additional 4 only by LigAmp (samples MP6, MP9, MP17, and MP3; designated with an asterisk in Table 1), suggesting that the latter four PanINs, harbored lower levels KRAS2 gene mutations.
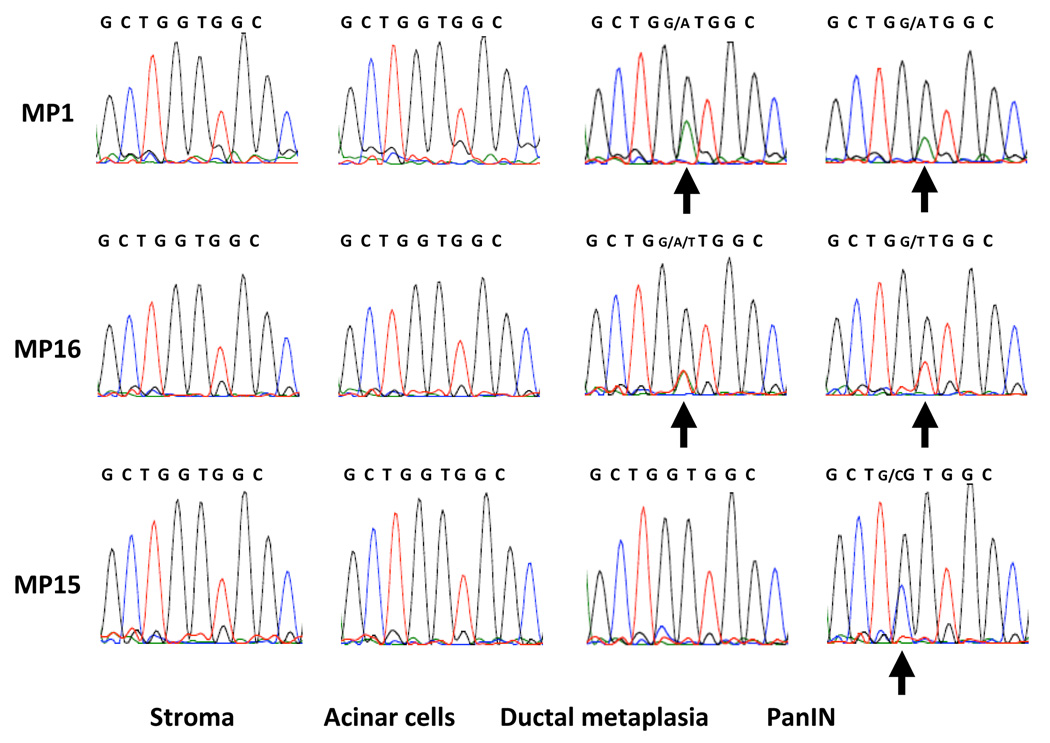
In 12 of the 14 lesions (86%) in which a KRAS2 gene mutation was identified in the PanIN, a KRAS2 gene mutation was also found in the associated ADM lesion, and in all 12 of these cases the same mutation was found in the PanIN lesion and in the associated focus of ADM (Group IA). These 12 mutations included GGT to GAT (5 cases), GGT to GTT (6 cases), and GGT to GAT/GTT (1 case). Representative KRAS2 sequences of lesion MP1 from the stroma, acinar cells, ADM and PanIN lesion are shown in Figure 3. Cases were reviewed before microdissection by at least two of us (CS, SMH, RHH) and photographed after microdissection to rule out cross contamination between the microdissected PanIN lesions and foci of ADM (Figure 2). In addition, for each set, special precautions were taken during microdissection, for example, samples were always collected in a sequence of the stroma cells, acinar cells, ductal metaplasia and PanIN to minimize the potential for cross contamination. Interestingly, in lesion MP16, a second mutation (GAT) was found in the associated ADM, in addition to the GTT mutation identified in the PanIN lesion (Figure 3).
Figure 3.
Representative KRAS2 gene sequences, from left-to-right, in the stroma cells, acinar cells, ADM and PanINs. Top row: Representative KRAS2 gene sequences from MP1. Note that the same KRAS2 gene mutation (arrows) is present in both the ADM and PanIN lesion. Middle row: Representative KRAS2 gene sequences from MP16. Note that two KRAS2 gene mutations (GAT and GTT, arrows) are present in the ADM, while only one mutation (GTT) is present in the PanIN. Bottom row: Representative KRAS2 gene sequences from MP15. Note that the KRAS2 gene mutation (CGT, arrow) is only present in the PanIN.
In the remaining 2 of the 14 PanINs in which a KRAS2 gene mutation was found in a PanIN, the associated foci of ADM were KRAS2 wild-type (Group IB, Table 1). The KRAS2 gene mutations in these 2 PanINs included GGT to GTT (1 of 2) and CGT (1 of 2). Representative KRAS2 DNA sequences from lesion MP15 are shown in Figure 3. In this case, a GGT to CGT mutation was identified in the PanIN. However, no mutations were detected in either the accompanying ADM or adjacent acinar and stromal cells.
The remaining 5 cases with a PanIN lesion (Group IC, Table 1) demonstrated no KRAS2 gene mutations in either the PanINs or the acinar-ductal metaplastic lesions.
Discussion
An understanding of the earliest precursors to invasive pancreatic cancer can be built by combining morphologic observations with careful molecular genetic studies. Such analyses, for example, have shown that PanIN lesions are a precursor to invasive ductal adenocarcinoma of the human pancreas (5). Genetically engineered mouse models in which genetic alterations are specifically targeted to selected cell populations can also be used to identify the cells, which when genetically manipulated, can give rise to a cancer (13, 14, 16–18). It is important that these two approaches to identifying precursors to invasive cancer are periodically reconciled such that the studies of human disease can benefit from insights gained in mouse models, and so that those studying mouse models can develop models that most accurately reflects the human condition (27).
The findings from a number of genetically engineered mouse models of pancreatic neoplasia have recently suggested that the acinar cell compartment in the pancreas can give rise to neoplasms with a ductal phenotype (16–18). For example, in one model the expression of an oncogenic KRAS transgene in acinar cells induced occasional low-grade PanINs (28), suggesting that acinar cells could be “cell of origin” of pancreatic ductal carcinoma. More recently, Zhu et al observed that ADM was the earliest prominent change to occur in LSL-KRASG12D/+/p48Cre+ mice, where metaplastic cells repressed acinar cell features and activated expression of the duct cell genes (18). They also demonstrated that ADM and early mPanIN (mouse PanIN) lesions in their mouse model exhibited comparable cellular and molecular properties. Similarly, another group has demonstrated that selective expression of an endogenous mutant KRAS oncogene in embryonic cells of acinar/centroacinar lineage resulted in a full spectrum of mPanIN-1 to mPanIN-3 and invasive carcinoma (17). In contrast, selective expression of oncogenic KRAS2 transgene in ductal lineages driven by the cytokeratin 19 promoter, failed to induce mPanINs or invasive carcinoma (15). These observations made in genetically engineered mouse models have led to the hypothesis that the acinar/centroacinar compartment and not ductal cells give rise to mouse ductal neoplasia.
These results from the mouse models have led to a re-evaluation of the early morphologic changes in human pancreatic ductal neoplasia. For example, it is now recognized that human PanIN lesions are often associated with foci of ADM. Detlefsen et al. documented a close association between lobular fibrosis and PanINs in elderly patients (21). The fibrotic foci shown in their report included ADM in addition to acinar atrophy and fibrosis. Similarly, Brune et al. examined PanIN lesions and parenchymal changes in the pancreas in patients having a strong family history of pancreatic cancer and found a close association of PanINs, even PanIN-1A, with atrophy of the lobular unit surrounding the duct containing the PanIN (20). Atrophic lobules exhibited a thinning of the acinar cells, loss of the apical granular cytoplasm and slight dilatation of the acinar lumen, namely ADM. While these investigators hypothesized that the PanINs developed first, producing small ductal obstructions, which, in turn, progress to lobulocentric atrophy and ADM. These same observations, interpreted in light of the observations made in the genetically engineered mouse models, could be interpreted as suggesting that ADM precedes the development of human PanIN lesions. Indeed, in the present study we observed that some foci of ADM arise in the absence of PanIN lesions, in areas where the associated pancreatic duct appears morphologically normal.
In the current study, we integrated the findings made in genetically engineered mouse models with the genetic changes in human acinar cells, foci of human ADM, and human PanINs. In all of the cases we examined, the acinar cells surrounding ADM lesions were KRAS2 gene wild-type. This finding suggests that acinar cells in human pancreata are unlikely “the cell of origin” for human pancreatic ductal adenocarcinoma, at least as defined by a clonal KRAS2 gene mutation. By contrast, three-fourths of the PanIN lesions harbored an activating point mutation in the KRAS2 gene, and these mutations were of the type seen in infiltrating ductal adenocarcinomas of the pancreas, consistent with the hypothesis that PanINs are precursor lesions that can progess to infiltrating adenocarcinoma (6). Of mutation bearing PanINs, 85% of their associated ADM lesions harbored a KRAS2 gene mutation. In contrast, when these lesions occurred in isolation, without an associated PanIN lesion, they were always KRAS2 wild-type. The only time we observed a KRAS2 gene mutation in a lesion of ADM was when the lesion was associated with a PanIN, and in all instances the same mutation was found in both the PanIN and in the lesion of ADM. The failure to find a KRAS2 gene mutation in an ADM lesion not associated with a PanIN, combined with the finding that the two lesions harbor the same mutation, suggests that the ADM lesions associated with a PanIN may represent retrograde extension of the PanIN into the smaller ducts. It is possible is that there are two distinct molecular mechanisms of ADM formation, one that occurs in isolation, has low mucin content and rarely involves mutations in KRAS2. The second ADM mechanism occurs within the same lobule as PanIN, contains mucin and generally bears the same KRAS2 gene mutation as the PanIN.
Our failure to demonstrate KRAS2 gene mutations in isolated ADM lesions suggests that a centroacinar origin for human pancreatic cancer is less likely, however we cannot rule out this possibility. Studies of genetically engineered mouse models have shown that centroacinar cells can give rise to pancreatic neoplasia, particularly after injury promotes differentiation into ductal cells. Most of the promoters used in the genetically engineered mouse models are expressed in centroacinar cells, and findings in mice with targeted PTEN gene deletion suggest that ductal metaplasia results from the expansion of centroacinar cells rather than transdifferentiation of acinar cells (29)..
Some conclusions can be made about the relationship between PanINs, their associated ADMs and KRAS2 gene mutations. First, while KRAS2 gene mutations occur commonly in PanIN (74%), mutation of this gene appears not to be essential for PanIN formation, as mutations were absent in 26% of lesions. Alternatively, activation of the KRAS2 pathway may be essential, but the pathway activation may be occurring through a less common KRAS2 mutation (e.g. codon 61) or a currently unidentified alternative pathway member in KRAS2 wild-type cases (11). Second, when a PanIN harbors a KRAS2 gene mutation, the vast majority of ADMs within the same lobule will bear the same mutation, suggesting that the two lesions derive from one another. The question of “who is the mother and who is the daughter?” cannot be definitively determined from these data. The occurrence of KRAS2 gene mutations detected in the PanIN-associated metaplastic acinar structures may be due to retrograde extension of the PanIN cells into the smaller ducts associated with acini, arising from physical backwards extension of PanIN cells into smaller ducts producing the ADM phenotype. It is also alternatively conceivable that ADMs are the initial lesion and that those with KRAS2 mutations, tend to progress to PanIN. The latter possibility is supported by the observation that 4 of ADM mutations could only be detected using the more sensitive LigAmp assay and were missed by standard DNA sequencing, suggesting that the mutations may be present in only a subpopulation of cells within the ADM. This is further supported by the existence of 2 mutant KRAS2 containing PanINs where their associated ADMs were KRAS2 wild-type. Alternatively, in the retrograde extension model, we would have to attribute this to incomplete extension or contamination of the microdissections with normal cells.
Although KRAS2 gene mutations were not identified in isolated acinar cells or in isolated ADM lesions in this study, it is possible that this result was obtained because the isolated lesions were early neoplastic lesions driven by something other than a KRAS2 gene mutation. While this is possible, the KRAS2 gene was chosen for this study because it is the first known gene targeted in the development of human pancreatic neoplasia, and because this is the very gene driving most of the genetically engineered mouse models that suggest the possibility of an acinar cell origin for pancreatic ductal neoplasia.
In summary, observations by a number of groups in several genetically engineered mouse models suggest that acinar cells, centroacinar cells, postulated stem cells or ADM can be responsible for the development of pancreatic ductal neoplasia. Our current data, however, indicate that acinar cells are most likely not the origin of human pancreatic neoplasia (Figure 4).
Figure 4.
Potential “cell of origin” of pancreatic ductal neoplasia in mouse models and human pancreata. Adapted and reprinted, with permission, from the Annual Review of Pathology: Mechanisms of Disease, Vol. 3, (c)2008 by Annual Reviews www.annualreviews.org(5). Vertical bars indicate the cell of origin as currently established. Backward arrows signify the remaining possibility that the PanIN precursor cell may originate in centroacinar cells.
Acknowledgements
We acknowledge Dr. Scott Kern for helpful discussions. This work was supported by the NIH SPORE (Specialized Program of Research Excellence) in Gastrointestinal Cancer Grant CA62924, the Michael Rolfe Foundation, Richard and Nancy Riordan, and the Sol Goldman Pancreatic Cancer Research Center.
References
- 1.Hulst SPL. Zur kenntinis der Genese des Adenokarzinoms und Karzinoms des Pankreas. Virchows Arch (B) 1905;180:288–316. [Google Scholar]
- 2.Cubilla AL, Fitzgerald PJ. Morphological lesions associated with human primary invasive nonendocrine pancreas cancer. Cancer Res. 1976;36:2690–2698. [PubMed] [Google Scholar]
- 3.Hruban RH, Adsay NV, Albores-Saavedra J, et al. Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol. 2001;25:579–586. doi: 10.1097/00000478-200105000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Brat DJ, Lillemoe KD, Yeo CJ, Warfield PB, Hruban RH. Progression of pancreatic intraductal neoplasias to infiltrating adenocarcinoma of the pancreas. Am J Surg Pathol. 1998;22:163–169. doi: 10.1097/00000478-199802000-00003. [DOI] [PubMed] [Google Scholar]
- 5.Maitra A, Hruban RH. Pancreatic Cancer. Annual Review of Pathology. 2008:157–188. doi: 10.1146/annurev.pathmechdis.3.121806.154305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hruban RH, van Mansfeld AD, Offerhaus GJ, et al. K-ras oncogene activation in adenocarcinoma of the human pancreas. A study of 82 carcinomas using a combination of mutant-enriched polymerase chain reaction analysis and allele-specific oligonucleotide hybridization. Am J Pathol. 1993;143:545–554. [PMC free article] [PubMed] [Google Scholar]
- 7.Hruban RH, Goggins M, Parsons J, Kern SE. Progression model for pancreatic cancer. Clin Cancer Res. 2000;6:2969–2972. [PubMed] [Google Scholar]
- 8.Moskaluk CA, Hruban RH, Kern SE. p16 and K-ras gene mutations in the intraductal precursors of human pancreatic adenocarcinoma. Cancer Res. 1997;57:2140–2143. [PubMed] [Google Scholar]
- 9.Tada M, Ohashi M, Shiratori Y, et al. Analysis of K-ras gene mutation in hyperplastic duct cells of the pancreas without pancreatic disease. Gastroenterology. 1996;110:227–231. doi: 10.1053/gast.1996.v110.pm8536861. [DOI] [PubMed] [Google Scholar]
- 10.Yanagisawa A, Ohtake K, Ohashi K, et al. Frequent c-Ki-ras oncogene activation in mucous cell hyperplasias of pancreas suffering from chronic inflammation. Cancer Res. 1993;53:953–956. [PubMed] [Google Scholar]
- 11.Jones S, Zhang X, Parsons DW, et al. Core Signaling Pathways in Human Pancreatic Cancers Revealed by Global Genomic Analyses. Science. 2008 doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hruban RH, Adsay NV, Albores-Saavedra J, et al. Pathology of genetically engineered mouse models of pancreatic exocrine cancer: consensus report and recommendations. Cancer Res. 2006;66:95–106. doi: 10.1158/0008-5472.CAN-05-2168. [DOI] [PubMed] [Google Scholar]
- 13.Hingorani SR, Wang L, Multani AS, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 14.Hingorani SR, Petricoin EF, Maitra A, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 15.Brembeck FH, Schreiber FS, Deramaudt TB, et al. The mutant K-ras oncogene causes pancreatic periductal lymphocytic infiltration and gastric mucous neck cell hyperplasia in transgenic mice. Cancer Res. 2003;63:2005–2009. [PubMed] [Google Scholar]
- 16.Murtaugh LC, Leach SD. A case of mistaken identity? Nonductal origins of pancreatic "ductal" cancers. Cancer Cell. 2007;11:211–213. doi: 10.1016/j.ccr.2007.02.020. [DOI] [PubMed] [Google Scholar]
- 17.Guerra C, Schuhmacher AJ, Canamero M, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 18.Zhu L, Shi G, Schmidt CM, Hruban RH, Konieczny SF. Acinar cells contribute to the molecular heterogeneity of pancreatic intraepithelial neoplasia. Am J Pathol. 2007;171:263–273. doi: 10.2353/ajpath.2007.061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carriere C, Seeley ES, Goetze T, Longnecker DS, Korc M. The Nestin progenitor lineage is the compartment of origin for pancreatic intraepithelial neoplasia. Proc Natl Acad Sci U S A. 2007;104:4437–4442. doi: 10.1073/pnas.0701117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brune K, Abe T, Canto M, et al. Multifocal neoplastic precursor lesions associated with lobular atrophy of the pancreas in patients having a strong family history of pancreatic cancer. Am J Surg Pathol. 2006;30:1067–1076. [PMC free article] [PubMed] [Google Scholar]
- 21.Detlefsen S, Sipos B, Feyerabend B, Kloppel G. Pancreatic fibrosis associated with age and ductal papillary hyperplasia. Virchows Arch. 2005;447:800–805. doi: 10.1007/s00428-005-0032-1. [DOI] [PubMed] [Google Scholar]
- 22.Leach SD. Mouse models of pancreatic cancer: the fur is finally flying! Cancer Cell. 2004;5:7–11. doi: 10.1016/s1535-6108(03)00337-4. [DOI] [PubMed] [Google Scholar]
- 23.Hruban RH, Takaori K, Klimstra DS, et al. An illustrated consensus on the classification of pancreatic intraepithelial neoplasia and intraductal papillary mucinous neoplasms. Am J Surg Pathol. 2004;28:977–987. doi: 10.1097/01.pas.0000126675.59108.80. [DOI] [PubMed] [Google Scholar]
- 24.Shi C, Eshleman SH, Jones D, et al. LigAmp for sensitive detection of single-nucleotide differences. Nat Methods. 2004;1:141–147. doi: 10.1038/nmeth713. [DOI] [PubMed] [Google Scholar]
- 25.Shi C, Fukushima N, Abe T, et al. Sensitive and quantitative detection of KRAS2 gene mutations in pancreatic duct juice differentiates patients with pancreatic cancer from chronic pancreatitis, potential for early detection. Cancer Biol Ther. 2008;7:353–360. doi: 10.4161/cbt.7.3.5362. [DOI] [PubMed] [Google Scholar]
- 26.Hruban RH, Pitman MB, Klimstra DS. AFIP Atlas of Tumor Pathology, Series IV. Tumors of the Pancreas: American Registry of Pathology. 2007 [Google Scholar]
- 27.Hruban RH, Rustgi AK, Brentnall TA, Tempero MA, Wright CV, Tuveson DA. Pancreatic cancer in mice and man: the Penn Workshop 2004. Cancer Res. 2006;66:14–17. doi: 10.1158/0008-5472.CAN-05-3914. [DOI] [PubMed] [Google Scholar]
- 28.Grippo PJ, Nowlin PS, Demeure MJ, Longnecker DS, Sandgren EP. Preinvasive pancreatic neoplasia of ductal phenotype induced by acinar cell targeting of mutant Kras in transgenic mice. Cancer Res. 2003;63:2016–2019. [PubMed] [Google Scholar]
- 29.Stanger BZ, Stiles B, Lauwers GY, et al. Pten constrains centroacinar cell expansion and malignant transformation in the pancreas. Cancer Cell. 2005;8:185–195. doi: 10.1016/j.ccr.2005.07.015. [DOI] [PubMed] [Google Scholar]