Abstract
Population isolates, such as Finland, have proved beneficial in mapping rare causative genetic variants due to a limited number of founders resulting in reduced genetic heterogeneity and extensive linkage disequilibrium. We have here used this special opportunity to identify rare alleles in autism by genealogically tracing 20 autism families into one extended pedigree with verified genealogical links reaching back to the 17th century. In this unique pedigree we performed a dense microsatellite marker genome-wide scan of linkage and linkage disequilibrium, and followed initial findings with extensive fine-mapping. We identified a putative autism susceptibility locus at 19p13.3, and obtained further evidence for previously identified loci at 1q23 and 15q11-13. Most promising candidate genes were TLE2 and TLE6 genes clustered at 19p13 and ATP1A2 at 1q23.
Introduction
Autism (AD [MIM 209850]) is a severe childhood-onset disorder characterized by impaired development in social interaction and communication as well as presence of stereotyped patterns of interests and behavior before the age of three years (1, 2). It is the most recognizable syndrome in a group of autism spectrum disorders (ASDs; or PDDs, pervasive developmental disorders), which include also Asperger syndrome (AS) and atypical forms of autism. The currently reported population prevalence for autism is 4-10/10 000, whereas the total prevalence for ASDs is reported as 10-60/10 000 (3-5). Awareness of ASDs has substantially increased during the past few years, and it is now recognized that when the early onset of the disorder is taken into account, the public health burden of autism in patient years is substantial.
Based on twin and family studies, predisposition to autism is strongly heritable. The estimated sibling risk of 2-4% indicates significant familial clustering of the disorder, whereas twin studies have suggested heritability estimates of over 90% (6-8). Nevertheless, only a few confirmed genetic disorders that may lead to an autistic phenotype are currently known. These include syndromes, such as Fragile X, and chromosomal aberrations, especially on chromosomes X and 15q11-13 (9-12). These and some more recent findings, such as identification of mutations in the neuroligin genes (12, 13), SHANK3 (14, 15), and CNTNAP2 (16-19) point out that rare high-penetrance mutations may be causative for the autistic phenotype. In addition, de novo DNA copy number variation (CNV) seems to influence autism predisposition (20-22). Yet, the predominant hypothesis is that a combination of multiple predisposing genetic and environmental factors is required in most cases. In fact, assuming multifactorial inheritance, it is estimated that potentially more than 15 individual loci each having a minor effect might be involved in the etiology of autism (23). Assuming such small effect variants, and thus extensive genetic heterogeneity, it is not surprising that the numerous genome-wide linkage scans performed to date have yielded poorly replicated findings and modest levels of significance. However, based on a recent meta-analysis of six separate genome-wide linkage scans comprising a total of 771 affected sib-pairs, especially the locus at 7q22-32 appears as a promising candidate for autism (24), as well as 11p12-p13, which was the single major locus identified recently by the Autism Genome Project Consortium (25). Genetic heterogeneity and contribution of numerous rare alleles most probably complicate also ongoing genome-wide association studies, which rely on association to common variants. This heterogeneity was recently highlighted in a study by Morrow et al. (26) where homozygosity mapping was carried out in autistic children of consanguineous marriages. All of the mutations identified in this study were associated to autism only within single families.
Due to the limited success in linkage-based genome-wide scans in autism, novel approaches are needed to reveal the molecular mechanism underlying ASDs. If the genetic background of autism reflects that of other complex diseases, massive study samples are required for genome-wide association studies which can be expected to expose common, most probably low impact alleles (27). On the other hand, linkage studies in large pedigrees are likely to contribute to our understanding of rare mutations with a high impact. Yet an alternative approach is to focus on population isolates, or preferably on further restricted sub-isolates with a well-established genealogy, in which a single (or few) causative variant(s) can be expected to be enriched (28). Such approaches focusing on the rare forms of common diseases have proved to be of high importance and revealed genes, which have led to a significant improvement in our understanding of molecular pathways underlying the disease. Further, there is an increasing number of examples of susceptibility genes initially mapped in rare families that have proved to be important also at the population level (29).
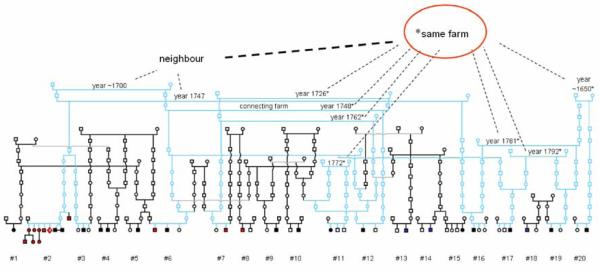
Population isolates, such as Finland, offer several well-recognized advantages for disease gene mapping. The results have been most striking in the mapping of rare Mendelian traits, but many of them hold also for mapping of common complex diseases. The benefits include for e.g. (i) shared common environment and culture of the study families, (ii) well-standardized diagnostic criteria and common training of clinicians, (iii) centralized health care records, (iv) opportunity to identify small sub-isolates or reconstruct large pedigrees based on population registers, and (v) reduced genetic heterogeneity at least in the cases where rare alleles confer significant increase in susceptibility to complex diseases (28, 30). Moreover, recent evidence verifies that population isolates exhibit substantially higher linkage disequilibrium (LD) around common alleles than outbred samples suggesting that relatively sparse marker maps might be sufficient for initial disease gene localization (31). Here, we describe an extended ASD pedigree constructed from 20 nuclear families (Figure 1) scattered all over the country by systematically examining the genealogy of all of the families in our nationwide ASD study sample. We carried out a dense microsatellite-based genome-wide scan in this unique pedigree and report a putative ASD susceptibility locus at 19p13.3 and further evidence for previously identified loci at 1q23 and 15q11-13.
Figure 1.
Extended ASD pedigree originating from Central Finland. The core families and links to the common ancestors from two neighbouring farms are marked in light blue. Individuals with infantile autism (n=17) are indicated in black, Asperger syndrome (n=14) in red, and pervasive developmental disorder not otherwise specified (PDD-NOS; n=3) in blue.
Results
We performed a dense genome-wide scan in the 20 nuclear families of the extended pedigree (naffected=33) with a set of 1109 microsatellites, which yielded an average intermarker distance of 3.43 cM. Individual families were analyzed separately, except in the three cases where two nuclear families can be connected as one family on the level of first or second cousins (see Figure 1). Two primary strategies were selected to analyze the data. Since the fundamental hypothesis behind the current study was that the observed genealogical links reflect identical-by-decent (IBD) sharing of the same ancestral susceptibility variant(s) in the current set of families, we wanted to maximally extract information of allele-sharing both within and across the families. Our primary approach was the LD+Linkage method of Pseudomarker analysis program, as described by Göring and Terwiliger (32). Both dominant and recessive Pseudomarker analyses were conducted. However, the structure of the pedigree and the obtained results support recessive inheritance due to which we mainly focused on the results of the “recessive Pseudomarker analysis”. The “dominant Pseudomarker analysis” is analogous to affected relative pair methods in large families, and it weights the sharing between parents affected with ASDs and their affected children more strongly than that between unaffected parents and affected children. In the “recessive Pseudomarker analysis” contributions of both parents are weighted equally. The Pseudomarker approach enables the combination of linkage and association evidence in various types of samples (family-based and case-control samples) in the same analysis (32). We also employed non-parametric multipoint linkage (NPL) analysis of Simwalk2 v. 2.91 software, which is especially suitable for complex pedigrees, to monitor allele-sharing within the families (linkage) (33). Of the five NPL statistics produced by Simwalk2, we chose to report the two that have been shown to be most powerful and best suitable for dominant and recessive traits (“BLOCKS”, referred here as “NPL_recessive” and “MAX-TREE”, referred here as “NPL_dominant”) (34). These analyses were additionally run using all known genealogical connections (see Figure 1) to the common ancestors, but this had no marked effect on the results (data not shown). In the initial scan, genotypes from 22 regionally matched controls were included in the analyses to improve power in LD analyses as well as to better estimate allele-frequencies in the linkage analysis. In the extended pedigree, family #2 includes a twin pair with severe infantile autism as well as 11 individuals with AS, of which four have a relatively mild AS phenotype. To avoid the dominance of this single family in the initial LD+Linkage analysis, all AS individuals in the family were excluded from the primary Pseudomarker analysis (designated as Set 1). The strategy of assigning these individuals as both unaffected (Set 1) and affected (Set 2) was employed also in the multipoint linkage analyses by Simwalk2 as well as in the follow-up and fine-map of the best loci.
Initial genome-wide scan
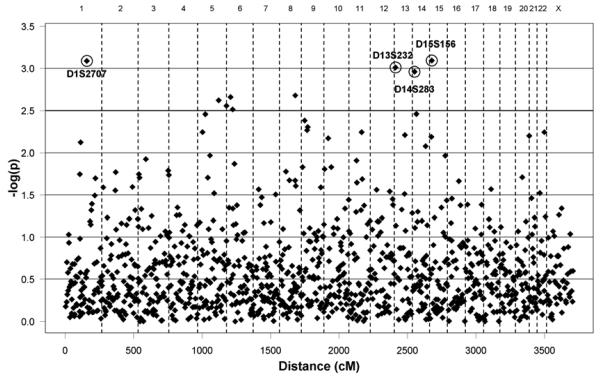
Summary of the genome-wide LD+Linkage results from the recessive Pseudomarker analysis in the extended pedigree (Set1) is given in Figure 2. Altogether nine loci exceeded the −log(p) value of 2.5 which was chosen based on the distribution of the results as an arbitrary cut-off for the selection of a reasonable number of follow-up loci (Table 1). For these nine loci, we additionally monitored the LD|Linkage value in Pseudomarker. This test of LD allowing, but not assuming, for linkage is used to demonstrate that some of the signals are obtained from haplotype sharing among individuals instead of linkage only. The best LD+Linkage p-values were observed with markers D1S2707 on 1q23.2 (p=0.00082, Set 1) and D15S156 on 15q12 (p=0.00081, Set 1). The evidence for D1S2707 was almost entirely attributable to LD (p=0.00079, LD!Linkage, Set 1). The flanking markers of D1S2707 (D1S1653 and D1S484, 4.5 cM proximally and 1.4 cM distally, respectively) yielded no evidence of either linkage or LD in the Pseudomarker analysis. On 15q12, by contrast, also D15S975 (2.1 cM proximally to D15S156) yielded a suggestive p-value in the Linkage+LD analysis (p=0.0065, Set 1), with most evidence for both markers again resulting from LD (p=0.02 to p=0.0009, LD|Linkage). In the dominant Pseudomarker analysis (Set 1), only one locus exceeded the −log(p)=2.5 cut-off. Results of the dominant analysis are given in Supplementary Material, Figure S1.
Figure 2.
Distribution of microsatellite marker LD+Linkage values of the initial genome-wide scan. Results produced by recessive Pseudomarker analysis in the extended pedigree (Set 1). The y-axis presents −log(p) values as a function of genetic distance (x-axis, in cM). Markers with −log(p)~3 are displayed.
Table 1.
Results of the initial genome-wide recessive Pseudomarker analysis.
| Ranking from screen |
Marker | Chromosomea | Genetic distanceb (cM) |
LD+Linkage [−log(p)] |
LD|Linkage [−log(p)] |
|---|---|---|---|---|---|
| 1 | D15S156 | 15q12 | 15.1 | 3.09 | 3.05 |
| 2 | D1S2707 | 1q23 | 156.1 | 3.09 | 3.10 |
| 3 | D13S232 | 13q12 | 8.7 | 3.01 | 2.85 |
| 4 | D14S283 | 14q11 | 14.7 | 2.96 | 2.52 |
| 5 | D8S1132 | 8q23 | ~113.1 | 2.68 | 2.84 |
| 6 | D6S1279 | 6p24 | ~30 | 2.66 | 1.59 |
| 7 | D5S2090 | 5q32 | 150.0 | 2.62 | 2.81 |
| 8 | D5S2006 | 5q35 | 205.7 | 2.56 | 2.74 |
| 9 | D6S422 | 6p22 | 42.8 | 2.51 | 2.58 |
All markers with LD+Linkage −log(p)>2.5 are displayed, together with the LD|Linkage values for these loci. Results produced by recessive Pseudomarker analysis (32) in the extended pedigree (Set1).
UCSC Human Genome Browser, May 2004 Assembly (http://genome.ucsc.edu)
From ptel based on deCODE Genetic map (70)
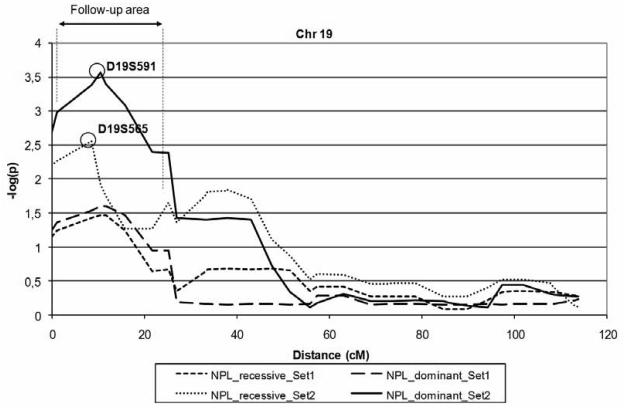
In addition to the nine loci identified in the recessive Pseudomarker analysis (Table 1), we identified one significant locus in the Simwalk2 analysis with non-parametric multipoint linkage of −log(p)=3.57 at D19S591 (Set 2, “NPL_dominant”; see Figure 4) located at 19p13.3. These ten loci were selected for the follow-up stage, as discussed below (see Figure 3 for a flow-chart of the study). The second most significant linkage in the Simwalk2 analysis was observed on chromosome 6 with −log(p)=2.15 (D6S958, Set 2, “NPL_dominant”) while the results for all other chromosomes were below −log(p)=1.5 (data not shown).
Figure 4.
Multipoint linkage results, chromosome 19. Results produced by non-parametric Simwalk2 analysis in the extended pedigree. All microsatellites and candidate genes analyzed in the follow-up and fine-map stages fall between markers D19S883 and D19S873 (follow-up area). The best markers of the Set 2 analysis are displayed. “NPL_dominant” equals to the Simwalk2 “MAX-TREE” statistic, while “NPL_recessive” equals to the “BLOCKS” statistics (34).
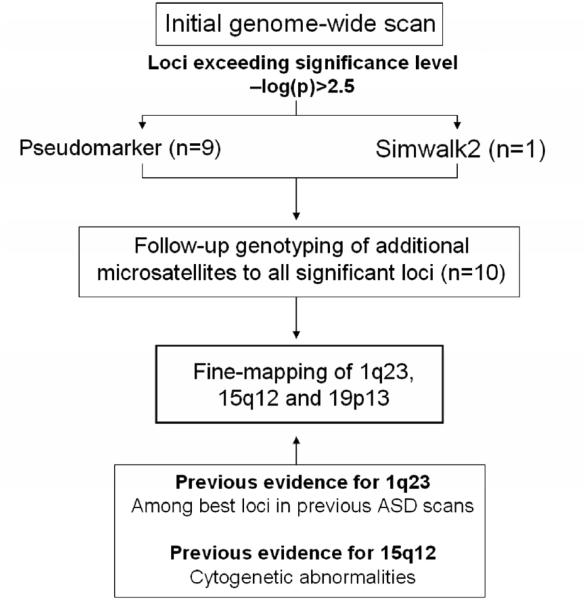
Figure 3.
Flow-chart illustrating the current study.
Follow-up stage and candidate gene analyses
Altogether 44 additional microsatellites from the ten genomic regions were analyzed in the extended pedigree in the follow-up stage. Detailed information of the follow-up markers in each locus is included in Supplementary Material, Table S1, whilst all follow-up results are reported in Supplementary Material, Table S2 and Figure S2. Two of the loci selected for follow-up (1q23 and 15q12) have been indicated in earlier autism studies. Two independent genome-wide screens for ASDs performed in Finnish families found evidence for 1q23, reporting highest LOD scores of 2.63 (D1S1653; ASD scan) and 3.58 (D1S484; AS scan) within 4.5cM of D1S2707 (35, 36). The extended pedigree of the current study overlaps with the previous ASD scan with four families (eight affected individuals); with the AS scan there is no overlap. The exclusion of the overlapping samples from the analyses was not considered because this would have decreased the already small sample size of the pedigree and broken up some of the genealogical links, resulting in a significant loss of information. The prior evidence for 15q11-13 locus in autism arises from cytogenetic studies demonstrating that some 1-3% of autism cases are caused by inverted maternal duplications of this region (10, 11, 37). 15q11-13 is also a well-known imprinted locus playing a key role in Angelman and Prader-Willi syndromes.
Based on this prior evidence and the results of the follow-up, candidate genes were chosen from the best loci at chromosomes 1q, 15q and 19p and analyzed further with SNP markers (Table 2). Candidates from 1q and 15q were analyzed in (i) the extended pedigree as in the primary screen, (ii) a sample of 97 Finnish families with infantile autism (naffected = 119), and (iii) a sample of 28 Finnish families with AS individuals (naffected = 119). Since no previous evidence exists for chromosome 19, the candidate genes at 19p13 were analyzed primarily in the extended pedigree, with only the best genes analyzed additionally in the nationwide autism and AS sample sets (Supplementary Material, Table S1). Complete fine-map results for all candidate genes are reported in Supplementary Material, Table S2.
Table 2.
Candidate genes at 1q23, 15q12 and 19p13.
| Gene | Chromosome | Position (bp)a | Size (bp) | Genotyped SNPs |
|---|---|---|---|---|
| KCNJ10 | 1q23 | 156821108-156853111 | 32 003 | 6 |
| KCNJ9 | 1q23 | 156864433-156872285 | 7852 | 2 |
| ATP1A2 | 1q23 | 156898621-156926445 | 27 824 | 5 |
| ATP1A4 | 1q23 | 156934436-156969836 | 35 400 | 7 |
| NOS1AP | 1q23 | 158771239-159069886 | 298 647 | 6 |
| RGS4 | 1q23 | 159770809-159778036 | 7227 | 5 |
| UBE3A | 15q12 | 23133489-23201746 | 68 257 | 10 |
| GABRB3 | 15q12 | 24342556-24570020 | 227 464 | 19 |
| GABRA5 | 15q12 | 24663151-24776749 | 113 598 | 6 |
| GABRG3 | 15q12 | 24799263-25451729 | 652 466 | 6 |
| PALM | 19p13 | 660002-699328 | 39 326 | 9 |
| GRIN3B | 19p13 | 951437-960723 | 9286 | 2 |
| EFNA2 | 19p13 | 1237168-1250944 | 13 776 | 2 |
| MBD3 | 19p13 | 1527678-1543652 | 15 974 | 4 |
| GNG7 | 19p13 | 2462218-2653663 | 191 445 | 21 |
| TLE6 | 19p13 | 2928562-2946176 | 17 614 | 7 |
| TLE2 | 19p13 | 2948637-2980022 | 31 385 | 11 |
| AES | 19p13 | 3003908-3013371 | 9463 | 8b |
| GNA15 | 19p13 | 3087230-3114741 | 27 512 | 8 |
| SH3GL1 | 19p13 | 4311368-4351471 | 40 104 | 4 |
| SEMA6B | 19p13 | 4493600-4509503 | 15 904 | 2 |
| NRTN | 19p13 | 5774818-5779335 | 4518 | 1 |
| PSPN | 19p13 | 6326305-6326860 | 556 | 1 |
UCSC Human Genome Browser, May 2004 Assembly (http://genome.ucsc.edu)
Includes one intergenic SNP
On 1q, six candidates were selected for further study: KCNJ9, KCNJ10, ATP1A2, and ATP1A4, which make up a syntenic rodent epilepsy locus around D1S2707 (38, 39), and RGS4 and NOS1AP (also known as CAPON) which are positional candidate genes also for schizophrenia (40-42) located 2.9 Mb and 1.9 Mb from D1S2707, respectively. A total of 31 SNPs were genotyped for these candidates. In the extended pedigree, significant evidence was detected at ATP1A2 with rs1016732 (p=0.00048, LD+Linkage, Set 2, recessive Pseudomarker), located just 14.7 kb from D1S2707, the best microsatellite of the initial scan. As with D1S2707, the evidence for rs1016732 was primarily attributable to sharing across the families (p=0.00055, LD|Linkage, Set2). The minor allele frequency of this SNP was 0.13 in affected individuals compared with 0.097 in controls. Encouragingly, also the four subsequent SNPs yielded suggestive evidence for LD+Linkage in the same analysis (from p=0.03 to p=0.006) which could be attributed to sharing across families as well. Similarly with the RGS4 gene, some evidence of sharing across families could be observed in the extended pedigree with two SNPs (p=0.01-0.03, LD|Linkage, Set1 and Set2, recessive Pseudomarker). Only marginal evidence of association or linkage was observed with any of the SNPs analyzed in the nationwide autism and AS families (best p-values ~ 0.01).
On 15q, two of the most promising markers in the initial scan, D15S156 and D15S975 are located either near (~129 kb distally) or within a GABAA receptor subunit gene cluster. Therefore, the three subunit genes, GABRB3, GABRG3, and GABRA5 were chosen as both positional and functional candidates. Also UBE3A, located 1.1 Mb away from the cluster, was included due to a previous report of association to ASDs (43) and its role in phenotypically related Angelman syndrome. With the 41 SNPs analyzed, LD+Linkage was detected in the extended pedigree with six SNPs from the GABAA cluster (from p=0.02 to p=0.0023, Set2, recessive Pseudomarker), with the most significant results again originating from four consecutive SNPs within the GABRB3 gene, yielding LD|Linkage from p=0.03 to p=0.00084 (best p-value rs7173713). Again, when analyzed in the nationwide study sample (autism and AS families), only modest evidence of LD+Linkage or LD|Linkage was seen at the region (from p=0.04 to p=0.002).
On 19p, we selected 13 biologically relevant candidate genes in the best multipoint linkage region for further analysis: PALM, GRIN3B, EFNA2, MBD3, GNG7, TLE6, TLE2, AES, GNA15, SH3GL1, SEMA6B, NRTN and PSPN (Table 2). Altogether 80 SNPs for these candidates were analyzed in the extended pedigree. Of these genes, TLE6, TLE2 and AES (also known as TLE5) are situated as a cluster 428 kb from D19S565 and 12.8 kb from D19S591, the two best markers of the initial scan (Figure 4 and Supplementary Material, Figure S3). These three genes belong to a TLE family of proteins homologous to the Drosophila Groucho protein which is involved in neurogenesis during embryonic development. Since the most significant linkage signal from the initial Simwalk2 analysis was detected in a Set2 analysis, where the multiple AS-individuals of family#2 markedly contribute to the linkage signal, we here focused on Set1 results to identify family#2-independent association signals. Interestingly, the most significant results at 19p13 - and in the whole study - were seen within the TLE6-TLE2-AES gene cluster with altogether eight consecutive SNPs yielding LD+Linkage p-values <0.04 in the same analysis (Set1) with dominant Pseudomarker, consistently with the original multipoint linkage. Of these, rs4806893 and rs216283 yielded the best results (both p=0.000078, LD+Linkage, Set 1, dominant Pseudomarker) together with rs216276 (p=0.00063). For five of the eight SNPs, there was also evidence of sharing across families (p-values from 0.00019 to 0.05, LD|Linkage, Set 1, dominant Pseudomarker). When comparing allele frequencies of these SNPs between affected individuals and controls, the minor allele of rs216276 was notably overrepresented in cases compared with controls (0.15 vs. 0.06). With rs4806893 and rs216283 the major allele was instead overrepresented (0.74 in cases vs. 0.54 in controls, both SNPs). The eight SNPs cover a region of 16.5 kb and are located mainly within TLE2 and the 3′UTR/intergenic region of both TLE2 and TLE6 (transcribed in reversed directions; see Supplementary Material, Figure S3 for details). Some evidence of LD+Linkage (p<0.01) in the extended pedigree was additionally seen with the GNA15 gene (best p=0.004, Set 1, dominant Pseudomarker). Due to the encouraging results with the TLE-cluster, we additionally genotyped the SNPs (n=26) for these genes in the nationwide autism and AS study samples. However, the analyses disclosed no comparable evidence of association in these study samples outside the extended pedigree (best LD+Linkage p=0.02; majority >0.05). In order to further investigate the variation at this specific locus we constructed haplotypes of the eight associating SNPs using Phase v2.1.1 program, separately for the cases in the extended pedigree and the regional controls. The distribution of the different haplotypes between cases and controls revealed three common haplotypes (Supplementary Material, Table S3, of which one was notably more frequent in the cases (59%) compared with controls (38%) suggesting the presence of a susceptibility variant on this haplotype. However, based on the distribution of the haplotype frequencies, no single haplotype could be expected to account for the entire association signal.
Discussion
Linkage disequilibrium (LD) has been efficiently used for disease gene identification in monogenic disorders in the Finnish population. As few as four affected individuals have been sufficient for disease gene localization due to extensive haplotype sharing around the disease-causing mutation (44, 45). The starting point for this study is in many ways analogous to these early examples: the Finnish small founder population as a whole shows extensive degree of LD, not to mention young sub-isolates, such as the region from which this extended ASD pedigree originates (31). Additionally, this pedigree contains strikingly many individuals with an ASD, indicating possible enrichment of causative variants and thus providing an ideal setting for genetic mapping based on linkage and haplotype sharing.
We obtained evidence of linkage at three loci in the extended pedigree. Our results provide additional support to previously reported ASD susceptibility loci at 1q23 and 15q11-13, and reveal an additional interesting locus at 19p13. Results at 19p13 were obtained with the Set2 analysis in which the signal mostly originates from the multiple individuals with AS in family#2. Also, the results at 19p13 were obtained with a model best suitable for dominant inheritance in both Simwalk2 and Pseudomarker. By contrast, the most significant results at 1q23 and 15q12 were obtained with a Set1 recessive Pseudomarker analysis. Due to the unique structure of the extended pedigree, it is important to be able to separate out the effects of linkage from the results, because linkage regions generally are much larger than LD regions. Therefore, using the test of LD allowing for linkage in Pseudomarker, it is possible to model only for the sharing between families and not within families. The test does not assume linkage, but instead subtracts out the information about linkage from the joint analysis of LD+Linkage, making it applicable in situations where the linkage signal might not be formally significant. Also, since the power of SNPs to detect linkage is smaller than with microsatellites, significant linkage cannot necessarily be expected. At all three loci, 1q23, 15q12 and 19p13, substantial evidence of sharing across families, or LD, was detected as expected, pointing to greater genetic homogeneity due to isolation. In the association study of regional candidates, the most plausible and interesting findings were ATP1A2 at 1q and the TLE gene cluster at 19p, both of which provided considerable evidence of association.
At 1q23, the most significant results were obtained with five consecutive SNPs spanning 25.8 kb of the ATP1A2 gene in the extended pedigree (best p=0.00055, LD|Linkage, Set1). Linkage to this locus has been detected in at least two previous ASD studies (35, 36) as well as in multiple schizophrenia studies (40, 46). ATP1A2 is one of the four genes that make up the syntenic seizure susceptibility locus (Szs1) in mouse, originally identified by quantitative trait locus (QTL) mapping (38, 39). Association between seizure susceptibility and idiopathic generalized epilepsy and KCNJ10, has since been detected in both in mice and humans (38, 39, 47). Association of this locus with ASDs is of interest since up to 30% of individuals with autism suffer from epilepsy (48).
Suggestive linkage at 19p13 has been reported in previous genome-wide scans for ASDs, but reported values have been modest (49-54). Therefore, to observe a multipoint - log(p) value of 3.57 (p=0.00029) at this locus with only 20 nuclear families, seems encouraging. In fact, at 19p13, all of the families in the extended pedigree show complete segregation with the trait (linkage), implying that this locus might contribute to the disease risk in this pedigree. In particular family#2 displays significant sharing across affected individuals (Supplementary Material, Figure S4), which explains in part the observed linkage signal.
In the fine-mapping stage, the most significant results in the whole study were observed within a cluster of three genes located just 12.8 kb away from D19S591, the best marker in the initial scan. Eight consecutive SNPs, located in the borderline of TLE6 and TLE2 genes (Supplementary Material, Figure S3), displayed significant LD+Linkage (p-values from 0.04 to 0.000078, Set1). With five of these SNPs, the signal was mostly attributable to sharing across families (p-values from 0.05 to 0.00019, LD|Linkage, Set1). Due to the lack of established methods to correct for multiple testing in multi-level gene-mapping studies, we have not attempted to correct for multiple testing or LD structure in our study. Taking the small sample size into account, the results would not remain significant if corrected for all markers and tests. However, the most significant association signal with TLE2 and TLE6 (p=0.000078) does remain significant if it is corrected for the total number of SNPs (n=152) and tests (dominant and recessive Pseudomarker analysis) performed (Bonferroni, p=0.024). The different tests performed by Pseudomarker (LD+Linkage and LD|Linkage) cannot be included in this correction since the tests are not independent and the Bonferroni correction assumes independence of tests. The genes, TLE6, TLE2 and AES (also known as TLE5), belong to the human TLE (transducin-like Enhancer of split) protein family that is extensively homologous with the Drosophila Groucho (Gro; http://flybase.bio.indiana.edu/) protein which, together with proteins of the Hairy/Enhancer of split (HES) family, is involved in neurogenesis during embryonic development as components of the Notch signalling pathway (55, 56). All of the members in the human groucho/TLE family share a conserved TLE_N (PF03920; http://pfam.sanger.ac.uk/) protein domain and act as transcriptional corepressors. They have been suggested to perform functions analogous to their Drosophila counterparts, that is, negatively regulating neuronal development and differentiation (57). Loss of function of Groucho and HES proteins, and other components of the Notch signalling pathway, results in the overproduction of central and peripheral neurons (58), which is of interest in regard of macrocephaly reported in ~20% of individuals with autism (59) and increased brain volume frequently observed in autistic cases (60). In Drosophila, Groucho is also known to interact with the conserved Engrailed protein (61), whose human homologue EN2 was recently associated with autism (62).
As a conclusion, on the basis of this study, we suggest that loci at 1q23, 15q12 and 19p13 are likely to contain genes that increase susceptibility to ASDs. In particular the results obtained with SNPs at 19p are promising with considerable linkage supporting the association evidence. In the next step, extensive sequencing of the candidates is required to reveal the complete allelic variability of the genes. However, the fact that none of the association results could be replicated outside the extended pedigree in the nationwide study samples indicates that the putative predisposing loci represent an enrichment of these loci in a small founder population. Since comparable information from single consanguineous autism families has provided useful also in other studies, as highlighted recently by Morrow et al (26), the results obtained with this pedigree should not be ignored simply because all of them do not reach genome-wide significance. We are currently working with detailed phenotypic analyses and a 317k SNP array to further characterize this unique ASD pedigree with the hope to eventually identify the specific genetic factors responsible for the disease phenotype in the pedigree. Such assumingly rare factors could provide new insight to the molecular mechanism of the autistic phenotype, as has been the case with some of the rare highpenetrance mutations identified in individuals with autism to date.
Materials and Methods
Study samples and genealogy
We first identified a total of 10 families with autism whose ancestors originated from a single small farm in a village of the late-settlement region of Central Finland some 5-10 generations ago. When we followed up all the ancestral trees back up to 12 generations, we were able to distinguish nine ancestors connecting the 10 autism families, which, most interestingly, were born on the same small farm 215-350 years ago. Local church and civil registers were utilized for information after year 1850 and the Finnish National Archives for the earlier periods in accordance with published criteria (63). It is thus probable that these families share one common ancestor, although the archives did not reach back enough to allow the identification of the founder couple. We were also able to link 10 additional nuclear families with ASDs to this core pedigree as well as to reveal additional genealogical links among the 20 nuclear families, as shown in Figure 1. The consanguinity of these autism families leading to the identified pedigree structure here is strikingly similar to the pedigrees we have uncovered in numerous rare recessive Mendelian disorders of the Finnish disease heritage, such as variant form of late infantile neuronal ceroid lipofuscinosis (vLINCL [MIM 256731]) (64) or infantile onset spinocerebellar ataxia (IOSCA [MIM 271245]) (65), thus providing an optimal setting for genetic mapping studies and identification of the disease gene(s) (66, 67). In total, our extended pedigree consists of 20 Finnish nuclear families with altogether 34 individuals affected with an ASD, of which 25 are males and 9 females. Of these, 17 are diagnosed with infantile autism, 14 with Asperger syndrome (AS), and three with a PDD not otherwise specified (PDD-NOS). Since two of the males affected with infantile autism are monozygotic twins, we have used only one of them in the analyses, making the total number of affected individuals 33. In all Pseudomarker analyses performed in the extended pedigree, we utilized genotype data from regionally matched controls to properly control for the diversity of alleles in the general population. The control samples were gathered from the same village where the families in the extended pedigree originate. In the initial scan we analyzed 22 controls and for the follow-up and fine-map stages the number of controls was increased to 93.
We followed the most promising loci emerging from the initial scan by genotyping regional SNP markers in the complete pedigree as well as in the nationwide sample collection of 238 familial autism and Asperger syndrome cases and their family members (Table 3), for which the diagnostic procedures have been described earlier (35, 68, 69). The carefully phenotyped autism study sample consists of 97 Finnish families with 119 affected individuals diagnosed with infantile autism according to the ICD-10 (1) and DSM-IV (2) criteria. Only families with at least one child with infantile autism were included, whilst families with associated medical conditions such as Fragile X syndrome or profound mental retardation were excluded. The AS sample contains 28 large Finnish pedigrees with 119 affected individuals. The pedigrees contain only AS cases fulfilling the ICD-10 criteria in multiple subsequent generations (Table 3). (It should be noted that in the extended pedigree four individuals in family #2 display notable AS-like features [here assigned as AS] but do not completely meet all of the ICD-10 criteria for AS.) Only individuals with normal overall cognitive development before the age of three were included in the AS sample. Of the 20 nuclear families in the extended pedigree, 16 are included in the current nationwide autism study sample, one in both autism and AS samples, and three in neither. This study has been approved by relevant ethical committees and informed written consent was received from all the participating families.
Table 3.
Description of the study sample.
| Study sample | Families | Affected individuals | Males | Females |
|---|---|---|---|---|
| Extended pedigree | 20a | 34b | 25 | 9 |
| Autism | 97 | 119 | 90 | 28 |
| Asperger syndrome (AS) | 28 | 119 | 74 | 45 |
Of which 16 included in the autism study sample, one in both autism and AS samples, and three in neither
Of which 33 used in analyses (a MZ twin left out)
Laboratory methods
The microsatellite markers of the initial scan (n=1109) were genotyped by standard procedures at deCODE Genetics Inc. (Reykjavik, Iceland). Follow-up microsatellites were genotyped with the ABI 3730 DNA sequencer, analyzed with GeneMapper v.3.0 software (Applera Corporation, Norwalk, CT, USA), and verified by two individuals independently. In all multipoint analyses, we used the deCODE high resolution genetic map (70). SNP markers in the fine-map stage were genotyped either with Sequenom's homogenous MassEXTEND (hME) and iPLEX technology using the Mass ARRAY Platform, as specified by manufacturer's instructions (Sequenom, San Diego, California, USA), or by fluorogenic 5′ nuclease allelic discrimination chemistry (TaqMan) with an ABI Prism 7900 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). All genotypes were checked for correct Mendelian transmission with PEDCHECK v.1.1 (71) and monitored for Hardy-Weinberg equilibrium. Markers accepted for analysis displayed a minimum genotyping success rate of 90%, with the majority of markers having a success rate of >95%. The borderline for the minor allele frequency (MAF) of SNPs was 5%, with most of the SNPs having MAF > 10%. Individuals were treated either as affected or unknown.
Supplementary Material
Acknowledgements
We wish to thank Drs Irma Järvelä, Reija Alen, Raija Vanhala, Raili Riikonen, Taina Nieminen-von Wendt, Ismo Makkonen and Mari Auranen for their contribution in collecting and characterizing the sample. Professor Joseph D Terwilliger is thanked for advice in statistical issues concerning Pseudomarker.
Funding
This work was supported by the Center of Excellence in Complex Disease Genetics of the Academy of Finland; Biocentrum Helsinki; Päivikki and Sakari Sohlberg Foundation; and Autism Speaks/Cure Autism Now [to T.Y.]
Abbreviations
- ASD
autism spectrum disorders
- AS
Asperger syndrome
- Ext ped
Extended pedigree
- PDD-NOS
pervasive developmental disorder not otherwise specified
Footnotes
Conflict of Interest Statement
The authors declare no conflicts of interest.
References
- 1.World Health Organization . The ICD-10 Classification of Mental and Behavioural Disorders. Diagnostic Criteria for Research. WHO; Geneva: 1993. [Google Scholar]
- 2.American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders (4th edn) (DSM-IV) 4 ed. APA; Washington, DC: 1994. [Google Scholar]
- 3.Chakrabarti S, Fombonne E. Pervasive developmental disorders in preschool children. Jama. 2001;285:3093–3099. doi: 10.1001/jama.285.24.3093. [DOI] [PubMed] [Google Scholar]
- 4.Charman T. The prevalence of autism spectrum disorders. Recent evidence and future challenges. Eur. Child Adolesc. Psychiatry. 2002;11:249–256. doi: 10.1007/s00787-002-0297-8. [DOI] [PubMed] [Google Scholar]
- 5.Yeargin-Allsopp M, Rice C, Karapurkar T, Doernberg N, Boyle C, Murphy C. Prevalence of autism in a US metropolitan area. Jama. 2003;289:49–55. doi: 10.1001/jama.289.1.49. [DOI] [PubMed] [Google Scholar]
- 6.Steffenburg S, Gillberg C, Hellgren L, Andersson L, Gillberg IC, Jakobsson G, Bohman M. A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J. Child Psychol. Psychiatry. 1989;30:405–416. doi: 10.1111/j.1469-7610.1989.tb00254.x. [DOI] [PubMed] [Google Scholar]
- 7.Bolton P, Macdonald H, Pickles A, Rios P, Goode S, Crowson M, Bailey A, Rutter M. A case-control family history study of autism. J. Child Psychol. Psychiatry. 1994;35:877–900. doi: 10.1111/j.1469-7610.1994.tb02300.x. [DOI] [PubMed] [Google Scholar]
- 8.Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol. Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- 9.Feinstein C, Reiss AL. Autism: the point of view from fragile X studies. J. Autism Dev. Disord. 1998;28:393–405. doi: 10.1023/a:1026000404855. [DOI] [PubMed] [Google Scholar]
- 10.Gillberg C. Chromosomal disorders and autism. J. Autism Dev. Disord. 1998;28:415–425. doi: 10.1023/a:1026004505764. [DOI] [PubMed] [Google Scholar]
- 11.Wassink TH, Piven J, Patil SR. Chromosomal abnormalities in a clinic sample of individuals with autistic disorder. Psychiatr. Genet. 2001;11:57–63. doi: 10.1097/00041444-200106000-00001. [DOI] [PubMed] [Google Scholar]
- 12.Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat. Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laumonnier F, Bonnet-Brilhault F, Gomot M, Blanc R, David A, Moizard MP, Raynaud M, Ronce N, Lemonnier E, Calvas P, et al. X-Linked Mental Retardation and Autism Are Associated with a Mutation in the NLGN4 Gene, a Member of the Neuroligin Family. Am. J. Hum. Genet. 2004;74:552–557. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsater H, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat. Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moessner R, Marshall CR, Sutcliffe JS, Skaug J, Pinto D, Vincent J, Zwaigenbaum L, Fernandez B, Roberts W, Szatmari P, et al. Contribution of SHANK3 Mutations to Autism Spectrum Disorder. Am. J. Hum. Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alarcon M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, Sebat J, Wigler M, Martin CL, Ledbetter DH, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am. J. Hum. Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, Rea A, Guy M, Lin S, Cook EH, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bakkaloglu B, O'Roak BJ, Louvi A, Gupta AR, Abelson JF, Morgan TM, Chawarska K, Klin A, Ercan-Sencicek AG, Stillman AA, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82:165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strauss KA, Puffenberger EG, Huentelman MJ, Gottlieb S, Dobrin SE, Parod JM, Stephan DA, Morton DH. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354:1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 20.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, et al. Association between Microdeletion and Microduplication at 16p11.2 and Autism. N Engl J Med. 2008 doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 22.Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, Gilliam TC, Nowak NJ, Cook EH, Jr., Dobyns WB, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 23.Risch N, Spiker D, Lotspeich L, Nouri N, Hinds D, Hallmayer J, Kalaydjieva L, McCague P, Dimiceli S, Pitts T, et al. A genomic screen of autism: evidence for a multilocus etiology. Am J Hum Genet. 1999;65:493–507. doi: 10.1086/302497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trikalinos TA, Karvouni A, Zintzaras E, Ylisaukko-oja T, Peltonen L, Jarvela I, Ioannidis JP. A heterogeneity-based genome search meta-analysis for autism-spectrum disorders. Mol Psychiatry. 2006;11:29–36. doi: 10.1038/sj.mp.4001750. [DOI] [PubMed] [Google Scholar]
- 25.Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, Mukaddes NM, Balkhy S, Gascon G, Hashmi A, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 28.Varilo T, Peltonen L. Isolates and their potential use in complex gene mapping efforts. Current Opinion in Genetics & Development. 2004;14:227–323. doi: 10.1016/j.gde.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 29.Peltonen L, Perola M, Naukkarinen J, Palotie A. Lessons from studying monogenic disease for common disease. Hum Mol Genet. 2006;15(Suppl 1):R67–74. doi: 10.1093/hmg/ddl060. [DOI] [PubMed] [Google Scholar]
- 30.Peltonen L, Palotie A, Lange K. Use of population isolates for mapping complex traits. Nat Rev Genet. 2000;1:182–190. doi: 10.1038/35042049. [DOI] [PubMed] [Google Scholar]
- 31.Service S, Deyoung J, Karayiorgou M, Roos JL, Pretorious H, Bedoya G, Ospina J, Ruiz-Linares A, Macedo A, Palha JA, et al. Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nat Genet. 2006;38:556–560. doi: 10.1038/ng1770. [DOI] [PubMed] [Google Scholar]
- 32.Goring HH, Terwilliger JD. Linkage analysis in the presence of errors IV: joint pseudomarker analysis of linkage and/or linkage disequilibrium on a mixture of pedigrees and singletons when the mode of inheritance cannot be accurately specified. Am. J. Hum. Genet. 2000;66:1310–1327. doi: 10.1086/302845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am. J. Hum. Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- 34.Lange EM, Lange K. Powerful allele sharing statistics for nonparametric linkage analysis. Hum. Hered. 2004;57:49–58. doi: 10.1159/000077389. [DOI] [PubMed] [Google Scholar]
- 35.Auranen M, Vanhala R, Varilo T, Ayers K, Kempas E, Ylisaukko-Oja T, Sinsheimer JS, Peltonen L, Jarvela I. A genomewide screen for autism-spectrum disorders: evidence for a major susceptibility locus on chromosome 3q25-27. Am. J. Hum. Genet. 2002;71:777–790. doi: 10.1086/342720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ylisaukko-Oja T, Nieminen-Von Wendt T, Kempas E, Sarenius S, Varilo T, Wendt Lv L, Peltonen L, Jarvela II. Genome-wide scan for loci of Asperger syndrome. Mol. Psychiatry. 2004;9:161–168. doi: 10.1038/sj.mp.4001385. [DOI] [PubMed] [Google Scholar]
- 37.Veenstra-VanderWeele J, Cook EH. Molecular genetics of autism spectrum disorder. Mol. Psychiatry. 2004 doi: 10.1038/sj.mp.4001505. [DOI] [PubMed] [Google Scholar]
- 38.Buono RJ, Lohoff FW, Sander T, Sperling MR, O'Connor MJ, Dlugos DJ, Ryan SG, Golden GT, Zhao H, Scattergood TM, et al. Association between variation in the human KCNJ10 potassium ion channel gene and seizure susceptibility. Epilepsy Res. 2004;58:175–183. doi: 10.1016/j.eplepsyres.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 39.Ferraro TN, Golden GT, Smith GG, Martin JF, Lohoff FW, Gieringer TA, Zamboni D, Schwebel CL, Press DM, Kratzer SO, et al. Fine mapping of a seizure susceptibility locus on mouse Chromosome 1: nomination of Kcnj10 as a causative gene. Mamm Genome. 2004;15:239–251. doi: 10.1007/s00335-003-2270-3. [DOI] [PubMed] [Google Scholar]
- 40.Brzustowicz LM, Hodgkinson KA, Chow EW, Honer WG, Bassett AS. Location of a major susceptibility locus for familial schizophrenia on chromosome 1q21-q22. Science. 2000;288:678–682. doi: 10.1126/science.288.5466.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brzustowicz LM, Simone J, Mohseni P, Hayter JE, Hodgkinson KA, Chow EW, Bassett AS. Linkage disequilibrium mapping of schizophrenia susceptibility to the CAPON region of chromosome 1q22. Am .J. Hum. Genet. 2004;74:1057–1063. doi: 10.1086/420774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams NM, Preece A, Spurlock G, Norton N, Williams HJ, McCreadie RG, Buckland P, Sharkey V, Chowdari KV, Zammit S, et al. Support for RGS4 as a susceptibility gene for schizophrenia. Biol. Psychiatry. 2004;55:192–195. doi: 10.1016/j.biopsych.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 43.Nurmi EL, Bradford Y, Chen Y, Hall J, Arnone B, Gardiner MB, Hutcheson HB, Gilbert JR, Pericak-Vance MA, Copeland-Yates SA, et al. Linkage disequilibrium at the Angelman syndrome gene UBE3A in autism families. Genomics. 2001;77:105–113. doi: 10.1006/geno.2001.6617. [DOI] [PubMed] [Google Scholar]
- 44.Nikali K, Suomalainen A, Terwilliger J, Koskinen T, Weissenbach J, Peltonen L. Random search for shared chromosomal regions in four affected individuals: the assignment of a new hereditary ataxia locus. Am. J. Hum. Genet. 1995;56:1088–1095. [PMC free article] [PubMed] [Google Scholar]
- 45.Peltonen L, Jalanko A, Varilo T. Molecular genetics of the Finnish disease heritage. Hum. Mol. Genet. 1999;8:1913–1923. doi: 10.1093/hmg/8.10.1913. [DOI] [PubMed] [Google Scholar]
- 46.Gurling HM, Kalsi G, Brynjolfson J, Sigmundsson T, Sherrington R, Mankoo BS, Read T, Murphy P, Blaveri E, McQuillin A, et al. Genomewide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q32.2, 5q33.2, and 8p21-22 and provides support for linkage to schizophrenia, on chromosomes 11q23.3-24 and 20q12.1-11.23. Am. J. Hum. Genet. 2001;68:661–673. doi: 10.1086/318788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lenzen KP, Heils A, Lorenz S, Hempelmann A, Hofels S, Lohoff FW, Schmitz B, Sander T. Supportive evidence for an allelic association of the human KCNJ10 potassium channel gene with idiopathic generalized epilepsy. Epilepsy Res. 2005;63:113–118. doi: 10.1016/j.eplepsyres.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 48.Gillberg C, Billstedt E. Autism and Asperger syndrome: coexistence with other clinical disorders. Acta Psychiatr Scand. 2000;102:321–330. doi: 10.1034/j.1600-0447.2000.102005321.x. [DOI] [PubMed] [Google Scholar]
- 49.McCauley JL, Li C, Jiang L, Olson LM, Crockett G, Gainer K, Folstein SE, Haines JL, Sutcliffe JS. Genome-wide and Ordered-Subset linkage analyses provide support for autism loci on 17q and 19p with evidence of phenotypic and interlocus genetic correlates. BMC Med Genet. 2005;6:1. doi: 10.1186/1471-2350-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu J, Nyholt DR, Magnussen P, Parano E, Pavone P, Geschwind D, Lord C, Iversen P, Hoh J, Ott J, et al. A genomewide screen for autism susceptibility loci. Am J Hum Genet. 2001;69:327–340. doi: 10.1086/321980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shao Y, Wolpert CM, Raiford KL, Menold MM, Donnelly SL, Ravan SA, Bass MP, McClain C, von Wendt L, Vance JM, et al. Genomic screen and follow-up analysis for autistic disorder. Am J Med Genet. 2002;114:99–105. doi: 10.1002/ajmg.10153. [DOI] [PubMed] [Google Scholar]
- 52.Philippe A, Martinez M, Guilloud-Bataille M, Gillberg C, Rastam M, Sponheim E, Coleman M, Zappella M, Aschauer H, Van Maldergem L, et al. Genome-wide scan for autism susceptibility genes. Paris Autism Research International Sibpair Study. Hum Mol Genet. 1999;8:805–812. doi: 10.1093/hmg/8.5.805. [DOI] [PubMed] [Google Scholar]
- 53.IMGSAC A full genome screen for autism with evidence for linkage to a region on chromosome 7q. International Molecular Genetic Study of Autism Consortium. Hum Mol Genet. 1998;7:571–578. doi: 10.1093/hmg/7.3.571. [DOI] [PubMed] [Google Scholar]
- 54.Buxbaum JD, Silverman J, Keddache M, Smith CJ, Hollander E, Ramoz N, Reichert JG. Linkage analysis for autism in a subset families with obsessive-compulsive behaviors: evidence for an autism susceptibility gene on chromosome 1 and further support for susceptibility genes on chromosome 6 and 19. Mol Psychiatry. 2004;9:144–150. doi: 10.1038/sj.mp.4001465. [DOI] [PubMed] [Google Scholar]
- 55.Miyasaka H, Choudhury BK, Hou EW, Li SS. Molecular cloning and expression of mouse and human cDNA encoding AES and ESG proteins with strong similarity to Drosophila enhancer of split groucho protein. Eur J Biochem. 1993;216:343–352. doi: 10.1111/j.1432-1033.1993.tb18151.x. [DOI] [PubMed] [Google Scholar]
- 56.Stifani S, Blaumueller CM, Redhead NJ, Hill RE, Artavanis-Tsakonas S. Human homologs of a Drosophila Enhancer of split gene product define a novel family of nuclear proteins. Nat Genet. 1992;2:119–127. doi: 10.1038/ng1092-119. [DOI] [PubMed] [Google Scholar]
- 57.Chen G, Courey AJ. Groucho/TLE family proteins and transcriptional repression. Gene. 2000;249:1–16. doi: 10.1016/s0378-1119(00)00161-x. [DOI] [PubMed] [Google Scholar]
- 58.Heitzler P, Bourouis M, Ruel L, Carteret C, Simpson P. Genes of the Enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signalling in Drosophila. Development. 1996;122:161–171. doi: 10.1242/dev.122.1.161. [DOI] [PubMed] [Google Scholar]
- 59.Fombonne E, Roge B, Claverie J, Courty S, Fremolle J. Microcephaly and macrocephaly in autism. J Autism Dev Disord. 1999;29:113–119. doi: 10.1023/a:1023036509476. [DOI] [PubMed] [Google Scholar]
- 60.Cody H, Pelphrey K, Piven J. Structural and functional magnetic resonance imaging of autism. Int J Dev Neurosci. 2002;20:421–438. doi: 10.1016/s0736-5748(02)00053-9. [DOI] [PubMed] [Google Scholar]
- 61.Jimenez G, Paroush Z, Ish-Horowicz D. Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes Dev. 1997;11:3072–3082. doi: 10.1101/gad.11.22.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benayed R, Gharani N, Rossman I, Mancuso V, Lazar G, Kamdar S, Bruse SE, Tischfield S, Smith BJ, Zimmerman RA, et al. Support for the homeobox transcription factor gene ENGRAILED 2 as an autism spectrum disorder susceptibility locus. Am. J. Hum. Genet. 2005;77:851–868. doi: 10.1086/497705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Varilo T. The age of the mutations in the Finnish disease heritage; a genealogical and linkage disequilibrium study. National Public Health Institute and University of Helsinki; Helsinki: 1999. [Google Scholar]
- 64.Varilo T, Savukoski M, Norio R, Santavuori P, Peltonen L, Jarvela I. The age of human mutation: genealogical and linkage disequilibrium analysis of the CLN5 mutation in the Finnish population. Am. J. Hum. Genet. 1996;58:506–512. [PMC free article] [PubMed] [Google Scholar]
- 65.Varilo T, Nikali K, Suomalainen A, Lonnqvist T, Peltonen L. Tracing an ancestral mutation: genealogical and haplotype analysis of the infantile onset spinocerebellar ataxia locus. Genome Res. 1996;6:870–875. doi: 10.1101/gr.6.9.870. [DOI] [PubMed] [Google Scholar]
- 66.Savukoski M, Klockars T, Holmberg V, Santavuori P, Lander ES, Peltonen L. CLN5, a novel gene encoding a putative transmembrane protein mutated in Finnish variant late infantile neuronal ceroid lipofuscinosis. Nat. Genet. 1998;19:286–288. doi: 10.1038/975. [DOI] [PubMed] [Google Scholar]
- 67.Nikali K, Suomalainen A, Saharinen J, Kuokkanen M, Spelbrink JN, Lonnqvist T, Peltonen L. Infantile onset spinocerebellar ataxia is caused by recessive mutations in mitochondrial proteins Twinkle and Twinky. Hum. Mol. Genet. 2005;14:2981–2990. doi: 10.1093/hmg/ddi328. [DOI] [PubMed] [Google Scholar]
- 68.Ylisaukko-oja T, Nieminen-von Wendt T, Kempas E, Sarenius S, Varilo T, von Wendt L, Peltonen L, Jarvela I. Genome-wide scan for loci of Asperger syndrome. Mol. Psychiatry. 2004;9:161–168. doi: 10.1038/sj.mp.4001385. [DOI] [PubMed] [Google Scholar]
- 69.Kilpinen H, Ylisaukko-Oja T, Hennah W, Palo OM, Varilo T, Vanhala R, Nieminen-von Wendt T, von Wendt L, Paunio T, Peltonen L. Association of DISC1 with autism and Asperger syndrome. Mol. Psychiatry. 2008;13:187–196. doi: 10.1038/sj.mp.4002031. [DOI] [PubMed] [Google Scholar]
- 70.Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, et al. A high-resolution recombination map of the human genome. Nat. Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 71.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am. J. Hum. Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.